Recurrent duplication of two key C4 genes during the emergence of a C4 cycle in the grass genus Alloteropsis is correlated with increases in transcript abundance
Keywords: Biochemical pathway, C4 photosynthesis, copy number variation, dosage effect, gene duplication, grasses, low-coverage sequencing
Abstract
The importance of gene duplication for evolutionary diversification has been mainly discussed in terms of genetic redundancy allowing neofunctionalization. In the case of C4 photosynthesis, which evolved via the co-option of multiple enzymes to boost carbon fixation in tropical conditions, the importance of genetic redundancy has not been consistently supported by genomic studies. Here, we test for a different role for gene duplication in the early evolution of C4 photosynthesis, via dosage effects creating rapid step changes in expression levels. Using genome-wide data for accessions of the grass genus Alloteropsis that recently diversified into different photosynthetic types, we estimate gene copy numbers and demonstrate that recurrent duplications in two important families of C4 genes coincided with increases in transcript abundance along the phylogeny, in some cases via a pure dosage effect. While increased gene copy number during the initial emergence of C4 photosynthesis probably offered a rapid route to enhanced expression, we also find losses of duplicates following the acquisition of genes encoding better-suited isoforms. The dosage effect of gene duplication might therefore act as a transient process during the evolution of a C4 biochemistry, rendered obsolete by the fixation of regulatory mutations increasing expression levels.
Introduction
C4 photosynthesis is a complex trait that results from the co-ordinated action of multiple biochemical and anatomical components to concentrate CO2 at the site of Rubisco, increasing photosynthetic efficiency under warm and dry conditions (Hatch, 1987; Sage, 2004). Despite its complexity, the C4 trait evolved multiple times independently in several groups of angiosperms (Sage et al., 2011). All enzymes required for the C4 pathway were present in non-C4 ancestors, where they were responsible for different, non-photosynthetic functions (Sage, 2004; Aubry et al., 2011). The evolution of C4 photosynthesis consequently required the co-option of these enzymes into new functions, followed by changes in their expression patterns and/or catalytic properties (Bläsing et al., 2000; Tausta et al., 2002; Gowik et al., 2004; Akyildiz et al., 2007; Christin et al., 2007; Hibberd and Covshoff, 2010; Huang et al., 2017). It has been hypothesized that this massive co-option was facilitated by gene duplication, with one of the duplicates acquiring the novel C4 function via neofunctionalization while the other continued to fulfil the ancestral function (Monson, 1999, 2003; Sage, 2004). However, recent genomic studies have not supported this hypothesis of genetic redundancy facilitating neofunctionalization, meaning that the genomic mechanisms enabling the acquisition of novel functions during C4 evolution remain largely unknown.
Most C4-related enzymes are encoded by multigene families, with numerous paralogues that emerged via multiple rounds of whole-genome and single-gene duplications during angiosperm diversification (Wang et al., 2009; Christin et al., 2013, 2015; Huang et al., 2017). However, the number of paralogues within each of these gene families does not differ significantly between C3 and C4 species (Williams et al., 2012; van den Bergh et al., 2014). Comparative genomics on a handful of grasses have identified duplicates that have been retained on branches leading to two C4 origins, but these did not encode enzymes necessarily involved in the C4 cycle (Emms et al., 2016). Indeed, investigations focusing on genes families with a known function in C4 photosynthesis indicate that the gain of a C4-specific function was generally not directly preceded by a gene duplication event (Christin et al., 2007, 2009; Wang et al., 2009). Although the creation of a large reservoir of ancient duplications might still be important (Monson, 2003), these various lines of evidence suggest that C4 evolution did not consistently involve duplication followed by neofunctionalization of one copy while the other retained the ancestral function. However, gene duplication might still have played a role in the initial emergence of C4 photosynthesis, via a combination of dosage effects and neofunctionalization.
Small-scale or whole-genome duplications are generally expected to increase transcript abundance through a gene dosage effect (Otto et al., 1986; Kondrashov et al., 2002; Conant and Wolfe, 2008; Conant et al., 2014). Instances of retention of duplicated genes due to a dosage effect on expression levels have been reported for a number of adaptive traits, which include insecticide resistance in the Culex mosquito (Mouchès et al., 1986), cold protection in Antarctic fishes (Chen et al., 2008), and nematode resistance in soybean (Cook et al., 2012). Positive selection on the dosage effect of newborn duplicates is predicted in cases where the protein products physically interact with molecules such as toxins or nutrients, or in cases in which proteins need rapid and constant production at high levels (Kondrashov et al., 2002; Kondrashov, 2012). The dosage effect of gene duplication might consequently be important for the establishment of a C4 cycle. Current models of C4 evolution hypothesize that a weak C4 cycle can first emerge using enzymes that have not been adapted to the C4 catalytic context (Sage, 2004; Heckmann et al., 2013; Christin and Osborne, 2014; Mallmann et al., 2014; Heckmann, 2016; Dunning et al., 2017). Gene duplications increasing the transcript abundance of C4-related genes in plants with a weak C4 cycle would increase the strength of the pathway, which is predicted to boost carbon assimilation and fitness (Heckmann et al., 2013; Mallmann et al., 2014), leading to the preferential retention of the duplicates. We propose here to test the hypothesis that gene duplications contributed to the initial emergence of a C4 biochemistry via dosage effects, with subsequent neofunctionalization. We capitalize on the diversity of C4 enzymes that evolved in the recent past within the grass genus Alloteropsis.
The Alloteropsis genus contains five species, four of which are C4, while the fifth, A. semialata, encompasses C4 as well as non-C4 populations with and without a weak C4 cycle (Ellis, 1974; Lundgren et al., 2016). The diversification of A. semialata took place during the last 3 million years (Lundgren et al., 2015), and only a few genes are markedly up-regulated in the C4 accessions compared with C3 populations (Dunning et al., 2017). In some cases, the identity of genes used for the C4 cycle differs among C4 populations of A. semialata, which is interpreted as the footprint of a gradual adaptation of C4 photosynthesis during the diversification of the group involving secondary gene flow among previously isolated populations (Olofsson et al., 2016; Dunning et al., 2017). This group therefore represents an outstanding system to investigate the small-scale processes that led to C4 photosynthesis, including the importance of genomic rearrangements such as duplications for C4 evolution.
Genome scans coupled with genome size estimates are used here to assess the gene content of accessions of the genus Alloteropsis varying in their photosynthetic type, testing (i) whether the copy number of genes encoding C4-related proteins varies among accessions of Alloteropsis; (ii) whether gene duplications coincide with the co-option of genes for a C4 function; and (iii) whether increases in gene copy number result from the duplication of genomic material or from retroposition events (i.e. insertion of retrotranscribed RNA into the genome; Kaessmann et al., 2009). In addition, we retrieve published transcriptomes for members of the Alloteropsis genus (Dunning et al., 2017) and associate them with newly generated high-coverage genome sequencing to test (iv) whether recently duplicated genes are expressed; (v) whether multiple copies all contribute to overall transcript abundance; and (vi) whether increases in copy number of C4-related genes along the phylogenetic tree were associated with increases in expression levels. This comparative analysis of gene copy numbers provides evidence for a potential role for recent gene duplications in physiological innovation through rapid and drastic changes of transcript abundance.
Materials and methods
Taxon sampling and genome data
A total of 20 genome-wide, low-coverage sequencing data sets of Alloteropsis J. Presl were retrieved from published studies (Table 1; Lundgren et al., 2015; Olofsson et al., 2016; NCBI accession no. SRP082653). These include two accessions of the C4A. angusta Stapf, one of the C4 species A. cimicina (L.) Stapf, and 17 of A. semialata (R. Br.) Hitchc. Among these 17 A. semialata, 12 are C4 individuals sampled across a broad geographical range from West Africa to Australia, and the five non-C4 include three individuals with a weak C4 cycle (‘C3+C4’ in Dunning et al., 2017; note that this term is equivalent to ‘type II C3–C4 intermediates’ sensuEdwards and Ku, 1987) and two C3 individuals from South Africa. Each of the genomic data sets consists of paired-end Illumina reads, with read lengths of 100, 125, or 150 bp (Table 1). In this study, the raw reads were filtered using the NGSQC Toolkit (Patel and Jain, 2012) to retain only high-quality sequences (i.e. >70% of read length with Phred quality >20), and to remove primer and adaptor contaminated reads. The genome size and ploidy level of some of the individuals analysed here were retrieved from previous studies that used the same accessions (Lundgren et al., 2015; Olofsson et al., 2016). Some accessions were only available as herbarium samples, preventing estimates of genome sizes or ploidy levels.
Table 1.
Genome data information
| ID | Species |
Carbon
isotope |
Genome size
(Gb/2Cx a )/ploidy |
Country |
Transcriptome sample
b |
Sequencing
batch c |
Sequencer |
Read
length |
Total nuclear
genome reads |
Organellar
reads (%)d |
Theoretical
coverage e |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cim1 | A. cimicina | C4 | – | Madagascar | ACIM | 2 | HiSeq 2500 | 100 | 20898025 | 2.0 | 0.95 |
| Ang1 | A. angusta | C4 | – | DRC | – | 5 | HiSeq 3000 | 150 | 14751007 | 2.6 | 1.01 |
| Ang2 | A. angusta | C4 | 1.95/2n | Uganda | – | 2 | HiSeq 2500 | 100 | 18665954 | 1.9 | 0.96 |
| RSA1 | A. semialata | C3 | – | South Africa | – | 1 | HiSeq 2500 | 100 | 14821009 | 0.8 | 0.67 |
| RSA2 | A. semialata | C3 | 1.80/2n | South Africa | RSA5 | 1 | HiSeq 2500 | 100 | 12524356 | 0.6 | 0.70 |
| TAN1 | A. semialata | C3+C4 | 1.88/2n | Tanzania | TAN1 | 2 | HiSeq 2500 | 100 | 18899157 | 4.0 | 1.01 |
| TAN2-A | A. semialata | C3+C4 | 2.19/2n | Tanzania | TAN2 | 2 | HiSeq 2500 | 100 | 20065838 | 4.2 | 0.92 |
| TAN2-Af | A. semialata | C3+C4 | 2.19/2n | Tanzania | TAN2 | 6 | HiSeq 2500 | 250 | 45774384 | 3.4 | 5.05 |
| TAN3 | A. semialata | C3+C4 | – | Tanzania | – | 3 | HiSeq 2500 | 125 | 35782290 | 1.6 | 2.03 |
| DRC1 | A. semialata | C4 | – | DRC | – | 5 | HiSeq 3000 | 150 | 33933832 | 3.6 | 2.31 |
| DRC2 | A. semialata | C4 | – | DRC | – | 4 | HiSeq 3000 | 150 | 23098686 | 3.1 | 1.57 |
| DRC3 | A. semialata | C4 | – | DRC | – | 3 | HiSeq 2500 | 125 | 28889427 | 6.4 | 1.64 |
| DRC4 | A. semialata | C4 | – | DRC | – | 5 | HiSeq 3000 | 150 | 14749392 | 4.0 | 1.01 |
| TAN4 | A. semialata | C4 | 2.01/2n | Tanzania | TAN4 | 2 | HiSeq 2500 | 100 | 18596076 | 3.2 | 0.93 |
| RSA3 | A. semialata | C4 | 5.22/6n | South Africa | RSA3 | 1 | HiSeq 2500 | 100 | 13824190 | 0.8 | 0.26 |
| KEN1 | A. semialata | C4 | – | Kenya | – | 3 | HiSeq 2500 | 125 | 25405608 | 4.9 | 1.44 |
| BUR1 | A. semialata | C4 | 1.95/2n | Burkina Faso | BUR1 | 1 | HiSeq 2500 | 100 | 13498418 | 0.9 | 0.69 |
| MAD1 | A. semialata | C4 | 2.05/2n | Madagascar | MAD1 | 1 | HiSeq 2500 | 100 | 16440692 | 1.8 | 0.80 |
| THA1 | A. semialata | C4 | – | Thailand | – | 2 | HiSeq 2500 | 100 | 16873534 | 2.1 | 0.77 |
| TPE1-3 | A. semialata | C4 | 1.87/2n | Taiwan | TPE1 | 2 | HiSeq 2500 | 100 | 15435339 | 4.8 | 0.83 |
| TPE1-10f | A. semialata | C4 | 1.87/2n | Taiwan | TPE1 | 7 | HiSeq 2500 | 250 | 169555422 | 3.4 | 21.92 |
| AUS1 | A. semialata | C4 | 2.20/2n | Australia | AUS1 | 1 | HiSeq 2500 | 100 | 11600487 | 0.8 | 0.53 |
a Genome size (Gb/2Cx)=total genome mass (pg)×0.978.
b Data retrieved from Dunning et al. (2017).
c Accessions with the same batch number were sequenced together.
d Percentage of reads mapping to chloroplast and mitochondrial genomes.
e Based on 2C genome size; after removing organellar reads; assuming a value of 2.2 Gb (maximum value of a diploid individual of Alloteropsis) for unknown genome sizes.
f Data set generated for this study. Other data sets were retrieved from either Lundgren et al. (2015) or Olofsson et al. (2016).
High-coverage sequencing data sets were generated here for two individuals to allow single nucleotide polymorphism (SNP) analyses (see below). This included one C3+C4 accession from Tanzania (TAN2) already sequenced at low coverage and one C4 accession from a population where another individual was sequenced at low coverage (TPE1; Table 1). For these two samples, 250 bp long paired reads were obtained with the Illumina technology.
The different sequence data sets were obtained from whole genomic DNA, so that reads can belong to any of the nuclear, chloroplast, and mitochondrial genomes. Reads from the two organellar genomes were identified by mapping the genomic data sets onto representative chloroplast and mitochondrial genomes using Bowtie2 (Langmead and Salzberg, 2012) with default parameters, and removed before analyses. Mitochondrial genomes were assembled de novo (Supplementary text S1 at JXB online) using the approach described in Lundgren et al. (2015), while chloroplast genomes were retrieved from Lundgren et al. (2015) and Olofsson et al. (2016). On average, 3% of the initial reads were removed because of their organellar origin (Table 1).
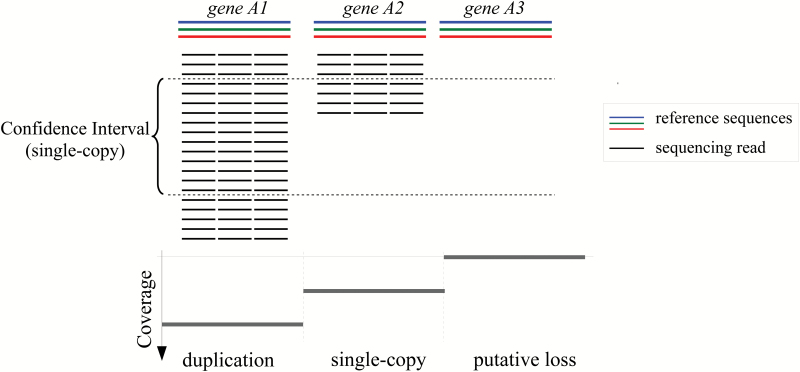
Mapping of reads on reference data sets
Gene copy numbers were estimated using a modified read depth approach (Alkan et al., 2009; Yoon et al., 2009; Teo et al., 2012). This strategy divides the genome into non-overlapping regions (bins) and uses the number of genomic reads mapped to each of these regions to estimate gene copy number. Bins receiving in some accessions more or fewer reads than expected under a null statistical model are considered copy number variants (Fig. 1). Given the current lack of a reference genome for any Alloteropsis species, genomic data were mapped to a reference data set consisting of coding sequences (CDSs) of A. cimicina and A. semialata, which was retrieved from the transcriptome study of Dunning et al. (2017). Briefly, this data set comprises groups of co-orthologues at the Panicoideae subfamily level, the group of grasses that includes the genus Alloteropsis. Each group of co-orthologues encompasses all the genes that are descended by speciation and/or gene duplication from a single gene in the common ancestor of Panicoideae. Only genes captured in one of the Alloteropsis transcriptomes and with co-orthologues in at least one of Sorghum bicolor and Setaria italica were included. Increases in copy number detected here therefore correspond to duplications that happened after the initial diversification of Panicoideae, about 30 million years ago. Manually curated alignments using longer transcripts of 23 gene families with a known function in C4 biochemistry (Bräutigam et al., 2011) and the gene encoding the Rubisco small subunit (rbcS) were added into the reference data set. These manually curated alignments improved read mapping accuracy in cases where paralogues with high sequence similarity were present, such as laterally acquired forms previously identified for phosphoenolpyruvate carboxylase (PEPC; ppc gene) and phosphoenolpyruvate carboxykinase (PCK; pck gene; Christin et al., 2012; Dunning et al., 2017). Overall, this genome-wide data set comprised 12688 groups of co-orthologues, belonging to 5589 gene families.
Fig. 1.
Read depth approach for gene copy number estimation. Duplications are inferred when the number of read counts expected for a determined gene is significantly higher than the expected read counts for single-copy genes, according to an underlying statistical model.
Genomic reads were mapped onto the genome-wide CDS data set using Bowtie2, with default parameters, randomly assigning reads mapped to multiple sequences to one of the top hits, and using the local alignment option. Reads were mapped as single-end reads to avoid false negatives when one of the reads mapped outside the CDS. The number of mapped reads (counts) per group of co-orthologues was obtained using SAMtools (Li et al., 2009) and used to compute gene copy number estimates as described below.
Estimates of copy numbers
Under the assumption that each site in the genome has an equal probability of being the first site of a given read, the expected read count (c) for any genomic region i of length L can be computed as:
| (1) |
where N is the total number of sequencing reads and G is the haploid genome size (in number of bases). Assuming the counts c is a random variable that follows a binomial distribution, with the total binomial trials being the total number of reads N, the probability of a region i being captured by one read is equivalent to the probability of success in each binomial trial, which is:
| (2) |
A well-known complication of quantitative genomic studies based on read depth is the sequencing bias linked to the GC content of the sequenced region, which is particular to sequencing approaches where library preparation includes PCR steps, as required for degraded DNA extracted from herbarium samples (Dohm et al., 2008; Aird et al., 2011; Benjamini and Speed, 2012; Teo et al., 2012). The relationship between sequencing depth and GC content can vary across sequencing runs (Benjamini and Speed, 2012), and previous studies have quantified this relationship using various metrics (Alkan et al., 2009; Bellos et al., 2012; Benjamini and Speed, 2012). In this study, preliminary analyses confirmed that the relationship varied among the different batches of library preparation and sequencing (Supplementary Fig. S1). The relationship between read counts and GC content was consequently estimated for each sample by using the counts of genes extracted from the genome-wide reference mapping. Read counts were normalized by gene length, and genes with no count or counts >1.5 times the median count were removed from this particular analysis, to enrich the data set with putative single-copy genes. These length-normalized counts were then expressed as a linear function of the mean GC content of the target genes (xi), so that:
| (3) |
The coefficients a and b were estimated individually for each genome data set using a linear model fit procedure in R (R Development Core Team, 2017). To homogenize the number of genes across GC content classes, 60 genes were randomly drawn from those present in each of nine equally spaced classes of GC content from 38% to 78%, and linear coefficients were calculated on the pooled subsample. Only genes longer than 700 bp were used here, since such long genes receive more reads and therefore provide more accurate copy number estimates. This procedure was repeated 100 times, providing a non-parametric estimate of variation for the coefficients. An approximate correction of the binomial probability of success in each trial (Equation 2) by the GC content was then obtained by substituting Equations 3 and 1 in Equation 2, so that:
| (4) |
Note that these new probabilities are independent of the genome size and can therefore be estimated for any sample. If E(ci) is the expected count when a target gene is present as a single copy, an estimate of the absolute number of copies ki can be obtained as:
| (5) |
The expected counts and confidence intervals for single-copy genes were computed using a binomial quantile function implemented in R, with a confidence level of 99% corrected for multiple comparisons using the Bonferroni method. Genes were considered duplicated if the counts were above the upper limit of this confidence interval, and single copy if the counts were within the confidence interval limits (inclusive). Although partial copies can exist following incomplete duplications, copy number estimates for duplicated genes were rounded up for follow-up analyses. Genes were considered absent when no read count was detected, provided the confidence intervals for the expected counts did not include zero. In such cases, and in cases where read counts were below the lower limit of the confidence interval, the genes were removed from the analysis, since accurate copy numbers could not be estimated.
Quantitative real-time PCR estimates of copy number
A number of concerns have been raised about the use of high-throughput sequencing data for genome analyses of structural diversity, such as copy number variants (Benjamini and Speed, 2012; Teo et al., 2012). In particular, the above-mentioned GC content bias and others resulting from the library preparations represent potential caveats. We consequently performed quantitative real-time PCR (qPCR) assays to confirm the accuracy of the copy numbers estimated from the genome data. The gene family encoding the key C4 enzyme phosphoenolpyruvate carboxylase (ppc genes) was selected for qPCR analyses since it included genes encompassing a wide range of copy numbers according to the read depth estimates (see the Results). Three paralogues (ppc_1P3, ppc_1P6, and ppc_1P7) were analysed in six individuals of A. semialata from a wide geographic and phylogenetic sampling (BUR1, RSA2, TAN2, TAN1, MAD1, and TPE1).
Alignments consisting of partial gene models of ppc groups of co-orthologues were assembled for Alloteropsis species using a genome-walking approach to include intron sequences, and were used as reference for primer design. Two pairs of primers per paralogue were designed to amplify 92–161 bp regions that include exon and intron sequences (except for one pair for ppc_1P7, which encompassed only exon sequences; Supplementary Table S1). The copy number estimated via qPCR consequently captured only putative duplications of genomic DNA, and excluded potential retroposition instances (Zhang, 2003; Kaessmann et al., 2009; Reams and Roth, 2015). To perform the assays, genomic DNA (gDNA) was isolated from fresh leaves of A. semialata individuals using the DNeasy Plant Kit (Qiagen), following the manufacturer’s instructions. SYBR green-based qPCRs were prepared using 1× Power SYBR green PCR Master Mix (Thermo Fisher Scientific), 0.25 µM of each primer, and 6.25 ng of gDNA in a total volume of 20 µl, with three technical replicates and non-template controls per reaction. Assays were carried out on a QuantStudio 12K Flex Real Time PCR instrument (Life Technologies) with an initial incubation of 10 min at 95 °C (Taq activation), followed by 40 cycles of 15 s at 95 °C (denaturation) and 60 s at 60 °C (annealing and extension). Amplification specificity was assessed via melting curves generated immediately after each assay, in which samples were incubated for 15 s at 95 °C and 60 s at 60 °C, followed by incremental temperature increases of 0.3 °C up to 95 °C. The melting temperature of the amplified fragments was then calculated based on their expected sequences and compared with the peak temperature values obtained from the melting curve assays. Baseline, threshold cycle, and PCR efficiency were determined using the LinRegPCR software v. 2016.0 (Ramakers et al., 2003). Samples with PCR efficiency <1.85 or >2.1 were excluded from the subsequent analysis. The Pfaffl method (Pfaffl, 2001) was used to correct for different PCR efficiencies across amplicon groups, and copy numbers of ppc genes were expressed relative to the mean of the two pairs of primers used for the ppc_1P7 gene.
Phylogenetic analyses of duplicated genes
To determine whether duplications of ppc and pck (see the Results) occurred before or after the diversification of A. semialata lineages, we assembled partial allele models by manually phasing polymorphisms using paired-end information. Ambiguous nucleotides were called for polymorphisms that could not be phased. Alleles of TPE1 and TAN2 were assembled using the high-coverage data, while raw transcriptome data of the genus Alloteropsis retrieved from Dunning et al. (2017) were used for the other accessions. Sequences were aligned using MAFFT v7.130b (Katoh and Standley, 2013), and phylogenetic trees were inferred using PhyML (Guindon and Gascuel, 2003) under a GTR+G model of nucleotide substitution, with 100 bootstrap pseudoreplicates.
Allele-specific expression analyses
The relative contribution of each allele/paralogue of pck and ppc to the overall transcript abundance was assessed and compared with their relative frequency in the genomes through the analysis of SNPs. Reads from the genome and transcriptome data sets were mapped to reference alignments of the ppc and pck gene families, and the read depth was determined for each SNP of each gene using Geneious v. 6.8 (Kearse et al., 2012). For each SNP, the abundance of the minor allele (defined on the transcriptome data as the variant base receiving fewer reads) was calculated as a proportion of the total read count for that site, for both transcriptome and genome data. Because the genomic frequency can vary among SNPs for multicopy genes (i.e. each variant can be present in any number of alleles up to twice the number of copies in a diploid individual), the contribution of different alleles to transcript abundance was evaluated via frequency correlations between transcriptome and genome data sets. Note that the polyploid individual was excluded from these analyses because of insufficient coverage to assess accurately polymorphisms among its high number of alleles.
Association between changes in copy number and transcript abundance
To test for an association between changes in copy number and changes in gene expression, transcript abundances in leaves were retrieved for 14 C4-related genes captured in a study of transcriptomes of the genus Alloteropsis grown in controlled conditions (Dunning et al., 2017). The average abundance between two biological replicates in reads per kilobase per million mapped reads (RPKM) is used here. Values were log10 transformed before analysis to homogenize variances. Accessions were considered for this analysis only if genome and transcriptome data were available for the same individual, or individuals from the same population, except in two cases (representing A. cimicina and the C3A. semialata) for which genome and transcriptome data were available for closely related individuals from different populations (Lundgren et al., 2015; Olofsson et al., 2016). Note that excluding these two individuals did not significantly alter the results. High-coverage sequence data were not used here to avoid pseudoreplication of some populations.
Homologous genes within a gene family do not represent independent data points as they result from events of gene duplication and/or speciation from a common ancestor. We consequently used phylogenetic generalized least squares (PGLS) under a Brownian model of evolution to test for correlated changes between gene copy number and transcript abundance using the R packages nlme and APE (Paradis et al., 2004). A Bonferroni correction was used to adjust significance levels for multiple testing. The sequence alignment of the respective gene family was extracted from the genome-wide data set generated from transcriptomes (see above), and the accessions with no associated genome data were removed. Bayesian trees were inferred from this alignment under a GTR+G+I substitution model using MrBayes v3.2.2 (Ronquist et al., 2012), with two parallel analyses running for 10000000 generations. After verifying the convergence of the runs, a consensus tree was generated using trees sampled after a burn-in period of 50%. The effect of topological uncertainty on the PGLS results was assessed by repeating the analysis using 100 independent trees sampled every 50000 generations after the burn-in period.
Results
Background distribution of gene copy numbers
Copy numbers were estimated for markers sampled across the genome for each accession, providing a background distribution of copy numbers per haploid chromosome set (Supplementary Fig. S2). Most genes were estimated as single copy, and the proportion of duplicated genes ranged from 9% to 28% across accessions, with 0.5–1.3% genes being absent (Table 2). The same copy numbers were estimated among individuals belonging to the same nuclear group, as previously defined in A. semialata (Olofsson et al., 2016), for 82% of the genes, on average. Although there was a weak positive correlation between coverage and the proportion of absent genes (R2=0.34, P=0.055), no significant association was found between coverage and the proportion of single-copy (R2=0, P=0.41) or duplicated genes (R2=0, P=0.53), which suggests that the inferred duplications reflect biological rather than methodological differences. Similar estimates were found moreover between individuals from the same population based on low- and high-coverage data sets (Supplementary Fig. S3), indicating that low-coverage sequencing provides an accurate assessment of gene copy number variation. The variation in genome size (Table 1) was not explained by differences in gene copy number, with correlations being non-significant for the proportion of both absent and duplicated genes.
Table 2.
Background distribution of gene copy numbers in Alloteropsis accessions
| Accession | Species | Metabolism | Total genes analyseda |
Proportions (%)b | ||
|---|---|---|---|---|---|---|
| Single-copy | Duplicated | Absent | ||||
| Cim1 | A. cimicina | C4 | 12057 | 89.4 (88.2–90.6) |
9.8 (8.6–11) |
0.9 (0.8–0.9) |
| Ang1 | A. angusta | C4 | 8966 | 83.9 (81.4–86.9) |
14.8 (11.7–17.4) |
1.2 (1.2–1.4) |
| Ang2 | A. angusta | C4 | 9700 | 84.2 (81.6–85.8) |
14.5 (12.8–17.1) |
1.3 (1.3–1.4) |
| RSA1 | A. semialata | C3 | 8935 | 83.8 (81.7–85.9) |
15.5 (13.4–17.6) |
0.7 (0.7–0.8) |
| RSA2 | A. semialata | C3 | 6996 | 86.4 (84.9–88) |
13.1 (11.3–14.6) |
0.5 (0.5–0.7) |
| TAN1 | A. semialata | C3+C4 | 11376 | 88.4 (87.5–89.3) |
10.8 (9.9–11.6) |
0.8 (0.8–0.9) |
| TAN2 | A. semialata | C3+C4 | 11221 | 86.1 (85.3–87.2) |
13.2 (12.1–14.1) |
0.7 (0.7–0.7) |
| TAN3 | A. semialata | C3+C4 | 12195 | 79.5 (77.7–82.2) |
19.9 (17.2–21.7) |
0.6 (0.6–0.6) |
| DRC1 | A. semialata | C4 | 12162 | 79 (76.4–81.3) |
20.4 (18.1–23) |
0.6 (0.6–0.6) |
| DRC2 | A. semialata | C4 | 11946 | 81.1 (78.5–83.1) |
18.3 (16.3–20.9) |
0.6 (0.6–0.6) |
| DRC3 | A. semialata | C4 | 11941 | 78.3 (75.3–80.7) |
21 (18.6–24) |
0.7 (0.7–0.7) |
| DRC4 | A. semialata | C4 | 11014 | 81.4 (79.1–83.9) |
17.9 (15.4–20.2) |
0.7 (0.6–0.7) |
| TAN4 | A. semialata | C4 | 11214 | 86.6 (85.6–87.3) |
12.6 (11.8–13.6) |
0.8 (0.8–0.8) |
| RSA3 | A. semialata | C4 | 10248 | 88.1 (86.3–89.4) |
11.2 (9.9–13.1) |
0.6 (0.6–0.7) |
| KEN1 | A. semialata | C4 | 10381 | 70.6 (64.1–76.5) |
28.4 (22.5–35) |
1 (1–1) |
| BUR1 | A. semialata | C4 | 9448 | 88.4 (87.4–89.5) |
10.9 (9.7–11.9) |
0.7 (0.7–0.8) |
| MAD1 | A. semialata | C4 | 10226 | 88.1 (86.7–89.1) |
11.2 (10.2–12.6) |
0.7 (0.7–0.7) |
| THA1 | A. semialata | C4 | 10926 | 87.5 (86–88.6) |
11.7 (10.6–13.3) |
0.8 (0.7–0.8) |
| TPE1 | A. semialata | C4 | 10730 | 88.5 (87.5–89.3) |
10.7 (9.9–11.7) |
0.8 (0.7–0.8) |
| AUS1 | A. semialata | C4 | 7174 | 88.3 (87–89.7) |
11 (9.6–12.3) |
0.7 (0.6–0.7) |
a After removing genes having confidence intervals for the expected read counts that included zero, and/or read counts between 1 and the lower limit of the confidence interval (see the Materials and methods).
b Percentage of single-copy, duplicated, or absent genes relative to the total number of genes analysed. Values are medians calculated from the resampling procedure used for the GC content correction, with the minimum and maximum values shown in parentheses.
Duplications of C4 protein-coding genes
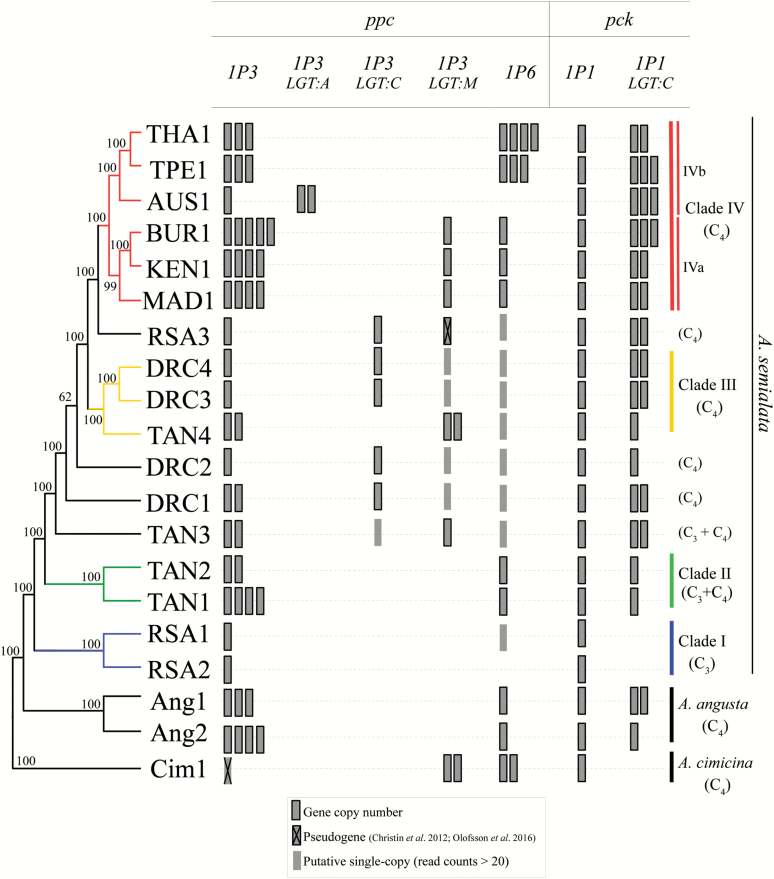
We estimated copy numbers for a total of 82 genes belonging to 23 gene families with some gene lineages encoding proteins known to be involved in the C4 pathway of some species. For 45 of these genes belonging to 19 families, at least one duplication was observed in the genus Alloteropsis (Supplementary Table S2). Putative ancient duplications (shared by A. semialata, A. angusta, and A. cimicina) include those for pyruvate kinase (pk_1P1) and NADP-dependent malic enzyme (nadpme_1P4). A number of genes have incurred independent duplications and/or secondary losses within A. semialata and A. angusta, including those for a tonoplast malate/fumarate transporter (tdt_1P2), in addition to those encoding phosphoenolpyruvate carboxylase (ppc_1P3) and phosphoenolpyruvate carboxykinase (pck_1P1_LGT:C). The pck_1P1_LGT:C gene was laterally acquired after the split between the C3 lineage and the lineage including C3+C4 and C4A. semialata, which now use it as part of their C4 cycle (Olofsson et al., 2016; Dunning et al., 2017), and subsequently duplicated only in the C4 group (Fig. 2). The ppc gene family has a particularly high diversity of copy numbers, which is especially marked for ppc_1P3 and ppc_1P6, both of which are used for the C4 cycle of some accessions (Dunning et al., 2017).
Fig. 2.
Copy number variation of selected genes of phosphoenolpyruvate carboxylase (ppc) and phosphoenolpyruvate carboxykinase (pck) in the Alloteropsis genus. LGT:A, C, and M are laterally acquired genes (Christin et al., 2012). Nuclear phylogeny of the Alloteropsis genus was modified from Olofsson et al. (2016), with bootstrap support values shown near nodes, and lineages indicated on the right. Copy number estimates are based on low-coverage genome data, and are rounded to the nearest integer.
The phylogenetic distribution of duplicates could be explained by different combinations of duplications and secondary gene losses (Fig. 2), but these scenarios can be distinguished based on gene trees. The multiple copies of pck_1P1_LGT:C retrieved from the C4A. semialata form a monophyletic clade, which is split into subgroups corresponding to African and Asian/Australian accessions (Supplementary Fig. S4). This pattern could be explained by independent duplications in each of the two groups or a duplication at their base followed by recombination or concerted evolution within each of the groups. The multiple copies of ppc_1P6 specific to TPE1 (and THA1; Fig. 2), which is the only accession to use this gene for its C4 pathway (Dunning et al., 2017), are very similar and cluster in the phylogeny (Supplementary Fig. S5), which supports the hypothesis of very recent duplications. The multiple ppc_1P3 copies of the C3+C4 and C4A. semialata form distinct, well-supported monophyletic groups and, within the C4 group, copies from the same accession tend to cluster despite a lack of resolution in some parts of the tree (Supplementary Fig. S6). This, again, suggests either independent duplications or concerted evolution following early duplications. Secondary losses of extra copies of ppc_1P3 and the complete loss of ppc_1P6 are inferred in the Australian accession (AUS1), which is the only accession carrying one of the laterally acquired ppc genes (ppc_1P3_LGT:A;Fig. 2).
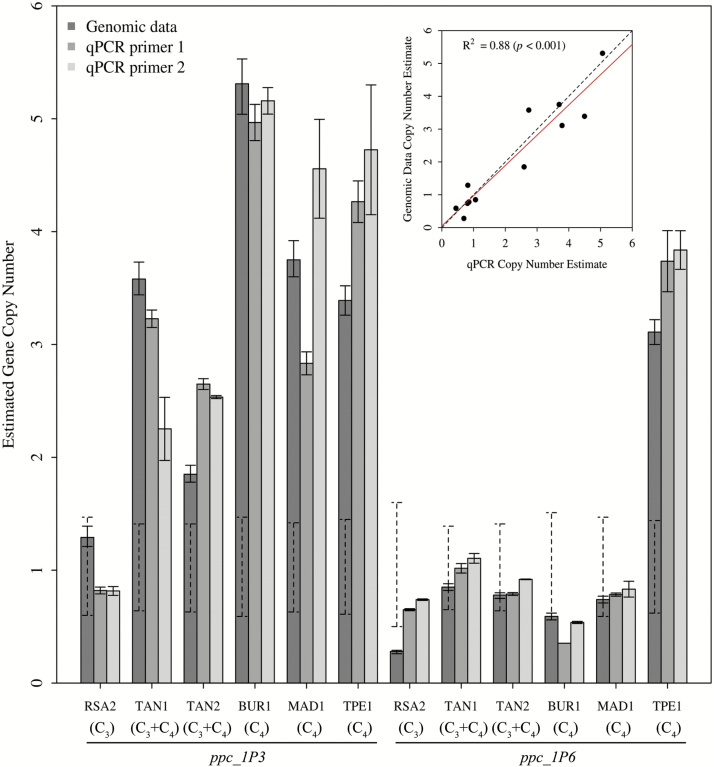
The copy numbers estimated for ppc_1P3 and ppc_1P6 from the genome data were significantly correlated with those estimated by qPCR (R2=0.88, P<0.001; Fig. 3). Since intronic regions were amplified in both pairs of primers used for the qPCR analysis, we conclude that the observed duplications correspond to duplications of genomic DNA. Differences in copy number of ppc_1P3 between different primer pairs may be explained by the existence of a polymorphism in a region amplified by one of the primers, which would prevent the amplification of one of the alleles. Analyses of sequence alignments confirmed this was the case for at least one individual (MAD1). Alternatively, it is also possible that in other accessions some of the duplicates are present as partial copies originating from illegitimate recombination.
Fig. 3.
Comparison of copy number estimates obtained from qPCR assays and from low-coverage genomic data for the genes ppc_1P3 and ppc_1P6 in six A. semialata accessions. Copy numbers are expressed relative to the ppc_1P7 gene. Error bars are SEs from 2–3 technical replicates for qPCR estimates, and non-parametric error estimates from the GC correction resampling procedure for the genomic estimates of copy number. Dashed lines on the genomic estimates indicate confidence intervals for single-copy genes. The upper panel indicates the correlation between qPCR estimates (mean value of both pairs of primers) and genomic estimates of copy number for ppc_1P3 and ppc_1P6, with the red line being the regression line and the dashed line the identity line.
Increases in transcript abundance associated with lineage-specific duplications
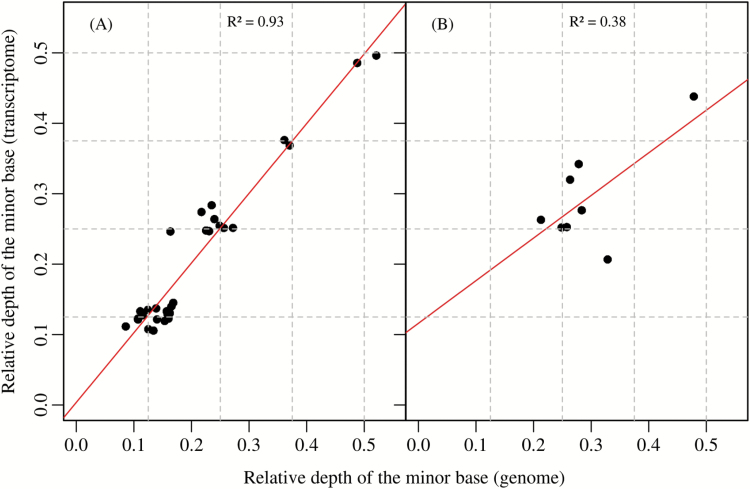
Our analyses of C4-related genes revealed remarkable variation in copy number of ppc and pck among Alloteropsis lineages. For each polymorphic site, the frequency of the minor variant was strongly correlated between high-coverage genome and transcriptome data sets across the eight copies of ppc_1P6 identified in TPE1 by the qPCR analysis (R2=0.93, P<0.001; Fig. 4). While the correlation between transcriptome and genome sequencing was also observed for ppc_1P3 of TPE1, it was weaker (R2=0.38, P=0.06; Fig. 4; Supplementary Table S4), which might stem from lower overall transcript abundance and a small number of SNPs increasing statistical noise, or variation in the transcript contribution of different copies. The association between genome and transcriptome SNP frequencies varied among the other samples (Supplementary Table S4), which reflects a combination of low genome coverage of individual variants, variants not shared among the individuals used for genome and transcriptome sequencing, and biased transcriptome contribution of different copies. Nonetheless, the analyses of ppc_1P3 and pck_1P1_LGT:C genes clearly show that multiple copies are expressed at consequent levels in the C3+C4 and C4 accessions, contributing to the elevated overall transcript levels of these genes in the C3+C4 and C4A. semialata (Supplementary Table S3; Dunning et al., 2017). Overall, the SNP analyses provide strong support for duplicates being equally expressed in some accessions (e.g. ppc_1P6 of TPE1), and show a widespread contribution of multiple copies to the elevated transcript abundance of ppc and pck genes.
Fig. 4.
Relative read depth of variants detected at polymorphic sites of (A) ppc_1P6 and (B) ppc_1P3 genes in the genome and transcriptome of a C4 individual of A. semialata (TPE1). Each data point is a polymorphic site and is expressed as the depth of the minor base relative to the total depth for that site. The red line is the linear regression between transcriptome and genome data . The data points cluster around frequencies of 0.125, 0.25, 0.375, and 0.5, as indicated by dashed grid lines, which correspond to one, two, three, and four alleles out of a total of eight alleles from four duplicates.
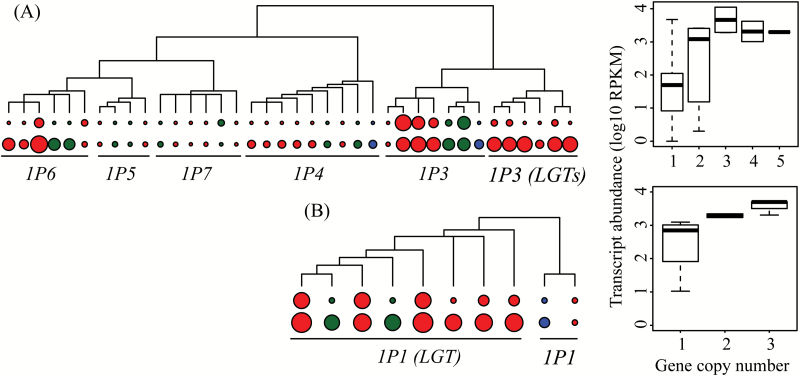
Finally, we tested whether the observed changes in copy number were statistically associated with changes in transcript abundance during the evolutionary diversification of the genus Alloteropsis. The conclusions of the statistical tests are robust to topological uncertainty (Supplementary Table S5), and we therefore discuss here only the results of the PGLS analyses based on the consensus tree (Table 3). Out of the 14 C4-related gene families for which transcript abundance was available in Dunning et al. (2017), 10 showed copy number variation among the accessions used for this analysis. We found a consistent positive association between changes in copy number and changes in transcript abundance that was significant after correction for multiple testing in two of them, ppc (P<0.001) and pck (P=0.002; Table 3; Fig. 5; Supplementary Fig. S7). In the case of ppc, these effects were mainly driven by a few copy number changes in ppc_1P3 and ppc_1P6 (Fig. 5A), which, along with the laterally acquired ppc genes (ppc_1P3_LGT:A, ppc_1P3_LGT:M, and ppc_1P3_LGT:C), are the most highly expressed copies of this gene family in the C4 accessions of the Alloteropsis genus (Dunning et al., 2017). For pck, the duplication of pck_1P1_LGT:C after the split between the C3+C4 and C4 lineages was tightly associated with increases in transcript abundance of this gene (Fig. 5B). Although the other eight families include, in some cases, genes varying in copy number and transcript abundance, the statistical association was not significant after taking the phylogeny into account. In addition, analyses of rbcS showed a decrease in abundance in C3+C4 and C4 accessions, which was associated with increases in gene copy numbers, highlighting processes other than dosage effects during the diversification of this gene family in terms of copy number and transcript abundance (Supplementary Fig. S8).
Table 3.
Association between changes in gene copy number and changes in transcript abundance of C4-related gene families in Alloteropsis
| Gene family |
Copy number
range |
Transcript abundance
range a |
P-value b |
|---|---|---|---|
| Alanine aminotransferase (ALA-AT) | 1–2 | 0–1838 | 0.08 |
| Aspartate aminotransferase (ASP-AT) | 1–2 | 9–2632 | 0.48 |
| Carbonic anhydrase (CA) | 1–3 | 3–13169 | 0.46 |
| Dicarboxylate transporter (DIT) | 1 | 0–342 | – |
| NAD-malate dehydrogenase (NAD-MDH) | 1–4 | 21–1528 | 0.11 |
| NAD-malic enzyme (NAD-ME) | 1–2 | 12–162 | 0.57 |
| NADP-malate dehydrogenase (NADP-MDH) | 1 | 15–3537 | – |
| NADP-malic enzyme (NADP-ME) | 1–3 | 0–5746 | 0.56 |
| PEP carboxykinase (PCK) | 1–3 | 11–5187 | 0.002 |
| PEP carboxylase (PEPC) | 1–5 | 0–11153 | < 0.001 |
| Pyruvate phosphate dikinase (PPDK) | 1–2 | 0–12796 | 0.82 |
| PEP-phosphate translocator (PPT) | 1–2 | 19–2593 | 0.62 |
| Sodium bile acid symporter (SBAS) | 1 | 17–7105 | – |
| Triosephosphate-phosphate translocator (TPT) | 1–2 | 8–3213 | – |
a In RPKM; retrieved from Dunning et al. (2017);
b P-values were obtained using a phylogenetic generalized least squares (PGLS) fitting under a Brownian model of character evolution; gene families lacking P-values do not show copy number variation, or contain representatives with no gene sequence available for the phylogenetic analysis.
Fig. 5.
Association between changes in gene copy number and transcript abundance for (A) phosphoenolpyruvate carboxylase (ppc) and (B) phosphoenolpyruvate carboxykinase (pck). For each gene in each accession, circles next to the tips of the gene phylogeny are proportional to the estimated gene copy number (top) and transcript abundance (log10 RPKM; bottom). Circles are coloured according to the photosynthetic type (blue=C3, green=C3+C4, red=C4). The boxplots on the right show the distribution of transcript abundances per class of copy numbers for each gene family.
Discussion
Recent gene duplications linked to physiological innovation via potential dosage effects
In this study, we used genome analyses to show that genes for ppc and pck recurrently increased in numbers during the evolution of C4 photosynthesis in the genus Alloteropsis (Fig. 2). These genes encode some of the few enzymes that reach very high levels in the C3+C4 and C4A. semialata (Ueno and Sentoku, 2006; Lundgren et al., 2016; Dunning et al., 2017), and increases in copy numbers statistically coincided with enhanced transcript abundance (Table 3; Fig. 5). One potential explanation for this pattern is that increased gene expression and high transcript abundance favoured frequent retroposition; that is, high transcription caused gene duplication (Kaessmann et al., 2009). However, if this were the case, we would expect that increased copy number would uniquely involve exon sequences, which is disproved by our qPCR results. Analyses of polymorphisms further demonstrate that the multiple copies contribute to the overall high transcript abundances, with at least in some cases an equal contribution from each copy (Fig. 4). We therefore conclude that duplication of genomic DNA directly contributed to the expression levels of these genes, via dosage effects. Modifications of the regulatory mechanisms during the diversification of land plants and grasses are probably responsible for the variation of transcript abundance observed among single-copy gene lineages, and recent duplications would then have quickly enhanced the transcript level associated with some of the ancestral gene (Fig. 5), which can reach consequent levels in the non-C4 ancestors (Moreno-Villena et al., 2018). Evidence for this mechanism was obtained here for only two genes, which encode proteins that are responsible for the initial fixation of atmospheric carbon into organic compounds and the release of CO2 to feed the C4 cycle, respectively. Three other enzymes show marked increases in transcript abundance in the C3+C4 and/or C4A. semialata (Dunning et al., 2017), without evidence of gene copy number increases (Table 3). Unsurprisingly, the proposed dosage effect therefore concerns only a subset of the C4 genes, but it probably played a key role first in the emergence of a weak C4 cycle in the C3+C4 accessions, and then in the strengthening of this cycle in the C4 accessions, which is predicted to impact positively on fitness (Heckmann et al., 2013; Mallmann et al., 2014; Bräutigam and Gowik, 2016). Our results therefore suggest that dosage effects contributed to physiological innovation in the studied taxa, in association with changes in the regulatory properties of genes encoding other enzymes.
Establishing the context of the duplications behind these increased copy numbers would require assembled genomes, but could involve unequal crossing over, chromosomal duplication, or the action of transposable elements (Zhang, 2003; Reams and Roth, 2015). Using high-coverage sequencing from genomic DNA or transcriptomes, we were able to assemble multiple copies of some ppc and pck genes in diploid accessions of A. semialata. While phylogenetic trees supported early duplications in some cases, the copies tended to group per accessions (Supplementary Figs S4–S6). The number of assembled copies was moreover below that estimated based on sequencing depth, suggesting that identical alleles exist. These patterns could be explained by recurrent gene duplications during the history of the Alloteropsis genus, or recombination, for example among tandem duplicates, leading to concerted evolution homogenizing the duplicated copies within geographically isolated lineages (Brown et al., 1972; Nei and Rooney, 2005).
Duplicates get lost after the acquisition of better-suited copies
At least three events of lateral gene transfers (LGTs) of ppc and one of pck occurred in the Alloteropsis genus (Christin et al., 2012; Olofsson et al., 2016), and some of the laterally acquired genes are expressed at high levels in the transcriptome of the accessions carrying such genes (Dunning et al., 2017). In most of these accessions, the vertically inherited copies of ppc and pck are strongly down-regulated, or not expressed at all (Dunning et al., 2017). Apart from the Southeast Asian clade, all C4 accessions of A. semialata studied here carry at least one laterally acquired ppc gene in their genomes. Interestingly, in this exception, multiple duplications of ppc_1P6 were retained and are associated with drastic changes in transcript abundance that are specific to this clade (Fig. 5). On the other hand, the presence of some LGT copies (ppc_1P3_LGT:C and ppc_1P3_LGT:A) coincides with the loss of the initial duplicates of the vertically inherited ppc_1P3 gene (Fig. 2; Olofsson et al., 2016). These findings indicate that, once a gene better suited for the C4 function is acquired, the selective pressure on the original copy is relaxed, leading over time to pseudogenization and/or gene loss.
With multiple copies of genes related to C4 metabolism, the chances that some of these copies will acquire C4 adaptive mutations increase. Our analyses indeed identified non-synonymous polymorphisms among multiple copies of some genes. In four cases, such substitutions on ppc generate amino acid changes that were recurrently selected in a number of other C4 grasses, suggesting that they adapt the protein for the C4 catalytic context (Christin et al., 2007). While not detectable with our approach, regulatory mutations, identified for other C4 groups (e.g. Gowik et al., 2004; Akyildiz et al., 2007), might similarly be present in only some of the multiple copies reported here. Genes that do not have the adaptive mutations can be lost via negative selection or drift, and those with the beneficial mutations are retained, leading to typical neofunctionalization. As reported here, the acquisition of more suitable gene versions, illustrated by the LGTs, can indeed relax the selection over duplicated copies that were once preserved via dosage selection, but from there on will be subjected to pseudogenization or eventually neofunctionalization. This suggests that during the course of evolution, fewer, more optimized genes are likely to remain, which would explain why more established C4 lineages are not enriched in C4-related genes (Williams et al., 2012; van den Bergh et al., 2014). The presence of multiple gene copies therefore probably contributes to the emergence of C4 photosynthesis via a combination of dosage effects and increased opportunities for neofunctionalization, both of which are evolutionarily transient.
Low-coverage sequencing correctly identified duplicates
Low-coverage genomic data sets are increasingly used for a wide range of population genomic (Buerkle and Gompert, 2013; Nicod et al., 2016; Olofsson et al., 2016) and phylogenetic studies (Bock et al., 2014; Dodsworth, 2015; Washburn et al., 2015). While such data sets are relatively cheap to obtain and can be generated from poorly conserved samples such as those from museum collections (Besnard et al., 2014; Silva et al., 2017), they come with their limitations. In particular, sequencing biases are inherent to the PCR steps involved in the sample preparation, and lead to over-representation of regions with specific GC contents (Benjamini and Speed, 2012; Ross et al., 2013; see the Materials and methods). It is therefore necessary to validate the results with independent evidence, provided here by qPCR. Slight variation between qPCR estimates and those based on low-coverage data confirmed that copy numbers inferred from read depths are in some cases under- or overestimated, as expected given both the low coverage and the difficulty in precisely correcting for the sequencing bias. However, the general patterns are correctly identified, as indicated by the similarity of estimates among closely related accessions, and by the strong agreement in the estimates based on low- and high-coverage data sets in cases where both were available for individuals from the same population (Supplementary Fig. S3). In addition, individual events of gene duplication inferred from low-coverage data are qualitatively correct, being in all cases confirmed by independent qPCR.
The intersection of different lines of evidence shows that our approach represents a valid strategy to infer patterns of copy number variation for a large number of non-model species. Some of the genomic data sets included here come from samples only available in herbarium collections, which were collected up to 60 years ago (Olofsson et al., 2016). In cases where living material is not available, low-coverage sequencing represents a valuable resource to shed light on not only the phylogenetic relationships, but also the genomic content of important taxa (Besnard et al., 2014), and, as shown here, variation in gene copy number. In the near future, the increasing availability of sequencing data sets for non-model species will offer multiple opportunities to track the genomic dynamics underlying a large array of physiological adaptations in a variety of taxa.
Conclusion
Using comparative genomics, we showed that the duplication of genes encoding two key enzymes required for C4 photosynthesis coincided with the co-option of these genes for the new metabolic pathway. Based on published transcriptome data, we propose that changes in copy number altered the expression levels via pure dosage effects, with duplication events representing major effect mutations that can rapidly double transcription levels of some genes, which might have contributed to the emergence of a weak C4 cycle in some plants. Once the C4 cycle was in place, selection could act to optimize it, which probably involved fixing beneficial mutations on individual genes, including substitutions and indels in both regulatory and coding sequences. The selection of better-suited isoforms apparently led to pseudogenization of the previous duplicates. We therefore suggest that gene copy number decreases as beneficial mutations in the promoter or coding sequences are fixed, in a process of neofunctionalization. The beneficial effects of gene duplication for physiological innovation are therefore likely to be transitory, with no footprint on longer evolutionary scales.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Relationship between length-normalized read count and GC content in the genomic data sets of accessions of the genus Alloteropsis.
Fig. S2. Background gene copy number distribution in accessions of the genus Alloteropsis. Copy numbers are expressed as observed read count divided by expected read count.
Fig. S3. Comparison between copy number estimates using high- and low-coverage data sets for individuals within the same population.
Fig. S4. Phylogenetic tree of pck genes in the genus Alloteropsis.
Fig. S5. Phylogenetic tree of ppc_1P6 genes in the genus Alloteropsis.
Fig. S6. Phylogenetic tree of ppc_1P3 genes in the genus Alloteropsis.
Fig. S7. Distribution of transcript abundance among classes of gene copy numbers for 12 C4-related gene families.
Fig. S8. Distribution of transcript abundance among classes of copy numbers for genes encoding the small unit of Rubisco (rbcS).
Table S1. List of primer sequences of ppc genes used for quantitative real-time PCR assays.
Table S2. List of duplicated genes of C4-related gene families within the genus Alloteropsis.
Table S3. Read depth of transcriptome and genome data for polymorphic sites of ppc and pck genes of accessions of the genus Alloteropsis.
Table S4. Association between read depth of transcriptome and genome data for polymorphic sites of ppc and pck genes of Alloteropsis accessions.
Table S5. Effect of phylogenetic tree on the phylogenetic generalized least squares (PGLS) analysis used to test for an association between changes in gene copy number and changes in transcript abundance.
Text S1. Mitochondrial genome contigs of Alloteropsis semialata (accession MAD1).
Acknowledgements
We thank Ilia Leitch and Oriane Hidalgo for comments on genome size results. MEB is supported by the Brazilian Research Council (CNPq) through a ‘Science without Borders’ scholarship (grant no. 201873/2014-1), LTD by an NERC grant (grant no. NE/M00208X/1), JJMV by a Royal Society Research Grant (grant no. RG130448), and PAC by a Royal Society University Research Fellowship (grant no. URF120119). The laboratory work was supported by the UK Natural Environment Research Council (NERC) Biomolecular Analysis Facility at the University of Sheffield. Library preparation and sequencing were carried out by Edinburgh Genomics, The University of Edinburgh. Edinburgh Genomics is partly supported through core grants from the NERC (R8/H10/56), MRC (MR/K001744/1), and BBSRC (BB/J004243/1).
References
- Aird D, Ross MG, Chen WS, Danielsson M, Fennell T, Russ C, Jaffe DB, Nusbaum C, Gnirke A. 2011. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biology 12, R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akyildiz M, Gowik U, Engelmann S, Koczor M, Streubel M, Westhoff P. 2007. Evolution and function of a cis-regulatory module for mesophyll-specific gene expression in the C4 dicot Flaveria trinervia. The Plant Cell 19, 3391–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Kidd JM, Marques-Bonet T, et al. 2009. Personalized copy number and segmental duplication maps using next-generation sequencing. Nature Genetics 41, 1061–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubry S, Brown NJ, Hibberd JM. 2011. The role of proteins in C3 plants prior to their recruitment into the C4 pathway. Journal of Experimental Botany 62, 3049–3059. [DOI] [PubMed] [Google Scholar]
- Bellos E, Johnson MR, Coin LJ. 2012. cnvHiTSeq: integrative models for high-resolution copy number variation detection and genotyping using population sequencing data. Genome Biology 13, R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Speed TP. 2012. Summarizing and correcting the GC content bias in high-throughput sequencing. Nucleic Acids Research 40, e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard G, Christin PA, Malé PJ, Lhuillier E, Lauzeral C, Coissac E, Vorontsova MS. 2014. From museums to genomics: old herbarium specimens shed light on a C3 to C4 transition. Journal of Experimental Botany 65, 6711–6721. [DOI] [PubMed] [Google Scholar]
- Bläsing OE, Westhoff P, Svensson P. 2000. Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyl-terminal part of the enzyme is a major determinant for C4-specific characteristics. Journal of Biological Chemistry 275, 27917–27923. [DOI] [PubMed] [Google Scholar]
- Bock DG, Kane NC, Ebert DP, Rieseberg LH. 2014. Genome skimming reveals the origin of the Jerusalem artichoke tuber crop species: neither from Jerusalem nor an artichoke. New Phytologist 201, 1021–1030. [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Gowik U. 2016. Photorespiration connects C3 and C4 photosynthesis. Journal of Experimental Botany 67, 2953–2962. [DOI] [PubMed] [Google Scholar]
- Bräutigam A, Kajala K, Wullenweber J, et al. 2011. An mRNA blueprint for C4 photosynthesis derived from comparative transcriptomics of closely related C3 and C4 species. Plant Physiology 155, 142–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Wensink PC, Jordan E. 1972. A comparison of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. Journal of Molecular Biology 63, 57–73. [DOI] [PubMed] [Google Scholar]
- Buerkle AC, Gompert Z. 2013. Population genomics based on low coverage sequencing: how low should we go?Molecular Ecology 22, 3028–3035. [DOI] [PubMed] [Google Scholar]
- Chen Z, Cheng CHC, Zhang J, et al. 2008. Transcriptomic and genomic evolution under constant cold in Antarctic notothenioid fish. Proceedings of the National Academy of Sciences, USA 105, 12944–12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Arakaki M, Osborne CP, Edwards EJ. 2015. Genetic enablers underlying the clustered evolutionary origins of C4 photosynthesis in angiosperms. Molecular Biology and Evolution 32, 846–858. [DOI] [PubMed] [Google Scholar]
- Christin PA, Boxall SF, Gregory R, Edwards EJ, Hartwell J, Osborne CP. 2013. Parallel recruitment of multiple genes into C4 photosynthesis. Genome Biology and Evolution 5, 2174–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christin PA, Edwards EJ, Besnard G, Boxall SF, Gregory R, Kellogg EA, Hartwell J, Osborne CP. 2012. Adaptive evolution of C4 photosynthesis through recurrent lateral gene transfer. Current Biology 22, 445–449. [DOI] [PubMed] [Google Scholar]
- Christin PA, Osborne CP. 2014. The evolutionary ecology of C4 plants. New Phytologist 204, 765–781. [DOI] [PubMed] [Google Scholar]
- Christin PA, Petitpierre B, Salamin N, Büchi L, Besnard G. 2009. Evolution of C4 phosphoenolpyruvate carboxykinase in grasses, from genotype to phenotype. Molecular Biology and Evolution 26, 357–365. [DOI] [PubMed] [Google Scholar]
- Christin PA, Salamin N, Savolainen V, Duvall MR, Besnard G. 2007. C4 photosynthesis evolved in grasses via parallel adaptive genetic changes. Current Biology 17, 1241–1247. [DOI] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC. 2014. Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Current Opinion in Plant Biology 19, 91–98. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nature Reviews Genetics 9, 938–950. [DOI] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, et al. 2012. Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338, 1206–1209. [DOI] [PubMed] [Google Scholar]
- Dodsworth S. 2015. Genome skimming for next-generation biodiversity analysis. Trends in Plant Science 20, 525–527. [DOI] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H. 2008. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Research 36, e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning LT, Lundgren MR, Moreno-Villena JJ, Namaganda M, Edwards EJ, Nosil P, Osborne CP, Christin PA. 2017. Introgression and repeated co-option facilitated the recurrent emergence of C4 photosynthesis among close relatives. Evolution 71, 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards EJ, Ku MB. 1987. Biochemistry of C3–C4 intermediates. In: Hatch MD, Boardman NK, eds. The biochemistry of plants: a comprehensive treatise, Vol 14: photosynthesis. New York: Academic Press, 275–325. [Google Scholar]
- Ellis R. 1974. Significance of the occurrence of both Kranz and non-Kranz leaf anatomy in the grass species Alloteropsis semialata. South African Journal of Science 70, 169–173. [Google Scholar]
- Emms DM, Covshoff S, Hibberd JM, Kelly S. 2016. Independent and parallel evolution of new genes by gene duplication in two origins of C4 photosynthesis provides new insight into the mechanism of phloem loading in C4 species. Molecular Biology and Evolution 33, 1796–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowik U, Burscheidt J, Akyildiz M, Schlue U, Koczor M, Streubel M, Westhoff P. 2004. cis-Regulatory elements for mesophyll-specific gene expression in the C4 plant Flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. The Plant Cell 16, 1077–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Systematic Biology 52, 696–704. [DOI] [PubMed] [Google Scholar]
- Hatch MD. 1987. C4 photosynthesis: a unique blend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta 895, 81–106. [Google Scholar]
- Heckmann D. 2016. C4 photosynthesis evolution: the conditional Mt. Fuji. Current Opinion in Plant Biology 31, 149–154. [DOI] [PubMed] [Google Scholar]
- Heckmann D, Schulze S, Denton A, Gowik U, Westhoff P, Weber AP, Lercher MJ. 2013. Predicting C4 photosynthesis evolution: modular, individually adaptive steps on a Mount Fuji fitness landscape. Cell 153, 1579–1588. [DOI] [PubMed] [Google Scholar]
- Hibberd JM, Covshoff S. 2010. The regulation of gene expression required for C4 photosynthesis. Annual Review of Plant Biology 61, 181–207. [DOI] [PubMed] [Google Scholar]
- Huang P, Studer AJ, Schnable JC, Kellogg EA, Brutnell TP. 2017. Cross species selection scans identify components of C4 photosynthesis in the grasses. Journal of Experimental Botany 68, 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. 2009. RNA-based gene duplication: mechanistic and evolutionary insights. Nature Reviews Genetics 10, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proceedings of the Royal Society B: Biological Sciences 279, 5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA, Rogozin IB, Wolf YI, Koonin EV. 2002. Selection in the evolution of gene duplications. Genome Biology 3, RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren MR, Besnard G, Ripley BS, et al. 2015. Photosynthetic innovation broadens the niche within a single species. Ecology Letters 18, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Lundgren MR, Christin PA, Escobar EG, Ripley BS, Besnard G, Long CM, Hattersley PW, Ellis RP, Leegood RC, Osborne CP. 2016. Evolutionary implications of C3–C4 intermediates in the grass Alloteropsis semialata. Plant, Cell and Environment 39, 1874–1885. [DOI] [PubMed] [Google Scholar]
- Mallmann J, Heckmann D, Bräutigam A, Lercher MJ, Weber AP, Westhoff P, Gowik U. 2014. The role of photorespiration during the evolution of C4 photosynthesis in the genus Flaveria. ELife 3, e02478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson RK. 2003. Gene duplication, neofunctionalization, and the evolution of C4 photosynthesis. International Journal of Plant Sciences 164, S43–S54. [Google Scholar]
- Monson RK. 1999. The origins of C4 genes and evolutionary pattern in the C4 metabolic phenotype. In: Sage RF, Monson RK, eds. C4 plant biology. San Diego: Academic Press, 377–410. [Google Scholar]
- Moreno-Villena JJ, Dunning LT, Osborne CP, Christin PA. 2018. Highly expressed genes are preferentially co-opted for C4 photosynthesis. Molecular Biology and Evolution 35, 94–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchès C, Pasteur N, Bergé JB, Hyrien O, Raymond M, de Saint Vincent BR, de Silvestri M, Georghiou GP. 1986. Amplification of an esterase gene is responsible for insecticide resistance in a California Culex mosquito. Science 233, 778–780. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annual Review of Genetics 39, 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicod J, Davies RW, Cai N, et al. 2016. Genome-wide association of multiple complex traits in outbred mice by ultra-low-coverage sequencing. Nature Genetics 48, 912–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsson JK, Bianconi M, Besnard G, et al. 2016. Genome biogeography reveals the intraspecific spread of adaptive mutations for a complex trait. Molecular Ecology 25, 6107–6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto E, Young JE, Maroni G. 1986. Structure and expression of a tandem duplication of the Drosophila metallothionein gene. Proceedings of the National Academy of Sciences, USA 83, 6025–6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLos One 7, e30619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience Letters 339, 62–66. [DOI] [PubMed] [Google Scholar]
- R Development Core Team 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reams AB, Roth JR. 2015. Mechanisms of gene duplication and amplification. Cold Spring Harbor Perspectives in Biology 7, a016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, Nusbaum C, Jaffe DB. 2013. Characterizing and measuring bias in sequence data. Genome Biology 14, R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage RF. 2004. The evolution of C4 photosynthesis. New Phytologist 161, 341–370. [DOI] [PubMed] [Google Scholar]
- Sage RF, Christin PA, Edwards EJ. 2011. The C4 plant lineages of planet Earth. Journal of Experimental Botany 62, 3155–3169. [DOI] [PubMed] [Google Scholar]
- Silva C, Besnard G, Piot A, Razanatsoa J, Oliveira RP, Vorontsova MS. 2017. Museomics resolve the systematics of an endangered grass lineage endemic to north-western Madagascar. Annals of Botany 119, 339–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tausta SL, Coyle HM, Rothermel B, Stiefel V, Nelson T. 2002. Maize C4 and non-C4 NADP-dependent malic enzymes are encoded by distinct genes derived from a plastid-localized ancestor. Plant Molecular Biology 50, 635–652. [DOI] [PubMed] [Google Scholar]
- Teo SM, Pawitan Y, Ku CS, Chia KS, Salim A. 2012. Statistical challenges associated with detecting copy number variations with next-generation sequencing. Bioinformatics 28, 2711–2718. [DOI] [PubMed] [Google Scholar]
- Ueno O, Sentoku N. 2006. Comparison of leaf structure and photosynthetic characteristics of C3 and C4Alloteropsis semialata subpsecies. Plant, Cell and Environment 29, 257–268. [DOI] [PubMed] [Google Scholar]
- van den Bergh E, Külahoglu C, Bräutigam A, Hibberd JM, Weber APM, Zhu XG, Schranz ME. 2014. Gene and genome duplications and the origin of C4 photosynthesis: birth of a trait in the Cleomaceae. Current Plant Biology 1, 2–9. [Google Scholar]
- Wang X, Gowik U, Tang H, Bowers JE, Westhoff P, Paterson AH. 2009. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biology 10, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn JD, Schnable JC, Davidse G, Pires JC. 2015. Phylogeny and photosynthesis of the grass tribe Paniceae. American Journal of Botany 102, 1493–1505. [DOI] [PubMed] [Google Scholar]
- Williams BP, Aubry S, Hibberd JM. 2012. Molecular evolution of genes recruited into C4 photosynthesis. Trends in Plant Science 17, 213–220. [DOI] [PubMed] [Google Scholar]
- Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. 2009. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Research 19, 1586–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. 2003. Evolution by gene duplication: an update. Trends in Ecology and Evolution 18, 292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.