The stabilization of microtubules by interaction with BRASSINOSTEROID INSENSITIVE2 (BIN2), a negative regulator of brassinosteroid responses, links brassinosteroid signaling to cytoskeletal dynamics and the regulation of plant development.
Keywords: BIN2, brassinosteroids, cytoskeleton, microtubule, pavement cell, tubulin protein
Abstract
Brassinosteroids (BRs), a group of plant steroid hormones, play important roles in regulating plant development. The cytoskeleton also affects key developmental processes and a deficiency in BR biosynthesis or signaling leads to abnormal phenotypes similar to those of microtubule-defective mutants. However, how BRs regulate microtubule and cell morphology remains unknown. Here, using liquid chromatography–tandem mass spectrometry, we identified tubulin proteins that interact with Arabidopsis BRASSINOSTEROID INSENSITIVE2 (BIN2), a negative regulator of BR responses in plants. In vitro and in vivo pull-down assays confirmed that BIN2 interacts with tubulin proteins. High-speed co-sedimentation assays demonstrated that BIN2 also binds microtubules. The Arabidopsis genome also encodes two BIN2 homologs, BIN2-LIKE 1 (BIL1) and BIL2, which function redundantly with BIN2. In the bin2-3 bil1 bil2 triple mutant, cortical microtubules were more sensitive to treatment with the microtubule-disrupting drug oryzalin than in wild-type, whereas in the BIN2 gain-of-function mutant bin2-1, cortical microtubules were insensitive to oryzalin treatment. These results provide important insight into how BR regulates plant pavement cell and leaf growth by mediating the stabilization of microtubules by BIN2.
Introduction
Brassinosteroids (BRs) are a group of steroid hormones that play important roles in regulating plant development, including vascular differentiation, senescence, stress responses, and photomorphogenesis (Clouse et al., 1996; Li and Chory, 1997). A deficiency in BR biosynthesis or signaling leads to dwarf phenotypes due to retarded plant growth (Li and Chory, 1997; Clouse and Sasse, 1998). BR regulates plant cell and organ growth by stimulating transverse microtubule organization (Catterou et al., 2001).
Many components of the BR signaling pathway have been identified. BR is perceived by the transmembrane serine/threonine protein kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1) (Li and Chory, 1997). The bri1 mutant is morphologically similar to BR biosynthetic mutants and is insensitive to exogenous BR application (Clouse et al., 1996; Li and Chory, 1997; Noguchi et al., 1999; Friedrichsen et al., 2000). BR binds to the extracellular domain of BRI1 and induces heterodimerization of BRI1 and BAK1, leading to their activation (Li et al., 2002; Nam and Li, 2002; Santiago et al., 2013). The binding of BRI1 to BAK1 is negatively regulated by BRI1 KINASE INHIBITOR1 (BKI1), a plasma membrane-associated protein (Wang and Chory, 2006). In the absence of BR, BKI1 binds to the kinase domain of BRI1 and prevents the interaction between BAK1 and BRI1 (Wang and Chory, 2006). The activation of BRI1 leads to the dissociation of BKI1 from the plasma membrane into the cytoplasm (Wang and Chory, 2006). BIN2, one of the ten glycogen synthase kinase 3 (GSK3)-like kinases in Arabidopsis, is another negative regulator of BR responses (Li et al., 2001; Li and Nam, 2002). BIN2 encodes a 380 aa protein that contains an N-terminal variable domain, a conserved kinase domain and a C-terminal domain. A highly conserved four-amino-acid TREE motif (threonine–arginine–glutamic acid–glutamic acid) in the C terminus is important for the activity of BIN2, and a mutation in the TREE motif affected the degradation of BIN2. In the absence of BR, BIN2 phosphorylates BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1), which are targeted for degradation by the proteasome (Yin et al., 2002, 2005).
The cytoskeleton plays important roles in regulating cell morphogenesis, intracellular transport, and cell division. Microtubules are formed by the polymerization of α- and β-tubulin dimers. The intrinsic polarity of microtubules provides them with a less-dynamic minus end and a more-dynamic plus end. During polymerization, tubulin molecules are positioned within a microtubule so that α-tubulin faces the slow-growing minus end and β-tubulin faces the fast-growing plus end. The dynamic instability of the cytoskeleton allows it to quickly alter plant growth and development to help the plant adapt to various conditions. Loss-of-function mutants of major microtubule components usually exhibit abnormal growth (Nakajima et al., 2004), and altering the dynamic instability of microtubules leads to right-handed helical growth (Abe and Hashimoto, 2005). For instance, the pilz mutant fails to form basic microtubule structures, and its growth is arrested at early stages of embryogenesis (Mayer et al., 1999). The fass mutant, which is unable to correctly form microtubule structures, shows defects in polarized cell growth and radial pattering (Torres-Ruiz and Jürgens, 1994). Plants carrying mutations in SPIRAL1 (SPR1) and SPR2, which participate in the organization of microtubule structure, show increased radial growth and shorter cells compared with wild-type (Furutani et al., 2000). Disrupting the microtubules results in abnormal cell shape and plant development. However, little is known about how cytoskeletal dynamics are regulated, although auxin was shown to induce the reorganization (Takahashi et al., 1995) and depolymerization of microtubules (Baluska et al., 1996).
The BR receptor mutant bri1 has round leaves, short petioles, and reduced apical dominance. BRI1-overexpressing plants exhibit right-handed helical growth, and bzr1-1D and bes1-D mutants have curly hypocotyls and leaves (Wang et al., 2002). The expression of TUB1 is affected in the BR-deficient mutant dim (Takahashi et al., 1995; Klahre et al., 1998). Another BR-defective mutant, bul1-1 (boule 1-1), exhibits reduced microtubule levels and a total lack of parallel microtubule organization, which can be rescued by BR treatment (Catterou et al., 2001). The similarity in phenotype between BR and cytoskeleton mutants and the ability of BR to rescue microtubule organization suggest that BR might regulate plant development by regulating cytoskeletal dynamics (Catterou et al., 2001). A recent study showed that BZR1 targets and up-regulates MDP40 (MICROTUBULE DESTABILIZING PROTEIN 40) to mediate hypocotyl cell elongation by influencing the orientation and stability of cortical microtubules (Wang et al., 2012). Brassinolide (BL, an active BR) treatment also alters the orientation of microtubules in hypocotyls and increases the sensitivity of microtubules to the microtubule-disrupting drug oryzalin by regulating the expression of MDP40. Although BR was shown to regulate plant development and hypocotyl elongation through regulating microtubule organization, little is known about how defective BR signaling leads to morphological defects in the leaves of these mutants due to abnormal formation of the cytoskeleton. While the process of leaf pavement cell development is more intricate than that of hypocotyls, whether the mechanism underlying BR-regulated microtubules in pavement cells is the same as that in hypocotyls remains to be investigated.
Microtubule dynamics are regulated by the acetylation, tyrosination, detyrosination, polyglutamylation, polyglycylation, palmitoylation, and phosphorylation of tubulins (LeDizet and Piperno, 1987; Eddé et al., 1991). Among these, protein phosphorylation is the most common and important post-translational modification. Protein phosphorylation plays important roles in the dynamic instability of microtubules. For instance, the phosphorylation of Ser172 in β-tubulin by cyclin-dependent kinase (Cdk1) can impair both its binding to GTP and the interaction between tubulin dimers (Fourest-Lieuvin et al., 2006). Furthermore, CASEIN KINASE 1-LIKE 6 (CKL6), which contains a tubulin-binding domain, plays a role in anisotropic cell growth and shape formation in Arabidopsis by regulating microtubule organization, possibly through the phosphorylation of tubulins (Ben-Nissan et al., 2008). A kinase-inactive mutant form of CKL6 induces altered cortical microtubule organization and anisotropic cell expansion, and calmodulin-dependent kinase can also phosphorylate tubulins and microtubule-associated proteins (Goldenring et al., 1983; Ben-Nissan et al., 2008).
In this study, we searched for BIN2-interacting proteins by liquid chromatography–tandem mass spectrometry (LC-MS/MS) of immunoprecipitated (IPed) proteins from BIN2-FLAG transgenic plants, finding that BIN2 interacted with all tubulin proteins. Whereas microtubule-associated mutants display an isotropically expanding cell phenotype, including distorted trichomes, swollen roots, short hypocotyls, and aberrant pavement cells (Mathur, 2004), the BR biosynthesis mutant det2 (deetiolated 2), the BR signaling-defective mutants bri1-5 and bin2-1, and a BKI1-overexpression line (BKI1-OX) all showed short hypocotyls, round leaf shapes, and abnormal pavement cells. We therefore reasoned that BR regulates pavement cell development through its effect on microtubules. We further confirmed that BIN2 interacts with tubulin proteins both in vitro and in vivo. Co-sedimentation assays indicated that BIN2 directly binds to microtubules. Treatment with the microtubule-disrupting drug oryzalin confirmed that BIN2 indeed inhibits microtubule disruption. Our findings suggest that BR regulates plant development and pavement cell development by mediating microtubule stabilization induced by BIN2.
Materials and methods
Plant materials and growth conditions
Arabidopsis plants, including wild-type (Col-0 and Ws-2), det2 (en2), bin2-1 (Col-0), the bin2-3 bil1 bil2 triple mutant (Ws-2), bri1-5 (Ws-2), BKI1 overexpression lines (Col-0), BRI1 overexpression lines (Col-0), sav2 (Col-0), and bes1-D (Col-0) were used in this study. Transgenic Arabidopsis plants expressing green fluorescent protein (GFP)-tagged α-tubulin crossed with BR mutants were used for observation of cortical microtubules. Seeds were grown on half-strength Murashige and Skoog (½MS) medium (pH 5.8) with 0.8% agar and 1% sucrose for 7 d at 22 °C. The plants were grown at 22 °C under a 16 h light–8 h dark photoperiod.
Microscopy and image analysis
Samples were imaged using a Leica TCS SP8 laser scanning confocal microscope and Spinning disk confocal microscope. Images were obtained with ×25 objective for propidium iodide (PI) staining and ×40 and ×60 objective for microtubule labelling. For the microtubule labelling, maximum Z-projections were used and the Z-step size was 1 μm. Images were captured by a Leica TCS SP8 at 488 and 561 nm laser excitation and 500–550 nm and 590–630 nm emission for GFP and PI staining. Images captured by a spinning disk confocal microscope were obtained with ×40 and ×60 objective. For the microtubule labelling, maximum Z-projections were used and the Z-step size was 1 μm. Images were captured at 488 nm laser excitation and 500–550 nm emission.
PI (5 μg ml−1)-stained cotyledon pavement cells from 7-day-old seedlings were imaged. ImageJ software was used to measure the lobe number, lobe length, neck width, perimeter, and area (http://rsb.info.nih.gov/ij). Circularity was analysed according to Zhang et al. (2011).
Microtubule alignment was quantified according to Gomez et al. (2016). At least 50 cells were used. The values were recorded and the significance was analysed using Student’s paired t-test.
BRZ, eBL, and bikinin treatment
For the BRZ (224047-41-0, Cayman Chemical), eBL (78821-43-9, Sigma-Aldrich), and bikinin (188011-69-1, Sigma-Aldrich) treatment assays, wild-type seeds were grown on ½MS medium with 1 μM BRZ, 1 μM eBL, or 30 μM bikinin for 7 d, followed by observation of pavement cells in the seedlings.
Plasmid construction and recombinant protein purification
For the glutathione S-transferase (GST) pull-down assays, BIN2, bin2-1, and BIN2K69R were cloned into the pGEX4T-1 vector, and TUBULIN genes were cloned into the pET28a vector. BIN2–GST, bin2-1–GST, BIN2K69R–GST, and TUA1–His, TUA3–His, TUA5–His, TUB3–His, TUB4–His, TUB6–His, TUB7–His, and TUB8–His were expressed in Escherichia coli cells (Rosetta strain) and purified using glutathione resin and Ni-NTA agarose.
Protein extraction from plants
Plants were ground to a fine powder in liquid nitrogen, solubilized with 2× extraction buffer (100 mM Tris–HCl pH 7.5, 300 mM NaCl, 1% Triton X-100, 10% glycerol, and protease inhibitor), separated on a 10% SDS-PAGE gel, transferred to a nitrocellulose membrane (Amersham Biosciences), and then detected with corresponding antibodies.
Pull-down and semi-in vivo pull-down assays
Purified proteins, including GST, BIN2–GST, BIN2K69R–GST, and bin2-1–GST were bound to 20 μl GST resin in binding buffer (1× PBS, pH 7.4, 0.1% Triton X-100) for 2 h at 4 °C. The beads were washed three times and incubated with purified tubulin-His proteins for 2 h at 4 °C for binding. Binding buffer (80 μl) and 20 μl 5× SDS loading buffer were added to the samples, which were boiled for 5 min and separated by 10% SDS-PAGE. His antibody (CWBIO, CW0286, monoclonal) and α-tubulin antibody (Beyotime, AT819, monoclonal) were used for detection, goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (CWBIO, CW0145) were used. An ECL Substrate Kit (Thermo Fisher Scientific, NCI4106) was used to detect the HRP-conjugated secondary antibodies. After incubated with ECL buffer (A:B, 1:1) for 2 min at room temperature, the polyvinylidene fluoride membrane was put into a cassette and the signals were visualized with X-ray film.
For the semi-in vivo pull-down assay, GST and BIN2–GST, BIN2K69R–GST, and bin2-1–GST were bound to 20 μl GST resin in binding buffer (1× PBS, pH 7.4, 0.1% Triton X-100) for 2 h at 4 °C. GST was used as a negative control. After three washes, tubulin proteins purified from porcine brain were added to the samples, followed by rebinding for 2 h at 4 °C. Tubulin antibody was used for detection.
Plants overexpressing TUA6-GFP were ground with liquid nitrogen and total protein was extracted with 2× extraction buffer (100 mM Tris–HCl pH 7.5, 300 mM NaCl, 1% Triton X-100, 10% glycerol, and protease inhibitor). Protein extracts were centrifuged at 20000 g for 10 min, and the resulting supernatant was incubated with prewashed BIN2–GST beads for 1 h at 4 °C. Beads were resuspended with 1× SDS-PAGE loading buffer and analysed by SDS-PAGE and immunoblotting.
In vitro kinase assay
For the phosphorylation assay, BIN2–GST or BIN2K69R–GST and tubulins from porcine brain were incubated in 24 μl reaction buffer (50 mM HEPES pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, 10 μM ATP, 1 μl [10 μCi] [γ-32P]ATP) for 1 h at 37 °C. The reaction was terminated by adding 6 μl 6×SDS loading buffer. The samples were separated by 10% SDS-PAGE and stained with Coomassie Brilliant Blue, followed by drying and autoradiography of the gel.
Microtubule binding assay
GST–BIN2, GST–BIN2K69R, and GST–bin2-1 were purified and protein concentrations determined using a Bio-Rad protein assay kit. Porcine brain tubulins were purified as described previously (Castoldi and Popov, 2003). The microtubule polymerization and co-sedimentation experiments were performed as previously described (Mao et al., 2005).
Recombinant GST–BIN2, GST–BIN2K69R, and GST–bin2-1 were co-sedimented with 5 mM Taxol-stabilized microtubules in PEMT buffer (100 mM PIPES, 1 mM EGTA, 1 mM MgCl2, and 20 μM Taxol, pH 6.9). After incubation at 25 °C for 30 min, the samples were centrifuged at 100000 g for 30 min at 25 °C. Pellets and supernatants were analysed by 10% SDS-PAGE and visualized by staining with Coomassie Brilliant Blue R250. GST was used as a negative control.
Oryzalin treatment
Cotyledon pavement cells from 7-day-old seedlings including wild-type, bin2-3 bil1 bil2, and bin2-1 with GFP–α-tubulin expressed in the background were used for observation. Oryzalin (3,5-dinitro-N4,N4-dipropylsulfanilamide, Ps-410, 19044-88-3, Sigma-Aldrich) was applied to whole plants at the indicated concentrations. Seedlings were treated with 15 μM oryzalin in tubes for 3 and 10 min, and then the seedlings were put on a glass slide and observed using confocal microscopy.
Real-time PCR analysis
Total RNA was extracted from seedlings using the Qiagen RNAprep plant kit. Primers for real-time PCR are listed in Supplementary Table S1 at JXB online. The UBOX gene was used as a control to normalize the level of total RNA. PCRs were performed with a Bio-Rad CFX96 real-time system.
Results
BR regulates microtubule organization and pavement cell development
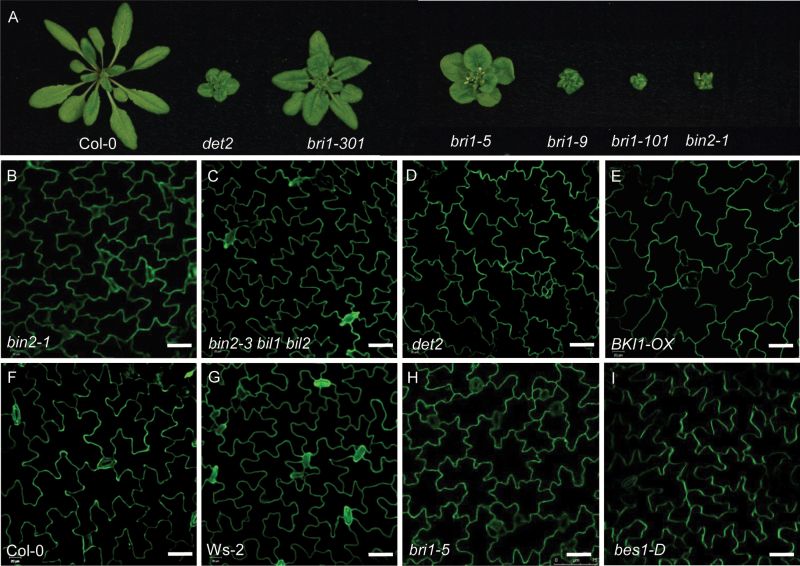
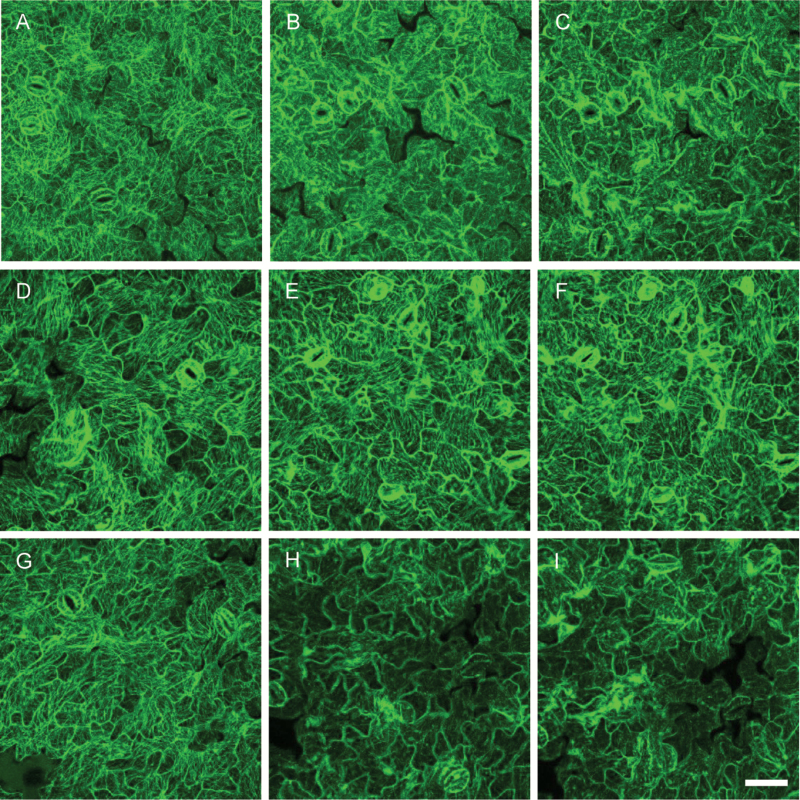
BR biosynthesis and signaling-deficient mutants have rounder leaves and shorter petioles than wild-type (Fig. 1A). To investigate the cellular mechanism underlying these phenotypes, we examined pavement cell shape in the BR mutants using propidium iodide (PI) staining and confocal microscopy. Wild-type mature pavement cells exhibited a ‘jigsaw puzzle’ shape, with lobes interdigitated with the necks of neighboring cells (Fig. 1F). By contrast, the BR mutants exhibited a smaller area of pavement cells and defective lobe formation (Fig. 1B–I). Quantitative analysis (Table 1) confirmed that pavement cell area, perimeter, and circularity value were smaller, with wider necks, in BR signaling and biosynthesis-deficient mutants (such as det2, bri1-5, bin2-1, and BKI1-Overexpression line (BKI1-OX)) compared with wild-type. By contrast, in BR signaling overexpression lines such as bes1-D, there were more lobes with pointed tips and the lobes were fewer in number. In the bin2-3 bil1 bil2 triple mutant, the area of the pavement cells was larger than in wild-type, with more lobes that were shorter in length. All tubulin-related mutants showed increased cell expansion, a phenotype similar to that of BR-related mutants such as bri1, BKI1-OX, det2, and bin2-1. Since the T-DNA insertion β-tubulin mutant tub1 (SALK_036755C) and the α-tubulin mutant tua6 (SALK_091096C) we ordered from the Arabidopsis Biological Resource Center (ABRC) are homozygous lethal, we used the β-tubulin mutant tub4 (also designated sav2) for analysis; sav2 was identified through a forward genetic screen and contains a point mutation in TUB4 (Yu et al., 2015). As shown in Supplementary Fig. S1, the sav2 mutant had short petioles and round leaves, like BR mutants. PI staining showed that sav2 pavement cells were also round, with wide necks (Supplementary Fig. S1). Quantitative analysis (Table 1) confirmed the phenotype of sav2 is similar to BR mutants.
Fig. 1.
BR mutants showed abnormal pavement cell shapes. (A) Phenotype of BR-related mutants. (B–I) Pavement cell shape of bin2-1 (B), bin2 bil1 bil2 triple mutant (C), det2 (D), BKI1-OX (E), Col-0 (F), Ws-2 (G), bri1-5 (H), and bes1-D (I). Cotyledons stained by propidium iodide. Bars in (B–I): 40 μm.
Table 1.
The size of cotyledon pavement cells of BR mutants
| Background | Lobe no. | Lobe length (μm) |
Neck width (μm) |
Perimeter (μm) |
Area (μm2) |
Circularity | |
|---|---|---|---|---|---|---|---|
| Col-0 | 9.1 ± 0.2 | 28.1 ± 4.8 | 31.2 ± 1.8 | 493.2 ± 14 | 7146 ± 382 | 0.37 ± 0.02 | |
| WS-2 | 10.1 ± 0.8 | 25.27 ± 0.2 | 25.20 ± 1.2 | 555 ± 11.3 | 5685 ± 324 | 0.23 ± 0.01 | |
| BKI1-OX | Col-0 | 13.3 ± 1.1**** | 14.2 ± 2.1**** | 35.1 ± 3.1**** | 381.4 ± 0.7**** | 4383 ± 269**** | 0.38 ± 0.02*** |
| BRI1-OX | Col-0 | 8.6 ± 0.7**** | 17.0 ± 1.6**** | 27.6 ± 0.4**** | 369.9 ± 4.4**** | 4657 ± 218**** | 0.45 ± 0.02**** |
| det2 | En2 | 8.5 ± 0.4**** | 11.7 ± 2.0**** | 37.01 ± 3.0**** | 358.3 ± 8.8**** | 5418 ± 98**** | 0.53 ± 0.01**** |
| bin2-1 | Col-0 | 7 ± 0.4**** | 12.8 ± 1.2**** | 27.65 ± 1.3**** | 328 ± 13.5**** | 3334 ± 174**** | 0.39 ± 0.02**** |
| bin2-3 bil1 bil2 | WS-2 | 14.5 ± 2**** | 16.5 ± 1.7**** | 22.71 ± 2.6**** | 502.1 ± 8.6**** | 5704 ± 215**** | 0.28 ± 0.01**** |
| bri1-5 | WS-2 | 12 ± 1.3**** | 14.3 ± 1.1**** | 29.5 ± 2.3**** | 398.9 ± 16.6**** | 5495 ± 266**** | 0.44 ± 0.02**** |
| bes1-D | En2 | 8.4 ± 0.37**** | 12.6 ± 1.1**** | 22.87 ± 2.2**** | 402.5 ± 7.8**** | 4920 ± 153**** | 0.38 ± 0.05*** |
| sav2 | Col-0 | 9.4 ± 0.8**** | 13.6 ± 0.13**** | 40.5 ± 3**** | 334 ± 81**** | 6278 ± 294**** | 0.71 ± 0.03**** |
n>63 for the lobe number, lobe length, and neck width data, and n>30 for the perimeter, area, and circularity measurements. The measurements were performed using ImageJ software (http://rsb.info.nih.gov/ij). t-Test: ***P<0.001, ****P<0.0001.
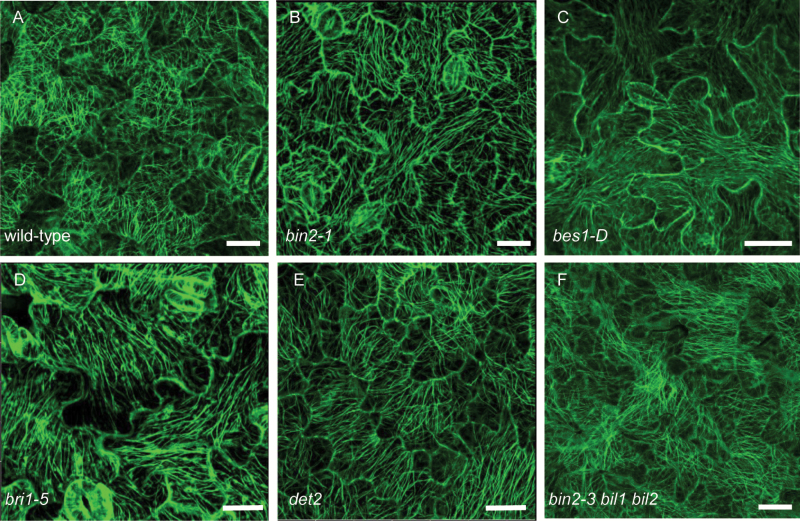
The abnormal pavement cell shape in the BR mutants and the similarity between BR mutants and tubulin mutants imply that microtubule arrangement or dynamics might be affected in the BR mutants. To clarify this, we examined the microtubule arrangement in BR mutants expressing GFP–tubulin. The TUBULIN gene expression levels were detected by real-time PCR (see Supplementary Fig. S2A) and tubulin protein levels were detected by western blotting with anti-GFP antibody (Supplementary Fig. S2B). As shown in Fig. 2, the microtubule organization was altered in the BR mutants. The Microtubule alignment was quantified according to Gomez et al. (2016) (Table 2). MTSD is the degree of deviation from the mean direction of the tubulin signal and reduces as microtubules become aligned with each other (Gomez et al., 2016). From Fig. 2A and Table 2, in wild-type mature pavement cells, the microtubules formed a randomly oriented array and were densely localized around the neck region, but in the BR mutants det2, bri1-5, bes1-D, bin2-1, and bin2-3 bil1 bil2, the microtubules were organized in nearly parallel arrays, with fewer microtubules localized around the neck than in wild-type (Fig. 2B–F; Table 2). These observations indicate that defects in BR biosynthesis and signaling result in an abnormal arrangement of microtubules.
Fig. 2.
BR mutants showed parallel arrangement of microtubules. (A–F) Cortical microtubules of cotyledon pavement cells in wild-type (A), bin2-1 (B), bes1-D (C), bri1-5 (D), det2 (E), and bin2-3 bil1 bil2 triple mutant (F) with a transgene expressing GFP–tubulin in the background for all lines. Bars: 20 μm. (This figure is available in color at JXB online.)
Table 2.
Microtubule organization of cotyledon pavement cells of BR mutants
| Wild-type | bri1-5 | det2 | bin2-1 | bin2-3 bil1 bil2 | bes1-D | eBL- treated wild-type |
BRZ- treated wild-type |
Bikinin-treated wild-type |
Bikinin-treated bin2-1 |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Random orientation |
87% | 14.9%**** | 13.4%**** | 11.3%**** | 16.4%**** | 17.2%**** | 6.7%**** | 5.4%**** | 6.3%**** | 9.4%**** |
| Paralleled orientation |
13% | 85.1%**** | 86.6%**** | 88.7%**** | 83.6%**** | 82.8%**** | 93.3%**** | 94.6%**** | 93.7%**** | 91.6%**** |
| MTSD (degrees) |
52.83 ± 0.76 | 14.9 ± 4.2**** | 25.9 ± 2.8**** | 17.7 ± 3.3**** | 23.2 ± 1.9**** | 24.6 ± 2.8**** | 24.0 ± 7.8**** | 14.7 ± 0.1**** | 34.3 ± 1.4**** | 16.9 ± 3**** |
MTSD is the degree of deviation from the mean direction of tubulin signal (Gomez et al. 2016). n>47 cells for each data point. t-Test: ****P<0.0001.
Inhibition of BIN2 activity affects the arrangement of microtubules
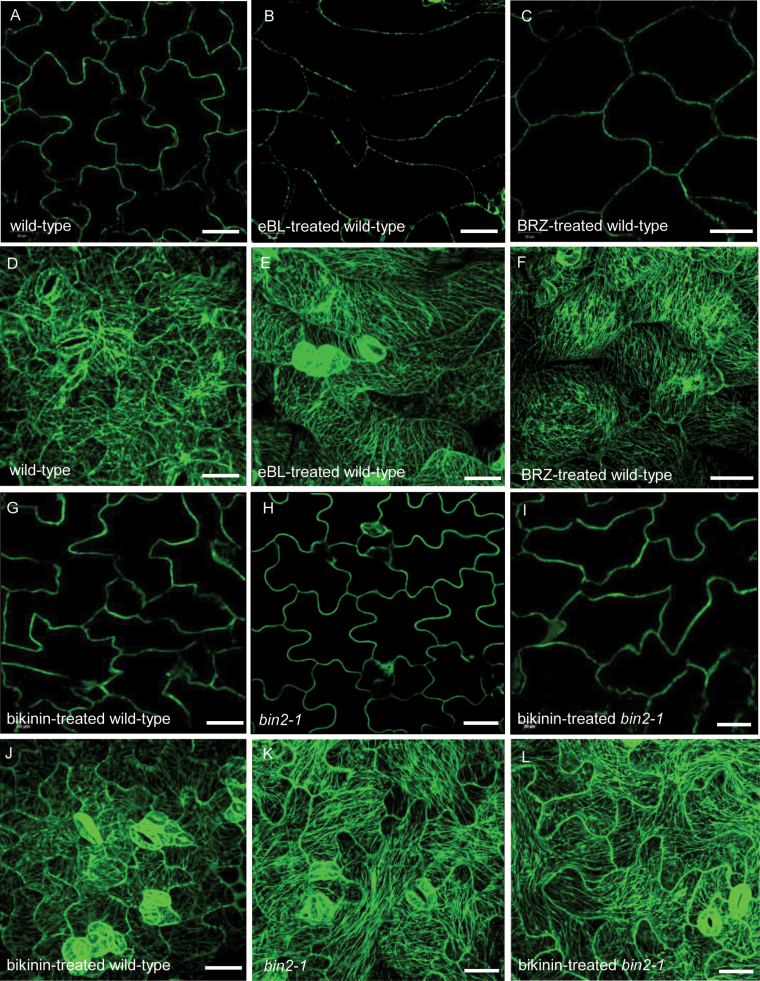
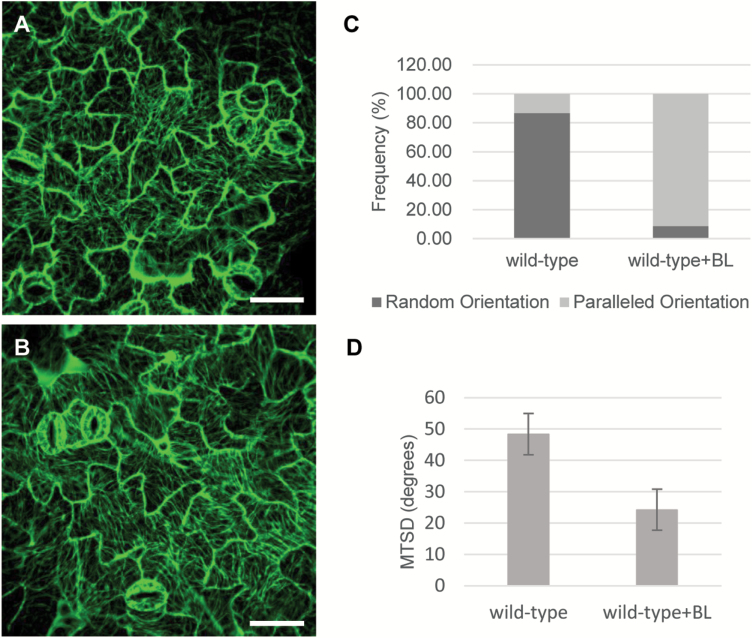
To investigate whether exogenously applied epi-brassinolide (eBL, an active BR) and brassinazole (BRZ, a BR biosynthesis inhibitor) affect microtubule arrangement, we grew wild-type seeds on ½MS medium containing 1 μM eBL or 1 μM BRZ for 7 d. As shown in Fig. 3 and Table 2, when treated with eBL, the wild-type pavement cells changed from jigsaw puzzle shaped to pointed tip shaped (Fig. 3B). By contrast, BRZ-treated pavement cells exhibited a swollen shape (Fig. 3C). Following eBL and BRZ treatment, the microtubule arrangement changed from random to parallel arrays (Fig. 3E, F). Both wild-type and bin2-1 plants treated with bikinin (an inhibitor of GSK3 kinases) showed similar phenotypes to those of eBL-treated plants (Fig. 3G–L). To detect whether short term treatment of eBL also leads to microtubule reorientation, we treat the wild-type with 10 μM eBL for 1 h. After treatment, the microtubule arrangement in wild-type changed from random to parallel arrays, which is the same as the result of 7 d treatment (Fig. 4A–C). Quantification of microtubule alignment showed that the MTSD changed from 48.26 to 24.16 degrees (Fig. 4D) (Gomez et al., 2016). These results indicate that BR regulates microtubule orientation and affects pavement cell shape directly. The inhibition of BIN2 activity also affects microtubule orientation. Taken together, these results suggest that BR regulates microtubule orientation through its effect on the activity of BIN2.
Fig. 3.
Treatment with eBL, BRZ, and bikinin led to abnormal pavement cell shape and arrangement of microtubules. (A–C, G–I) Pavement cell shape of wild-type (A), eBL-treated wild-type (B), BRZ-treated wild-type (C), bikinin-treated wild-type (G), bin2-1 (H), and bikinin-treated bin2-1 (I) cotyledons with a transgene expressing GFP–tubulin in the background for all lines. (D–F, J–L) Cortical microtubules of cotyledon pavement cells in wild-type (D), eBL-treated wild-type (E), BRZ-treated wild-type (F), bikinin-treated wild-type (J), bin2-1 (K), and bikinin-treated bin2-1 (L) with a transgene expressing GFP–tubulin in the background for all lines. Bar: 40 μm. (This figure is available in color at JXB online.)
Fig. 4.
One hour treatment with eBL led to microtubule reorientation. (A) Cortical microtubules of cotyledon pavement cells in wild-type. (B) Cortical microtubules of cotyledon pavement cells in wild-type with eBL treated for 1 h. (C) Frequency of microtubule orientation in pavement cells of wild-type and eBL-treated wild-type. (D) Quantification of MT alignment. Bar: 40 μm. (This figure is available in color at JXB online.)
BIN2 interacts with tubulin proteins in vitro and in vivo
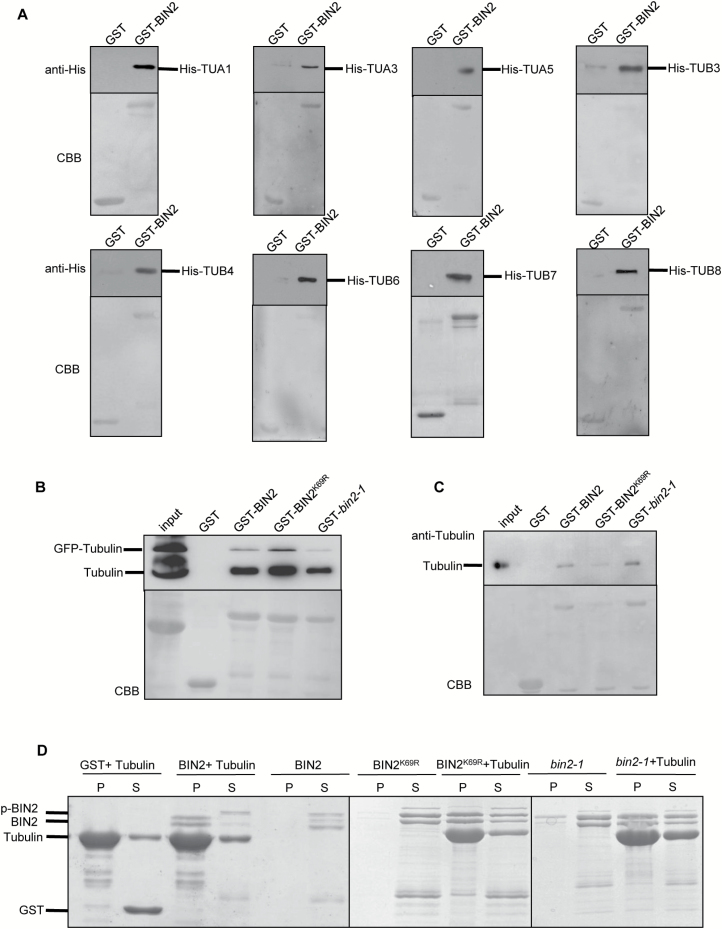
To identify proteins that interact with BIN2, we performed LC-MS/MS analysis of proteins that immunoprecipitated with BIN2, using extracts from p35S-BIN2::FLAG transgenic plants. The results indicated BIN2 may interact with tubulin proteins. To confirm that BIN2 interacts with tubulin proteins, we conducted pull-down assays. To that end, we cloned the full-length coding sequence of BIN2 in the pGEX-4T vector and TUBULIN genes in the pET28a vector and separately transformed these two constructs into E. coli. GST pull-down assays indicated that BIN2 directly interacts with TUA1, TUA3, TUA5, TUB3, TUB4, TUB6, TUB7, and TUB8 in vitro (Fig. 5A).
Fig. 5.
BIN2 interacts with tubulins in vitro and semi-in vivo. (A) BIN2 directly interacts with tubulins in vitro by pull-down assay. Tubulin–His was pulled down by GST–BIN2 immobilized on glutathione resin and analysed by western blotting using anti-His antibody. GST was used as a negative control. GST–BIN2 and tubulin–His were expressed and purified from E. coli. The data presented are a western blot (top) and a Coomassie-stained gel (bottom). (B) The interaction between BIN2 and tubulins was detected by a semi-in vivo pull-down assay. GST–BIN2 was expressed and purified from E. coli. Protein extracts from plants overexpressing tubulin–GFP were used for pull-down assays. Tubulin–GFP and tubulin were pulled down by GST–BIN2, GST–BIN2K69R and GST–bin2-1 immobilized on glutathione resin and analysed by western blotting using anti-α-tubulin antibody. GST was used as a negative control. The data presented are a western blot (top) and a Coomassie-stained gel (bottom). (C) The interaction between BIN2 and tubulins was tested by semi-in vivo pull-down assays. Tubulins were pulled down by GST–BIN2, GST–BIN2K69R, and GST–bin2-1 immobilized on glutathione resin and analysed by western blotting using anti-tubulin antibody. GST was used as a negative control. GST–BIN2 was expressed and purified from E. coli and tubulin was purified from porcine brain. The data presented are a western blot (top) and a Coomassie-stained gel (bottom). (D) The interaction between BIN2 and microtubules was detected by co-sedimentation assays. Recombinant GST–BIN2, GST–BIN2K69R, and GST–bin2-1 proteins were co-sedimented with 5 mM Taxol microtubules. Recombinant proteins appeared mainly in the supernatant (S) after centrifugation in the absence of microtubules and co-sedimented with microtubules into the pellets (P) in the presence of microtubules. GST was used as a negative control. (This figure is available in color at JXB online.)
To investigate whether BIN2 interacts with tubulin proteins in vivo, we extracted tubulin proteins from plants overexpressing tubulin–GFP, finding that BIN2 interacted with these proteins (Fig. 5B). Because tubulin proteins are dimers comprising α-tubulin and β-tubulin, we purified dimerized tubulin proteins from porcine brain (Fig. 5C) and found that BIN2 interacted with these dimers.
To investigate whether the kinase activity of BIN2 is important for its binding to tubulin proteins, we performed a semi-in vivo pull-down assay and found that BIN2–GST, the kinase-dead form BIN2K69R, and activated bin2-1 all interacted with tubulin proteins extracted from plants overexpressing tubulin–GFP (Fig. 5B) and with tubulin proteins purified from porcine brain (Fig. 5C).
The expression patterns of α-tubulin, TUA1, TUA3, and TUA5, and β-tubulin, TUB3, TUB4, TUB6, TUB7, and TUB8, are similar to that of BIN2 at various stages of development (see Supplementary Fig. S3A–F). This also indicated BIN2 interacted with tubulin proteins in various organ to modulate development.
BIN2 binds microtubules in vitro
Although we determined that BIN2 could bind tubulin proteins, we lacked evidence that it directly affects microtubules. Therefore, we performed a high-speed co-sedimentation assay to investigate whether BIN2 binds directly to microtubules. The full-length coding sequences of BIN2, BIN2K69R, and bin2-1 were cloned separately into the pGEX-4T vector. Recombinant proteins GST–BIN2, GST–BIN2K69R, and GST–bin2-1 were incubated individually with pre-formed Taxol-stabilized microtubules (5 mM) at room temperature for 20 min. After centrifugation at 100000 g, the supernatants and pellets were subjected to SDS-PAGE. Before the addition of microtubules, BIN2, BIN2K69R, and bin2-1 were only present in the supernatant (Fig. 5D). After incubation and centrifugation, BIN2, BIN2K69R, and bin2-1 were detected in the pellet, indicating that all three proteins directly bound to and co-sedimented with microtubules (Fig. 5D).
The bin2-3 bil1 bil2 triple mutant is more sensitive to the microtubule-disrupting drug oryzalin than wild-type and bin2-1
Both the bin2-1 and the bin2-3 bil1 bil2 mutants exhibited an abnormal microtubule arrangement, prompting us to investigate the relationship between this phenotype and the stability of cortical microtubules. To investigate microtubule stability, we examined the microtubule arrangement in bin2-1 and bin2-3 bil1 bil2 plants expressing GFP–tubulin and treated with the microtubule-disrupting drug oryzalin. As shown in Fig. 6A, D, G, before treatment the microtubules were arranged in a more parallel orientation in bin2-1 (Fig. 6D) and bin2-3 bil1 bil2 (Fig. 6G) than in wild-type (Fig. 6A). After 3 min of treatment with 15 μM oryzalin, the number of cortical microtubules in pavement cells differed between wild-type, bin2-1, and bin2-3 bil1 bil2 plants. The microtubules were disrupted in bin2-3 bil1 bil2 pavement cells (Fig. 6H) and weakly disrupted in wild-type pavement cells (Fig. 6B), while the microtubules in bin2-1 pavement cells were almost normal (Fig. 6E). When we increased the treatment time to 10 min, the microtubules in bin2-3 bil1 bil2 (Fig. 6I) and wild-type pavement cells were almost completely disrupted (Fig. 6C), while microtubules in bin2-1 were almost normal (Fig. 6F). These results indicate that the binding of BIN2 has a stabilizing effect on microtubules.
Fig. 6.
Microtubules are hypersensitive to treatment with oryzalin in bin2-3 bil1 bil2 cells but more resistant to oryzalin in bin2-1 cells. Cortical microtubules were observed in the cotyledon pavement cells of the wild-type (A–C), bin2-1 (D–F), and bin2-3 bil1 bil2 (G–I) plants after treatment with 0 (A, D, G) or 15 μM oryzalin (B, E, H) for 3 min or 15 μM oryzalin for 10 min (C, F, I). Bar in (I): 50 μm. (This figure is available in color at JXB online.)
Discussion
In this study, we provided several lines of evidence showing that BR plays important roles in regulating microtubule arrangement and stability via BIN2. First, pavement cell shape was dramatically altered in BR-defective mutants and in wild-type plants grown on ½MS medium containing eBL, bikinin, or BRZ for 7 d. Seven-day treatment and 1 h treatment of eBL led to microtubule reorientation demonstrating that GSK3-like kinase, BIN2, or its upstream components are involved in this regulation. Second, BIN2 directly interacted with tubulin proteins, suggesting that it directly regulates microtubule organization. Third, BIN2 binds microtubules, suggesting that it directly regulates microtubule stability and arrangement. Finally, the bin2-1 mutant was insensitive to the microtubule-disrupting drug oryzalin whereas the bin2-3 bil1 bil2 triple mutant was hypersensitive to oryzalin, indicating that BIN2 regulates pavement cell growth and leaf development by stabilizing microtubules.
Microtubules are formed by the polymerization of α-tubulin and β-tubulin dimers. Polymerization-biased dynamic instability at the plus end and slow depolymerization at the minus end contribute to cortical microtubule dynamics. Dynamic microtubules become organized into cortical microtubule arrays. The suppression of cortical microtubule dynamics and organization results in abnormal cell morphogenesis. Interference with the microtubule cytoskeleton results in pavement cells with defects in lobe formation. In the current study, we found that BR biosynthesis-defective mutants had round leaves, as well as smaller and fewer lobes in their pavement cells compared with wild-type, while BR overexpression lines showed right-handed helical growth in leaves and spindle-like pavement cells. The microtubule-related mutants such as sav2 had round, small rosette leaves and round and wide pavement cells, which is similar to the phenotype of BR-defective mutants (see Supplementary Fig. S1). The similarity between BR mutants and microtubule-related mutants suggests that BR might mediate pavement cell growth through its effect on cortical microtubules. We also found that BIN2 interacted with both α-tubulin and β-tubulin (Fig. 5). Thus, it is highly likely that the binding of tubulins by BIN2 affects tubulin dimerization and microtubule organization.
We also performed an in vitro kinase assay to investigate whether BIN2 can phosphorylate tubulin proteins (see Supplementary Fig. S4). GST–BIN2 and GST– BIN2K69R were incubated separately with dimerized tubulins extracted from porcine brain for 1 h at 37 °C. Autoradiography showed that BIN2 could phosphorylate these proteins, but the kinase-dead form of BIN2 (BIN2K69R) could not. Therefore, while both BIN2 and BIN2K69R bind tubulin proteins, only BIN2 can phosphorylate these proteins. Although we did not determine which tubulin protein is phosphorylated by BIN2 and which phosphorylation site is responsible for the phosphorylation and the resulting phenotype, our findings suggest that the interaction and phosphorylation of these proteins affect microtubule organization and stability.
Using a co-sedimentation assay, we confirmed the binding between BIN2 and microtubules. To investigate the binding mechanism in vivo, we treated bin2-1 and bin2-3 bil1 bil2 triple mutant plants with the microtubule-disrupting drug oryzalin. The bin2-3 bil1 bil2 mutant showed hypersensitivity to oryzalin treatment, while bin2-1 was insensitive. Many microtubule-binding proteins in plants have been reported, such as MAP65 (Mao et al., 2005), MAP18 (Wang et al., 2007), and MDP25 (Li et al., 2011). Microtubule-binding proteins are considered to be microtubule stabilizers or destabilizers depending on their effect on microtubule stability. Among these, MAP65 is a microtubule stabilizer, whereas MAP18 and MDP25 are microtubule destabilizers. A MAP18 overexpression line (Wang et al., 2007), in which cortical microtubules were destabilized, had a similar response to oryzalin treatment to that of the bin2-3 bil1 bil2 triple mutant, indicating that BIN2 is also a microtubule stabilizer. BR likely mediates pavement cell growth through its effect on the interaction with tubulin proteins and the stabilization of microtubules by BIN2.
BIN2 is expressed in various organs, suggesting that BIN2 plays a general role in regulating microtubule dynamics. Treatment with the microtubule-disrupting drug oryzalin leads to left-skewing root growth. The microtubule-binding protein mutant spr1 shows a right-skewing root growth phenotype; the direction of skewing is reversed by treatment with 3 µM microtubule depolymerizing drug propyzamide (Furutani et al., 2000; Sedbrook et al., 2004). The α-tubulin mutant lefty and a MAP18 overexpression line both exhibit left-skewing root growth due to inhibited tubulin polymerization. Therefore, the stabilization and disruption of cortical microtubules both lead to root growth defects. To investigate the role of BIN2 in root growth, we compared the roots of bin2-1 and bin2-3 bil1 bil2 with those of wild-type. When treated with 0.1 µM oryzalin, bin2-1 roots were significantly shorter than those of wild-type and skewed to the right, whereas bin2-3 bil1 bil2 roots were longer than wild-type and skewed to the left (see Supplementary Fig. S5). These observations indicate that BIN2 is a microtubule-stabilizing factor that plays an important role in regulating plant growth and development.
BR regulates both hypocotyl elongation and pavement cell development. Hypocotyl growth is polarized growth, whereas pavement cell development is a much more intricate process involving a combination of localized tip growth and diffuse growth to produce interlocking cells with complex shapes. Pavement cell morphogenesis is controlled by the countersignaling of two pathways with opposite effects (Fu et al., 2005). In addition to BR, auxin regulates plant growth in various ways depending on the developmental stage and plant organ (Nick et al., 1992). In roots and hypocotyls, auxin treatment alters the arrangement of microtubules towards a longitudinal direction (Chen et al., 2014), but in shoots, auxin treatment leads to a transverse microtubule orientation (Sassi et al., 2014). Therefore, BR may regulate microtubule stability differently in pavement cells than in hypocotyls. BZR1, a transcription factor in the BR signal pathway, can target and up-regulate MDP40 to mediate hypocotyl cell elongation by influencing the orientation and stability of cortical microtubules (Wang et al., 2012). In the current study, we found that in pavement cells, BR regulates microtubule dynamics through direct binding to BIN2. We also examined the microtubule arrangements in hypocotyls of bin2-1 and bin2-3 bil1 bil2 triple mutants and found the microtubules in bin2-1 and bin2-3 bil1 bil2 triple mutants were oblique to the long expansion axis of the cell while in wild-type the microtubules were transversely reoriented to the long expansion axis of the cell in the upper regions of hypocotyls (see Supplementary Fig. S6). While BIN2 can phosphorylate BZR1 directly, the abnormality of microtubule arrangements in bin2-1 and bin2-3 bil1 bil2 triple mutants may be caused by the abnormal BZR1. Although the mechanism uncovered in the current study differs from that described by Wang et al. (2012), the effect of BR on microtubules is the same. Both studies indicated that BR regulates plant growth by destabilizing microtubules.
Auxin modulates the interdigitated growth of leaf pavement cells and cell expansion through an ABP1-related pathway that activates ROP2 and ROP6 (Xu et al., 2010). Auxin activates ROP6 to modulate the association of RIC1 with microtubules and ROP2 to activate RIC4, promoting the assembly of cortical actin microfilaments required for localized outgrowth (Fu et al., 2005). Gibberellic acid (GA) also affects cortical microtubule orientation. The application of GA induces a transverse cortical microtubule array organization through the interaction between DELLA and the prefoldin proteins PFD3 and PFD5 (Locascio et al., 2013; Vineyard et al., 2013). DELLAs interact with PFD3 and PFD5, causing them to be retained in the nucleus to inhibit α-tubulin and β-tubulin heterodimerization. When GA levels are high, degraded DELLAs release PFD3 and PFD5 into the cytoplasm to promote tubulin dimerization (Locascio et al., 2013). Ethylene inhibits etiolated hypocotyl elongation through the activity of EIN3, which directly targets and up-regulates the expression of the microtubule-stabilizing protein WDL5 (Sun et al., 2015). Most phytohormones regulate plant growth through their effects on microtubules, suggesting that there is crosstalk among hormonal signaling pathways. The relationship between these phytohormones and how they regulate the cytoskeleton represents an exciting and fertile area of research. Our finding that BR regulates pavement cell formation through its effect on the binding and stabilization of microtubules by BIN2 contributes to our understanding of this important topic.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. The tub4 mutant shows an abnormal rosette leaf phenotype and pavement cell shape.
Fig. S2. The relative expression levels of TUBULIN gene and protein levels of tubulin in BR mutants.
Fig. S3. Expression patterns of TUBULIN genes.
Fig. S4. BIN2 phosphorylates tubulins in vitro.
Fig. S5. The bin2-1 and bin2-3 bil1 bil2 mutants exhibit a skewed-root phenotype when treated with 0.1 μM oryzalin.
Fig. S6. The cortical microtubule array is altered in hypocotyl epidermal cells of bin2-1 and bin2-3 bil1 bil2 mutants.
Table S1. Primers used for real-time PCR.
Acknowledgements
We are grateful to Professor Yi Tao for the kind gift of sav2 seeds. This work was supported by the National Natural Science Foundation of China (31600219) and the China Postdoctoral Science Foundation.
References
- Abe T, Hashimoto T. 2005. Altered microtubule dynamics by expression of modified α-tubulin protein causes right-handed helical growth in transgenic Arabidopsis plants. The Plant Journal 43, 191–204. [DOI] [PubMed] [Google Scholar]
- Baluska F, Barlow PW, Volkmann D. 1996. Complete disintegration of the microtubular cytoskeleton precedes its auxin-mediated reconstruction in postmitotic maize root cells. Plant & Cell Physiology 37, 1013–1021. [DOI] [PubMed] [Google Scholar]
- Ben-Nissan G, Cui W, Kim DJ, Yang Y, Yoo BC, Lee JY. 2008. Arabidopsis casein kinase 1-like 6 contains a microtubule-binding domain and affects the organization of cortical microtubules. Plant Physiology 148, 1897–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi M, Popov AV. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expression and Purification 32, 83–88. [DOI] [PubMed] [Google Scholar]
- Catterou M, Dubois F, Schaller H, Aubanelle L, Vilcot B, Sangwan-Norreel BS, Sangwan RS. 2001. Brassinosteroids, microtubules and cell elongation in Arabidopsis thaliana. I. Molecular, cellular and physiological characterization of the Arabidopsis bul1 mutant, defective in the Δ7-sterol-C5-desaturation step leading to brassinosteroid biosynthesis. Planta 212, 659–672. [DOI] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H et al. 2014. Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516, 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. 1996. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiology 111, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. 1998. BRASSINOSTEROIDS: Essential regulators of plant growth and development. Annual Review of Plant Physiology and Plant Molecular Biology 49, 427–451. [DOI] [PubMed] [Google Scholar]
- Eddé B, Rossier J, Le Caer JP, Berwald-Netter Y, Koulakoff A, Gros F, Denoulet P. 1991. A combination of posttranslational modifications is responsible for the production of neuronal α-tubulin heterogeneity. Journal of Cellular Biochemistry 46, 134–142. [DOI] [PubMed] [Google Scholar]
- Fourest-Lieuvin A, Peris L, Gache V, Garcia-Saez I, Juillan-Binard C, Lantez V, Job D. 2006. Microtubule regulation in mitosis: tubulin phosphorylation by the cyclin-dependent kinase Cdk1. Molecular Biology of the Cell 17, 1041–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. 2000. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiology 123, 1247–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z. 2005. Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120, 687–700. [DOI] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masukawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T. 2000. The SPIRAL genes are required for directional control of cell elongation in Aarabidopsis thaliana. Development 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Goldenring JR, Gonzalez B, McGuire JS Jr, DeLorenzo RJ. 1983. Purification and characterization of a calmodulin-dependent kinase from rat brain cytosol able to phosphorylate tubulin and microtubule-associated proteins. The Journal of Biological Chemistry 258, 12632–12640. [PubMed] [Google Scholar]
- Gomez JM, Chumakova L, Bulgakova NA, Brown NH. 2016. Microtubule organization is determined by the shape of epithelial cells. Nature Communications 7, 13172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. 1998. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. The Plant Cell 10, 1677–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. 1987. Identification of an acetylation site of Chlamydomonas α-tubulin. Proceedings of the National Academy of Sciences, USA 84, 5720–5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J. 1997. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. 2002. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295, 1299–1301. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. 2001. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiology 127, 14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang X, Qin T et al. 2011. MDP25, a novel calcium regulatory protein, mediates hypocotyl cell elongation by destabilizing cortical microtubules in Arabidopsis. The Plant Cell 23, 4411–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. 2002. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. [DOI] [PubMed] [Google Scholar]
- Locascio A, Blázquez MA, Alabadí D. 2013. Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Current Biology 23, 804–809. [DOI] [PubMed] [Google Scholar]
- Mao T, Jin L, Li H, Liu B, Yuan M. 2005. Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiology 138, 654–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur J. 2004. Cell shape development in plants. Trends in Plant Science 9, 583–590. [DOI] [PubMed] [Google Scholar]
- Mayer U, Herzog U, Berger F, Inzé D, Jürgens G. 1999. Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. European Journal of Cell Biology 78, 100–108. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Furutani I, Tachimoto H, Matsubara H, Hashimoto T. 2004. SPIRAL1 encodes a plant-specific microtubule-localized protein required for directional control of rapidly expanding Arabidopsis cells. The Plant Cell 16, 1178–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KH, Li J. 2002. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212. [DOI] [PubMed] [Google Scholar]
- Nick P, Schäfer E, Furuya M. 1992. Auxin redistribution during first positive phototropism in corn coleoptiles: microtubule reorientation and the Cholodny-Went theory. Plant Physiology 99, 1302–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. 1999. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiology 121, 743–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J, Henzler C, Hothorn M. 2013. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892. [DOI] [PubMed] [Google Scholar]
- Sassi M, Ali O, Boudon F et al. 2014. An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Current Biology 24, 2335–2342. [DOI] [PubMed] [Google Scholar]
- Sedbrook JC, Ehrhardt DW, Fisher SE, Scheible WR, Somerville CR. 2004. The Arabidopsis SKU6/SPIRAL1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. The Plant Cell 16, 1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ma Q, Mao T. 2015. Ethylene regulates the arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiology 169, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Gasch A, Nishizawa N, Chua NH. 1995. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes & Development 9, 97–107. [DOI] [PubMed] [Google Scholar]
- Torres-Ruiz RA, Jürgens G. 1994. Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967–2978. [DOI] [PubMed] [Google Scholar]
- Vineyard L, Elliott A, Dhingra S, Lucas JR, Shaw SL. 2013. Progressive transverse microtubule array organization in hormone-induced Arabidopsis hypocotyl cells. The Plant Cell 25, 662–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chory J. 2006. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122. [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T. 2012. Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. The Plant Cell 24, 4012–4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M. 2007. Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. The Plant Cell 19, 877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J et al. 2002. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Developmental Cell 2, 505–513. [DOI] [PubMed] [Google Scholar]
- Xu T, Wen M, Nagawa S et al. 2010. Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143, 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. 2005. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. 2002. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191. [DOI] [PubMed] [Google Scholar]
- Yu J, Qiu H, Liu X, Wang M, Gao Y, Chory J, Tao Y. 2015. Characterization of tub4(P287L), a β-tubulin mutant, revealed new aspects of microtubule regulation in shade. Journal of Integrative Plant Biology 57, 757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Halsey LE, Szymanski DB. 2011. The development and geometry of shape change in Arabidopsis thaliana cotyledon pavement cells. BMC Plant Biology 11, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.