Abstract
Plants are known for their capacity to regenerate organs, such as shoot, root and floral organs. Recently, a number of studies contributed to understanding the mechanisms of shoot and root regeneration. However, the mechanisms underlying floral organ regeneration are largely unknown. In this study, we established a carpel regeneration system in which two types of carpels were induced by exogenous cytokinin. For type I, all the floral organs in the regenerated inflorescence were transformed into carpels. For type II, carpels were generated directly from callus. The transcript level of AGAMOUS (AG), the carpel identity gene, was up-regulated during carpel induction. The expression signals of AG were detected in the initiating carpel primordia and regenerating carpels, and co-localized with those of two Type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), ARR1 and ARR10. Repression of either AG or type-B ARRs reduced carpel regeneration. Binding analyses showed that ARR1 and ARR10 directly bound to transcriptional regulatory regions of AG and positively regulated its expression. In addition, the expression of type-B ARRs overlapped with that of AG in the floral primordia in planta. Defects in type-B ARRs reduced the number of carpels. The results indicate that type-B ARRs control carpel regeneration through activating AG expression. Our results provide new information for understanding the mechanism of carpel formation.
Keywords: AGAMOUS, Arabidopsis, ARR, Carpel, Cellular reprogramming, Regeneration
Introduction
High cellular plasticity confers a powerful capacity to regenerate organs in plants (Ikeuchi et al. 2016, Shang et al. 2016). Under proper culture conditions, the balance between auxin and cytokinin promotes cellular reprogramming and determines organ regeneration (Iwase et al. 2017). A high cytokinin to auxin ratio leads to shoot regeneration, while a high auxin to cytokinin ratio specifies root formation (Skoog and Miller 1957). The whole plant body can be derived from successive formation of shoot and root (Duclercq et al. 2011). In addition to shoot and root regeneration, reproductive organs can be induced directly from callus cells. In some plant species, such as Hyacinthus orientalis, all types of floral organs including the tepal, stamen and carpel can be regenerated from the plant tissue by different levels of plant hormones (Lu et al. 1988, Lu 2003, Wu et al. 2003, An et al. 2004, Xu et al. 2004). These regenerative systems shed new light on understanding organ initiation and promote the progress of biological breeding. The regeneration of these organs is correlated to functions of transcription factors, which play critical roles in regulating floral organ development (Li et al. 2002, Xu et al. 2004). However, the molecular mechanisms remain largely unknown.
Most flowers consist of four types of floral organs which are controlled by the class-A, -B and -C organ identity genes (Coen and Meyerowitz 1991). In Arabidopsis, AGAMOUS (AG), encoding members of the MADS family of transcription factors, is classified as the class-C gene that is required for stamen and gynoecium determination (Yanofsky et al. 1990). Moreover, the floral homeotic gene AG required for floral meristem determinacy is activated by WUSCHEL (WUS) expression in the early floral meristem at floral stage 3 (Lenhard et al. 2001, Lohmann et al. 2001, Liu et al. 2011). Expression of AG is also mediated by LEAFY (LFY) and APETALA1 (AP1) synergistically, which controls the transition of meristem activity from indeterminate to determinate (Weigel and Meyerowitz 1993). In ag mutants, organ development does not terminate with the formation of fourth-whorl organs, but continues indeterminately (Huang et al. 2017). Thus, ag-1 mutant flowers have an indeterminate number of whorls containing only sepals and petals, resulting in a flower-within-flower phenotype (Bowman et al. 1989, Bowman et al. 1991). Our previous work indicates that HAG1, a homolog of AG in H. orientalis, is required for the determination of floral organ types, which is mediated by auxin and cytokinin (Li et al. 2002).
Auxin and cytokinin determine organ regeneration through their signaling components (Duclercq et al. 2011, Iwase et al. 2017). In Arabidopsis, auxin regulates shoot meristem formation through AUXIN RESPONSE FACTOR3 (ARF3) repressing the expression of the ATP/ADP ISOPENTENYLTRANSFERASE5 gene, which encodes the key enzyme of cytokinin biosynthesis (Cheng et al. 2013). Interestingly, cytokinin signaling components, type-B ARABIDOPSIS RESPONSE REGULATORs (ARRs), activate the expression of WUS encoding a key regulator for stem cell specification (Meng et al. 2017, Wang et al. 2017, Zhang et al. 2017). Here, we described that the carpels or carpel-like organs were induced with a high level of cytokinin through AG expression promoted by its signal transduction factors type-B ARRs . Our study provides new information for understanding the mechanism of carpel formation.
Results
Exogenous cytokinin induces carpel regeneration
We have described the inflorescence regeneration system previously (Cheng et al. 2010). Pistils at stage 10 were used as explants and were first cultured on a callus-inducing medium (CIM) for 15 d to generate callus. The calli were then transferred onto an inflorescence-inducing medium (IIM) containing 2.0 mg l–1 zeatin (ZT) and 0.01 mg l–1 IAA to induce inflorescence regeneration. To induce carpel regeneration, we established a system by adjusting the hormonal concentration in IIM. Carpel regeneration occurred at high frequency when the ZT concentration was increased to 20 mg l–1. The medium containing 20 mg l–1 ZT and 0.01 mg l–1 IAA was thus defined as the carpel-inducing medium (CaIM).
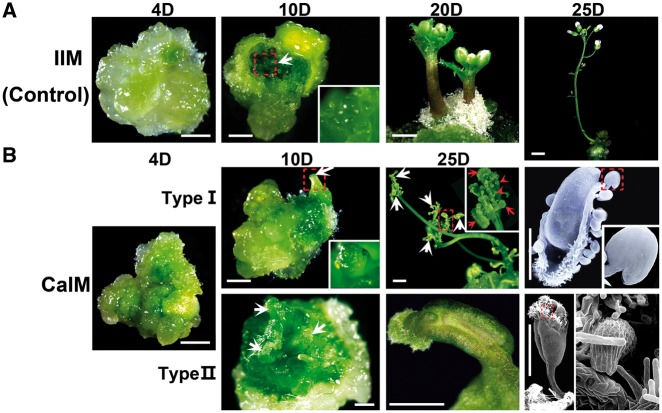
Two types of carpels (type I and II) regenerated in our carpel induction system (Fig. 1A, B; Supplementary Fig. S1). Inflorescence primordia on callus were observed at 10 d induction on CaIM (CaIM10). At CaIM25, flower-like structures were generated from the inflorescence (Fig. 1B; Supplementary Fig. S2). The inflorescences regenerated on IIM formed normal lateral organs. In contrast, in the inflorescences regenerated on CaIM, the lateral organs (including cauline leaves, sepals, petals and stamens) were transformed into carpels; these were designated as type I carpels (Fig. 1B). Type II carpels regenerated directly from the callus (Fig. 1B). Carpel primordia were visible on the surface of the callus at CaIM10 and further developed into mature carpels at CaIM25. The frequency of type I and II regeneration was 92.9% and 57.5%, respectively (Supplementary Fig. S3). In 50.4% of the examined calli, both types were observed. Although some of the regenerated carpels were unfused, type I and II carpels had normal stigma and ovule structures (Fig. 1B). Carpels also regenerated from callus cultured on CaIM without IAA, but at a lower frequency than that on CaIM with IAA (data not shown). These results indicate that a high concentration of exogenous cytokinin induces carpel regeneration.
Fig. 1.
Morphology of regenerated carpels induced by cytokinin. (A) Inflorescence regeneration procedure as control. Calli were cultured on inflorescence induction medium (IIM) for 4, 10, 20 and 25 d. The white arrow indicates the inflorescence meristem initiation site. (B) Carpel regeneration procedures. Calli were cultured on carpel induction medium (CaIM) for 4, 10, 20 and 25 d. For type I regeneration (upper), inflorescence regeneration initiated at 10 CaIM. Carpels were generated at floral organ primordia of regenerated inflorescences. For type IIregeneration (lower), carpels initiated directly from calli. White arrows indicate regenerated carpels; red arrows indicate formation of ovules. Scanning electron microscope images show structures of ovules (upper) and stigma (lower) of regenerated carpels. Dotted boxes indicate regions magnified in insets. Scale bars = 2 mm. ‘D’ indicates the number of days of culture on IIM or CaIM.
AG is required for cellular reprogramming during carpel regeneration
To study the molecular mechanisms underlying carpel regeneration, we detected the transcript levels of genes involved in floral organ development. The AG transcript level was up-regulated at CaIM4, and further increased at CaIM6, compared with that at CaIM0 (Supplementary Fig. S4). The AG transcript levels at CaIM4 and CaIM6 were significantly higher than those at IIM4 and IIM6, respectively.
We further traced the expression pattern of AG during carpel regeneration using pAG::AG-GFP lines (Ji et al. 2011). In type I carpel regeneration, the inflorescence meristem was initiated at CaIM6. The green fluorescent protein (GFP) signals became detectable at the peripheral region of the regenerated meristems, indicating the formation of carpel primordia (Fig. 2A). At CaIM10, AG expression was detected in the carpel primordia and regenerating carpels. The expression pattern of AG was confirmed using pAG::GUS transgenic lines, and similar results were obtained. However, in the normal inflorescence regenerated on IIM as a control, AG expression was only observed in the central region of flower primordia (Fig. 2C). In type II carpel regeneration, AG expression signal was detected in the initiating carpel primordia at CaIM6, and was distributed throughout the regenerated carpel thereafter (Fig. 2B). The expression pattern of AG was further confirmed by in situ hybridization and pAG::GUS transgenic lines (Fig. 2B). These results show that AG expression is involved in cellular reprogramming during carpel initiation.
Fig. 2.
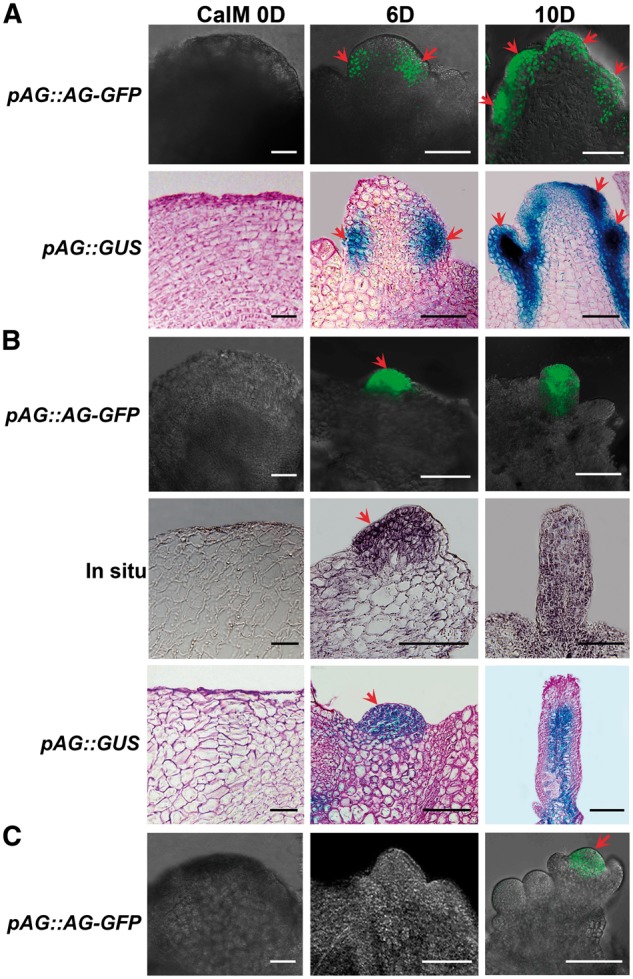
AG expression patterns during carpel regeneration. (A) Expression patterns of pAG::AG-GFP and pAG::GUS during type I carpel regeneration. Calli were cultured in CaIM for 0, 6 and 10 d. Red arrows indicate carpel primordial and regenerating carpels. (B) Distribution pattern of pAG::AG-GFP and pAG::GUS, and in situ hybridization of AG during type II regeneration. Calli were cultured on CaIM for 0, 6 and 10 d. Red arrows indicate carpel primordia. (C) Expression pattern of pAG::AG-GFP during inflorescence induction as control. The red arrow indicates GFP signals in the central region of the floral primordium. Calli were cultured on IIM for 0, 6 and 10 d. Scale bars = 100 μm. ‘D’ indicates the number of days of culture on IIM or CaIM.
Next, we tested whether AG is required for carpel regeneration. Because we could not obtain pistil explants from the ag mutant, we used artificial microRNAs (ams) driven by an ethanol-inducible promoter to knock down the transcripts of AG temporally (Leibfried et al. 2005, Zhao et al. 2010). Without exogenous ethanol, the seedling and flower phenotypes of am-AG transgenic lines were indistinguishable from those of the wild type (Supplementary Fig. S5A, C). After ethanol induction for 12 h, the transcript level of AG was significantly reduced (Supplementary Fig. S5B). An ethanol treatment of the transgenic line after bolting led to a phenotype similar to that of the ag mutant (Supplementary Fig. S5C). Thus, we used the am-AG line for carpel induction. When ethanol was added to the CaIM, the carpel initiation exhibited a 2 d delay, and reached the regenerative frequency of 82.1% at CaIM25, demonstrating that exogenous ethanol has little effect on carpel induction (Fig. 3). However, the carpel regeneration was inhibited in the am-AG line compared with the control. In addition, leaf-like structures were generated (Fig. 3). The results indicate that AG function is required for cellular reprogramming and subsequent carpel regeneration.
Fig. 3.
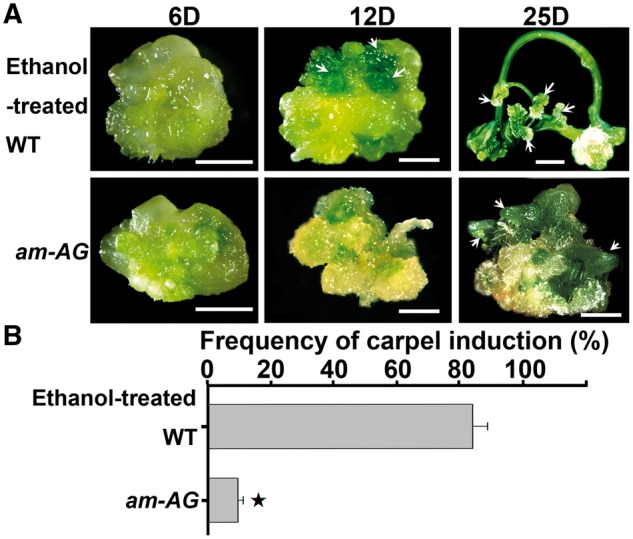
AG is required for carpel regeneration. (A) Carpel induction from wild-type and am-AG transgenic calli. Calli were cultured on CaIM for 6, 12 and 25 d. Arrows indicate initiation of carpel regeneration and regenerating carpels from wild-type callus, and regeneration of leaf-like structures from am-AG transgenic callus. Scale bars = 2 mm. ‘D’ indicates the number of days of culture on CaIM. (B) Frequency of carpel regeneration in (A). Error bars indicate tjhe SDs of three biological replicates. The star indicates P < 0.01 (Student’s t-test).
Type-B ARRs are involved in the regulation of carpel regeneration
Because carpel regeneration is induced by exogenous cytokinin, we determined whether cytokinin signaling plays a role in this process. Our previous study showed that ARR1 and ARR10 are required for shoot regeneration (Meng et al. 2017). Thus, we analyzed the spatiotemporal expression pattern of ARR1 during carpel regeneration using pARR1::ARR1-GFP reporter lines (Fig. 4A). At the beginning of CaIM culture, ARR1 signals were distributed evenly at the edge of the callus. During type I carpel regeneration, the signals were restricted to the peripheral region of the regenerated inflorescence meristem at CaIM6, and distributed in carpel primordia and regenerating carpels at CaIM10. Similarly, during type II carpel regeneration, the signals were restricted to the carpel primordium at CaIM6, and subsequently to the regenerating carpel. These results demonstrate that the expression pattern of ARR1 is similar to that of AG.
Fig. 4.
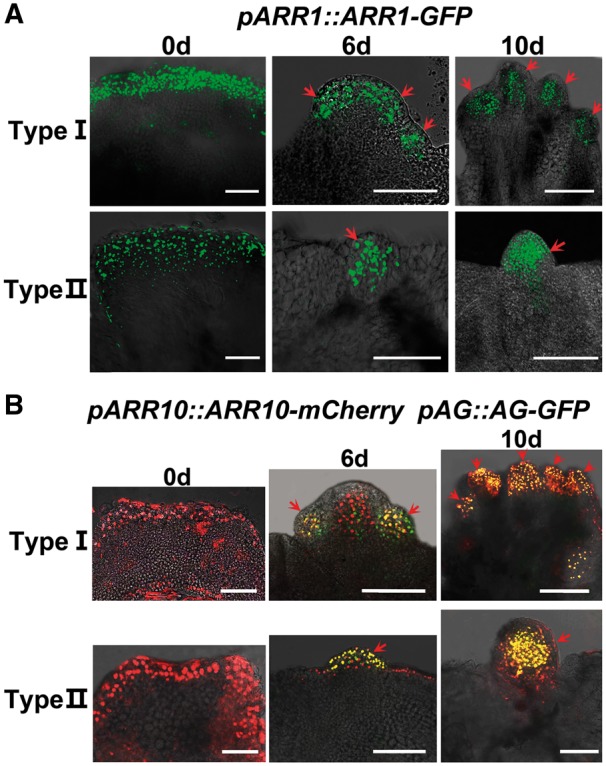
Overlapping expression patterns of type-B ARRs and AG during carpel regeneration. (A) Expression of pARR1::ARR1-GFP during carpel regeneration. (B) Expression patterns of pARR10::ARR10-mCherry (red) and pAG::AG-GFP (green) in the double reporter line. Expression signals of ARR10 overlapped with that of AG in carpel primordia and regenerating carpels (yellow). Calli were cultured on CaIM for 0, 6 and 10 d. Red arrows indicate carpel primordial and regenerating carpels. Scale bars = 100 μm. ‘D’ indicates the number of days of culture on CaIM.
To examine the relationship between the expression patterns of type-B ARR genes and AG further, we generated pARR10::ARR10-mCherry; pAG::AG-GFP double reporter lines (Fig. 4B). The expression pattern of ARR10 was similar to that of ARR1 at the beginning of CaIM culture. During both type I and type II carpel regeneration, the expression signals of ARR10 overlapped with those of AG in carpel primordia and regenerating carpels, suggesting that type-B ARR genes are involved in AG function.
Next, we determined whether type-B ARRs play a role in carpel regeneration. We used transgenic lines expressing an artificial microRNA to target specifically ARR1 and ARR10 under an ethanol-inducible promoter. Callus formation was not affected in am-ARR1/10 lines. However, when am-ARR1/10 callus was transferred onto CaIM to which ethanol had been added, the frequency of carpel regeneration was significantly decreased compared with that of the control, confirming that type-B ARRs are required for carpel regeneration (Fig. 5A, B). The results were further confirmed using arr1 10 double and arr1 10 12 triple mutants which also display highly reduced carpel induction (Fig. 5B). Quantitative reverse transcription–PCR (qRT–PCR) and our previous study revealed that the reduction of carpel regeneration is related to the decreased transcript levels of ARR1 and ARR10 (Supplementary Fig. S6) (Meng et al. 2017).
Fig. 5.
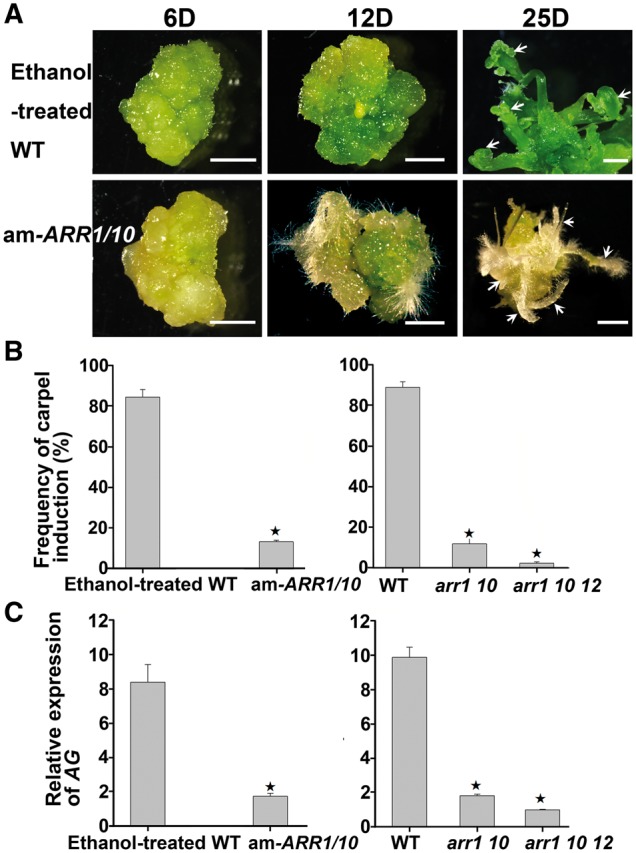
Type-B ARRs are involved in carpel regeneration. (A) Carpel induction from wild-type and am-ARR1/10 transgenic calli. Calli were cultured on CaIM for 6, 12 and 25 d. Arrows indicate initiation of carpel regeneration and regenerating carpels from wild-type callus, and root regeneration from am-ARR1/10 transgenic callus. Scale bars = 2 mm. ‘D’ indicates the number of days of culture on CaIM. (B) Compared with the wild type, am-ARR1/10 transgenic lines as well as arr1 10 double and arr1 10 12 triple mutants showed reduced frequencies of carpel regeneration. (C) Compared with the wild type, am-ARR1/10 transgenic lines as well as arr1 10 double and arr1 10 12 triple mutants showed reduced transcriptional levels of AG. (B, C) Error bars indicate the SDs of three biological replicates. The star indicates P < 0.01 (Student’s t-test).
ARR1 and ARR10 directly regulate AG expression
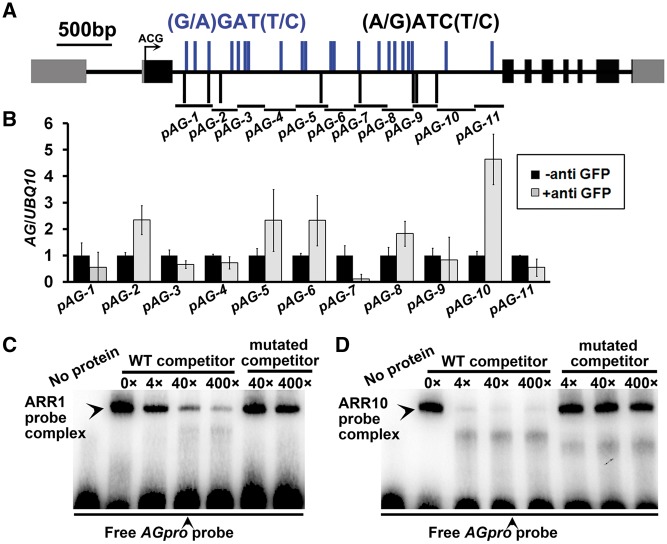
The overlapping expression patterns of type-B ARR genes and AG prompted us to test whether type-B ARRs regulate AG expression. To this end, we detected the transcriptional levels of AG using qRT–PCR and found an obvious reduction in the arr1 10 and arr1 10 12 mutants, indicating that type-B ARRs play a positive regulatory role in AG transcription (Fig. 5C). To test whether type-B ARRs directly regulate AG expression, we performed chromatin immunoprecipitation (ChIP) analyses using pARR1::ARR1-GFP transgenic explants at CaIM10. A previous study showed that sequences in the second intron of AG are sufficient for its proper expression pattern, while a 744 bp fragment within this region confers the carpel-specific expression pattern (base pairs 2,495–3,239, downstream of the ACG start codon; Deyholos and Sieburth 2000). We analyzed fragments covering the second intron in the ChIP analyses (Fig. 6A), and found that the fragment pAG-10 (base pairs +2,644 to + 2,911, downstream of the ACG start codon, containing part of the region sufficient for carpel-specific expression) was strongly enriched (Fig. 6B).
Fig. 6.
ARR1 and ARR10 bind to the transcriptional regulatory region of AG. (A) Diagram of the AG genomic region. The arrow marks the translation start site, black boxes indicate coding sequences, gray boxes indicate untranslated regions and black bold lines indicate intron/intergenic regions. pAG-1 to pAG-10 indicate positions of fragments used in ChIP-qPCR analyses. Blue and the black bars indicate type-B ARR-binding elements (G/A)GAT(T/C) and (A/G)ATC(T/C), respectively. (B) ChIP analyses showing an association between ARR1 and the transcriptional regulatory region of AG. (C) EMSAs confirming binding of ARR1 and ARR10 to the transcriptional regulatory region of AG. Arrowheads above indicate band shifts (complexes of ARR proteins and probe DNA). Arrows below indicate free probes. Non-labeled oligonucleotides were used as competitors. Mutated competitors were generated by replacing two base pairs in ARR-binding elements (GATC/T to CTTC/T).
The direct binding of ARR1 and ARR10 to fragment pAG-10, which contains two type-B ARR-binding elements, was examined by electrophoretic mobility shift assays (EMSAs). An oligonucleotide (base pairs +2,686 to + 2,736, downstream of the ACG start codon) including these two type-B ARR-binding elements was biotin labeled and used as a probe. Both ARR1 and ARR10 produced clear band shifts with the probe (Fig. 6C, D). Moreover, the addition of excess unlabeled competitor probes effectively reduced the amounts of shifted bands, while mutated probes could not compete in the association with the ARR proteins. These results confirm that ARR1 and ARR10 directly associate with AG transcriptional regulatory regions to regulate its expression positively.
Type-B ARRs regulate carpel initiation in planta
Finally, we studied whether the type-B ARRs regulate carpel initiation in planta. We first visualized the expression patterns of ARR1, ARR10 and AG using pARR1::ARR1-GFP, pAG::AG-GFP and pARR10::ARR10-mCherry; pAG::AG-GFP reporter lines. The expression signals of ARR1, ARR10 and AG overlapped in the central region of flower primordia, suggesting that there is a regulatory relationship among these proteins (Fig. 7A). Next, we determined whether defects in ARR genes affect floral organogenesis. We found that 15.71% of the arr1 10 12 pistils and 5.72% of the pistils in the am-ARR1/10/12 lines contained only one carpel (Fig. 7B). Therefore, compared with the wild type, the arr1 10 12 triple mutant and am-ARR1/10/12 lines produced fewer carpels per flower, but did not show changes in other floral organs including stamens, petals and sepals (Fig. 7C). These results indicate that type-B ARRs are required for carpel initiation in planta.
Fig. 7.
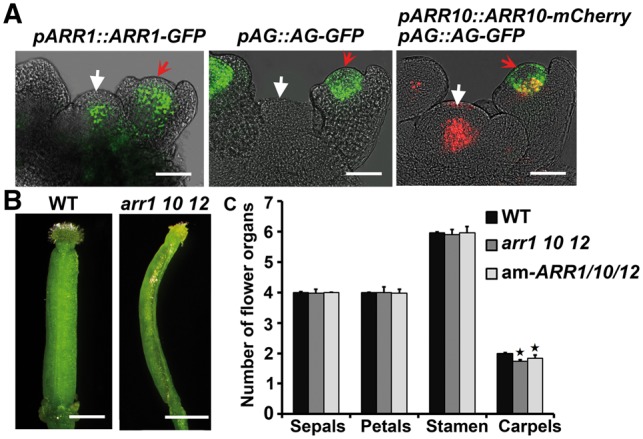
ARRs regulate carpel initiation in planta. (A) Expression pattern of pARR1::ARR1-GFP (green), pAG::AG-GFP (green), pARR10::ARR10-mCherry (red) and pAG::AG-GFP (green) double reporters in inflorescence meristems and floral primordia. White arrows indicate inflorescence meristems and red arrows indicate floral primordia at stage 3. ARR10 and AG signals overlapped in the central regions of the floral primordium (yellow). Scale bars = 50 μm. (B) Pistil (15.71%) from the arr1 10 12 triple mutant carpels formed only one carpel. Scale bars = 1 mm. (C) Compared with the wild type, arr1 10 12 triple mutant and am-ARR1/10 transgenic lines showed a reduced number of carpels, but not other floral organs. Error bars indicate the SDs of three biological replicates. The star indicates P < 0.01 (Student’s t-test).
Discussion
Hormone-induced ectopic expression of organ identity genes is critical for cellular reprogramming during plant regeneration
Plants are able to regenerate the whole body through establishing shoot and root apical meristems in vitro (Ikeuchi et al. 2016). Also, different types of floral organs can be induced in vitro, independent of the regeneration of the plant body (Li et al. 2002, An et al. 2004, Xu et al. 2004, Guan et al. 2006, Wu et al. 2008, Cheng et al. 2010). In recent years, significant progress has been made towards understanding specification of apical meristems (Liu et al. 2014, Kareem et al. 2015, Iwase et al. 2017, Meng et al. 2017, Zhang et al. 2017). However, the mechanisms underlying organ regeneration remained largely unknown. In the present study, we established a carpel regeneration system using a medium containing a high concentration of cytokinin. Culturing callus on CaIM with cytokinin alone was sufficient to induce carpel regeneration, but at a lower frequency, which may result from the attenuated callus proliferation without auxin. The results showed that type-B ARRs activated the expression of AG, the carpel identity gene, thus triggering the cellular reprogramming that specifies carpel formation.
Recent studies have shown that during shoot regeneration, type-B ARRs directly activate the expression of WUS, which encodes a key factor for maintaining shoot stem cells, and reprogram callus cells into the shoot stem cell niche (Meng et al. 2017, Zhang et al. 2017). For root regeneration, auxin directly induces the expression of WUSCHEL-RELATED HOMEOBOX 11 (WOX11) and WOX12, which act redundantly to up-regulate LATERAL ORGAN BOUNDARIES DOMAIN16 (LBD16) and LBD29, and promote the conversion from procambium cells to root founder cells (Chen et al. 2014, Liu et al. 2014, Sang et al. 2016). Therefore, it seems that the ectopic expression of organ identity genes induced by hormonal signaling is the critical step for plant organ regeneration. Thus, it is possible to control organ regeneration by mediating expression of organ identity genes.
Type-B ARRs regulates carpel initiation through mediating specific AG expression
AG is required for the determination of the stamen and gynoecium in planta (Yanofsky et al. 1990, Alvarez-Buylla et al. 2010). Overexpression of AG in Arabidopsis or Hyacinthus resulted in carpelloid sepals and stamioid petals, suggesting that the ectopic function of AG promotes the formation of both stamen and carpel structures (Mizukami and Ma 1992, Mizukami and Ma 1997, Li et al. 2002). However, in our system, AG expression induced by exogenous cytokinin led to the regeneration of carpels, and not staminoid structures. It has been shown that different regions in the second intron of AG confer its stamen-specific or carpel-specific expression patterns (Deyholos and Sieburth 2000). In developing flower primordia in planta, AG mRNA was detected in the third and fourth whorls before the initiation of stamen and carpel primordia, respectively, indicating that spatially specific expression of AG is sufficient for the initiation of stamen and carpel (Smyth et al. 1990, Yanofsky et al. 1990, Drews et al. 1991). Our ChIP and EMSA results revealed that during carpel induction, ARR1 and ARR10 specifically associated with the regulatory region sufficient for carpel-specific expression (Fig. 6). These results suggest that type-B ARRs regulate the carpel-specific expression of AG, which is required for carpel regeneration.
Previous studies have revealed the relationship between cytokinin and carpel initiation. For example, in the ckx3 ckx5 double mutant, the endogenous cytokinin content and the number of carpels were increased (Bartrina et al. 2011). In rice, mutations in LONELY GUY, which encodes a cytokinin-activating enzyme, led to abortion of pistil initiation (Kurakawa et al. 2007). In maize, the spikelet meristem gives rise to two florets; the lower one aborts early and cannot generate a pistil. Expression of the cytokinin-synthesizing ISOPENTENYL-TRANSFERASE gene under an Arabidopsis senescence-inducible promoter resulted in the formation of a functional pistil in the lower floret (Young et al. 2004). These findings demonstrate that cytokinin plays positive roles in regulating carpel initiation. However, the mechanisms underpinning this process remain elusive.
In this study, we found that the expression areas of ARR1 and ARR10 overlapped with that of AG during flower development. Mutations in ARR1, ARR10 and ARR12 gave rise to flowers with only one carpel, suggesting that type-B ARRs regulate carpel initiation through mediating AG expression in planta (Fig. 7). Our results demonstrate that the information derived from in vitro systems can help to understand mechanisms of biological processes in planta. Moreover, although defects in ARR1 and ARR10 significantly repressed carpel regeneration (Fig. 5), carpel initiation remained normal. Similar trends have also revealed previously that double mutations in type-B ARR genes suppressed shoot regeneration but did not affect shoot development in planta (Meng et al. 2017, Zhang et al. 2017). We thus suggest that in vitro regeneration is more sensitive to defects in cytokinin signaling than shoot development in planta.
Relationship between explant types and regenerated organs
Besides stage 10 pistils, other explants, including petals, sepals, leaves and roots, were also tested for their ability to regenerate carpels. However, after 25 d of culture on CaIM, only calli derived from pistils gave rise to regenerated carpels (data not shown). Similar results were also obtained in our previous studies, indicating that explant type is important to regenerate the same types of organs (Li et al. 2002, An et al. 2004, Xu et al. 2004, Guan et al. 2006, Wu et al. 2008, Cheng et al. 2010).
The transcriptional activities of key regulatory genes are always under spatiotemporal epigenetic regulation (He et al. 2011). For instance, histone H3 lysine 27 trimethylation (H3K27me3), a transcriptional repressive epigenetic modification marker, is highly enriched at the WUS locus in mature leaves, where WUS is not expressed (Li et al. 2011, Liu et al. 2011). The WUS locus exhibits a higher level of H3K27me3 and a lower level of H3 acetylation (the transcriptionally active modification) in early-stage than in late-stage leaf axils, correlating well with WUS activation and axillary meristem formation (Wang et al. 2017). The epigenetic regulation of AG expression has been suggested previously. In METHYLTRANSFERASE1 antisense transgenic plants, ectopic AG transcripts were detected in leaves, indicating that DNA methylation repressed AG expression (Finnegan et al. 1996). Histone acetylation has also been suggested to be involved in the regulation of AG expression (Bertrand et al. 2003). It is possible that during carpel regeneration, AG is under a permissive epigenetic environment in pistils, where it is easier for ARR proteins to activate its transcription and drive carpel formation. Furthermore, epigenetic regulation has been found to be cell division dependent in some cases, because cell division can dilute the inhibitory modifications (Sun et al. 2014, Wang et al. 2017, Zhang et al. 2017). This is consistent with our finding that carpels regenerated either from floral organ primordia or from callus, both of which contain actively dividing cells.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Wassilewskija (Ws) was used as the wild type in this study. The T-DNA insertion mutants arr1-2 (At3g16857, CS6368), arr10-1 (At4g31920, CS6369) and arr12-1 (At2g25180, CS6978) were obtained from the Arabidopsis Biological Resource Center. Double and triple mutants of ARR genes were derived by crossing. The transgenic lines of am-arr1/10 and am-arr1/10/12, and the reporter lines of pARR1::ARR1-GFP and pARR10::ARR10-mCherry have been described previously (Meng et al. 2017). To clear the genetic background, pAG::AG-GFP (kindly provided by Dr. Xuemei Chen, University of California at Riverside) was crossed with wild-type Ws for at least five generations. The pTFL1::GUS marker lines have been described previously (Guan et al. 2006).
Surface-sterilized Arabidopsis seeds were sown on 0.8% Phytoagar plates [half-strength Murashige and Skoog (MS) salts, 1% sucrose, pH 5.7]. After vernalization at 4 °C for 4 d, seedlings were grown under sterile conditions or in soil at 20–22 °C, under long-day conditions (16 h 100 μmol m–2 s–1 white light and 8 h dark).
Plant regeneration
For the inflorescence and carpel induction, pistils from flowers at stages 9 and 10 were used as explants. Pistils were excised and transferred onto CIM: MS medium containing 2.5 mg l–1 2,4-D, 0.4 mg l–1 6-benzyladenine and 0.8% (w/v) agarose. After 25 d of culture on CIM, the calli were transferred onto medium containing 0.01 mg l–1 IAA and 2.0 mg l–1 ZT for inflorescence induction, or medium containing 0.01 mg l–1 IAA and 20.0 mg l–1 ZT for carpal induction. For ethanol induction, am-AG or am-ARR1/10 callus was incubated on CaIM to which ethanol was added to a final concentration of 0.05% (v/v). Wild-type callus cultured on the same medium was used as control.
Plasmid construction
Artificial microRNAs targeting AG were designed using WMD3-Designer and were cloned into an ethanol-inducible vector (Leibfried et al. 2005, Zhao et al. 2010) to produce the pAlcA:am-AG constructs. The pAG::GUS vector, a 3,597 bp DNA fragment containing the second intron of AG, was PCR-amplified using the primers pAG-HindIII-F and pAG-KpnI-R, and was cloned into the pBI121 expression vector. The sequences of all primers are listed in Supplementary Table S1.
Scanning electron microscopy
The structures of the regenerated carpels were observed with a JSM-6610LV scanning electron microscope (JEOL) and photographed as previously described (Dai et al. 2014).
Histochemical GUS assay
For β-glucuronidase (GUS) staining, calli cultured for various periods on CIM or CaIM were harvested and fixed in 90% acetone on ice for 15 min, and were transferred into GUS staining buffer [50 mM NaPO4 (pH 7.2) 2 mM X-Gluc (Sigma-Aldrich), 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6], vacuum infiltrated, and incubated overnight at 37 °C. The calli were photographed using a stereomicroscope (SZX-16, Olympus) equipped with an Olympus DP72 digital camera. For anatomical analysis, stained calli were sectioned at 8 μm, stained with 0.2% ruthenium red and photographed using a stereomicroscope (BX-5, Olympus) equipped with an Olympus DP71 digital camera.
Confocal microscopy
For confocal microscopy, samples were prepared as described previously (Cheng et al. 2013, Meng et al. 2017). Images were captured under a TCS SP5II confocal laser scanning microscope (Leica) with a ×40 oil objective. Excitation and detection windows set-ups for GFP and mCherry were as described (Cheng et al. 2013, Meng et al. 2017).
qRT–PCR analyses
To examine the transcript levels of genes involving floral organ development, calli after 0, 4 and 6 d incubation on IIM or CaIM were obtained (Supplementary Fig. S4). To test the regulation of AG expression by type-B ARRs, calli derived from the wild type, am-ARR1/10 lines as well as arr1 10 double and arr1 10 12 triple mutants after 6 d incubation on CaIM were used (Fig. 5C). To examine the transcriptional levels of ARR1 and ARR10, callus derived from arr1 10 double mutants after 6 d incubation on CaIM was used (Supplementary Fig. S6). To detect the effectiveness of artificial microRNA against AG, the inflorescences before and after 12 h ethanol treatment were harvested from am-AG lines (Supplementary Fig. S5C). Total RNA was extracted from tissues using TRI Reagent (Sigma-Aldrich). The full-length cDNA was generated with the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). qRT–PCR was carried out on a Chromo4 real-time PCR system (Bio-Rad) using SuperReal PreMix Plus (Tiangen) with gene-specific primers. The gene transcript levels in each sample were normalized to that of the housekeeping gene TUBULIN2. Values shown are the mean ± SD of three biological replicates. The primers are listed in Supplementary Table S1.
In situ hybridization
The calli cultured for 0, 6 and 10 d on CaIM were collected and fixed in FAA (10% formaldehyde, 5% acetic acid and 50% alcohol) at 4 °C overnight., and then cut into 8 μm sections. RNA probes were synthesized and labeled in vitro, and the hybridized signals were detected as previously described (Zhao et al. 2006). Photographs were taken using a BX-51 microscope (Olympus) equipped with a DP71 digital camera (Olympus).
ChIP assays
The calli of pARR1::ARR1-GFP lines cultured on CaIM for 10 d were used as ChIP materials. The chromatin extract was immunoprecipitated using anti-GFP (Sigma-Aldrich). The ChIP experiments were performed as previously described (Cheng et al. 2013, Meng et al. 2017). In all of the experiments, three biological replicates were performed. The primers and oligonucleotides sequences are listed in Supplementary Tables S1 and S2.
Electrophoretic mobility shift assays
To construct plasmids for the expression of recombinant ARR1 and ARR10 proteins, the full-length coding sequences of the two genes were amplified and cloned into the pGEX-4T-1 vector, which was expressed in the Escherichia coli BL21 (DE3) cell line to produce glutathione S-transferase (GST)-tagged ARR proteins. The ARR1/10–GST proteins were purified using Glutathione Sepharose 4B (GE Healthcare). Double-stranded oligonucleotide probes were synthesized and labeled with biotin at the 5' end. The EMSAs were performed using a LightShift Chemiluminescent EMSA kit (Thermo). The competition experiments were carried out with different amounts of non-labeled probes. Mutated competitors were generated by replacing two base pairs in the ARR binding elements (GATC/T to CTTC/T). The oligonucleotides of the probes are listed in Supplementary Table S2.
Accession numbers
Sequence data from this study can be found in the EMBL/GenBank database and/or the Arabidopsis Genome Initiative database under the following accession numbers: AG (At4g18960), ARR1 (At3g16857), ARR10 (At4g31920), ARR12 (At2g25180), TFL1 (At5g03840), LFY (At5g61850), AP1 (At1g69120), AP2 (At4g36920), AP3 (At3g54340), TUBULIN2 (At5g62690) and UBQ10 (AT4G05320).
Supplementary Data
Supplementary data are available at PCP online.
Funding
This work was supported by the National Natural Sciences Foundation of China [31570281, 9121730 and 90917015] and Funds of Shandong ‘Double Tops’ Program [SYL2017XTTD13].
Supplementary Material
Acknowledgments
We thank the ABRC for providing the plant materials. We thank Dr. Xuemei Chen (University of California at Riverside) and Dr. Zhong Zhao (University of Science and Technology of China) for providing materials.
Disclosures
The authors of have no conflicts of interests to declare.
Glossary
Abbreviations
- AG
AGAMOUS
- am
artificial microRNA
- AP
APETALA
- ARR
Arabidopsis Response Regulator
- CaIM
carpel-inducing medium
- ChIP
chromatin immunoprecipitation
- CIM
callus-inducing medium
- EMSA
electrophoretic mobility shift assay
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- GUS
β-glucosidase
- IIM
inflorescence-inducing medium
- LBD
LATERAL ORGAN BOUNDARIES DOMAIN
- LFY
LEAFY
- qRT–PCR
quantitative reverse transcription–PCR
- WUS
WUSCHEL
- ZT
zeatin
References
- Alvarez-Buylla E.R., Benítez M., Corvera-Poiré A., Chaos Cador A., de Folter S., Gamboa, de Buen A., et al. (2010) Flower development. Arabidopsis Book 8: e0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Y.R., Li X.G., Su H.Y., Zhang X.S. (2004) Pistil induction by hormones from the callus of Oryza sativa in vitro. Plant Cell Rep. 23: 448–452. [DOI] [PubMed] [Google Scholar]
- Bartrina I., Otto E., Strnad M., Werner T., Schmülling T. (2011) Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 23: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand C., Bergounioux C., Domenichini S., Delarue M., Zhou D.X. (2003) Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 278: 28246–2851. [DOI] [PubMed] [Google Scholar]
- Bowman J.L., Drews G.N., Meyerowitz E.M. (1991) Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 3: 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J.L., Smyth D.R., Meyerowitz E.M. (1989) Genes directing flower development in Arabidopsis. Plant Cell 1: 37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Qu Y., Sheng L., Liu J., Huang H., Xu L. (2014) A simple method suitable to study de novo root organogenesis. Front. Plant Sci. 5: 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Wang L., Sun W., Zhang Y., Zhou C., Su Y.H., et al. (2013) Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161: 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Zhu S.S., Gao X.Q., Zhang X.S. (2010) Cytokinin and auxin regulates WUS induction and inflorescence regeneration in vitro in Arabidopsis. Plant Cell Rep. 29: 927–33. [DOI] [PubMed] [Google Scholar]
- Coen E., Meyerowitz E.M. (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37. [DOI] [PubMed] [Google Scholar]
- Dai X.R., Gao X.Q., Chen G.H., Tang L.L., Wang H., Zhang X.S. (2014) ABNORMAL POLLEN TUBE GUIDANCE1, an endoplasmic reticulum-localized mannosyl transferase homolog of GLYCOSYLPHOSPHATIDYLINOSITOL10 in yeast and PHOSPHATIDYLINOSITOL GLYCAN ANCHOR BIOSYNTHESIS B in human, is required for Arabidopsis pollen tube micropylar guidance and embryo development. Plant Physiol. 165: 1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyholos M.K., Sieburth L.E. (2000) Separable whorl-specific expression and negative regulation by enhancer elements within the AGAMOUS second intron. Plant Cell 12: 1799–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G.N., Bowman J.L., Meyerowitz E.M. (1991) Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65: 991–1002. [DOI] [PubMed] [Google Scholar]
- Duclercq J., Sangwan-Norreel B., Catterou M., Sangwan R.S. (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci. 16: 597–606. [DOI] [PubMed] [Google Scholar]
- Finnegan E.J., Peacock W.J., Dennis E.S. (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc. Natl. Acad. Sci. USA93: 8449–8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C.M., Zhu S.S., Li X.G., Zhang X.S. (2006) Hormone-regulated inflorescence induction and TFL1 expression in Arabidopsis callus in vitro. Plant Cell Rep. 25: 1133–7. [DOI] [PubMed] [Google Scholar]
- He G., Elling A.A., Deng X.W. (2011) The epigenome and plant development. Annu. Rev. Plant Biol. 62: 411–435. [DOI] [PubMed] [Google Scholar]
- Huang Z., Shi T., Zheng B., Yumul RE., Liu X., You C., et al. (2017) APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol. 215: 1197–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. (2016) Plant regeneration: cellular origins and molecular mechanisms. Development 143: 1442–1451. [DOI] [PubMed] [Google Scholar]
- Iwase A., Harashima H., Ikeuchi M., Rymen B., Ohnuma M., Komaki S., et al. (2017) WIND1 promotes shoot regeneration through transcriptional activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell 29: 54–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji L., Liu X., Yan J., Wang W., Yumul R.E., Kim Y.J., et al. (2011) ARGONAUTE10 and ARGONAUTE1 regulate the termination of floral stem cells through two microRNAs in Arabidopsis. PLoS Genet. 7: e1001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kareem A., Durgaprasad K., Sugimoto K., Du Y., Pulianmackal A.J., Trivedi Z.B., et al. (2015) PLETHORA genes control regeneration by a two-step mechanism. Curr. Biol. 25: 1017–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., et al. (2005) WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 438: 1172–1175. [DOI] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jurgens G., Laux T. (2001) Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105: 805–814. [DOI] [PubMed] [Google Scholar]
- Li Q.Z., Li X.G., Bai S.N., Lu W.L., Zhang X.S. (2002) Isolation of HAG1 and its regulation by plant hormones during in vitro floral organogenesis in Hyacinthus orientalis L. Planta 215: 533–540. [DOI] [PubMed] [Google Scholar]
- Li W., Liu H., Cheng Z.J., Su Y.H., Han H.N., Zhang Y., et al. (2011) DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 7: e1002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Sheng L., Xu Y., Li J., Yang Z., Huang H., et al. (2014) WOX11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell. 26: 1081–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Dinh T.T., Li D., Shi B., Li Y., Cao X., et al. (2014) AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 80: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kim Y.J., Müller R., Yumul R.E., Liu C., Pan Y., et al. (2011) AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J.U., Hong R.L., Hobe M., Busch M.A., Parcy F., Simon R., et al. (2001) A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105: 793–803. [DOI] [PubMed] [Google Scholar]
- Lu W., Enomoto K., Fukunaga Y., Kuo C. (1988) Regeneration of tapales, stamens and ovules in explants from perianth of Hyacinthus orientalis L. Importance of explant age and exogenous hormones. Planta 175: 478–484. [DOI] [PubMed] [Google Scholar]
- Lu W.L. (2003) Control of in vitro regeneration of individual reproductive and vegetative organs in Dracaena fragrans cv. massangeana Hort.—regularities of the direct regeneration of individual organs in vitro. Acta Bot. Sin. 45: 1453–1464. [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., et al. (2017) Type-B ARABIDOPSIS RESPONSE REGULATORs specify the shoot stem cell niche by dual regulation of WUSCHEL. Plant Cell 29: 1357–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukami Y., Ma H. (1992) Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 71: 119–131. [DOI] [PubMed] [Google Scholar]
- Mizukami Y., Ma H. (1997) Determination of Arabidopsis floral meristem identity by AGAMOUS. Plant Cell 9: 393–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y.L., Cheng Z.J., Zhang X.S. (2016) Endogenous auxin biosynthesis and de novo root organogenesis. J. Exp. Bot. 67: 4011–4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B., Xu C., Zhang X., Cao H., Xin W., Hu Y. (2016) Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc. Natl. Acad. Sci. USA 113: 5101–5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoog F., Miller C.O. (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp. Soc. Exp. Biol. 11: 118–130. [PubMed] [Google Scholar]
- Smyth D.R., Bowman J.L., Meyerowitz E.M. (1990) Early flower development in Arabidopsis. Plant Cell 2: 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Looi L.S., Guo S., He Z., Gan E.S., Huang J., et al. (2014) Timing mechanism dependent on cell division is invoked by Polycomb eviction in plant stem cells. Science 343: 1248559. [DOI] [PubMed] [Google Scholar]
- Wang J., Tian C., Zhang C., Shi B., Cao X., Zhang T.Q., et al. (2017) Cytokinin signaling activates WUSCHEL expression during axillary meristem initiation. Plant Cell 29: 1373–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D., Meyerowitz E.M. (1993) Activation of floral homeotic genes in Arabidopsis. Science 261: 1723–1726. [DOI] [PubMed] [Google Scholar]
- Wu B.H., Zheng Y.L., Liu D.C., Zhou Y.H., Yan Z.H. (2003) Unisexual pistillate flower regeneration in immature embryo culture of wheat. Acta Bot. Sin. 45: 452–459. [Google Scholar]
- Wu X.Q., Li X.G., Zhang X.S. (2008) Molecular analysis of hormone-regulated petal regeneration in Petunia. Plant Cell Rep. 27: 1169–1176 [DOI] [PubMed] [Google Scholar]
- Xu H.Y., Li X.G., Li Q.Z., Bai S.N., Lu W.L., Zhang X.S. (2004) Characterization of HoMADS 1 and its induction by plant hormones during in vitro ovule development in Hyacinthus orientalis L. Plant Mol Biol. 55: 209–220. [DOI] [PubMed] [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Dews G., Feldmann K., Meyerowitz E.M. (1990) The protein encoded by the Arabidopsis homeotic gene AGAMOUS resembles transcription factors. Nature 346: 35–39. [DOI] [PubMed] [Google Scholar]
- Young T.E., Geisler-Lee J., Gallie D.R. (2004) Senescence-induced expression of cytokinin reverses pistil abortion during maize flower development. Plant J. 38: 910–922. [DOI] [PubMed] [Google Scholar]
- Zhang T.Q., Lian H., Zhou C.M., Xu L., Jiao Y., Wang J.W. (2017) A two-step model for de novo activation of WUSCHEL during plant shoot regeneration. Plant Cell 29: 1073–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.Y., Cheng Z.J., Zhang X.S. (2006) Overexpression of TaMADS1, a SEPALLATA-like gene in wheat, causes early flowering and the abnormal development of floral organs in Arabidopsis. Planta 223: 698–707. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Andersen S.U., Ljung K., Dolezal K., Miotk A., Schultheiss S.J., et al. (2010) Hormonal control of the shootstem-cell niche. Nature 465: 1089–1092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.