Abstract
Due to the antimicrobial activity of flavonoids, it has been suggested that they may provide a possible alternative to antibiotics to stimulate productivity and reduce the environmental load of ruminant agriculture. We hypothesised that an extract of liquorice, rich in prenylated isoflavonoids and particularly glabridin, might potentially improve the efficiency of nitrogen utilisation and reduce methane production in the rumen. When added to a long-term rumen simulating fermentor (RUSITEC), liquorice extract at 1 g L−1 decreased ammonia production (−51%; P < 0.001) without affecting the overall fermentation process. When added at 2 g L−1, decreases in not only ammonia production (−77%; P < 0.001), but also methane (−27%; P = 0.039) and total VFA production (−15%; P = 0.003) were observed. These effects in fermentation were probably related to a decrease in protozoa numbers, a less diverse bacteria population as well as changes in the structure of both the bacterial and archaeal communities. The inclusion of an isoflavonoid-rich extract from liquorice in the diet may potentially improve the efficiency of the feed utilisation by ruminants.
Keywords: glabridin, isoflavonoids, liquorice, methane, rumen fermentation
An isoflavonoid-rich liquorice extract could potentially be used to boost productivity and decrease the environmental burden of livestock production due to its effects on rumen microbial communities.
INTRODUCTION
Since the ban of antibiotics as growth-promoting feed additives by the European Union in 2006, plant extracts and plant secondary metabolites have been considered as alternatives to manipulate rumen fermentation to boost productivity and decrease the environmental burden of livestock production (Hart et al.2008). Among plant secondary metabolites, flavonoids have recently gained interest because of their wide range of biological activities, particularly antimicrobial properties (Oskoueian, Abdullah and Oskoueian 2013).
Flavonoids are polyphenolic compounds consisting of a 15-carbon skeleton in which two benzene rings are linked via a heterocyclic pyran ring (Kumar and Pandey 2013). According to substitution pattern variations, flavonoids can be classified into different subclasses, providing an extremely diverse range of derivatives (Wang, Li and Bi 2018). Depending on their chemical structure, flavonoids can then have different antimicrobial effects (Wang, Li and Bi 2018) that ultimately determine the extent in which rumen fermentation can be altered (Oskoueian, Abdullah and Oskoueian 2013).
The effect of flavonoids on rumen fermentation has not been extensively evaluated (Patra et al.2017). In addition and taking into account their great variability in structure (over 9000 different compounds identified; Wang, Li and Bi 2018), only a small number of flavonoid-rich plant extracts or pure compounds have been tested so far. Some flavonoids, or the derivatives produced by microbial degradation in the rumen, have been reported to affect rumen microbial activity causing, amongst other effects, a decrease in methane production (Oskoueian, Abdullah and Oskoueian 2013; Kim et al.2015; Ma et al.2017). Furthermore, flavonoids have been shown to be effective in attenuating the effects of excessive grain feeding on rumen pH (Balcells et al.2012; de Nardi et al.2014). However, to our knowledge, a detailed characterisation of the changes in rumen microbial communities associated with the effects of flavonoids on rumen fermentation has not yet been published. In this study, we tested an extract of liquorice, rich in prenylated isoflavonoids and particularly glabridin (Asl and Hosseinzadeh 2008), for its effect in in vitro batch culture and its long-term effect on rumen fermentation and methanogenesis whilst also characterising its effect on bacterial and methanogen communities.
MATERIAL AND METHODS
Liquorice extract
Liquorice extract was obtained from the dried roots of liquorice (Glycyrrhiza glabra; 40 g) after extraction with 95% ethanol (2 L for 2 h) at 45°C and then concentrating under reduced pressure to give a 95% ethanol extract (1.20 g). The extract was further purified on silica gel eluted with ethyl acetate–methanol gradients (19 : 1; 9 : 1; 2 : 1) and finally with methanol. Nuclear magnetic resonance analysis revealed that glabridin was the major flavonoid in the extract, with five other related flavonoids found. Fitness R Us Ltd (Kiryat Shmona, Israel) provided the extract as Licogen powder (Batch No: 14090023PWDR). Liquorice powder is sold as a natural antioxidant, flavouring powder and phytoestrogen for menopausal women.
Measurement of protozoal activity
The effect of liquorice extract on protozoal activity was measured in vitro as the breakdown of [14C]-labelled bacteria by rumen protozoa as described by Wallace and McPherson (1987). Isotope-labelled bacteria were obtained by growing Streptococcus bovis ES1 in Wallace and McPherson media (Wallace and McPherson 1987) containing [14C] leucine (1.89 μCi/7.5 mL tube) as the sole nitrogen source, for 24 h. Cultures were centrifuged (3000 g, 15 min), supernatant discarded and pellets re-suspended in 7 mL of simplex-type salt solution (STS; Williams and Coleman 1992) containing 12C-leucine (5 mM). This process was repeated three times to prevent re-incorporation of released [14C] leucine by bacteria.
Rumen digesta was obtained from four rumen-cannulated Holstein–Frisian cows fed at maintenance level (composed of perennial ryegrass hay and concentrate at 67 : 33 on a DM basis). Animal procedures were carried out in accordance with the Animal Scientific Procedures Act 1986, and protocols were approved by the Aberystwyth University Ethical Committee. Rumen digesta was obtained before the morning feeding and strained through two layers of muslin and diluted with STS (1 : 1) containing 12C-leucine (5 mM). Diluted rumen fluid (7.5 mL) was then incubated with labelled bacteria prepared as described above (0.5 mL) in tubes containing no additive (control) or 0.25, 0.5, 1 or 2 g L−1 of liquorice extract. Incubations were carried out at 39°C under a stream of CO2, and tubes were sampled at time 0 and at 1 h intervals up to 5 h using a syringe with a 19 gauge needle. Samples (0.5 mL) were acidified (by adding 0.125 mL of 25% trichloroacetic acid (wt/vol) and centrifuged (13 000 g, 5 min). Supernatant (0.200 mL) was diluted with 2 mL of OptiPhase HiSafe 2 scintillation fluid (Perkin Elmer, Seer Green, UK) to determine the radioactivity released by liquid-scintillation spectrometry (Hidex 300 SL, Lablogic Systems Ltd, Broomhill, UK). Bacterial breakdown at each incubation time was expressed as the percentage of the acid-soluble radioactivity released relative to the total radioactivity present in the initial labelled bacteria (Wallace and McPherson 1987).
In vitro batch cultures
To measure the short term effect of liquorice extract on fermentation parameters, 24 h in vitro incubations were carried out. The experimental design consisted of a control (no additive) and liquorice extract added at 0.5, 1 or 2 g L−1. The experiment was conducted in quadruplicate, using rumen fluid from the same four cannulated cows. Rumen contents were sampled before the morning feeding, filtered through a double layer of muslin and diluted 1 : 2 in artificial saliva solution (Menke and Steingass 1988). Aliquots (30 mL) of the diluted strained rumen fluid were added anaerobically to 120 mL Wheaton bottles containing 0.3 g of diet composed of ryegrass hay and barley (40 : 60), previously ground to pass through a 1-mm2 mesh screen. Bottles were sealed and incubated at 39°C receiving a gentle mix before sampling at 24 h.
Fermentation pattern, in terms of pH, ammonia and VFA was determined after 24 h of the incubation. A subsample (4 mL) was diluted with 1 mL of deproteinising solution (200 mL L−1 orthophosphoric acid containing 20 mmol L−1 of 2-ethylbutyric acid as an internal standard) for the determination of VFA using gas chromatography, as described by Stewart and Duncan (1985). Another subsample (1 mL) was diluted with 0.250 mL of 25% trichloroacetic acid (wt/vol) for analysis of ammonia using a colourimetric method (Weatherburn 1967).
Rumen simulation technique
The rumen simulation technique (RUSITEC; Czerkawski and Breckenridge 1977) was used to study the effect of a control diet alone or supplemented with liquorice extract at 1 g L−1 (liquorice 1; 0.66 g d−1, 3.3% inclusion rate in DM) or 2 g L−1 (liquorice 2; 1.32 g d−1, 6.6% inclusion rate in DM), doses that were selected based on the results obtained in the 24 h batch culture trial described above. The experimental diet was the same one used in the batch culture trial (40 : 60, ryegrass hay and barley grounded to pass through 1 mm2 sieve size).
Rumen digesta was obtained from four rumen-cannulated Aberdale x Texel sheep, fed at maintenance level (diet composed of perennial ryegrass hay and concentrate at 67 : 33 on DM basis). Animal procedures were carried out in accordance with the Animal Scientific Procedures Act 1986, and protocols were approved by the Aberystwyth University Ethical Committee. Rumen digesta was obtained before the morning feeding, strained through two layers of muslin and stored anaerobically at 39°C.
The trial consisted of a single incubation period using 12 vessels that were considered as experimental units. Each dietary treatment was randomly allocated to the vessels that were inoculated with rumen fluid from four different sheep (four replicates). Vessels had an effective volume of 800 mL and were kept at 39°C under permanent vertical agitation.
On day 1, vessels were inoculated with strained rumen fluid mixed with artificial saliva (McDougall 1948) and demineralised water in a 1 : 1 : 1 ratio. Then, artificial saliva was continuously infused at a rate of 660 mL d−1 (dilution rate of 3%/h) using a multichannel peristaltic pump (Watson–Marlow 200 series, Cornwall, UK). Squeezed rumen solids (20 g FM) were placed in nylon bags (110 × 60 mm, pore size 100 μm2) and incubated in each vessel for 1 day to provide solid-associated bacteria, while experimental feed (20 g DM) was supplied in a second bag. On subsequent days, the feed bag that had remained 2 days in each vessel was squeezed, returning the liquid to the vessel, and discarded; a new bag, containing 20 g DM was then inserted to the vessel.
The trial lasted for 18 days, using the first 12 days for adaptation and the last 6 for sampling. Dry matter degradation, total gas and methane production and outflow of fermentation products were measured on days 13, 14, 15 and 16. Nylon bags were collected, rinsed with cold water for 20 min, and DM disappearance after 48 h incubation was calculated from the loss in weight. The residue was then analysed for organic matter (OM), nitrogen (N), Neutral-detergent (NDF) and Acid-detergent fibre (ADF) to determine nutrient disappearance. Fermentation gases were collected in gas-tight bags (TECOBAG 5L, PETP/AL/PE-12/12/75, Tesseraux container GmbH, Germany) to measure total gas and methane production. Daily production of ammonia and VFA were measured in the overflow flasks with 10 mL of saturated HgCl2 (diluted 1 : 5) added to stop the fermentation.
To describe diurnal changes in the fermentation pattern, on days 17 and 18 the content of the vessels was sampled (25 mL) by aspiration at 0, 2, 4 and 8 h after feeding. The pH was immediately recorded, and five subsamples were collected as follows: for microbial characterisation and enzymatic activity, 16 mL were collected and immediately frozen in liquid N prior to long-term storage at −80°C. For VFA determination, 1.6 mL of sample was diluted with 0.4 mL of deproteinising solution (200 mL L−1 orthophosphoric acid containing 20 mmol L−1 of 2-ethylbutyric acid as an internal standard). For ammonia analysis, 0.8 mL of sample was diluted with 0.2 mL of trichloro-acetate (25% wt:vol). For lactate determination, 1 mL sample was collected and snapped frozen in liquid N prior to long-term storage at −80°C. For protozoa counts, 0.5 mL of sample were added to 0.5 mL of saline formaline solution (4% formaldehyde and 0.9% NaCl in distilled water) and stored at room tempreture.
Sample analyses
For feed analysis, DM and OM content were determined by drying in an oven at 105°C for 24 h and heating at 550°C for 6 h in a muffle furnace, respectively. Nitrogen concentration was measured by the Dumas combustion method (Elementar analyser, Vario MAX cube, Hanau, Germany). For NDF and ADF determination, the automated fibre analyser (ANKOM 2000, Macedon, USA) was used. Methane concentration was determined by directly injecting 0.5 mL of gas sample into a gas chromatograph (ATI Unicam 610 Series, Cambridge, UK) fitted with a 40 cm Porapak N metal packed column (Agilent, Cheshire, UK) and flame ionisation detector. Ammonia and VFA concentrations in vessels and overflows were determined as described by Weatherburn (1967) using an automated spectrophotometer (ChemWell T,Astoria Pacific, Oregon, USA) and Stewart and Duncan (1985) using gas chromatography, respectively. Protozoa were quantified by optical microscope following the procedure described by Dehority (1993) and adapted by de la Fuente, Skirnisson and Dehority (2006). Concentrations of L-lactate and D-Lactate were measured using the Enzytec D/L-Lactic Acid kit (r-biopharm, Darmstadt, Germany); total lactate was calculated as the sum of both. Enzymatic activities in vessels content were measured according to the procedure described by Giraldo et al. (2008) and Belanche at al. (2016). Endoglucanase (EC 3.2.1.4.), xylanase (EC 3.2.1.8.) and amylase activities (EC 3.2.1.1.) were measured in triplicate and expressed as mmol of sugar released from the corresponding substrates in 1 min per gram DM of sample (or gram of protein).
DNA extraction and quantitative PCR
Genomic DNA was extracted from vessel samples withdrawn at different time points. Freeze-dried samples (25 mg DM) were bead beaten in 4% SDS lysis buffer for 45 s, and DNA was extracted using a CTAB/Chloroform method (adapted from Yu and Morrison 2004).
Concentration and quality of genomic DNA was assessed by spectrophotometry (Nanodrop ND-100, Thermo Scientific, USA). Absolute concentrations of DNA from total bacteria, methanogens and fungi were determined by qPCR and serial dilutions of their respective standards (10−1–10−5) as previously described (Belanche et al.2012, 2016). Quantitative PCR (qPCR) was conducted in triplicate using a LightCycler 480 System (Roche, Mannheim, Germany).
Ion torrent next generation sequencing
Rumen bacteria and methanogenic archaea communities were studied using Next Generation Sequencing (NGS) (de la Fuente et al.2014). For bacterial profiling, amplification of the V1–V2 hypervariable regions of the 16S rRNA gene was carried out using bacterial primers (27F and 357R) followed by Ion Torrent adaptors. For methanogens profiling, amplification of the V2-V3 hypervariable region of the 16S rRNA gene was performed using archaeal primers (86F and 519R) also followed by Ion Torrent adaptors.
Forward primers were barcoded with 10 nucleotides to allow sample identification. PCR was carried out on a 25 μL reaction containing DNA template (1 μL), 0.2 μL reverse primer, 1 μL forward primer, 5 μL buffer (PCR Biosystems Ltd., London, UK), 0.25 μL bio HiFi polymerase (PCR Biosystems) and 17.6 μL molecular grade water. Amplification conditions for bacteria and methanogens were 95°C for 1 min, and then 22 cycles of 95°C for 15 s, 55°C for 15 s and 72°C for 30 s. To assess quality of amplifications, resultant amplicons were visualised on a 1% agarose gel. PCR products were then purified using Agencourt AMpure XP beads (Beckman Coulter Inc., Fullerton, USA), and DNA concentration was determined using an Epoch Microplate Spectrophotometer fitted with a Take 3 Micro-Volume plate (BioTek, Potton, UK) to enable equimolar pooling of samples with unique barcodes.
Libraries were further purified using the EGel system with 2% agarose gel (Life Technologies Ltd., Paisley, UK). Purified libraries were assessed for quality and quantified on an Agilent 2100 Bioanalyzer with High Sensitivity DNA chip (Agilent Technologies Ltd., Stockport, UK). Library preparation for NGS sequencing was carried out using the Ion Chef system (Life Technologies UK Ltd) and the Ion PGM HiQ Chef kit, and sequencing using the Ion Torrent Personal Genome Machine (PGM) system on an Ion PGM Sequencing 316 Chip v2 BC. Due to the lower abundance of methanogens than total bacteria, methanogens library was sequenced using a smaller chip (Ion PGM Sequencing 314 Chip v2).
Following sequencing, data were processed as previously described (de la Fuente et al.2014). Briefly, sample identification numbers were assigned to multiplexed reads using the MOTHUR software package. Data were denoised by removing low-quality sequences, sequencing errors and chimeras (quality parameters: maximum 10 homopolymers, qaverage 13, qwindow 25, for archaea the qwindow was set at 30, and erate = 1; Chimera check, both de novo and database driven using Uchime).
Sequences were clustered into OTUs using the Uparse pipeline at 97% identity. Bacterial taxonomic information on 16S rRNA gene sequences was obtained by comparing against Ribosomal Database Project-II (Wang et al2007), while the methanogens were compared with the RIM-DB database (Seedorf et al.2014). The number of reads per sample was normalised to the sample with the lowest number of sequences. To exclude potential bacterial sequences from the methanogens dataset, methanogens sequences were blasted with the Ribosomal Database Project-II, and those annotations that matched with bacterial sequences were removed. Raw sequences reads from the bacterial and methanogens libraries were deposited at the EBI Short Read Archive of the European Nucleotide Archive (accession number PRJEB22945 and PRJEB22960, respectively).
Statistical analysis
Linear regression was conducted to model the relationship between the percentage of radioactivity released (relative to the 14C-bacterial inoculum) and the time (from 0 h to 5 h), as well as its correlation coefficient. The slope of this trend-line indicated the bacterial degradation rate (as % h−1) by the rumen protozoa and ultimately their activity. Trend line slopes, 24 h fermentation parameters, daily productions of VFA and ammonia in the RUSITEC system together with nutrient disappearance and methane data were analysed statistically by randomised block ANOVA, with individual cows/sheep as a blocking term. For the rates of bacterial degradation and 24 h fermentation parameters, polynomial contrasts were also used to determine linear (L) and/or quadratic (Q) responses to the treatments. Rumen fermentation and qPCR data in the RUSITEC were analysed using a repeated-measurements procedure (REML) including the different time-points (0, 2, 4 and 8). The effect of treatment, time and treatment × time interaction on the relative abundance of different bacteria, and archaea taxa was analysed by split plot ANOVA (three treatments × four time points). P values were adjusted for multiple testing using the method proposed by Benjamini and Hochberg (1995) to decrease the false discovery rate. When effects were detected, treatment means were compared by Fisher's protected LSD test. Findings with P < 0.05, P < 0.10 when applying Benjamini and Hochberg (1995) correction, were regarded statistically significant. Genstat 15th Edition (VSN International, Hemel Hempstead, UK) was used.
Permutation multivariate analysis of variance (PERMANOVA) was used to determine overall significant differences in bacterial and archaea community and was performed in PRIMER 6 and PERMANOVA + (versions 6.1.18 and 1.0.8, respectively; Primer-E, Ivybridge, UK). Abundance percentage data were subjected to square root transformation and Bray–Curtis distance matrices calculated. PERMANOVA was carried out using default settings with 9999 unrestricted permutations, and the Monte Carlo P value was calculated. Analysis of Similarity (ANOSIM) was carried out in PRIMER 6 and PERMANOVA + using the Bray–Curtis distance matrix calculated above. This analysis was used to provide a metric of the degree of divergence between communities as given by the R statistic.
To calculate the contribution of environmental data on bacteria and archaea communities, distance-based linear modelling was used to calculate which environmental variables had a significant correlation with the community data. Significant variables were used in distance-based redundancy analysis (dbRDA) (Legendre and Anderson 1999) as implemented in PRIMER 6 and PERMANOVA+.
RESULTS
Acute antiprotozoal activity and effect on fermentation parameters (in vitro batch incubations)
Bacterial degradation by protozoa increased linearly (R2 > 0.99) over the 5 h incubation with the control treatment. Increasing levels of liquorice extract resulted in a linear and quadratic decrease (P < 0.001) in the breakdown of bacteria by protozoa (Table S1, Supporting Information). Whereas the rate of bacterial breakdown was not affected by the addition of 0.25 g L−1 of the flavonoid-rich extract, it was reduced by 55.6% (P < 0.001) in the presence of 0.5 g L−1. Doses of 1 and 2 g L−1 of liquorice extract caused a dramatic reduction in protozoa activity (P < 0.001) with no bacterial breakdown observed.
Based on these results, doses of 0.5, 1 and 2 g L−1 of the extract were tested over 24 h in in vitro incubations (Table 1). Liquorice extract added at 1 and 2 g L−1 of the incubation caused only a moderate decrease in pH, albeit significant (P = 0.001), compared with the control. No effect on the concentration of total VFA or on the molar proportion of acetate was observed at any of the concentrations tested (P > 0.05). Doses of 1 and 2 g L−1 resulted in a decrease in ammonia concentration (P = 0.010) and in an increase in the molar proportion of propionate (P = 0.002). A reduction in the molar proportions of butyrate (P = 0.013) and branched chain volatile fatty acids (BCVFA) (P = 0.024) was only observed with the highest dose of liquorice.
Table 1.
Effect of liquorice extract at 0.5, 1 and 2 g L−1 on pH, NH3-N and VFA profile in ruminal digesta after 24 h of incubation.
| Dose (g L−1) | |||||||
|---|---|---|---|---|---|---|---|
| 0 | 0.5 | 1 | 2 | SED | P | Contrast | |
| pH | 6.44b | 6.44b | 6.37a | 6.35a | 0.017 | 0.001 | L*** |
| NH3-N (mg L−1) | 20.5c | 19.5bc | 18.2ab | 16.2a | 1.00 | 0.010 | L** |
| Total VFAs (mmo L−1) | 77.5 | 77.7 | 79.4 | 77.9 | 4.10 | 0.965 | – |
| Molar proportions | |||||||
| Acetate | 59.2 | 59.0 | 59.6 | 57.1 | 3.14 | 0.858 | – |
| Propionate | 17.4a | 18.0ab | 19.8b | 22.9c | 1.03 | 0.002 | L*** |
| Butyrate | 12.0b | 11.8b | 11.4b | 9.42a | 0.666 | 0.013 | L** |
| BCVFA | 2.49b | 2.42b | 2.34b | 1.95a | 0.149 | 0.024 | L** |
a–cMeans with different superscript differ (P < 0.05); L: linear response; **P < 0.01; ***P < 0.001. BCVFA = Branched chain volatile fatty acids.
Feed degradability and fermentation pattern (rumen simulation technique)
Because the batch culture experiment showed no effect on fermentation with liquorice added at 0.5 g L−1, only doses of 1 and 2 g L−1 were further tested in the RUSITEC system.
The addition of liquorice to the diet did not have any detrimental effect on feed disappearance after 48 h of incubation (Table 2), although a trend (P = 0.069) to decreased OM disappearance was observed with the highest dose of liquorice tested. When liquorice was added at 1 g L−1, no negative effects on fermentation were observed, whilst ammonia production decreased (−51%; P < 0.001). The addition of 2 g L−1, however, had a strong effect decreasing total VFA concentration (P = 0.014), shifting fermentation towards propionate (P = 0.012) at the expense of acetate (P = 0.003), as well as dramatically decreasing ammonia production (−77%; P < 0.001). Although total gas production was not affected by the inclusion of 2 g L−1 liquorice in the diet, methane production decreased (P < 0.05) by 35% (Table 3). Theoretical metabolic hydrogen production based on the VFA stoichiometry (Moss, Jouany and Newbold 2000) was also lower (P = 0.002) with 2 g L−1 of the flavonoid-rich extract.
Table 2.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) on feed disappearance in the RUSITEC system.
| Treatment | C | L1 | L2 | SED | P |
|---|---|---|---|---|---|
| Disappearance (%) | |||||
| DM | 43.4 | 43.4 | 42.2 | 1.42 | 0.653 |
| OM | 47.2 | 45.4 | 42.3 | 1.67 | 0.069 |
| N | 43.2 | 43.3 | 43.3 | 1.77 | 0.998 |
| NDF | 38.7 | 37.0 | 37.2 | 1.01 | 0.257 |
| ADF | 44.2 | 43.5 | 43.2 | 0.76 | 0.451 |
Table 3.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) on fermentation products and methanogenesis in the RUSITEC system.
| Treatment | C | L1 | L2 | SED | P |
|---|---|---|---|---|---|
| Fermentation products (mmol d−1) | |||||
| Total VFA | 33.7b | 34.1b | 28.5a | 1.056 | 0.003 |
| Acetate | 17.2b | 17.0b | 12.8a | 0.795 | 0.003 |
| Propionate | 3.55a | 4.06a | 4.94b | 0.313 | 0.012 |
| Butyrate | 8.08 | 7.95 | 7.56 | 0.333 | 0.332 |
| BCVFA | 3.51b | 3.74b | 0.482a | 0.213 | <0.001 |
| Ammonia | 1.37c | 0.674b | 0.315a | 0.106 | <0.001 |
| Gas emissions | |||||
| Total gas (L d−1) | 1.20 | 1.23 | 1.30 | 0.069 | 0.384 |
| Methane (mM) | 3.92b | 3.58b | 2.37a | 0.175 | <0.001 |
| Methane (mmol d−1) | 4.67b | 4.35b | 3.06a | 0.445 | 0.024 |
| Methane (mmol gDOM−1) | 0.510b | 0.492b | 0.374a | 0.043 | 0.039 |
| 2H produced (mmol d−1) | 70.2b | 69.8b | 60.8a | 1.59 | 0.002 |
a and bMeans with different superscript differ (P < 0.05). BCVFA = Branched chain volatile fatty acids.
The study of the fermentation pattern in the vessel over a 24 h period (days 17 and 18; Table S2, Supporting Information) showed the same differences between treatments as those described when studying the daily fermentation products in the overflow. Concentrations of D-, L- and total lactate were unaffected by the treatments. Sampling time had a strong effect on fermentation parameters with decreased ammonia concentration and increased propionate and butyrate concentrations after feeding (P < 0.01). D-, L- and total lactate concentrations decreased (P < 0.001) in samples taken after feeding.
Absolute and relative enzymatic activities (Table 4) increased (P < 0.05) when liquorice was added at 2 g L−1. Sampling time also had an effect, with decreased xylanase and endoglucanase activities after feeding (P < 0.01). Quantitative PCR revealed decreases in only the relative abundance of anaerobic fungi (P < 0.001) with 2 g L−1 liquorice. Protozoa concentration decreased (P < 0.001) in vessels fed liquorice, with the highest dose having a stronger effect. The addition of liquorice, at all the doses tested, caused the elimination of the holotrich protozoa.
Table 4.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) and the sampling time (0, 2, 4 and 8 h after feeding) on rumen enzymatic activity and microbial numbers in the RUSITEC system.
| Treatment | P-Value | Time after feeding | P-Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | L1 | L2 | SED | Trt | 0 h | 2 h | 4 h | 8 h | SED | T | TrtxT | |
| Absolute enzymatic activity (mmol of sugar gDM−1 min−1) | ||||||||||||
| Amylase | 0.060a | 0.069a | 0.138b | 0.016 | 0.005 | 0.083 | 0.097 | 0.092 | 0.083 | 0.011 | 0.435 | 0.464 |
| Xylanase | 0.081a | 0.086a | 0.103b | 0.006 | 0.034 | 0.123b | 0.083a | 0.083a | 0.071a | 0.006 | <0.001 | 0.166 |
| Endoglucanase | 0.062 | 0.067 | 0.074 | 0.004 | 0.097 | 0.085b | 0.064a | 0.062a | 0.060a | 0.003 | <0.001 | 0.031 |
| Relative enzymatic activity (mmol of sugar gProtein−1 min−1) | ||||||||||||
| Amylase | 0.306a | 0.315a | 0.664b | 0.031 | <0.001 | 0.405 | 0.444 | 0.401 | 0.463 | 0.061 | 0.567 | 0.391 |
| Xylanase | 0.42a | 0.396a | 0.493b | 0.022 | 0.012 | 0.606b | 0.392a | 0.357a | 0.390a | 0.039 | <0.001 | 0.437 |
| Endoglucanase | 0.326a | 0.314a | 0.356b | 0.010 | 0.013 | 0.421b | 0.304a | 0.267a | 0.337a | 0.030 | 0.006 | 0.940 |
| Microbial numbers | ||||||||||||
| Bacteria (log copies gDM−1) | 11.4 | 11.4 | 11.4 | 0.033 | 0.093 | 11.3 | 11.4 | 11.4 | 11.4 | 0.029 | 0.068 | 0.135 |
| Methanogens (log copies gDM−1) | 9.66a | 9.82b | 9.61a | 0.051 | 0.016 | 9.83c | 9.73b | 9.62a | 9.62a | 0.032 | <0.001 | 0.182 |
| Anaerobic fungi (log copies gDM−1) | 7.99b | 7.47b | 4.71a | 0.277 | <0.001 | 7.04 | 6.75 | 6.69 | 6.41 | 0.250 | 0.160 | 0.690 |
| Protozoa (log cells mL−1) | ||||||||||||
| Total | 3.57c | 2.89b | 2.30a | 0.114 | <0.001 | |||||||
| Holotrichs | 2.91b | 0a | 0a | 0.062 | <0.001 | |||||||
| Entodinomorphs | 3.46c | 2.89b | 2.30a | 0.126 | <0.001 | |||||||
a–cMeans with different superscript differ (P < 0.05). Trt: treatment; T: time.
Bacterial 16S rRNA gene sequencing
Quality filtering resulted in 1 684 022 high-quality sequences (320 bp long) that clustered in 1811 different OTUs with 6195 reads per sample after normalisation.
Permutational analysis of variance (Table 5) showed a strong effect of both doses of liquorice on the structure of the bacterial community (P = 0.0001). However, no effect of time was observed (P = 0.986). Pairwise comparison showed that the structure of the bacterial community differed between control and liquorice treatments (P = 0.0001) and between liquorice 1 and liquorice 2 treatments (P = 0.001). This was confirmed by ANOSIM (P = 0.001), with the greatest differences found between control and liquorice 2 and liquorice 1 and liquorice 2 treatments.
Table 5.
Effects of the supplementation with liquorice extract (1 and 2 g L−1) on the structure of the bacterial communities in the rumen simulating fermenter RUSITEC.
| Bacteria | Archaea | |||
|---|---|---|---|---|
| P(MC) (Permanova) | R-value (ANOSIM) | P(MC) (Permanova) | R-value (ANOSIM) | |
| Treatment effect | 0.0001 | 0.981 | 0.0001 | 0.744 |
| Pairwise comparison | ||||
| Control vs Liquorice 1 | 0.0001 | 0.924 | 0.0001 | 0.573 |
| Control vs Liquorice 2 | 0.0001 | 1 | 0.0001 | 0.971 |
| Liquorice 1 vs liquorice 2 | 0.0001 | 1 | 0.0001 | 0.867 |
| Time effect | 0.9863 | −0.161 | 0.993 | −0.194 |
| Pairwise comparison | ||||
| t0 vs t2 | 0.9179 | −0.163 | 0.943 | −0.208 |
| t0 vs t4 | 0.82 | −0.149 | 0.948 | −0.125 |
| t0 vs t8 | 0.4681 | −0.087 | 0.578 | −0.146 |
| t2 vs t4 | 0.9986 | −0.26 | 0.999 | −0.240 |
| t2 vs t8 | 0.8258 | −0.167 | 0.935 | −0.212 |
| t4 vs t8 | 0.9309 | −0.146 | 0.891 | −0.222 |
To detect possible correlations between the structure of the bacterial community and rumen fermentation parameters, a dbRDA was performed. The primary axis accounted for 55.4% of the variation, and a clear separation by treatment was observed (Fig. 1). Ammonia and BCVFA concentrations in the vessel (P < 0.001) and bacterial richness (P = 0.024) were positively and negatively correlated to the structure of the bacterial community of control and liquorice 2 samples, respectively.
Figure 1.
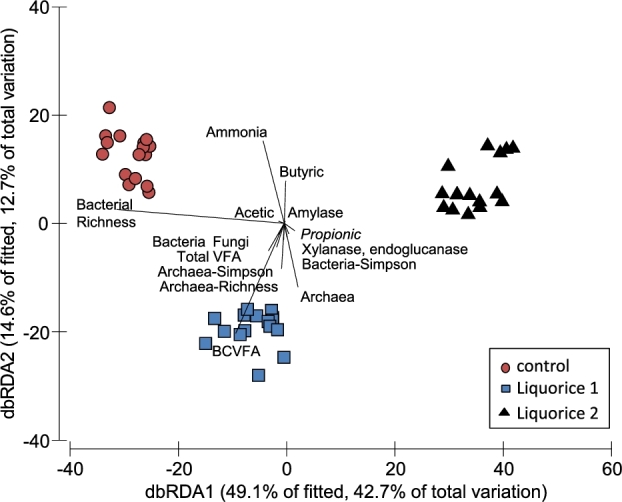
DbRDA illustrating the relationship between the structure of the bacterial community (based on Bray–Curtis distance matrices calculated of normalised and transformed abundance data) with the rumen fermentation pattern and microbial numbers and diversity in the RUSITEC system.
Regarding bacterial diversity (Table 6), the addition of liquorice decreased Shannon and Simpson indexes (P = 0.001 and P < 0.001, respectively) with the highest dose of liquorice having a stronger effect. Bacterial richness also decreased (P < 0.001) in the presence of liquorice extract as compared with the control.
Table 6.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) and the sampling time (0, 2, 4 and 8 h after feeding) on the structure of the bacteria and methanogen communities in the RUSITEC system.
| Treatment | P-Value | Time after feeding | P-Value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | L1 | L2 | SED | Trt | 0 h | 2 h | 4 h | 8 h | SED | T | TrtxT | |
| Bacteria | ||||||||||||
| Richness | 497c | 387b | 231a | 23.8 | <0.001 | 387 | 369 | 387 | 343 | 12.6 | 0.018 | 0.160 |
| Simpson index | 0.959b | 0.944b | 0.923a | 0.008 | 0.010 | 0.942 | 0.944 | 0.952 | 0.931 | 0.006 | 0.039 | 0.577 |
| Shannon index | 4.35c | 3.95b | 3.48a | 0.115 | <0.001 | 3.98 | 3.94 | 4.04 | 3.76 | 0.067 | 0.012 | 0.454 |
| Archaea | ||||||||||||
| Richness | 18.9 | 20.8 | 20.3 | 0.885 | 0.155 | 19.0 | 19.6 | 21.1 | 20.3 | 0.616 | 0.042 | 0.550 |
| Simpson index | 0.525 | 0.725 | 0.645 | 0.090 | 0.162 | 0.648 | 0.629 | 0.637 | 0.612 | 0.021 | 0.369 | 0.516 |
| Shannon index | 1.26 | 1.77 | 1.49 | 0.212 | 0.135 | 1.54 | 1.51 | 1.53 | 1.45 | 0.053 | 0.296 | 0.315 |
a–cMeans with different superscript differ (P < 0.05). Trt: treatment; T: time.
No differences in bacterial abundances because of the addition of liquorice were observed at phylum (Table 7; P > 0.1) or family level (Table S3, Supporting Information; P > 0.1). At genera level (Table 8), only changes in less abundant genera were detected. The greatest change observed was the increased amount of Rikenella with liquorice as compared to the control treatment (P corrected value = 0.186).
Table 7.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) and the sampling time (0, 2, 4 and 8 h after feeding) on relative abundance of bacteria phyla present at an average of more than 0.5% (false discovery rate for Benjamini–Hochberg: 0.25).
| Treatment | Time | SED | Uncorrected P | Benjamini–Hochberg P-Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | L1 | L2 | T0 | T2 | T4 | T8 | Trt | T | TrtxT | Trt | T | TrtxT | Trt | T | TrtxT | |
| Proteobacteria | 0.181 | 0.218 | 0.179 | 0.200 | 0.189 | 0.166 | 0.217 | 0.034 | 0.020 | 0.045 | 0.478 | 0.118 | 0.307 | 0.598 | 0.393 | 0.512 |
| Bacteroidetes | 0.482 | 0.473 | 0.517 | 0.470 | 0.496 | 0.502 | 0.495 | 0.032 | 0.029 | 0.054 | 0.406 | 0.623 | 0.472 | 0.580 | 0.811 | 0.590 |
| Firmicutes | 0.257 | 0.257 | 0.259 | 0.265 | 0.256 | 0.268 | 0.242 | 0.043 | 0.025 | 0.057 | 0.999 | 0.629 | 0.263 | 0.999 | 0.811 | 0.512 |
| Spirochaetes | 0.021 | 0.017 | 0.015 | 0.021 | 0.019 | 0.020 | 0.011 | 0.003 | 0.003 | 0.005 | 0.213 | 0.029 | 0.213 | 0.456 | 0.145 | 0.512 |
| unclassified | 0.021 | 0.009 | 0.007 | 0.013 | 0.011 | 0.014 | 0.012 | 0.007 | 0.003 | 0.009 | 0.217 | 0.649 | 0.251 | 0.456 | 0.811 | 0.512 |
| Tenericutes | 0.012 | 0.013 | 0.012 | 0.013 | 0.013 | 0.013 | 0.010 | 0.003 | 0.003 | 0.005 | 0.900 | 0.580 | 0.359 | 0.999 | 0.811 | 0.513 |
| Verrucomicrobia | 0.012 | 0.001 | 0.000 | 0.004 | 0.004 | 0.005 | 0.005 | 0.004 | 0.003 | 0.006 | 0.049 | 0.813 | 0.905 | 0.430 | 0.813 | 0.905 |
| Fibrobacteres | 0.006 | 0.003 | 0.001 | 0.005 | 0.004 | 0.003 | 0.001 | 0.002 | 0.001 | 0.002 | 0.086 | 0.020 | 0.104 | 0.430 | 0.145 | 0.512 |
| Synergistetes | 0.001 | 0.004 | 0.006 | 0.003 | 0.004 | 0.005 | 0.002 | 0.002 | 0.001 | 0.003 | 0.228 | 0.389 | 0.627 | 0.456 | 0.811 | 0.697 |
| Elusimicrobia | 0.006 | 0.003 | 0.001 | 0.004 | 0.003 | 0.003 | 0.003 | 0.003 | 0.002 | 0.004 | 0.356 | 0.746 | 0.047 | 0.580 | 0.813 | 0.470 |
Trt: treatment; T:time.
Table 8.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) and the sampling time (0, 2, 4 and 8 h after feeding) on relative abundance of bacteria genera present at an average of more than 0.2% (false discovery rate for Benjamini–Hochberg: 0.25).
| Treatment | Time | SED | Uncorrected P | Benjamini-Hochberg P-Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | L1 | L2 | T0 | T2 | T4 | T8 | Trt | T | TrtxT | Trt | T | TrtxT | Trt | T | TrtxT | |
| Ruminobacter | 0.054 | 0.055 | 0.035 | 0.035 | 0.035 | 0.047 | 0.076 | 0.016 | 0.027 | 0.043 | 0.400 | 0.355 | 0.764 | 0.777 | 0.880 | 0.882 |
| Prevotella | 0.250 | 0.274 | 0.277 | 0.264 | 0.279 | 0.280 | 0.246 | 0.035 | 0.030 | 0.057 | 0.721 | 0.606 | 0.343 | 0.901 | 0.880 | 0.828 |
| Unclassified | 0.226 | 0.203 | 0.192 | 0.216 | 0.198 | 0.219 | 0.195 | 0.018 | 0.023 | 0.039 | 0.237 | 0.551 | 0.205 | 0.631 | 0.880 | 0.828 |
| Christensenella | 0.018 | 0.012 | 0.017 | 0.014 | 0.011 | 0.016 | 0.023 | 0.007 | 0.006 | 0.011 | 0.624 | 0.245 | 0.273 | 0.901 | 0.880 | 0.828 |
| Anaerovorax | 0.021 | 0.004 | 0.012 | 0.013 | 0.013 | 0.012 | 0.013 | 0.009 | 0.011 | 0.018 | 0.228 | 0.991 | 0.401 | 0.631 | 0.991 | 0.828 |
| Vampirovibrio | 0.002 | 0.009 | 0.003 | 0.003 | 0.004 | 0.005 | 0.007 | 0.002 | 0.004 | 0.006 | 0.029 | 0.683 | 0.882 | 0.186 | 0.880 | 0.882 |
| Selenomonas | 0.010 | 0.017 | 0.014 | 0.013 | 0.019 | 0.011 | 0.012 | 0.005 | 0.004 | 0.008 | 0.467 | 0.196 | 0.539 | 0.830 | 0.880 | 0.828 |
| Roseburia | 0.003 | 0.006 | 0.003 | 0.005 | 0.006 | 0.003 | 0.002 | 0.004 | 0.003 | 0.007 | 0.731 | 0.600 | 0.401 | 0.901 | 0.880 | 0.828 |
| Paraprevotella | 0.042 | 0.044 | 0.039 | 0.057 | 0.029 | 0.050 | 0.029 | 0.024 | 0.025 | 0.045 | 0.975 | 0.547 | 0.570 | 0.975 | 0.880 | 0.828 |
| Treponema | 0.011 | 0.012 | 0.012 | 0.014 | 0.013 | 0.011 | 0.010 | 0.003 | 0.003 | 0.005 | 0.937 | 0.481 | 0.222 | 0.975 | 0.880 | 0.828 |
| Anaeroplasma | 0.007 | 0.010 | 0.011 | 0.009 | 0.010 | 0.009 | 0.009 | 0.002 | 0.002 | 0.004 | 0.241 | 0.890 | 0.602 | 0.631 | 0.949 | 0.828 |
| Subdivision5_ genera_ incertae_sedis | 0.009 | 0.002 | 0.001 | 0.006 | 0.005 | 0.005 | 0.002 | 0.005 | 0.003 | 0.007 | 0.276 | 0.529 | 0.792 | 0.631 | 0.880 | 0.882 |
| Fibrobacter | 0.005 | 0.003 | 0.002 | 0.006 | 0.004 | 0.003 | 0.002 | 0.002 | 0.001 | 0.003 | 0.413 | 0.059 | 0.156 | 0.777 | 0.880 | 0.828 |
| Acidaminococcus | 0.019 | 0.013 | 0.018 | 0.022 | 0.016 | 0.016 | 0.013 | 0.001 | 0.004 | 0.006 | 0.008 | 0.163 | 0.799 | 0.186 | 0.880 | 0.882 |
| Solobacterium | 0.002 | 0.003 | 0.002 | 0.002 | 0.003 | 0.002 | 0.002 | 0.000 | 0.001 | 0.001 | 0.259 | 0.604 | 0.864 | 0.631 | 0.880 | 0.882 |
| Pyramidobacter | 0.003 | 0.003 | 0.004 | 0.003 | 0.005 | 0.003 | 0.003 | 0.002 | 0.002 | 0.003 | 0.970 | 0.427 | 0.670 | 0.975 | 0.880 | 0.833 |
| Anaerovibrio | 0.020 | 0.017 | 0.017 | 0.015 | 0.020 | 0.017 | 0.021 | 0.008 | 0.007 | 0.014 | 0.919 | 0.690 | 0.198 | 0.975 | 0.880 | 0.828 |
| Streptococcus | 0.057 | 0.033 | 0.040 | 0.044 | 0.038 | 0.043 | 0.048 | 0.010 | 0.016 | 0.026 | 0.111 | 0.887 | 0.433 | 0.507 | 0.949 | 0.828 |
| Pseudobutyrivibrio | 0.015 | 0.017 | 0.018 | 0.017 | 0.018 | 0.015 | 0.016 | 0.006 | 0.007 | 0.012 | 0.856 | 0.955 | 0.356 | 0.975 | 0.986 | 0.828 |
| Succinivibrio | 0.121 | 0.145 | 0.139 | 0.134 | 0.152 | 0.118 | 0.137 | 0.027 | 0.026 | 0.048 | 0.670 | 0.603 | 0.252 | 0.901 | 0.880 | 0.828 |
| Succiniclasticum | 0.021 | 0.025 | 0.024 | 0.020 | 0.026 | 0.024 | 0.021 | 0.005 | 0.009 | 0.014 | 0.732 | 0.852 | 0.621 | 0.901 | 0.949 | 0.828 |
| Coprococcus | 0.003 | 0.005 | 0.006 | 0.004 | 0.005 | 0.005 | 0.005 | 0.001 | 0.001 | 0.002 | 0.142 | 0.285 | 0.323 | 0.568 | 0.880 | 0.828 |
| Phocaeicola | 0.009 | 0.004 | 0.006 | 0.007 | 0.005 | 0.006 | 0.008 | 0.001 | 0.003 | 0.005 | 0.044 | 0.692 | 0.582 | 0.235 | 0.880 | 0.828 |
| Lactobacillus | 0.014 | 0.017 | 0.016 | 0.019 | 0.013 | 0.012 | 0.019 | 0.003 | 0.005 | 0.008 | 0.552 | 0.392 | 0.436 | 0.901 | 0.880 | 0.828 |
| Rikenella | 0.012 | 0.022 | 0.021 | 0.013 | 0.019 | 0.019 | 0.022 | 0.003 | 0.005 | 0.009 | 0.027 | 0.341 | 0.336 | 0.186 | 0.880 | 0.828 |
| Sphaerochaeta | 0.004 | 0.004 | 0.006 | 0.005 | 0.005 | 0.005 | 0.004 | 0.001 | 0.001 | 0.002 | 0.396 | 0.539 | 0.879 | 0.777 | 0.880 | 0.882 |
| Asteroleplasma | 0.002 | 0.002 | 0.003 | 0.002 | 0.003 | 0.003 | 0.002 | 0.001 | 0.001 | 0.002 | 0.583 | 0.884 | 0.677 | 0.901 | 0.949 | 0.833 |
| Candidatus Endomicrobium | 0.001 | 0.003 | 0.005 | 0.002 | 0.003 | 0.004 | 0.004 | 0.002 | 0.002 | 0.004 | 0.188 | 0.715 | 0.599 | 0.631 | 0.880 | 0.828 |
| Eubacterium | 0.003 | 0.004 | 0.004 | 0.004 | 0.004 | 0.003 | 0.003 | 0.000 | 0.001 | 0.001 | 0.023 | 0.054 | 0.290 | 0.186 | 0.880 | 0.828 |
| Butyricimonas | 0.002 | 0.003 | 0.003 | 0.002 | 0.003 | 0.002 | 0.002 | 0.001 | 0.001 | 0.002 | 0.631 | 0.488 | 0.573 | 0.901 | 0.880 | 0.828 |
| Mucinivorans | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 | 0.002 | 0.003 | 0.001 | 0.001 | 0.002 | 0.786 | 0.629 | 0.409 | 0.932 | 0.880 | 0.828 |
| Anaerocella | 0.012 | 0.009 | 0.028 | 0.012 | 0.016 | 0.014 | 0.023 | 0.005 | 0.009 | 0.015 | 0.018 | 0.604 | 0.501 | 0.186 | 0.880 | 0.828 |
Trt: treatment; T:time.
Methanogens 16S rRNA gene sequencing
Quality filtering and removal of bacterial sequences resulted in 370 221 high-quality methanogen sequences (average length of 380 bp) that were clustered in to 33 unique OTUs with 3733 sequences per sample after normalisation.
Permutational analysis of variance (Table 5) showed an effect of liquorice addition on the structure of the archaeal community (P = 0.0001), but no effect of time was observed (P = 0.993). Pairwise comparison showed differences in the structure of the archaea community between control and liquorice treatments (P = 0.0001) and between liquorice 1 and liquorice 2 treatments (P = 0.001). ANOSIM analysis also showed these differences (P = 0.001), with the largest separation detected between the archaeal communities corresponding to control and liquorice 2 treatments. dbRDA (Fig. 2) primary axes displayed 71.3% of the variation and a separation between treatments. Several variables (total and molar proportions of VFA, ammonia and archaea diversity and richness) were positively correlated (P < 0.001) with the structure of the archaeal population in vessels corresponding to the liquorice 2 treatment.
Figure 2.
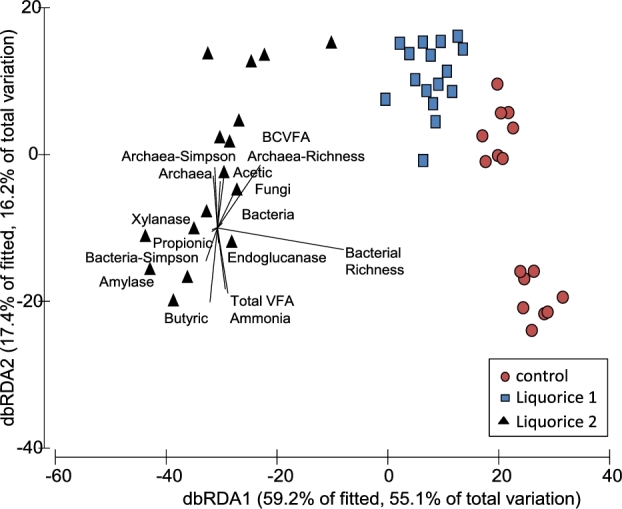
DbRDA illustrating the relationship between the structure of the archaeal community (based on Bray–Curtis distance matrices calculated of normalised and transformed abundance data) with the rumen fermentation pattern and microbial numbers and diversity in the RUSITEC system.
Contrary to the effects on bacterial community structure, methanogens richness and diversity was unaffected by the addition of liquorice (Table 6; P > 0.05). Based on the RIM-DB database, three families (Methanomassiliicoccaceae, Methanosarcinaceae and Methanobacteriaceae) made up the archaeal population in this experiment. The addition of liquorice extract influenced the abundance of the main methanogen groups with the highest dose having a stronger effect (Table 9). When added at 2 g L−1, the flavonoid-rich extract dramatically decreased Methanomassiliicoccus Group 12 (P = 0.035) and it also reduced Group 3a (P = 0.092). On the contrary, the highest dose of liquorice promoted an increased in the abundance of Methanomassiliicoccus Group 10 (P = 0.035), Methanobrevibacter (P = 0.092) and Methanosphaera (P = 0.053).
Table 9.
Effect of supplementing a control diet (C) with liquorice extract (L1 and L2, 1 and 2 g L−1, respectively) and the sampling time (0, 2, 4 and 8 h after feeding) on relative abundance of archaea genera present at an average of more than 0.2% (false discovery rate for Benjamini–Hochberg: 0.25).
| Treatment | Time | SED | Uncorrected P | Benjamini-Hochberg P-Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | L1 | L2 | T0 | T2 | T4 | T8 | Trt | T | TrtxT | Trt | T | TrtxT | Trt | T | TrtxT | |
| Methanomassiliicoccus G12 | 0.588 | 0.171 | 0.009 | 0.231 | 0.256 | 0.269 | 0.268 | 0.109 | 0.012 | 0.111 | 0.005 | 0.034 | 0.327 | 0.035 | 0.170 | 0.483 |
| Methanomassiliicoccus G9 | 0.222 | 0.423 | 0.258 | 0.260 | 0.310 | 0.299 | 0.335 | 0.164 | 0.026 | 0.168 | 0.470 | 0.114 | 0.337 | 0.470 | 0.380 | 0.483 |
| Methanomassiliicoccus G10 | 0.002 | 0.018 | 0.309 | 0.126 | 0.115 | 0.112 | 0.086 | 0.068 | 0.011 | 0.070 | 0.007 | 0.017 | <0.001 | 0.035 | 0.170 | < 0.011 |
| Methanomassiliicoccus G11 | 0.023 | 0.099 | 0.001 | 0.042 | 0.043 | 0.041 | 0.038 | 0.057 | 0.004 | 0.057 | 0.267 | 0.522 | 0.452 | 0.297 | 0.522 | 0.483 |
| Methanomassiliicoccus G3a | 0.039 | 0.012 | 0.001 | 0.013 | 0.018 | 0.018 | 0.019 | 0.012 | 0.003 | 0.012 | 0.042 | 0.252 | 0.140 | 0.092 | 0.407 | 0.483 |
| Methanomassiliicoccus G3b | 0.017 | 0.024 | 0.001 | 0.020 | 0.011 | 0.013 | 0.012 | 0.011 | 0.005 | 0.013 | 0.180 | 0.260 | 0.483 | 0.225 | 0.407 | 0.483 |
| Methanomassiliicoccus G8 | 0.023 | 0.056 | 0.004 | 0.041 | 0.024 | 0.021 | 0.024 | 0.017 | 0.013 | 0.026 | 0.062 | 0.326 | 0.464 | 0.103 | 0.407 | 0.483 |
| Methanimicrococcus | 0.002 | 0.040 | 0.004 | 0.023 | 0.014 | 0.014 | 0.011 | 0.020 | 0.009 | 0.024 | 0.176 | 0.425 | 0.402 | 0.225 | 0.472 | 0.483 |
| Methanobrevibacter | 0.083 | 0.151 | 0.354 | 0.218 | 0.186 | 0.192 | 0.189 | 0.086 | 0.019 | 0.091 | 0.046 | 0.324 | 0.112 | 0.092 | 0.398 | 0.466 |
| Methanosphaera | 0.0004 | 0.006 | 0.059 | 0.025 | 0.021 | 0.021 | 0.020 | 0.015 | 0.002 | 0.016 | 0.016 | 0.155 | 0.482 | 0.053 | 0.388 | 0.483 |
Trt: treatment; T:time.
DISCUSSION
Flavonoids have received interest as promising alternatives to antibiotics in ruminant feeding because of their antimicrobial activity (Cheng et al.2014). Indeed, in vitro studies have shown that flavonoid-rich plant extracts reduce methane production in the rumen (Bodas et al.2008; Patra and Saxena 2010; Oskoueian, Abdullah and Oskoueian 2013) that has been associated with its effect on the methanogen (Patra and Saxena 2010) and protozoal populations (Kim et al.2015). Furthermore, recent in vivo studies have reported changes in the bacteria community as a consequence of supplementing the diet with flavonoids (de Nardi et al.2016; Kasparovska et al.2016; Zhan et al.2017).
Liquorice, the root of the Glycyrrhiza species, has long been used worldwide in herbal medicine and as a natural sweetener (Asl and Hosseinzadeh 2008; Damle 2014). More than 20 triterpenoids and nearly 300 flavonoids have been isolated from liquorice (Wang et al.2015). Glycyrrhizin, a triterpenoid saponin, is considered as the bioactive constituent of liquorice (Asl and Hosseinzadeh 2008). However, it has been shown that many biological activities of liquorice, including estrogenic, anti-cancer, anti-microbial, skin whitening and metabolic syndrome preventive, could be ascribed to its isoflavonoid constituents (Vaya, Belinky and Aviram 1997). Isoflavonoids (3-phenyl benzopyrans) differ from other classes of flavonoids due to their greater structural variability, their presence mainly in free form, rather than as a glycoside, and by the greater frequency of isoprenoid substitution (Munke, Viswanathan and Phadatare 2011).
Our results showed that liquorice extract had a strong antiprotozoal effect in vitro when measuring protozoal activity based on the amount of released [14C] from labelled bacteria. Incubations for 24 h revealed that doses of 1 and 2 g L−1 decreased ammonia concentration by 11% and 21% and increased propionate molar proportion by 14% and 32%, respectively, without impairing the overall fermentation process. The highest dose of liquorice tested also decreased butyrate molar proportions by 21%. Stoichiometrically, and based on the equation of Moss, Jouany and Newbold (2000), the shift in the fermentation pattern observed with 1 and 2 g L−1 of liquorice extract should have resulted in a reduction in methane of 3% and 13%, respectively. When these doses of isoflavonoid-rich extract were tested for their long-term effects in the RUSITEC system, similar effects on fermentation, without negative effects on nutrient digestibility were observed. The addition of 2 g L−1 of the extract also caused a decrease in methane per gram of disappeared OM (−27%). A decrease in total VFA and a substantial shift in the fermentation pattern from acetate towards propionate was observed, leading to a decrease in the theoretical metabolic hydrogen production (−13%).
Despite the inherent difficulty of maintaining high numbers of protozoa in the RUSITEC system (Hillman, Williams and Lloyd 1991), protozoal numbers in our study were reasonable (3775 cells mL−1 for the control), allowing the assessment of the effect of the treatments on the protozoal community. The effects of our liquorice extract on methane emission could have been associated with a decreased protozoa population (−79% and −94% for doses of 1 and 2 g L−1, respectively) since protozoa provide hydrogen as a reducing substrate to methanogens (Newbold et al.2015). The elimination of holotrich protozoa, which play a disproportionate role in supporting methanogenesis (Newbold et al.2015), would be in line with the reduction in methane reported. Although 2 g L−1 of liquorice also caused a great reduction in anaerobic fungi, which together with protozoa plays a significant role in the degradation of ingested plant cellulosic fibres, the digestibility of the fibre resulted unaffected. Possibly an increase in bacterial activity, as reflected in a greater xylanase activity with liquorice, might have compensated for the decrease in protozoal and fungal activity. The greatest effect observed in the presence of liquorice was the reduction in ammonia production (−51 and −77% with 1 and 2 g L−1 of liquorice, respectively) that could have also been related to the decrease in protozoa as they are involved in the turnover of bacterial protein due to their predatory activity (Newbold et al.2015). It is also possible that the formation of isoflavonoids-protein complexes could have reduced the availability of nitrogen to rumen microorganisms, as has been previously reported for other polyphenolic compounds (Ozdal, Capanoglu and Altay 2013). Isoflavonoids may also have other effects on rumen fermentation and microbial activities: some authors have suggested that derivatives from the microbial degradation of flavonoids can be used as alternative carbon source for rumen microbial activities (McSweeney et al.2001; Smith, Zoetendal and Mackie 2005; Ouskoueian et al.2013), whilst others have proposed that flavonoids could act as a hydrogen sink via cleavage of ring structures and reductive dihydroxylation (Becker et al.2013).
Although the total number of bacteria were unaffected by the addition of liquorice, the isoflavonoid-rich extract promoted a less diverse bacterial community. ANOSIM analysis showed that the bacterial community structure was highly separated between treatments. Only changes in the relative abundance of less abundant genera were however observed. The greatest change was observed for Rikenella that are thought to be involved in structural carbohydrates degradation (Pitta et al.2010). Its increase in the presence of liquorice would be in line with the observed increase in xylanase activity. Contrary to previous studies (Oskoueian, Abdullah and Oskoueian 2013; Seradj et al.2014), no major effects on archaea numbers were observed with the addition of liquorice extract. The isoflavonoid-rich extract did not significantly affect archaea diversity. Liquorice extract had an effect on the structure of the methanogen community that differed between treatments, although not to the same extent as that of the bacterial communities. A shift in the methanogen community towards one less effective in producing methane could be suggested to explain differences in methane emissions. Although it has been reported that methane emission can be related to the concentration of archaea in rumen digesta (Wallace et al.2014), it seems that it is the metabolic activity of individual species rather than the number of archaea what is essential for the level of methane production (Shi et al.2014). Methanomassiliicoccus Group12 and Group 3a were replaced by Methanomassiliicoccus Group 10, Methanosphaera and Methanobrevibacter. Methanobrevibacter, theoretically less active in methane production (Kang et al.2013) increased by 0.27 log units with 2 g L−1 of the extract, as compared to the control. This observation was also reported by Belanche et al. (2016) when using ivy saponins in RUSITEC.
Liquorice extract added at 1 g L−1 decreased ammonia production without affecting the overall fermentation process. When added at 2 g L−1, decreases in not only ammonia production but also methane and total VFA production were observed. These effects in fermentation were probably related to decreases in protozoa numbers, a less diverse bacteria population as well as changes in the structure of both bacteria and archaea communities. The inclusion of an isoflavonoid-rich extract from liquorice in the diet could potentially improve the efficiency of the feed utilisation by ruminants. While we speculate that the observed effects could be attributed to the high content of isoflavonoids, and particularly glabridin, the contribution of other phytochemical to the reported effects cannot be ruled out.
Supplementary Material
Supplementary data are available at FEMSEC online.
Acknowledgements
The Authors thank SE Girdwood, R Hill and HJ Worgan at Aberystwyth University for their technical assistance. Thanks to A Marson (New-Food Innovation Ltd) for selecting the liquorice extract.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This work was financed by the Innovate UK project “Ivy for ruminants” Ref:101091. CJN acknowledges the support of the Biotechnology and Biological Sciences Research Council, UK via grant number BB/J0013/1.
REFERENCES
- Asl MN, Hosseinzadeh H. 2008. Review of pharmacological effects of glycyrrhiza sp. and its bioactive compounds. Phytother Res 2008;22:709–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells J, Aris A, Serrano A et al. Effects of an extract of plant flavonoids (Bioflavex) on rumen fermentation and performance in heifers fed high-concentrate diets1. J Anim Sci 2012;90:4975–84. [DOI] [PubMed] [Google Scholar]
- Becker PM, van Wikselaar PG, Franssen MCR et al. Evidence for a hydrogen-sink mechanism of (+) catechin-mediated emission reduction of the ruminant greenhouse gas methane. Metabolomics 2014;10:179–89. [Google Scholar]
- Belanche A, Doreau M, Edwards JE et al. Shifts in the rumen microbiota due to the type of carbohydrate and level of protein ingested by dairy cattle are associated with changes in rumen fermentation. J Nutr 2012;142:1684–92. [DOI] [PubMed] [Google Scholar]
- Belanche A, Pinloche E, Preskett D et al. Effects and mode of action of chitosan and ivy fruit saponins on the microbiome, fermentation and methanogenesis in the rumen simulation technique. FEMS Microbiol Ecol 2016;92:1 DOI: 10.1093/femsec/fiv160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate— a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 1995;57:289–300. [Google Scholar]
- Bodas R, Lopez S, Fernandez M et al. In vitro screening of the potential of numerous plant species as antimethanogenic feed additives for ruminants. Anim Feed Sci Technol 2008;145:245–58. [Google Scholar]
- Cheng G, Hao H, Xie S et al. Antibiotic alternatives: the substitution of antibiotics in animal husbandry?. Front Microbiol 2014;5:217 DOI: 10.3389/fmicb.2014.00217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerkawski JW, Breckenridge G. Design and development of a long-term rumen simulation technique (RUSITEC). Br J Nutr 1977;38:371–84. [DOI] [PubMed] [Google Scholar]
- de la Fuente G, Belanche A, Girwood SE et al. Pros and cons of ion-torrent next generation sequencing versus terminal restriction fragment length polymorphism T-RFLP for studying the rumen bacterial community. PLoS One 2014;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente G, Skirnisson K, Dehority BA. Rumen ciliate fauna of Icelandic cattle, sheep, goats and reindeer. Zootaxa 2006;1377:47–60. [Google Scholar]
- de Nardi R, Marchesini G, Plaizier JC et al. Use of dicarboxylic acids and polyphenols to attenuate reticular pH drop and acute phase response in dairy heifers fed a high grain diet. BMC Vet Res 2014;10:277 DOI: 10.1186/s12917-014-0277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nardi R, Marchesini G, Li S et al. Metagenomic analysis of rumen microbial population in dairy heifers fed a high grain diet supplemented with dicarboxylic acids or polyphenols. BMC Vet Res 2016;12:29 DOI 10.1186/s12917-016-0653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damle M. 2014. Glycyrrhiza glabra (Liquorice)—a potent medicinal herb. Int J Herbal Med 2014;2:132–6. [Google Scholar]
- Dehority BA. Laboratory Manual for Classification and Morphology of Ruminal Ciliate Protozoa. Boca Raton, Florida: CRC Press, 1993. [Google Scholar]
- Giraldo LA, Tejido ML, Ranilla MJ et al. Influence of direct-fed fibrolytic enzymes on diet digestibility and ruminal activity in sheep fed a grass hay-based diet. J Anim Sci 2008;86:1617–23. [DOI] [PubMed] [Google Scholar]
- Hart KJ, Yanez-Ruiz DR, Duval SM et al. Plant extracts to manipulate rumen fermentation. Anim Feed Sci Technol 2008;147:8–35. [Google Scholar]
- Hillman K, Williams AG, Lloyd D. Evaluation of matrices in the rumen simulation technique (RUSITEC) for the maintenance of ciliate protozoa. Lett Appl Microbiol 1991;12:129–32. [Google Scholar]
- Kang SH, Evans P, Morrison M et al. Identification of metabolically active proteobacterial and archaeal communities in the rumen by DNA- and RNA-derived 16S rRNA gene. J Appl Microbiol 2013;115:644–53. [DOI] [PubMed] [Google Scholar]
- Kasparovska J, Pecinkova M, Dadakova K et al. Effects of isoflavone-enriched feed on the rumen microbiota in dairy cows. PLoS One 2016;11:4 DOI: 10.1371/journal.pone.0154642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim ET, Guan LL, Lee SJ et al. Effects of flavonoid-rich plant extracts on in vitro ruminal methanogenesis, microbial populations and fermentation characteristics. Asian-Australas J Anim Sci 2015;28:530–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Pandey AK. Chemistry and biological activities of flavonoids: an overview. Sci World J 2013, 162750 DOI: 10.1155/2013/162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Anderson MJ. Distance-based redundancy analysis: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 1999;69:1–24. [Google Scholar]
- Ma T, Chen DD, Tu Y et al. 2017. Dietary supplementation with mulberry leaf flavonoids inhibits methanogenesis in sheep. Anim Sci J 2017;88:72–78. [DOI] [PubMed] [Google Scholar]
- McDougall EI. Studies on ruminant saliva. 1. The composition and output of sheep's saliva. Biochem J 1948;43:99–109. [PMC free article] [PubMed] [Google Scholar]
- McSweeney CS, Palmer B, McNeill DM et al. Microbial interactions with tannins: nutritional consequences for ruminants. Anim Feed Sci Technol 2001;91:83–93. [Google Scholar]
- Menke KH, Steingass H. Estimation of the energetic feed value obtained from chemical analysis and gas production using rumen fluid. Anim Res Dev 1988;28:7–55. [Google Scholar]
- Moss AR, Jouany JP, Newbold CJ. Methane production by ruminants: its contribution to global warming. Ann Zootech 2000;49:231–53. [Google Scholar]
- Munke AP, Viswanathan V, Phadatare AG. Structure pre-requisites for isoflavones as effective antibacterial agents. Phcog Rev 2011;5:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbold CJ, de la Fuente G, Belanche A et al. The role of ciliate protozoa in the rumen. Front Microbiol 2015;61313 DOI: 10.3389/fmicb.2015.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskoueian E, Abdullah N, Oskoueian A. Effects of flavonoids on rumen fermentation activity, methane production, and microbial population. BioMed Res Int 2013, 349129 DOI: 10.1155/2013/349129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdal T, Capanoglu E, Altay F. A review on protein–phenolic interactions and associated changes. Food Res Int 2013;51:954–70. [Google Scholar]
- Patra AK, Saxena J. A new perspective on the use of plant secondary metabolites to inhibit methanogenesis in the rumen. Phytochemistry 2010;71:1198–222. [DOI] [PubMed] [Google Scholar]
- Patra A, Park T, Kim M et al. Rumen methanogens and mitigation of methane emission by anti-methanogenic compounds and substances. J Anim Sci Biotechnol 2017;8:13 DOI: 10.1186/s40104-017-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitta DW, Pinchak WE, Dowd SE et al. 2010. Rumen bacterial diversity dynamics associated with changing from bermudagrass hay to grazed winter wheat diets. Microb Ecol 2010;59:511–22. [DOI] [PubMed] [Google Scholar]
- Seedorf H, Kittelmann S, Henderson G et al. RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J 2014;2:e494 DOI: 10.7717/peerj.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seradj AR, Abecia L, Crespo J et al. The effect of Bioflavex® and its pure flavonoid components on in vitro fermentation parameters and methane production in rumen fluid from steers given high concentrate diets. Anim Feed Sci Technol 2014;197:85–91. [Google Scholar]
- Shi W, Moon CD, Leahy SC et al. Methane yield phenotypes linked to differential gene expression in the sheep rumen microbiome. Genome Res 2014;24:1517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AH, Zoetendal E, Mackie RI. Bacterial mechanisms to overcome inhibitory effects of dietary tannins. Microb Ecol 2005;50:197–205. [DOI] [PubMed] [Google Scholar]
- Stewart CS, Duncan SH. The effect of avoparcin on cellulolytic bacteria of the ovine rumen. J Gen Microbiol 1985;131:427–35. [Google Scholar]
- Vaya J, Belinky PA, Aviram M. Antioxidant constituents from licorice roots: isolation, structure elucidation and antioxidative capacity toward LDL oxidation. Free Radic Biol Med 1997;23:302–13. [DOI] [PubMed] [Google Scholar]
- Wallace RJ, McPherson CA. Factors affecting the rate of breakdown of bacterial protein in rumen fluid. Br J Nutr 1987;58:313–23. [DOI] [PubMed] [Google Scholar]
- Wallace RJ, Rooke JA, Duthie CA et al. Archaeal abundance in post-mortem ruminal digesta may help predict methane emissions from beef cattle. Sci Rep 2015;4:5892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM et al. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl Environ Microbiol 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yang R, Yuan B et al. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharmaceutica Sinica B 2015;5:310–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TY, Li Q, Bi KS. Bioactive flavonoids in medicinal plants: structure, activity and biological fate. Asian J Pharm Sci 2018;13:12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weatherburn MW. Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 1967;39:971–4. [Google Scholar]
- Williams AG, Coleman GS. The Rumen Protozoa. New York: Springer-Verlag, 1992. [Google Scholar]
- Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques 2004;36:808–12. [DOI] [PubMed] [Google Scholar]
- Zhan J, Liu M, Su X et al. Effects of alfalfa flavonoids on the production performance, immune system, and ruminal fermentation of dairy cows. Asian-Australas J Anim Sci 2017;30:1416–24. DOI: 10.5713/ajas.16.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSEC online.


