Abstract
Background and Aim
Family history is the strongest risk factor for developing Crohn’s disease [CD] or ulcerative colitis [UC]. We investigated whether the proximity of relationship with the affected relative and concordance for type of inflammatory bowel disease [IBD] modifies the effect of family history on phenotype and disease severity.
Method
This cross-sectional study included patients with a confirmed diagnosis of IBD in a clinical registry. Family history of IBD was assessed by a questionnaire ascertaining presence of disease in a first-first-degree, second-second-degree or distant relative. Our primary outcomes were disease phenotype as per the Montreal classification and severity measured by need for immunomodulator, biologic, or surgical therapy. Genotyping was performed on the Immunochip and faecal samples were subjected to 16S rRNA microbiome sequencing.
Results
Our study included 2136 patients with IBD [1197 CD, 939 UC]. Just under one-third [32%] of cases ere familial IBD [17% first-degree, 21% second-degree]. Familial IBD was diagnosed at an earlier age, both in CD [26 vs 28 years, p = 0.0006] and UC [29 vs 32 years, p = 0.01]. Among CD patients, a positive family history for CD was associated with an increased risk for complicated disease in the presence of an affected family member (odds ratio [OR] 1.48, 95% confidence interval [CI] 1.07–2.03). However, this effect was significant only for first-degree relatives [OR 1.82, 95% CI 1.19–2.78].
Conclusions
A family history of CD in first-degree relatives was associated with complicated CD. Family history discordant for type of IBD or in distant relatives did not influence disease phenotype or natural history.
Keywords: Family history, inflammatory bowel disease, complicated disease, age at diagnosis, concordant
1. Introduction
Inflammatory bowel diseases [IBD] are complex immune-mediated diseases affecting over 1.6 million individuals in the USA, 2.2 million in Europe, and thousands more worldwide.1,2 Though the exact pathogenic mechanisms behind IBD are yet to be fully defined, the current hypothesis suggests that IBD develops at the interface of predisposing genetic variations, immunological alterations, shifts in the gut microbiome, and external environmental influences.
Genetic predisposition is one of the strongest factors influencing the development of IBD. Genome-wide association studies have identified over 200 distinct single nucleotide polymorphisms [SNPs] predisposing to the development of Crohn’s disease [CD] or ulcerative colitis [UC].3 Between 8% and 20% of patients with CD or UC will have an affected close relative.4–6 In the general population, a history of IBD in a first- or second-degree family member increases risk of incident disease 8-fold and 2-fold, respectively.7 Many studies have examined the impact of family history on disease risk, but fewer have examined if this history influences disease course in the index patient. Familial IBD may differ from sporadic IBD in having a stronger genetic predisposition and shared common environmental influences, which may, in turn, lead to similarities in gut microbial composition. Previous studies have demonstrated concordance in age at diagnosis, disease location and behaviour, and need for IBD-related surgery in affected family members,4,8,9 but have been limited by small sample sizes and lack of examination of the genetics or microbial composition underlying such phenotypes. In addition, previous studies often failed to examine the impact of proximity of the relationship of the affected family member or concordance for type of IBD.
Identifying the impact of family history on the natural history of IBD may also have prognostic implications, allowing for risk stratification and prediction of disease-related complications. Consequently, we performed this study using a large prospective cohort with the following aims: [1] to define the impact of family history on the clinical characteristics and natural history of IBD, stratifying by proximity of relationship and concordance for type of IBD; and [2] to compare the genetics and microbiome composition in a subset of patients with familial and sporadic IBD.
2. Methods
2.1. Study cohort
This study included patients recruited in a prospective registry, the Prospective Registry for IBD Study at Massachusetts General Hospital [PRISM]. All adult patients aged 18 years and older with a confirmed diagnosis of CD, UC, or IBD-unspecified [IBDU], seeking care at the Massachusetts General Hospital [MGH] Crohn’s and Colitis Center, were eligible for inclusion in the cohort as described in our previous publications.10–12 After provision of informed consent, patients completed an enrolment interview with a trained research coordinator, where information about demographics, disease characteristics including age at diagnosis, and phenotype according to the Montreal classification [location and behaviour in CD, extent in UC] were obtained and confirmed by medical record review. Information was also obtained about current and past medical treatments and need for IBD-related surgery.
A family history of IBD was assessed by a detailed questionnaire ascertaining presence of either CD or UC in a first-degree [parent, child, sibling], second-degree [grandparent, uncle, aunt], or distant relative. Patients were considered to have familial IBD in the presence of CD or UC in any relative. Patients with no reported family history were considered to have sporadic IBD. Patients who had the same type of IBD as their affected family member were considered to have a concordant family history, andthose with the other type of IBD were labelled as having a discordant family history.
Our primary end point in both CD and UC was severe disease defined as needing IBD-related surgery. In CD, our co-primary outcome was complicated disease defined as the presence of stricturing [B2] or penetrating disease [B3] or perianal involvement.
2.2. Genotyping
All consenting patients provided 10 mL blood from which genomic DNA was extracted for genotyping. Patients were genotyped on the Illumina Immunochip, a custom-chip designed to perform fine mapping of over 150000 loci relevant to immune-mediated diseases, at the Broad Institute.13 We extracted data on 201 distinct IBD-risk single nucleotide polymorphisms [SNPs] reported, and calculated a weighted genetic risk score [GRS] as described previously.12,14 For each of the risk alleles associated with either CD, UC, or both, we determined each patient to have wild type [scored as 0], heterozygous [scored as 1], or homozygous [scored as 2] variants. Odds ratios for strength of association of each of the alleles with CD, UC, or both were obtained from a recent multicentre consortium publication.3 A weighted genetic risk score was then constructed which was the cumulative sum of the natural logarithm of this odds ratio multiplied by the allele frequency.15 This yielded three separate risk scores that quantified magnitude of genetic predisposition towards developing any IBD, CD, or UC, respectively.
2.3. Sequencing of the microbiome
DNA was extracted from the faecal samples using the Qiagen AllPrep MiniKit [Qiagen, Valencia, CA, USA], incorporating bead-beating at several steps to improve homogenisation, as described previously.16 Specimens were then sent to the Broad Institute [Cambridge, MA] to generate a 16S DNA profile targeting the V4 region of the SSU rRNA gene via the Illumina HiSeq 2000 platform, with 100 bp paired-end reads, targeting ~2 Gbp per sample. Raw sequence data were demultiplexed and quality filtered using the computational pipeline QIIME.17 Operational taxonomic unit [OTU] tables were built using the ‘pick_closed_reference_otus.py’ command in QIIME. The ‘biome’ output file was then imported into R using the ‘Phyloseq’ statistical package for further analysis.18 Taxonomy was assigned using the ‘Greengenes’ reference database clustered at 97%.19
2.4. Statistical analysis
Institutional review board approval was obtained from the Partners Healthcare Human Subjects Research committee. Analysis of clinical covariates was performed using Stata 13.1 [StataCorp, College Station, TX]. Continuous variables were summarised using means and standard deviations, and categorical variables were expressed as proportions and compared using the chi square test. We first performed univariate analysis examining the association between family history and each of our study outcomes. Subsequently, multivariable models were constructed adjusting for disease-specific covariates including duration of disease, behaviour and extent of involvement, education, and employment status. Adjusted regression models were used to estimate adjusted odds ratios [OR] and 95% confidence intervals [CI]. A two-sided p-value < 0.05 indicated independent statistical significance. Separate analysis was performed examining the association with history of IBD in a first-degree relative alone, second-degree relative, or more distant relative, and concordance for type of IBD.
Genetic analysis was performed using Plink v1.07.20 A total of 185 [out of 201] SNPs passed our threshold of HardyWeinberg p < 0.0001 and a call rate > 95%. Individuals with genotyping success rates < 90% were excluded. First, we compared the weighted GRS between familial and sporadic IBD, by proximity to affected relative, and concordance for type of IBD. In an exploratory analysis, we then examined if specific SNPs were differentially distributed between familial and sporadic IBD, adopting a p-value threshold of 0.01 for nominal significance, and of 0.0003 adjusting for multiple comparisons.
Microbiome analysis was conducted using the ‘Phyloseq’ statistical package in R. Microbial alpha diversity was calculated on unfiltered data using the Shannon diversity index, and stratified by relevant covariates. First, we compared the diversity of the microbiome in familial with sporadic IBD, using the Student’s t test. We then stratified by measures indicating proximity to the affected relative [for example, comparing those with IBD and an affected first-degree relative with those with sporadic IBD] and degree of concordance [between those with a concordant family history compared with those with sporadic IBD]. Differential abundance testing for each of these comparisons was done using the MaAsLin pipeline.16 Briefly, MaAsLin performs a per-feature differential abundance testing of all microbes [OTUS] by regressing the relative abundance of each feature in a linear model. The relative abundances were arcsin-square-root-transformed to approximate homoscedasticity when applying linear models. We limited our analysis to only those features that were both prevalent and abundant, with mean abundance > 0.01% in at least 40% of the samples. Using ANOVA type III [tests of fixed effects], p-values of associations of each OTU were computed and subjected to Benjamini–Hochberg false discovery rate [FDR] correction with a cutoff of 0.25.21 All analyses adjusted for whether there was clinically active disease [HarveyBradshaw index > 4 or simple clinical colitis activity index > 2] at the time of the stool sample collection.
3. Results
3.1. Study population
Our study included 2094 patients with IBD [1, 97 CD, 857 UC] with a mean age of 41 years. Just over half the cohort were women [52%]. One-third of patients had a family history of IBD [32%]; 17% had an effected first-degree relative and 21% had an affected second-degree relative. In 69% of patients with a family history, there was concordance for type of IBD which was similar when the index diagnosis was CD or UC.
Patients with familial IBD were similar to those with sporadic IBD in age, type of IBD, and smoking status, but were likely to have higher education status [Table 1]. Patients with familial IBD had a younger age at diagnosis than those with sporadic IBD, both for CD [25.5 vs 28.4 years, p = 0.0006] and UC [29.4 vs 31.8 years, p = 0.01]. In CD, this association was noted irrespective of the concordance for type of IBD [concordant family history: 26 vs 28 years, p = 0.03; discordant family history 25 vs 28 years, p = 0.02]. However in UC, an earlier age of diagnosis was only noted when the affected family member had UC [29 vs 32 years, p = 0.01] but not CD [30 vs 31 years, p = 0.70].
Table 1.
Characteristics of the study population.
| Characteristic | Familial IBD [n = 677]* | Sporadic IBD [n = 1,407]** | p-Value |
|---|---|---|---|
| Mean age [SD] [in years] | 39.8 [15.2] | 41.0 [15.1] | 0.070 |
| Female sex, n [%] | 294 [43.4] | 705 [49.8] | 0.006 |
| Mean age at diagnosis [SD] [in years] | 27.1 [12.5] | 29.9 [14.5] | < 0.001 |
| Mean duration of IBD [SD] [in years] | 12.9 [11.8] | 11.0 [10.3] | < 0.001 |
| Smoking status | 0.106 | ||
| Never, n [%] | 440 [66.2] | 901 [65.5] | |
| Past, n [%] | 171 [25.7] | 393 [28.6] | |
| Current, n [%] | 54 [8.1] | 82 [6.0] | |
| High level education, n [%] | 475 [72.1] | 907 [66.7] | 0.015 |
| Employment status, n [%] | 537 [80.6] | 1,104 [79.5] | 0.543 |
| Type of IBD | 0.125 | ||
| Crohn’s disease, n [%] | 397 [58.6] | 776 [54.8] | |
| Ulcerative colitis, n [%] | 280 [41.4] | 641 [45.2] | |
| CD location, n [%] | 0.100 | ||
| Ileal [L1] | 92 [25.6] | 171 [25.0] | |
| Colon [L2] | 75 [20.9] | 183 [26.8] | |
| Ileocolon [L3] | 192 [53.5] | 330 [48.3] | |
| CD behaviour, n [%] | 0.019 | ||
| Inflammatory [B1] | 163 [44.3] | 365 [51.7] | |
| Stricturing [B2] | 76 [20.7] | 150 [21.3] | |
| Penetrating [B3] | 129 [35.1] | 191 [27.1] | |
| CD perianal disease, n [%] | 102 [27.7] | 183 [25.9] | 0.527 |
| UC extent, n [%] | 0.155 | ||
| Limited colitis | 37 [16.0] | 77 [13.5] | |
| Pancolitis | 194 [84.0] | 493 [86.5] | |
| Therapy | |||
| Surgery, n [%] | 205 [30.3] | 364 [25.7] | 0.027 |
| Biologics, n [%] | 326 [48.2] | 653 [46.1] | 0.382 |
| Immunomodulators, n [%] | 416 [61.7] | 848 [60.7] | 0.642 |
CD, Crohn’s disease; UC, ulcerative colitis; IBD, inflammatory bowel diseases; SD, standard deviation. Bold values represent a p-value < 0.05.
*Patients in the presence of CD or UC in any relative.
**Patients with no reported family history of CD or UC.
3.2. Complicated Crohn’s disease
On univariate analysis, CD patients with a family history were more likely to have complicated disease [56% vs 48%, OR 1.35, 95% CI 1.05–1.73]. However, differences were noted based on concordance for type of IBD. Upon adjustment for disease location, age at diagnosis, and duration of disease, CD patients with a concordant family history were more likely to have complicated disease [OR 1.48, 95% CI 1.07–2.03], particularly if the affected member was a first-degree relative [OR 1.82, 95% CI 1.19–2.78] [Table 2]. Among first-degree relatives, the association was more striking in the presence of CD in a sibling [p = 0.008] than parent [p = 0.41] or child [p = 0.42]. The presence of CD in a second-degree relative [OR 1.20, 95% CI 0.85–1.69], UC in a first- [OR 0.64, 95% CI 0.37–1.10] or second-degree relative [OR 0.88, 95% CI 0.56–1.37] did not modify risk of complicated CD.
Table 2.
Likelihood of complicated Crohn’s disease, by proximity and concordance of affected family member.
| Affected relative | Odds ratio [95% confidence interval] a |
|---|---|
| Any family history [close or distant] | 1.48 [1.07–2.03] |
| First-degree relative with CD | 1.82 [1.19–2.78] |
| Second-degree relative with CD | 1.17 [0.79–1.72] |
| Any relative with UC [discordant] | 0.66 [0.42–1.02] |
CD, Crohn’s disease; UC, ulcerative colitis.
aAdjusted for disease location, age at diagnosis, and duration of disease.
3.3. IBD-related surgery
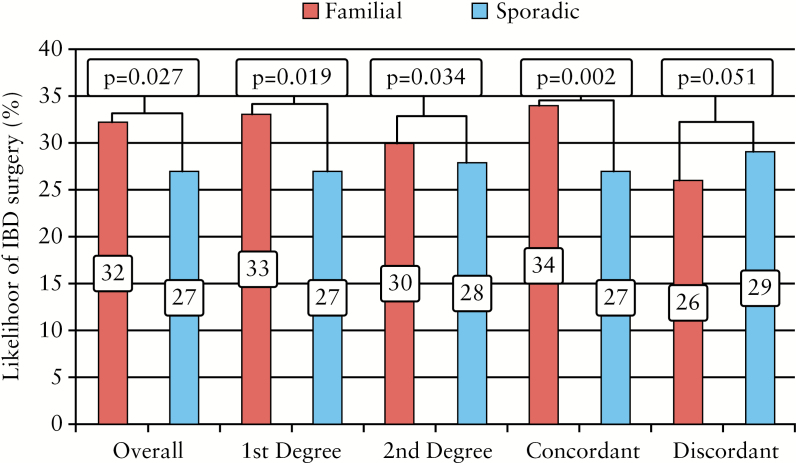
Patients with familial IBD were more likely to require IBD-related surgery than those with sporadic IBD [32% vs 27% p = 0.027] [Figure 1]. This result was more striking when the affected family member was a first-degree relative [33% vs 27%, p = 0.019] but not for a second-degree relative [30% vs 28%, p = 0.34] [Figure 1]. The association was also stronger when the family history was concordant for type of IBD [34% vs 27%, p = 0.002] than discordant [26% vs 29%, p = 0.51]. In both CD and UC, multivariable analysis adjusting for disease behaviour in addition to location, age at diagnosis, and disease duration, yielded no statistically significant association between family history and need for IBD-related surgery [CD: 1.04, 95% CI 0.75–1.43; UC: 1.10, 95% CI 0.67–1.79] suggesting that the influence of family history on IBD surgery was mediated through effect on age of onset and disease behaviour. We found no differences in need for biologic or immunosuppressive therapy based on the presence of family history of IBD or concordance for type of IBD [Supplementary Table 1, available as Supplementary data at ECCO-JCC online]. In stratifying by number of biologics, there was also no difference between the two groups on the number of patients receiving 0, 1, 2, 3, or 4 biologics for their IBD [data not shown].
Figure 1.
Likelihood of disease-related surgery among patients with Crohn’s disease, stratified by concordance and proximity of affected family member.
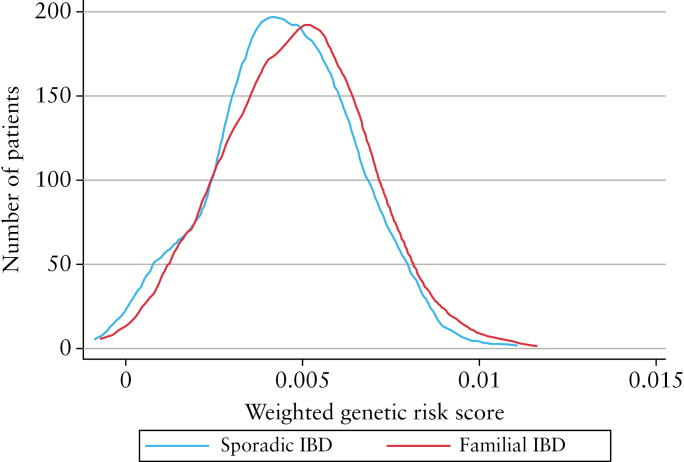
3.4. Genetic analysis
Genotype data for calculation of the weighted GRS were available in 1277 patients [796 CD, 481 UC or IBD-U]. Patients with familial IBD had a stronger genetic predisposition to develop IBD than those with sporadic IBD [p = 0.006] [Figure 2]. However, this difference was only in those with an affected first-degree [p = 0.004] but not second-degree relative [p = 0.35]. The greater genetic predisposition was also noted only in those with a concordant family history [p = 0.03] than when there was discordance for type of IBD [p = 0.19]. In CD, five SNPs were differently distributed between sporadic and familial CD, with a p-value < 0.01 [Supplementary Table 2, available as Supplementary data at ECCO-JCC online]. One SNP [rs1569328], coding for FOS [FOS proto-oncogene, AP-1 transcription factor subunit], a regulator of TGF-β mediated signaling, achieved significance above the false discovery threshold, with a minor allele frequency of 18% in familial IBD compared with 10% in sporadic disease [OR 1.934, p = 1.27 x 10–5]. This achieved a p-value of 0.0012 in CD patients with a concordant [18% vs 11%] but not discordant family member [17% vs 12%, p = 0.16]. None of the SNPs met a p-value threshold of 0.01 for differential distribution between sporadic and familial disease among those with UC.
Figure 2.
Comparison of inflammatory bowel disease genetic risk score between familial and sporadic disease.
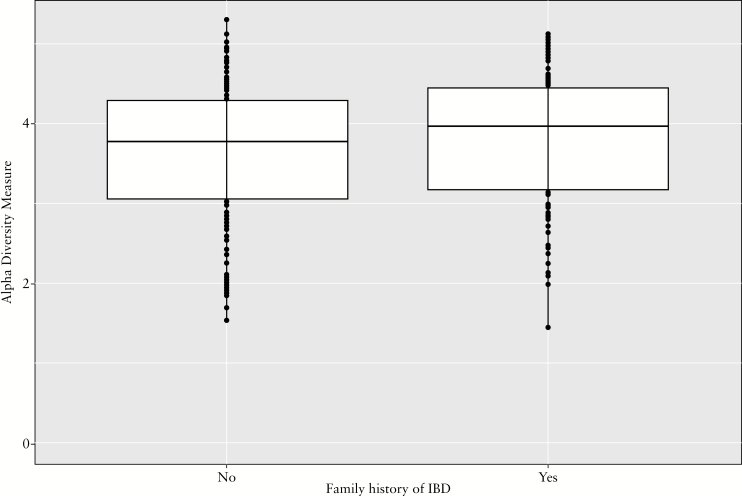
3.5. Microbiome analysis
Stool was available for 16S rRNA microbiome analysis from 268 patients. IBD patients with a positive family history had a trend towards greater alpha diversity than sporadic IBD [p = 0.05, Figure 3]. We noted some differences in the microbiome between sporadic and familial IBD, and by proximity to affected relative. Patients with familial IBD had greater abundance of Ruminococcaceae when compared with sporadic IBD [q = 0.18] [Supplementary Table 3, available as Supplementary data at ECCO-JCC online]. Among those with an involved first-degree relative, two families─Lachnospiraeceae and Erysipelotrichaceae [species Eubacterum dolichum] ─were more abundant in those with a family history, whereas the genus Streptococcus was less common than in those with sporadic IBD.
Figure 3.
Difference in gut microbial diversity between familial and sporadic inflammatory bowel disease.
4. Discussion
Inflammatory bowel diseases are complex and heterogeneous in their natural history, and are multifactorial in origin, with genetics and consequently family history being a strong risk factor for incident disease. Little is known about whether familial IBD differs from sporadic IBD in clinical characteristics, genetic architecture, or the gut microbiome. Using a large prospective cohort, we identified positive family history to be associated with earlier onset, complicated CD behaviour, and need for IBD-related surgery. Also important in our findings is that the association with family history depends on proximity of relationship to the index patient and on concordance for type of IBD. We also describe a higher genetic burden in familial IBD and association with specific genetic polymorphisms in CD, but not UC. In addition, certain microbial species were differentially abundant in the stool between familial and sporadic IBD.
It is widely accepted that the strongest risk factor for developing IBD is the presence of an affected first-degree relative, with weaker influences of more distant relations.4,22,23 The proportion with a positive family history in our study is similar to that noted in other studies,24–29 as is the concordance for type of IBD.24 A key clinical observation from our study is that a positive family history was associated with stricturing or penetrating CD and an earlier age of diagnosis, and through these influences, greater need for IBD-related surgery. This effect was notable primarily in those with an affected first-degree relative and when the family member had CD. Among first-degree relatives, the association was stronger when the affected family member was a sibling, consistent with previous studies demonstrating that siblings are at highest risk to develop IBD owing to being most genetically similar.4,25
There are limited data on whether having an affected family member influences disease course in the index patient. Consistent with our study, Henriksen et al. found that familial disease had a younger age at diagnosis than sporadic disease among those with CD, but not UC.24 A meta-analysis by Childers et al., which included 71 studies of UC, reported that a positive family history was more common in those with a younger age at diagnosis,30 a finding also supported by a cohort from Korea.31
The effect of family history on disease outcomes has yielded more conflicting results. A population-based cohort from Norway,24 a small study of 181 Jewish CD patients28 and a larger cohort of Finnish IBD patients,29 found no effect of family history on age at diagnosis, disease complications, or need for surgery. In contrast to these findings and consistent with our observations of the association between family history and disease severity, two larger studies from Korea noted more anti-TNF use31,32 and higher risk for CD-related surgery31 in those with a positive family history, compared with sporadic disease. A similarly large study by Trier Moller et al., using the national registry in Denmark, also identified higher rates of major surgery in familial compared with sporadic CD and a shorter time to needing anti-TNF therapy in both familial CD and UC.33 One reason for the differing results, in addition to the lack of statistical power in smaller studies, is that the effect of family history may be more nuanced and modified by proximity of relationship to the index patient and concordance for type of IBD, none of which have been examined in detail before.
Fewer studies have examined if familial IBD differs genetically from sporadic IBD and favours specific pathways. Previous studies have focused on either one or a few specific SNPs.34–36 In a large Dutch study by Weersma et al., higher genetic burden, defined as the number of variants in five CD-specific SNPs, was associated with diagnosis before the age of 40 years, more complicated disease, and need for IBD-related surgery.35 Greater genetic predisposition has been linked to both early age at diagnosis15 and ileal involvement, the strongest predictor of disease behaviour, which is supportive of our findings of a higher genetic risk score in familial compared with sporadic IBD.27 There were no specific SNPs that were more commonly affected in familial UC; however in CD, variants at five SNPs including one at the FOS gene, were nominally more common in familial disease. FOS plays a role in TGF-β signalling, which is important for the development of fibrosis in CD. Thus, the more common occurrence of FOS polymorphisms in familial CD may contribute to the higher risk of stricturing/penetrating phenotype in this cohort.
Gut microbial composition may predict progression of CD37 and determine response to therapy.38 Several studies have demonstrated that healthy relatives of patients with CD or UC demonstrate dysbiotic microbial profiles,39,40 but whether the microbiome of familial IBD differs from sporadic IBD has not been examined previously. Some of the differences in the microbiome between familial and sporadic IBD could contribute towards more complicated disease in the former. We found that a member of the Ruminococcus family was more common in individuals with a family history of CD. Interestingly, in a paediatric inception cohort the abundance of Ruminococcus was associated with the development of stricturing complications, consistent with our observation of the association of family history with complicated CD.41 Other members belonging to Ruminococcaceae, in particular R. gnavus, was more abundant in patients with a discordant family history, whereas Dorea spp. [belonging to the family lachnospiraeceae] was less common in those with an affected second-degree relative. In other cohorts, R. gnavus has been associated with CD disease activity, whereas Dorea spp. have been inversely associated with active disease.42 Thus, distinct microbial profiles of familial IBD, influenced by shared genetic or environmental influences, may modify disease course in the index patient.
There are several limitations to this study. First, our study was at a tertiary referral centre which may be biased towards more severe disease when compared with a population-based cohort. Assessment of family history was done by a detailed questionnaire, and we were unable to review records of family members to confirm diagnosis or ascertain IBD phenotype in relatives. However, the proportion with an affected family member in our cohort was similar to other studies, supporting the generalisability of our findings. Third, information was also not available on time-varying covariates such as proximal extension of colitis or response to specific therapies. Our preliminary data, suggesting some differences in disease phenotype, lay the groundwork for examining whether familial IBD differs from sporadic IBD in parameters such as response or lack thereof to specific therapies and progression of disease. Fourth, information on genetics and the microbiome was available only in a small subset of patients, limiting our statistical power. Larger cohorts are essential to more robustly define the similarities and differences between familial and sporadic IBD.
In conclusion, this large prospective cohort study provides evidence of earlier onset of disease in patients with familial IBD compared with sporadic cases of IBD. Furthermore, a family history of CD in first-degree [but not second-degree] relatives was associated with complicated CD, most strongly in those with an affected sibling and with concordance for type of IBD. This effect may be mediated through shared genetic risk factors and through differences in the microbiome. Our findings also provide useful data for risk stratification and determination of prognosis and disease course. Further studies in larger cohorts are essential to shed important light on the pathogenesis of these complex diseases.
Funding
This work is supported by the National Institutes of Health [NIH] [P30 DK043351], to the Center for Study of Inflammatory Bowel Diseases. ANA is supported in part by grants from the National Institutes of Health [K23 DK097142, R03 DK112909] and the Crohn’s and Colitis Foundation.
Conflict of Interest
ANA has served on scientific advisory boards for Abbvie, Takeda, and Merck.
Supplementary Data
Supplementary data are available at ECCO-JCC online.
Supplementary Material
References
- 1. Loftus EV., Jr Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 2004;126:1504–17. [DOI] [PubMed] [Google Scholar]
- 2. Crohn’s and Colitis Foundation of America. The Facts About Inflammatory Bowel Disease http://www.ccfa.org/assets/pdfs/updatedibdfactbook.pdf Accessed May 29, 2017.
- 3. Liu JZ, van Sommeren S, Huang H et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peeters M, Nevens H, Baert F et al. . Familial aggregation in Crohn’s disease: increased age-adjusted risk and concordance in clinical characteristics. Gastroenterology 1996;111:597–603. [DOI] [PubMed] [Google Scholar]
- 5. Roth MP, Petersen GM, McElree C et al. . Familial empiric risk estimates of inflammatory bowel disease in Ashkenazi Jews. Gastroenterology 1989;96:1016–20. [DOI] [PubMed] [Google Scholar]
- 6. Weterman IT, Peña AS. Familial incidence of Crohn’s disease in The Netherlands and a review of the literature. Gastroenterology 1984;86:449–52. [PubMed] [Google Scholar]
- 7. Moller FT, Andersen V, Wohlfahrt J, Jess T. Familial risk of inflammatory bowel disease: a population-based cohort study 1977-2011. Am J Gastroenterol 2015;110:564–71. [DOI] [PubMed] [Google Scholar]
- 8. Bayless TM, Tokayer AZ, Polito JM II, Quaskey SA, Mellits ED, Harris ML. Crohn’s disease: concordance for site and clinical type in affected family members–potential hereditary influences. Gastroenterology 1996;111:573–9. [DOI] [PubMed] [Google Scholar]
- 9. Satsangi J, Grootscholten C, Holt H et al. . Clinical patterns of familial inflammatory bowel disease. Gut 1996;38:738–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borren NZ, Conway G, Tan W et al. . Distance to specialist care and disease outcomes in inflammatory bowel disease. Inflamm Bowel Dis 2017;23:1234–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conway G, Velonias G, Andrews E et al. . The impact of co-existing immune-mediated diseases on phenotype and outcomes in inflammatory bowel diseases. Aliment Pharmacol Ther 2017;45:814–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barber GE, Yajnik V, Khalili H et al. . Genetic markers predict primary non-response and durable response to anti-TNF biologic therapies in Crohn’s disease. Am J Gastroenterol 2016;111:1816–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther 2011;13:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ananthakrishnan AN, Cagan A, Cai T et al. . Common genetic variants influence circulating vitamin D levels in inflammatory bowel diseases. Inflamm Bowel Dis 2015;21:2507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ananthakrishnan AN, Huang H, Nguyen DD et al. . Differential effect of genetic burden on disease phenotypes in Crohn’s disease and ulcerative colitis: analysis of a North American cohort. Am J Gastroenterol 2014;109:395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morgan XC, Tickle TL, Sokol H et al. . Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012;13:R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caporaso JG, Kuczynski J, Stombaugh J et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McDonald D, Price MN, Goodrich J et al. . An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 2012;6:610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Purcell S, Neale B, Todd-Brown K et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kasen S, Ouellette R, Cohen P. Mainstreaming and postsecondary educational and employment status of a rubella cohort. Am Ann Deaf 1990;135:22–6. [DOI] [PubMed] [Google Scholar]
- 22. Cho JH, Weaver CT. The genetics of inflammatory bowel disease. Gastroenterology 2007;133:1327–39. [DOI] [PubMed] [Google Scholar]
- 23. Orholm M, Munkholm P, Langholz E et al. . Familial occurrence of inflammatory bowel disease. N Engl J Med 1991;324:84–8. [DOI] [PubMed] [Google Scholar]
- 24. Henriksen M, Jahnsen J, Lygren I, Vatn MH, Moum B; IBSEN Study Group Are there any differences in phenotype or disease course between familial and sporadic cases of inflammatory bowel disease? Results of a population-based follow-up study. Am J Gastroenterol 2007;102:1955–63. [DOI] [PubMed] [Google Scholar]
- 25. Yang H, McElree C, Roth MP et al. . Familial empirical risks for inflammatory bowel disease: differences between Jews and non-Jews. Gut 1993;34:517–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang PQ, Hu J, Al Kazzi ES et al. . Family history and disease outcomes in patients with Crohn’s disease: a comparison between China and the United States. World J Gastrointest Pharmacol Ther 2016;7:556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cleynen I, Boucher G, Jostins L et al. . Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ben-Horin S, Avidan B, Yanai H et al. . Familial clustering of Crohn’s disease in Israel: prevalence and association with disease severity. Inflamm Bowel Dis 2009;15:171–5. [DOI] [PubMed] [Google Scholar]
- 29. Halme L, Turunen U, Heliö T et al. . Familial and sporadic inflammatory bowel disease: comparison of clinical features and serological markers in a genetically homogeneous population. Scand J Gastroenterol 2002;37:692–8. [DOI] [PubMed] [Google Scholar]
- 30. Childers RE, Eluri S, Vazquez C et al. . Family history of inflammatory bowel disease among patients with ulcerative colitis: a systematic review and meta-analysis. J Crohns Colitis 2014;8:1480–97. [DOI] [PubMed] [Google Scholar]
- 31. Hwang SW, Kwak MS, Kim WS et al. . Influence of a positive family history on the clinical course of inflammatory bowel disease. J Crohns Colitis 2016;10:1024–32. [DOI] [PubMed] [Google Scholar]
- 32. Chung SH, Park SJ, Lee HS et al. . Similar clinical characteristics of familial and sporadic inflammatory bowel disease in South Korea. World J Gastroenterol 2014;20:17120–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trier Moller F, Andersen V, Andersson M, Jess T. Hospital admissions, biological therapy, and surgery in familial and sporadic cases of inflammatory bowel disease: a population-based cohort study 1977-2011. Inflamm Bowel Dis 2015;21:2825–32. [DOI] [PubMed] [Google Scholar]
- 34. Connelly TM, Berg AS, Harris L III, Brinton D, Deiling S, Koltun WA. Genetic determinants associated with early age of diagnosis of IBD. Dis Colon Rectum 2015;58:321–7. [DOI] [PubMed] [Google Scholar]
- 35. Weersma RK, Stokkers PC, van Bodegraven AA et al. ; Dutch Initiative on Crohn and Colitis [ICC] Molecular prediction of disease risk and severity in a large Dutch Crohn’s disease cohort. Gut 2009;58:388–95. [DOI] [PubMed] [Google Scholar]
- 36. Ananthakrishnan AN, Xavier RJ. How does genotype influence disease phenotype in inflammatory bowel disease?Inflamm Bowel Dis 2013;19:2021–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kugathasan S, Denson LA, Walters TD et al. . Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389:1710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ananthakrishnan AN, Luo C, Yajnik V et al. . Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 2017;21:603–10.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hedin C, van der Gast CJ, Rogers GB et al. . Siblings of patients with Crohn’s disease exhibit a biologically relevant dysbiosis in mucosal microbial metacommunities. Gut 2016;65:944–53. [DOI] [PubMed] [Google Scholar]
- 40. Ijaz UZ, Quince C, Hanske L et al. . The distinct features of microbial ‘dysbiosis’ of Crohn’s disease do not occur to the same extent in their unaffected, genetically-linked kindred. PLoS One 2017;12:e0172605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Olbjørn C SM,. Thiis-Evensen E. Faecal microbiota profiles at diagnosis in paediatric inflammatory bowel disease: prediction of disease severity and subsequent need of biologic therapy. In: proceedings of the Annual Meeting of the European Society for Paediatric Gastroenterology, Hepatology and Nutrition; May 13, 2017; Prague [Google Scholar]
- 42. Mondot S, Lepage P, Seksik P et al. ; GETAID Structural robustness of the gut mucosal microbiota is associated with Crohn’s disease remission after surgery. Gut 2016;65:954–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.