Abstract
Background
Circular RNAs (circRNAs) are RNA transcripts that are widespread in the eukaryotic genome. Recent evidence indicates that circRNAs play important roles in tissue development, gene regulation, and carcinogenesis. However, whether circRNAs encode functional proteins remains elusive, although translation of several circRNAs was recently reported.
Methods
CircRNA deep sequencing was performed by using 10 pathologically diagnosed glioblastoma samples and their paired adjacent normal brain tissues. Northern blotting, Sanger sequencing, antibody, and liquid chromatograph Tandem Mass Spectrometer were used to confirm the existence of circ-FBXW7 and its encoded protein in in two cell lines. Lentivirus-transfected stable U251 and U373 cells were used to assess the biological functions of the novel protein in vitro and in vivo (five mice per group). Clinical implications of circ-FBXW7 were assessed in 38 pathologically diagnosed glioblastoma samples and their paired periphery normal brain tissues by using quantitative polymerase chain reaction (two-sided log-rank test).
Results
Circ-FBXW7 is abundantly expressed in the normal human brain (reads per kilobase per million mapped reads [RPKM] = 9.31). The spanning junction open reading frame in circ-FBXW7 driven by internal ribosome entry site encodes a novel 21-kDa protein, which we termed FBXW7-185aa. Upregulation of FBXW7-185aa in cancer cells inhibited proliferation and cell cycle acceleration, while knockdown of FBXW7-185aa promoted malignant phenotypes in vitro and in vivo. FBXW7-185aa reduced the half-life of c-Myc by antagonizing USP28-induced c-Myc stabilization. Moreover, circ-FBXW7 and FBXW7-185aa levels were reduced in glioblastoma clinical samples compared with their paired tumor-adjacent tissues (P < .001). Circ-FBXW7 expression positively associated with glioblastoma patient overall survival (P = .03).
Conclusions
Endogenous circRNA encodes a functional protein in human cells, and circ-FBXW7 and FBXW7-185aa have potential prognostic implications in brain cancer.
CircRNAs are characterized by “exon skipping” or “direct back-splicing,” which results in the formation of covalently closed-loop structures with no 5ʹ to 3ʹ polarity and that are widely expressed throughout the eukaryotic transcriptome (1–3). Because they have no PolyA tails and have relatively low expression, few circRNAs have been reported in the last decade; previously reported circRNAs include DCC and SRY (4,5). Because of advances in deep sequencing and computational approaches, many circRNAs have been identified in a variety of species (6–8). Previously considered transcriptional errors or side products, circRNAs have recently been shown to play critical roles in gene regulation, neural development, and carcinogenesis (9–11). Specifically, circRNAs have been verified as “microRNA (miRNA) sponges,” harboring multiple miRNAs and functioning as miRNA inhibitors (12,13). Nevertheless, few circRNAs contain perfect miRNA trapping sites, raising the question of whether circRNAs have additional unknown functions (3,14).
Current studies have revealed that some so-called “non-coding RNAs (ncRNAs),” including pre-mRNAs and miRNAs, can generate functional peptides in vivo (15–17). Interestingly, synthetic circRNAs can also encode peptides or proteins (18,19). Furthermore, a computational analysis–based database suggests the coding potential of human circRNAs (20). Just lately, two endogenous circRNAs, circZNF609 and circMbl, were reported to be translatable, and another report showed circRNAs can be translated driven by N6-methyladenosine, further supporting the coding ability of circRNAs (21–23).
In this study, we generated deep RNA sequencing data from 10 glioblastoma samples and their paired adjacent normal tissues and identified approximately 31 000 circRNA candidates. We focused on the most abundantly and differentially expressed circRNAs and matched them with circRNADb (http://reprod.njmu.edu.cn/circrnadb). We characterized the circular form of the FBXW7 gene, a well-characterized tumor-suppressive E3 ligase that encodes a novel 185-amino acid protein in human cells, which we termed “FBXW7-185aa.” We investigated FBXW7-185aa expression and its potential role as a tumor suppressor in vitro and in vivo. Clinal samples and patient data were also used to investigate the relationship between expression and patient outcome.
Methods
Human Cancer and Normal Tissues
All glioma (n = 100, random World Health Organization [WHO] grade glioma; n = 38, glioblastoma and their paired periphery normal brain tissues) and normal brain tissues (n = 100) from traumatic decompression patients were collected from the Department of Neurosurgery at The 1st Affiliated Hospital of Sun Yat-sen University. The human materials were obtained with informed consent, and the study was approved by the Clinical Research Ethics Committee.
RNA Fluorescence In Situ Hybridization
Cells were incubated at 37 °C in a solution containing 50% formamide, 2× SSC, 0.25 mg/mL Escherichia coli transfer RNA, 0.25 mg/mL salmon sperm DNA (Life Technologies, Carlsbad, CA), 2.5 mg/mL BSA (Roche, Indianapolis, IN), and fluorescently labeled junction probe at 125 nM (Generay, Shanghai, China). After 12 hours, the cells were washed and mounted in ProLong Gold (Life Technologies, Carlsbad, CA) and left overnight at room temperature. The probe for circ-FBXW7 was listed in Supplementary Table 1 (available online).
Animal Care and Ethics Statement
Four-week-old female BALB/c-nu mice were purchased from the Laboratory Animal Center of Sun Yat-sen University. Mice were housed in a temperature-controlled (22 °C) andlight-controlled pathogen-free animal facility with free access to food and water. All experimental protocols concerning the handling of mice were approved by the institutional animal care and use committee of Sun Yat-sen University.
Statistical Analysis
Experimental data are represented as the average ± SD of a minimum of three biological replicates. The Student’s two-tailed unpaired t test was used to determine statistical significance of in vitro experiments. The log-rank test or Gehan-Breslow-Wilcoxon test was used to determine the statistical differences of the survival data. All statistical tests were two-sided, and a P value of less than .05 was considered statistically significant.
Detailed methods are described in the Supplementary Materials (available online).
Results
Different CircRNAs Expression Patterns in Human Glioblastoma and Adjacent Normal Tissues
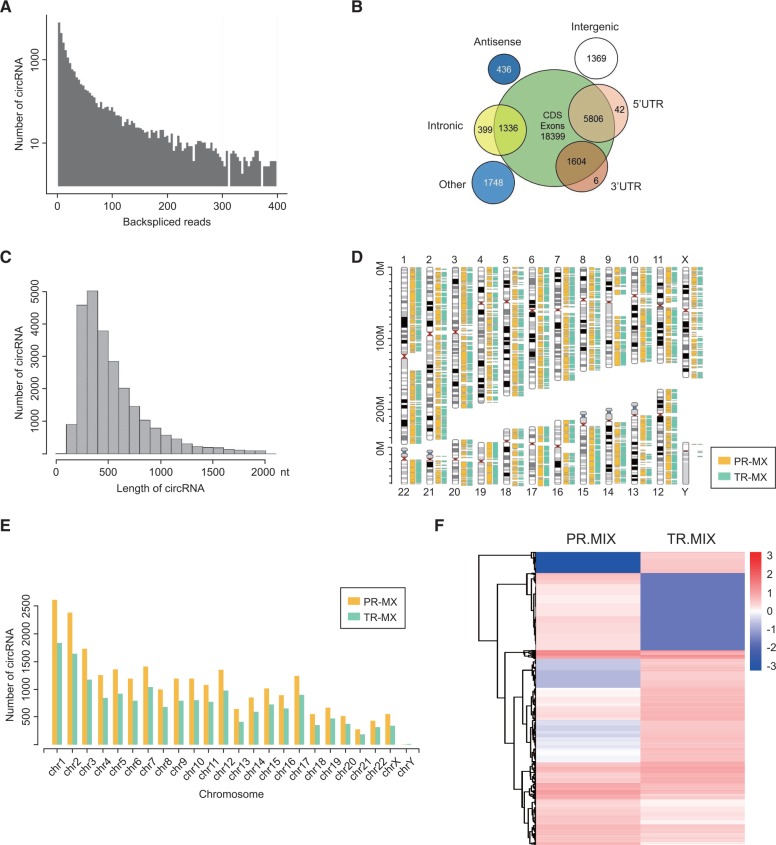
To generate a circRNA profiling database, we performed RNA-seq analyses of ribosomal RNA-depleted total RNA from clinical glioblastoma tissues and their paired adjacent normal tissues. Ten human glioblastoma samples were combined for the tumor group (TR-MIX), and their periphery normal tissues were combined for the normal group (PR-MIX). Tumor and normal RNA were sequenced on an Illumina Hiseq 2500. The reads obtained were mapped to reference ribosomal RNA (Bowtie2, http://bowtie-bio.sourceforge.net/bowtie2/) and reference genome (TopHat2, http://ccb.jhu.edu/software/tophat/) (24,25), and 20 mers from both ends of the unmapped reads were extracted and aligned to the reference genome to find unique anchor positions within the splice site. Anchor reads that aligned in the reverse orientation (head-to-tail) indicated circRNA splicing and were then subjected to find_circ (https://omictools.com/find-circ-tool) to identify circRNAs (12). A candidate circRNA was called if it was supported by at least two unique back-spliced reads in one sample. A total of 31 145 circRNAs were identified by this approach, 6442 of which were matched in circBase (http://www.circbase.org/) (Figure 1A) (26). We next annotated these identified candidates using the RefSeq database (27). Most of these circRNAs originated from protein coding exons and others aligned with introns, 5ʹ-UTR, 3 ʹ-UTR, or antisense sequences (Figure 1B). The majority of the identified circRNAs were less than 1500 nucleotides (nt) (Figure 1C). The chromosome distribution of identified circRNAs showed no obvious difference between cancerous and normal groups, while the total expression of circRNAs in the cancerous group was downregulated (Figure 1, D and E). As shown in Figure 1F, the cancerous and tumor groups had differential circRNA expression patterns. We focused on the candidates that had the greatest differential expression between the cancerous and normal groups and matched them with circRNADb (Supplementary Table 2, available online). Among these specific candidates, novel_circ_022705, which was formed by circularization of exon 3 and exon 4 of the FBXW7 gene (RPKM = 9.31), attracted our attention (Figure 2A).
Figure 1.
Profiling of circular RNAs in human glioblastoma and adjacent normal tissues. A) RNA-seq read abundance distribution of identified circular RNAs (circRNAs). x-axis: the back-spliced read numbers of circRNAs detected in RNA-seq. y-axis: the abundance of circRNAs classified by different read numbers. The majority of called circRNAs in the study were supported by more than 10 reads. B) Venn plot showing the number of circRNAs derived from different genomic regions. C) Length distribution of the identified circRNAs. x-axis: the length of circRNAs detected in this study. y-axis: the abundance of circRNAs classified by different lengths. D) The distribution of identified circRNAs in chromosomes. The yellow and cyan bars represent the location of detected circRNAs within different chromosomes in normal and tumor samples, respectively. E) Numbers of identified circRNAs in different chromosomes. The yellow and cyan bars represent the numbers of circRNAs within different chromosomes detected in normal and tumor samples, respectively. F) Heat map of all differentially expressed circRNAs between normal and tumor tissues. PR-MIX = tumor peripheral RNA mix; TR-MIX = tumor RNA mix.
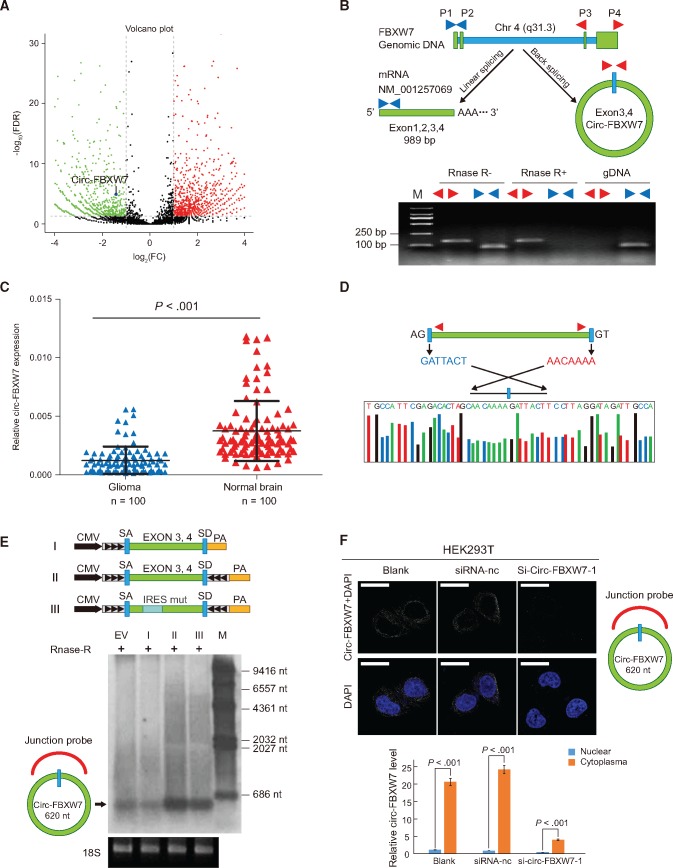
Figure 2.
Characterization of circ-FBXW7 as a circular RNA in vitro and in vivo. A) In the volcano plot, the green, red, and black points represent downregulated, upregulated, and no statistically significant difference circular RNAs (circRNAs) in the TR-MIX group compared with the PR-MIX group, respectively. x-axis: log2 ratio of circRNA expression levels between normal and tumor tissues. y-axis: the false discovery rate value (-log10 transformed) of circRNAs. The blue dot indicates novel_circ_022705 (circ-FBXW7). B) Upper panel, illustration of the annotated genomic region of FBXW7, the putative different RNA splicing forms, and the validation strategy for circular exon 3 and 4 (circ-FBXW7). Convergent (blue) and divergent (red) primers were designed to amplify the linear or back-splicing products. Lower panel: total RNA from 293T cells with or without RNase-R treatment were subjected to polymerase chain reaction (PCR). C) Expression levels of circ-FBXW7 in 100 randomly selected glioma samples and 100 normal brain tissues were detected by the junction primers (P < .001). Values are the average ± SD of three independent experiments. The Student’s two-tailed unpaired t test was used to determine statistical significance between the normal and glioma groups. D) Sanger sequencing following PCR conducted using the indicated divergent flanking primers confirmed the “head-to-tail” splicing of circ-FBXW7 in 293T cells. E) Upper panel: illustration of the synthetic circRNA expression plasmid. I: exons 3 and 4 of the FBXW7 gene were cloned between splicing acceptor (SA) and splicing donor (SD) sequences with upstream flanking repeat sequences (black arrows) (sequences showed in Supplementary Table 1, available online). II: exon 3 and 4 sequences of the FBXW7 gene were cloned between SA and SD with both sides of flanking repeat sequences. III: compared with vector II, the putative internal ribosomal entrance site sequences were mutated (as shown in Supplementary Figure 2, available online). Lower panel: empty vector and above-mentioned vectors were transfected into 293T cells. After 24 hours of transfection, total RNA was treated with RNase-R and subjected to Northern blotting using circ-FBXW7 junction probes. F) Upper panel: fluorescence in situ hybridization with junction-specific probes indicates the cellular localization of circ-FBXW7. Scale bars = 5 μM. Lower panel: circ-FBXW7 was detected in different cell fractions. Nuclear and cytoplasmic RNA was extracted, and junction primers were used for circ-FBXW7 detection. U6 was used as internal control of nuclear RNA, and GAPDH was used as internal control for cytoplasmic RNA. Values are the average ± SD of three independent experiments. The Student’s two-tailed unpaired t test was used to determine the statistical significance between nuclear and cytoplasm circ-FBXW7 expression. EV = transection of empty vector; M = nucleotide marker; NC = negative control.
Identification of Circ-FBXW7 as a Circular RNA
To verify that exons 3 and 4 of the FBXW7 gene formed an endogenous circRNA, we designed convergent and divergent primers that specifically amplified the canonical or back-spliced forms of FBXW7 (Figure 2B, upper panel). Polymerase chain reaction (PCR) after reverse transcription with the divergent primers (red) detected circ-FBXW7, which is resistant to digestion by RNase-R (28). These divergent primers could not amplify any products in genomic DNA, indicating that this result was not due to a PCR artifact or genomic rearrangement. In contrast, convergent primers (blue) amplified the PCR products from linear FBXW7 mRNA, which disappeared after RNase-R digestion (Figure 2B, lower panel). We verified the RNA-seq data using spanning junction primers to detect circ-FBXW7 expression in 100 glioma samples (random WHO grades) and 100 normal brains (from internal decompression patients) by using quantitative PCR (q-PCR). Consistent with the RNA-seq analysis, circ-FBXW7 expression was lower in glioma than normal brains (Figure 2C). Next, Sanger sequencing was used to confirm the circ-FBXW7 junction (Figure 2D). We found that the back-splicing formed the 620 nt circ-FBXW7 instead of the 1227 nt circ-FBXW7 as reported in Circbase (hsa_circ_0001451), most likely due to intronic sequence shearing (29). Endogenous circ-FBXW7 was further detected using a junction-specific probe in Northern blotting (Figure 2E, EV). The synthetic circRNA expression vector successfully overexpressed circ-FBXW7, induced by side flanking repeat sequences with splicing acceptor (SA) and donor (SD) (30), which was not observed for a control vector lacking the downstream flanking sequences (Figure 2E, lanes I and II). To demonstrate circ-FBXW7 cellular localization, we conducted FISH analysis. The junction probe detected abundant cytoplasmic circ-FBXW7 expression in 293T cells. When small interference RNA (siRNA) specifically designed to target the junction was transfected, circ-FBXW7 expression was inhibited compared with that of the control siRNA (Figure 2F, upper panel). q-PCR analysis of different cell fractions confirmed that circ-FBXW7 is predominantly located in the cytoplasm (Figure 2F, lower panel).
Evaluation of the Coding Ability of Circ-FBXW7
As circ-FBXW7 was matched in circRNADb, we next analyzed the putative open reading frame (ORF) in circ-FBXW7. There was a potential spanning junction ORF that encoded a 185 aa protein in circ-FBXW7 (Figure 3A). Conservation analysis showed that this ORF is highly conserved among different species, implying that this ORF is translatable (Supplementary Figure 1, A–C, available online). An internal ribosomal entrance site (IRES) is required for 5ʹ-cap independent translation (31). To test the putative IRES activity in circ-FBXW7, we used a dual-luciferase vector system. Full-length or truncated putative circ-FBXW7 IRES sequences (–120 bp from the ORF) (Supplementary Figure 1, D and E, available online) were cloned between the Rluc and Luc reporter genes. Rluc and Luc reporters with independent start and stop codons were directly connected in the empty vector. A luciferase assay showed that the full-length circ-FBXW7 IRES induced the highest Luc/Rluc activity compared with the truncated or mutated IRES. In contrast, empty vectors could not induce Luc activation (Figure 3B). These results indicated that circ-FBXW7 IRES could induce 5ʹ-cap independent translation. Next, we established a set of vectors to confirm that circ-FBXW7 is translatable in human cells. As shown in the Figure 3C upper panel, the junction of endogenous circ-FBXW7 is inside the ORF (endo-circ-FBXW7). To add a FLAG tag, we moved the junction to the stop codon of the ORF and separated the FLAG sequence to both sides, with side flanking sequences as well as SA and SD (circ-FBXW7-FLAG). For a negative control, downstream flanking sequences were deleted (linear-FL-FBXW7-AG). For a positive control, the linearized ORF plus FLAG tag was cloned in a linear vector (FBXW7-FLAG). We transfected these plasmids into 293T cells and detected their potential translated products. As shown in the Figure 3C lower panel, the FLAG tag antibody only detected an approximately 22 kDa protein in circ-FBXW7-FLAG- and FBXW7-FLAG-tranfected cells, indicating that the circ-FBXW7-FLAG vector is translated. The antibody specifically targeting the putative circ-FBXW7 translated protein (we termed it “FBXW7-185aa”) (antigen design shown in Figure 3C, middle) could detect endogenous FBXW7-185aa in blank and linear-FL-FBXW7-AG-transfected cells, while the circ-FBXW7-FLAG- and FBXW7-FLAG-transfected cells showed obvious overexpression of FBXW7-185aa. We next used liquid chromatograph Tandem Mass Spectrometer to characterize the amino acid sequences of FBXW7-185aa in circ-FBXW7-transfected 293T cells. As FBXW7-185aa is formed by the “spanning junction ORF,” the identified distinctive amino acids in this region further suggested that this novel protein was encoded by circ-FBXW7 (Figure 3, C and D).
Figure 3.
Evaluation of the coding ability of circ-FBXW7. A) Upper panel: the putative open reading frame (ORF) in circ-FBXW7. Note that the circ-FBXW7 junction is inside the ORF. Lower panel: the sequences of the putative ORF are shown in green, internal ribosomal entrance site (IRES) sequences are shown in purple, and specific amino acid sequences of FBXW7-185aa are shown in yellow. B) The putative IRES activity in circ-FBXW7 was tested. Upper panel: IRES sequences in circ-FBXW7 or its different truncations were cloned between Rluc and Luc reporter genes with independent start and stop codons. Lower panel: the relative luciferase activity of Luc/Rluc in the above vectors was tested. Values are the average ± SD of three independent experiments. C) Upper left: vector set for detecting circ-FBXW7 encoded protein. Endo-circ-FBXW7: illustration showing how endogenous circ-FBXW7 formed; the circular junction is inside the ORF and formed the unique sequences shown in yellow. Circ-FBXW7-FLAG: exon 3 and exon 4 of FBXW7 were cloned between splicing acceptor, splicing donor, and side flanking repeat sequences (black arrows); the circular junction was moved to the stop codon of the ORF. FLAG tag was divided to both sides (light and dark blue). Circularization of this vector could form the same circular RNA as endogenous circ-FBXW7, except with a FLAG tag added behind the ORF. Linear-FL-FBXW7-AG: circ-FBXW7-FLAG vector lacking the downstream flanking repeat sequence. FBXW7-185aa-FLAG: the 185 aa ORF with FLAG tag was directly cloned inside a linear expression vector. Middle: the putative FBXW7-185aa amino acid sequences and the antigen used to produce the FBXW7-185aa antibody. The red amino acids were distinctly formed by the circ-FBXW7 junction. Lower: FLAG tag antibody and FBXW7-185aa antibody were used to detect FBXW-185aa expression in 293T cells transfected with the above vectors. D) Total proteins from circ-FBXW7 or control plasmid-transfected 293T cells were separated via SDS-PAGE (left). FBXW7-185aa overexpression was confirmed by immunoblotting (upper right). The differential gel bands between 17 kDa and 26 kDa were extracted and subjected to liquid chromatograph Tandem Mass Spectrometer. Among the liquid chromatograph Tandem Mass Spectrometer results, two FBXW7-185aa junction-specific peptides were identified (lower right).
Effects of FBXW7-185aa on Glioma Cells and the Molecular Mechanism
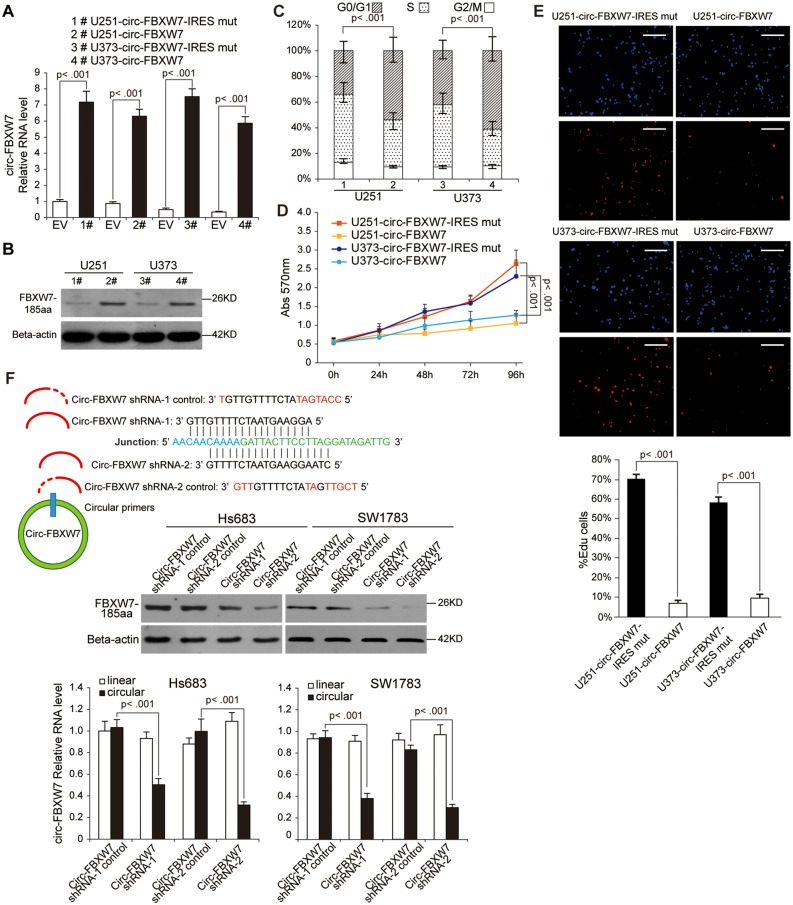
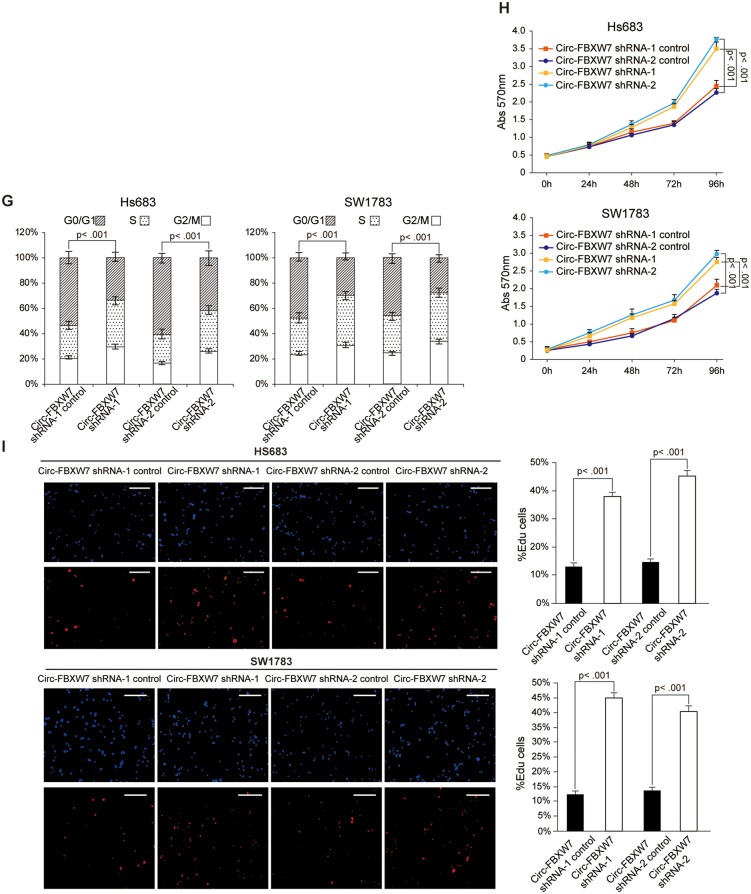
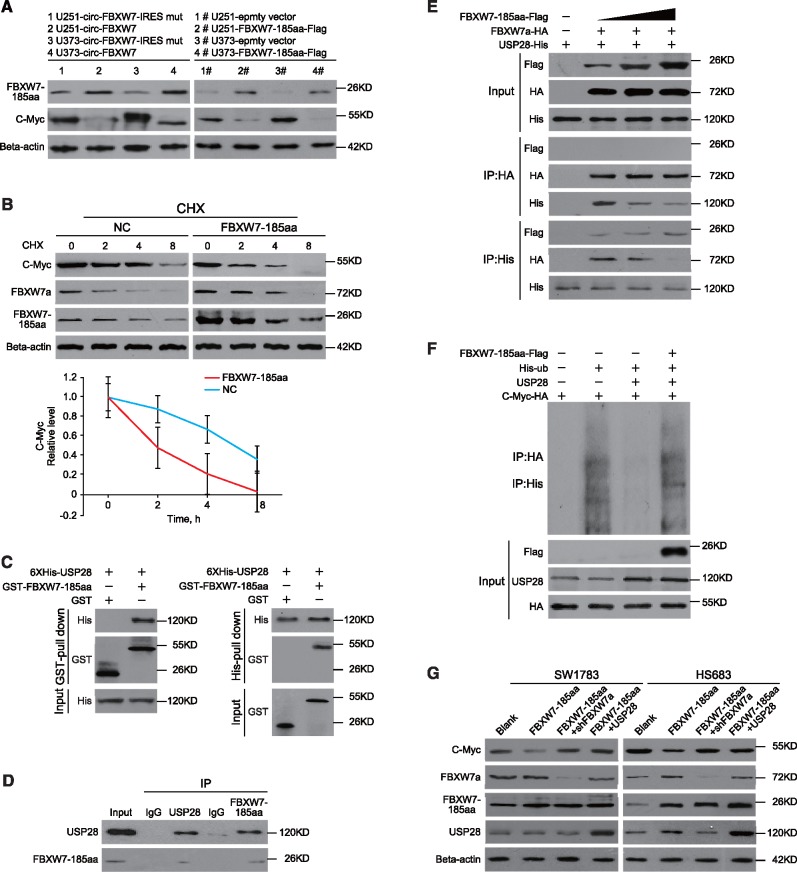
To further explore the biological functions of circ-FBXW7, we established stable circ-FBXW7-overexpressing U251 and U373 cells using the vectors described in Figure 2E, lane II. The IRES-mutated vector, in which the IRES sequences were mutated but the vector could form similar circRNAs as circ-FBXW7, was used as a negative control (Figure 2E, lane III; Supplementary Figure 1E, available online). Additionally, to exclude the possibility that the biological functions were induced by circ-FBXW7 instead of FBXW7-185aa, we used the linear FBXW7-FLAG vector described in Figure 3C to establish the FBXW7-185aa stably overexpressing U251 and U373 cells. By using circ-FBXW7 junction primers, we showed that both circ-FBXW7 and circ-FBXW7-IRES mut vectors were successfully overexpressed (Figure 4A). The FBXW7-FLAG vector did not induce circ-FBXW7 overexpression (Supplementary Figure 2A, available online). Meanwhile, instead of the circ-FBXW7-IRES mut vector, only circ-FBXW7 and FBXW7-FLAG vector overexpression could elevate FBXW7-185aa expression (Figure 4B;Supplementary Figure 2B, available online). We next tested the cell cycle and the cell proliferation rate in these cells. Circ-FBXW7 and FBXW7-FLAG stably overexpressing U251 and U373 cells exhibited a massive G1 phase arrest compared with their control cells (Figure 4C;Supplementary Figure 2C, available online). Additionally, the viability of circ-FBXW7 and FBXW7-FLAG stably overexpressing U251 and U373 cells was reduced compared with their control cells (Figure 4, D and E; Supplementary Figure 2D, available online). However, knocking down circ-FBXW7 by two junction-specific shRNAs in Hs683 and SW1783 anaplastic astrocytoma cells decreased the expression of FBXW7-185aa (Figure 4F) and increased cell cycle acceleration and cell viability (Figure 4, G–I). These results indicate that FBXW7-185aa, but not circ-FBXW7, could induce cell cycle arrest and reduce proliferation in glioma cells.
Figure 4.
Tumor-suppressive functions of FBXW7-185aa in glioma cell lines. A) circ-FBXW7 expression was detected by using junction primers in U251 and U373 cells stably expressing circ-FBXW7 or circ-FBXW7 internal ribosomal entrance site (IRES) mut plasmids. B) FBXW7-185aa expression was detected in circ-FBXW7 or circ-FBXW7 IRES mut plasmid stably overexpressing U251 and U373 cells. C) Cell cycle was determined in circ-FBXW7 or circ-FBXW7 IRES mut plasmid stably overexpressing U251 and U373 cells. D) The growth curve was determined by MTT assay in circ-FBXW7 or circ-FBXW7 IRES mut plasmid stably overexpressing U251 and U373 cells. E) Left panel: circ-FBXW7 or circ-FBXW7-IRES mut overexpressing U251 and U373 cells were treated with 10 µM EdU for two hours, then detected with Andy Fluor 594 azide (red); cells were counterstained with DAPI (blue). Scale bar = 50 μM. Right panel: quantification of left panel. The percentage of EdU-incorporated cells was counted in 200 cells. F) Two spanning junction shRNAs were designed for circ-FBXW7 and stably expressed in Hs683 and SW1783 anaplastic astrocytoma cell lines. The RNAi effect was determined by quantitative polymerase chain reaction using junction primers for circ-FBXW7. FBXW7-185aa expression was detected in circ-FBXW7 shRNA stably expressing Hs683 and SW1783 cells and their control cells. G) The cell cycle was determined in sh-circ-FBXW7 stably expressing Hs683 and SW1783 cells and their control cells. H) The growth curve was determined by MTT assay of sh-circ-FBXW7 stably expressing Hs683 and SW1783 cells and their control cells. I) Left panel: circ-FBXW7 stable knockdown Hs683 and SW1783 cells and their respective control cells were treated with 10 µM EdU for two hours, then detected with Andy Fluor 594 azide (red); cells were counterstained with DAPI (blue). Scale bar = 50 μM. Right panel: quantification of left panel. The percentage of EdU-incorporated cells was counted in 200 cells. In each panel of this figure, values are the average ± SD of three independent experiments. The Student’s two-tailed unpaired t test was used to determine statistical significance in/between indicated groups.
FBXW7α, the most abundant isoform of the FBXW7 gene, is a well-defined E3 ligase (32). FBXW7α could target c-Myc, a key regulator of tumorigenesis, for ubiquitination-induced degradation (33). FBXW7-185aa did not change FBXW7α mRNA or protein levels (Supplementary Figure 2E, available online). However, we found that the expression of c-Myc was also decreased in both circ-FBXW7- and FBXW7-185aa-overexpressing cells (Figure 5A). Meanwhile, c-Myc mRNA levels remained unchanged (Supplementary Figure 2F, available online). Given the above evidence, we inferred that FBXW7-185aa may also contribute to c-Myc protein stability. A half-life test confirmed that FBXW7-185aa could de-stabilize c-Myc compared with the negative control. Notably, FBXW7-185aa was more stable than FBXW7α (Figure 5B). As FBXW7-185aa only contained the 5ʹ terminal of the FBXW7 pre-mRNA (Supplementary Figure 3A, available online), it could not directly interact with c-Myc though the C terminal WD40 domains, in contrast to FBXW7α (33). However, the de-ubiquitinating enzyme USP28 was reported to bind to c-Myc through interaction with the FBXW7α N terminal, sequentially stabilizing c-Myc (34,35). We inferred that FBXW7-185aa may destabilize c-Myc through USP28. As expected, FBXW7-185aa interacts with USP28 in vitro and in vivo (Figure 5, C and D). Compared with FBXW7α, FBXW7-185aa has a higher binding affinity to USP28. Increased FBXW7-185aa could competitively free FBXW7α from USP28 in cancer cells (Figure 5E). In vivo and in vitro ubiquitination assays showed that FBXW7-185aa overexpression antagonized the de-ubiquitination of c-Myc induced by USP28 and increased c-Myc ubiquitination. However, FBXW7-185aa alone could not ubiquitinate c-Myc (Figure 5F;Supplementary Figure 3B, available online). In Hs683 and SW1783 cells, overexpression of FBXW7-185aa could not decrease c-Myc if FBXW7α was knocked down, indicating that FBXW7-185aa regulated c-Myc through FBXW7α. Furthermore, overexpressing USP28 inhibited FBXW7-185aa-induced c-Myc turnover (Figure 5G). In FBXW7α/circ-FBXW7-knockout Hs683 and SW1783 cells, the addition of FBXW7α could induce c-Myc degradation, but addition of FBXW7-185aa alone did not have the same effect (Supplementary Figure 4, available online). The above evidence collectively elucidates that FBXW7-185aa competitively interacted with USP28 and “freed” FBXW7α to degrade c-Myc.
Figure 5.
The molecular mechanism of FBXW7-185aa in suppressing glioma tumorigenesis. A) FBXW7-185aa and c-Myc expression were detected by immunoblotting of circ-FBXW7 or circ-FBXW7 internal ribosomal entrance site mut overexpressed U251 and U373 cells. B) Upper panel: FBXW-185aa or control plasmid-transfected U251 cells were treated with cycloheximide. Total lysates at the indicated time points were collected, and indicated proteins were determined by immunoblotting. Lower panel: semiquantification of c-Myc protein levels in the above immunoblotting. Values are the average ± SD of three independent experiments. C) GST-tagged FBXW7-185aa and 6XHis-tagged USP28 were expressed in Escherichia coli. Purified His-tagged-USP28 and GST-tagged-FBXW7-185aa were subjected to in vitro GST or His pull-down assays. The pull-down lysates were subjected to immunoblotting. D) In vivo interaction of USP28 and FBXW7-185aa was detected by immunoprecipitation in SW1783 cells. E) HA-tagged-FBXW7α and His-tagged-USP28 were cotransfected with increasing doses of FLAG-tagged-FBXW7-185aa. Immunoprecipitation was performed using anti-HA or anti-His antibody, followed by immunoblotting using the indicated antibodies. F) FLAG-tagged-FBXW7-185aa, His-tagged-Ubiquitin, USP28 and HA-tagged-Myc were transfected into 293T cells at the indicated combinations. Immunoprecipitation was performed using anti-HA antibody, and the ubiquitination level of c-Myc was determined by immunoblotting. G) FBXW7-185aa-overexpressing Hs683 or SW1783 cells were stably transfected with sh-FBXW7α or USP28 plasmids. After 72 hours, c-Myc, FBXW7α, FBXW7-185aa, and USP28 levels were detected by immunoblotting. All statistical tests were two-sided. CHX = cycloheximide.
Clinical Implications of Circ-FBXW7 and FBXW-185aa
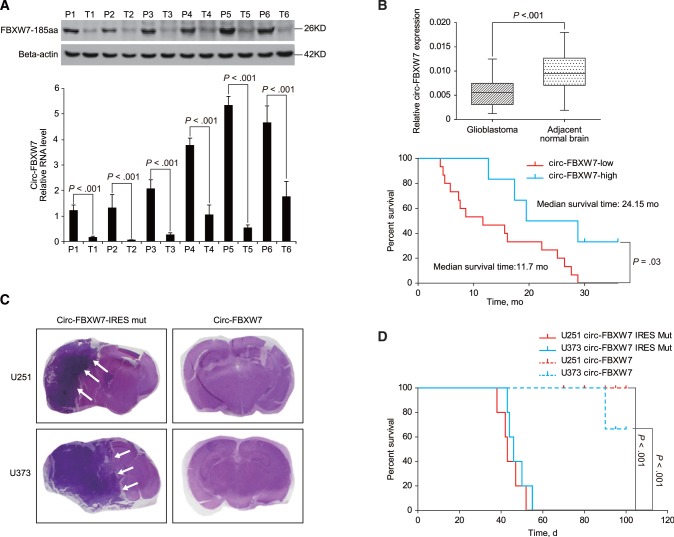
To explore the potential clinical implications, we detected FBXW7-185aa expression in several established glioma cell lines. NHA and 293T cells exhibited strong FBXW7-185aa expression, and SW1783 and Hs683 grade III glioma cell lines had reduced FBXW7-185aa expression. U251 and U373, which are glioblastoma cell lines, exhibited the lowest expression of FBXW7-185aa. The corresponding circ-FBXW7 had a similar expression pattern as FBXW7-185aa (Supplementary Figure 5A, available online). Additionally, FBXW7-185aa and circ-FBXW7 were decreased in six randomly selected glioblastoma patient samples compared with their paired tumor-adjacent tissues (P < .001) (Figure 6A;Supplementary Figure 5, B and C, available online). Next, the expression of circ-FBXW7 was detected in 38 pairs of cancerous and adjacent noncancerous tissues derived from glioblastoma patients (36). In glioblastoma tissues, circ-FBXW7 expression was lower compared with that in paired adjacent noncancerous tissues (P < .001) (Figure 6B, left). Meanwhile, glioblastoma patients with higher circ-FBXW7 (eight in 38 patients, 21.1%) had an increased total survival time compared with those with low circ-FBXW7 expression (median survival time in circ-FBXW7-high group is 24.2 months and in the circ-FBXW7-low group is 11.7 months, P = .03) (Figure 6B, right). As frequently seen FBXW7α mutations did not affect circ-FBXW7 or FBXW7-185aa (Supplementary Figure 6, available online), we assumed that circ-FBXW7 and FBXW7-185aa are independent prognostic markers for glioblastoma. Finally, circ-FBXW7 stably overexpressing U251 and U373 cells exhibited a much lower tumorigenicity compared with control cells as implantation of circ-FBXW7-overexpressing U251 and U373 cells reduced the xenograft brain tumor formation in nude mice (Figure 6C). Additionally, circ-FBXW7 stably overexpressing U251 and U373 cell-implanted mice lived longer than the control mice (five mice per group, P < .001) (Figure 6D).
Figure 6.
Clinical implications of circ-FBXW7 and FBXW-185aa. A) Six randomly selected glioblastoma patient samples and their paired adjacent normal brain tissues were collected. Circ-FBXW7 and FBXW7-185aa were detected by quantitative polymerase chain reaction (q-PCR) and immunoblotting, respectively. Values are the average ± SD of three independent experiments. The Student’s two-tailed unpaired t test was used to determine statistical significance between indicated groups. B) Upper panel: circ-FBXW7 levels in cancerous and adjacent normal tissues were detected in a cohort of 38 glioblastoma patients by q-PCR. Lower panel: patients in the cohort were divided into two groups according to relative circ-FBXW7 expression. The overall survival time of each group was calculated. The P value between the circ-FBXW7-high and -low groups is .03 (log-rank test) or .04 (Gehan-Breslow-Wilcoxon test). C) Circ-FBXW7 or circ-FBXW7-internal ribosomal entrance site mut plasmid stably overexpressing U251 and U373 cells were intracranially injected into nude mice; 5 × 105 cells were used per mouse, and each group contained five mice. Mice were killed, and the brain tissues were collected if clinical symptoms, such as emaciation, disorientation, or hemiplegia, were shown. At day 100, all mice were killed, and hematoxylin and eosin (HE) staining was performed on all collected brain tissues. Representative HE staining of brain sections in each experimental group is shown. Tumors are indicated by white arrows. D) The survival time in each experimental group was calculated. The survival time of mice injected with circ-FBXW7-overexpressed glioblastoma multiforme cells was longer than the mice injected with corresponding control cells (n = 5 mice per group, P < .001, Log rank test). All statistical tests were two-sided.
Discussion
CircRNAs are novel RNA molecules with different biological functions and pathological implications (10,37,38). Among these multiple functions, “miRNA sponge” represents the most conspicuous function (12,13,39). However, the general roles of circRNAs remain elusive (3,14). Interestingly, artificial circRNA was shown to be translatable in eukaryotic cells (18). Current evidence also shows that other types of so-called “non-coding RNA” can initiate protein synthesis, raising the question of whether endogenous circRNAs can encode proteins in mammalian cells (15–17). In this study, by matching the RNA deep sequencing data with circRNADb, we found that circ-FBXW7 encoded a novel protein that suppresses glioma cell proliferation and the cell cycle in vitro and in vivo. Translated from the spanning junction ORF formed by the covalent connection of exon 3 and exon 4 of the FBXW7 gene, FBXW7-185aa had distinct amino acid sequences compared with its related linear mRNA translated proteins. As a constituent of the Skp1-Cul1-F Box (SCF) ubiquitin ligase, linear mRNA encoding FBXW7 was reported to degrade key cellular regulators, including c-Myc, cyclin E1, c-Jun, and Notch1 (40). Thus far, three FBXW7 isoforms, including FBXW7α, β, and γ, have been reported. These isoforms are highly conserved at the C terminal, whereas different N terminals are driven by isoform-specific promoters (41). Although FBXW7 isoforms have diverse cellular localizations, evidence has shown that they are functionally identical as the C-terminal F-Box and WD-40 domains are common among them (41,42). These isoforms can be expressed upon specific signaling and execute precise cellular functions.
Although shorter than the above three isoforms, FBXW-185aa also inhibited the expression of c-Myc, thus decreasing cellular proliferation and inhibiting cell cycle acceleration. However, as FBXW7-185aa did not contain the conservative WD-40 domains, it could not degrade c-Myc through direct interaction-induced ubiquitination. Instead, FBXW7-185aa binds to USP28, which is a de-ubiquitinating enzyme that stabilizes c-Myc by inhibiting FBXW7α activity. FBXW7-185aa competitively interacts with USP28 and acts as a tumor-suppressive “decoy” that prevents USP28 binding to FBXW7α, thus antagonizing USP28-induced c-Myc stabilization. FBXW7 mutations have been commonly identified in various types of human cancers (43). These mutations lead to dominant-negative or premature termination of FBXW7 with a loss of tumor-suppressive functions (40,44,45). Given the critical role of the FBXW7 gene, we hypothesized that the identical functions of FBXW7 isoforms, as well as shorter and more stable circ-FBXW7-translated FBXW7-185aa, may serve as “multiple safe assurances” to control cellular proliferation by this critical regulator.
This study was limited to cultured cells and xenograft animal models. The clinical significance of circ-FBXW7 and FBXW7-185aa may also need to be addressed in larger cohorts. Furthermore, although the tumor-suppressive role is definitive, the clinical application of FBXW7-185aa still has a long way to go. Nevertheless, we provide clear evidence that circRNA encodes functional protein in vivo. Although circRNAs have been reported as important regulators during critical biological processes, no translatable circRNAs or their products have been reported during human carcinogenesis. Our findings expand upon the current understanding of circRNAs and imply that the coding potential of circRNAs is largely underestimated. Specifically, evidence from circ-FBXW7 and FBXW7-185aa strongly suggests that circRNAs and their translated proteins may have a role in glioma carcinogenesis as well as in patient clinical prognosis.
Funding
This work was supported by the National Key Research and Development Program of China (2016YFA0503000), the National Natural Science Foundation of China (81370072 and 81572477), the Science and Technology Project of Guangdong Province (S2013050014535, 2014A030313183, 2016A050502017), Guangzhou Science and Technology Program (201704020068), and the US National Cancer Institute (grants R01CA157933 and R01CA172233).
Notes
The funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
There are no potential conflicts of interest to disclose.
Supplementary Material
References
- 1. Salzman J, Gawad C, Wang PL et al. , Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;72:e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jeck WR, Sorrentino JA, Wang K et al. , Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;192:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jeck WR, Sharpless NE.. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;325:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nigro JM, Cho KR, Fearon ER et al. , Scrambled exons. Cell. 1991;643:607–613. [DOI] [PubMed] [Google Scholar]
- 5. Capel B, Swain A, Nicolis S et al. , Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;735:1019–1030. [DOI] [PubMed] [Google Scholar]
- 6. Zhang XO, Wang HB, Zhang Y et al. , Complementary sequence-mediated exon circularization. Cell. 2014;1591:134–147. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Y, Zhang XO, Chen T et al. , Circular intronic long noncoding RNAs. Mol Cell. 2013;516:792–806. [DOI] [PubMed] [Google Scholar]
- 8. Danan M, Schwartz S, Edelheit S et al. , Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2012;407:3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Guarnerio J, Bezzi M, Jeong JC et al. , Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;1652:289–302. [DOI] [PubMed] [Google Scholar]
- 10. You X, Vlatkovic I, Babic A et al. , Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;184:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Z, Huang C, Bao C et al. , Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;223:256–264. [DOI] [PubMed] [Google Scholar]
- 12. Memczak S, Jens M, Elefsinioti A et al. , Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;4957441:333–338. [DOI] [PubMed] [Google Scholar]
- 13. Hansen TB, Jensen TI, Clausen BH et al. , Natural RNA circles function as efficient microRNA sponges. Nature. 2013;4957441:384–388. [DOI] [PubMed] [Google Scholar]
- 14. Guo JU, Agarwal V, Guo H et al. , Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;157:409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nelson BR, Makarewich CA, Anderson DM et al. , A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science. 2016;3516270:271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lauressergues D, Couzigou JM, Clemente HS et al. , Primary transcripts of microRNAs encode regulatory peptides. Nature. 2015;5207545:90–93. [DOI] [PubMed] [Google Scholar]
- 17. Magny EG, Pueyo JI, Pearl FM et al. , Conserved regulation of cardiac calcium uptake by peptides encoded in small open reading frames. Science. 2013;3416150:1116–1120. [DOI] [PubMed] [Google Scholar]
- 18. Chen CY, Sarnow P.. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;2685209:415–417. [DOI] [PubMed] [Google Scholar]
- 19. Wang Y, Wang Z.. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;212:172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen X, Han P, Zhou T et al. , circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pamudurti NR, Bartok O, Jens M et al. , Translation of circRNAs. Mol Cell. 2017;661:9–21, e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Legnini I, Di Timoteo G, Rossi F et al. , Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;661:22–37, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Y, Fan X, Mao M et al. , Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;275:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Langmead B, Salzberg SL.. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;94:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kim D, Pertea G, Trapnell C et al. , TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;144:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Glazar P, Papavasileiou P, Rajewsky N.. circBase: A database for circular RNAs. RNA. 2014;2011:1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pruitt KD, Tatusova T, Brown GR et al. , NCBI reference sequences (RefSeq): Current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40(database issue):D130–D135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suzuki H, Zuo Y, Wang J et al. , Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;348:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Starke S, Jost I, Rossbach O et al. , Exon circularization requires canonical splice signals. Cell Rep. 2015;101:103–111. [DOI] [PubMed] [Google Scholar]
- 30. Liang D, Wilusz JE.. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;2820:2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mokrejs M, Vopalensky V, Kolenaty O et al. , IRESite: The database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res. 2006;34(database issue):D125–D130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Koepp DM, Schaefer LK, Ye X et al. , Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;2945540:173–177. [DOI] [PubMed] [Google Scholar]
- 33. Yada M, Hatakeyama S, Kamura T et al. , Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;2310:2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Popov N, Herold S, Llamazares M et al. , Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;619:2327–2331. [DOI] [PubMed] [Google Scholar]
- 35. Popov N, Wanzel M, Madiredjo M et al. , The ubiquitin-specific protease USP28 is required for MYC stability. Nat Cell Biol. 2007;97:765–774. [DOI] [PubMed] [Google Scholar]
- 36. Zhang N, Wu X, Yang L et al. , FoxM1 inhibition sensitizes resistant glioblastoma cells to temozolomide by downregulating the expression of DNA-repair gene Rad51. Clin Cancer Res. 2012;1821:5961–5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Du WW, Yang W, Liu E et al. , Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;446:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rybak-Wolf A, Stottmeister C, Glazar P et al. , Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;585:870–885. [DOI] [PubMed] [Google Scholar]
- 39. Zheng Q, Bao C, Guo W et al. , Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. King B, Trimarchi T, Reavie L et al. , The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell. 2013;1537:1552–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Welcker M, Clurman BE.. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;82:83–93. [DOI] [PubMed] [Google Scholar]
- 42. Bai C, Sen P, Hofmann K et al. , SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;862:263–274. [DOI] [PubMed] [Google Scholar]
- 43. Akhoondi S, Sun D, von der Lehr N et al. , FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;6719:9006–9012. [DOI] [PubMed] [Google Scholar]
- 44. Aydin IT, Melamed RD, Adams SJ et al. , FBXW7 mutations in melanoma and a new therapeutic paradigm. J Natl Cancer Inst. 2014;1066:dju107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Trinquand A, Tanguy-Schmidt A, Ben Abdelali R et al. , Toward a NOTCH1/FBXW7/RAS/PTEN-based oncogenetic risk classification of adult T-cell acute lymphoblastic leukemia: A Group for Research in Adult Acute Lymphoblastic Leukemia study. J Clin Oncol. 2013;3134:4333–4342. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.