Abstract
Wildlife species are critical for both feeding vectors and serving as reservoirs of zoonotic vector-borne pathogens. Transmission pathways leading to disease in humans or other target taxa might be better understood and managed given a complete understanding of the relative importance of different reservoir species in nature. Using the conceptual framework of “reservoir potential,” which considers elements of both reservoir competence and vector-host contact, we review the wildlife reservoirs of Trypanosoma cruzi in the southern United States, where many species of triatomine vectors occur and wildlife maintain enzootic cycles that create a risk of spillover to humans, domestic dogs, and captive nonhuman primates that may develop Chagas disease. We reviewed 77 published reports of T. cruzi infection in at least 26 wildlife species across 15 southern states. Among the most well-studied and highly infected reservoirs are raccoon (Procyon lotor), woodrat (Neotoma spp.), and opossum (Didelphis virginiana), with aggregate overall infection prevalences of 36.4, 34.7, and 22.9%, respectively. Just over 60% of studies utilized methods from which an infectiousness index could be generated and show that raccoons and striped skunk (Mephitis mephitis) are among the most infectious wildlife hosts. Triatomine-host contact has sparsely been quantified in the southern United States, but 18 of the 24 host species previously identified to have been fed upon by triatomines are wildlife. Future studies to parameterize the reservoir potential model, especially to quantify wildlife infectiousness, vector-host contact, and the epidemiological importance of parasite strains maintained by wildlife, could open new doors for managing enzootic cycles and reducing T. cruzi spillover risk.
Keywords: bloodmeal analysis, Chagas disease, reservoir, triatomine vector, Trypanosoma cruzi, wildlife
Introduction
Identifying and characterizing reservoirs of vector-borne zoonotic pathogens is critical for disease management interventions that aim to dampen transmission in natural disease cycles to reduce spillover to humans. However, these pathogens are usually maintained in complex transmission cycles involving diverse vertebrate taxa and multiple arthropod vector species. Additionally, these systems are often heterogeneous across space and time, creating challenges for characterizing the wild reservoirs of vector-borne zoonoses (Box 1). The purpose of our review is to provide a framework and highlight gaps in knowledge for the evaluation of candidate wildlife reservoirs of Trypanosoma cruzi, agent of Chagas disease (American trypanosomiasis), in the United States, although the approach we use is broadly applicable to any multihost vector-borne pathogen. T. cruzi is maintained in a complex multihost transmission system at enzootic levels in the southern United States. Despite the first report 100 years ago (Kofoid and Donat 1933; Kofoid and McCulloch 1916), the transmission cycles in the United States and relative importance of different reservoir species have been relatively understudied. In the United States, the disease poses a major threat to the health of domestic dogs and captive nonhuman primates (Dorn et al. 2012; Kjos et al. 2008), and autochthonous transmission has been demonstrated in humans as well (Bern et al. 2011; Curtis-Robles et al. 2017; Garcia et al. 2015; N. C. Woody and H. B. Woody 1955). Domestic dogs are key reservoirs of T. cruzi in South America where the parasite is transmitted in domestic cycles with vector species that colonize the home (Gürtler and Cardinal 2015). However, given ecological differences in T. cruzi vectors and transmission cycles in the southern United States (discussed below), we pose that wild species are critical for maintaining sylvatic transmission cycles and as a source of spillover to target hosts. Here, we outline a framework for assessing the relative importance of reservoir species, which will aid in the development of interventions that limit spillover to humans and domestic animals.
Box 1. Challenges in characterizing wildlife species as reservoirs of zoonotic pathogens.
- Diagnostic limitations
- Diagnostic tests are rarely validated for wildlife species and species-specific controls are largely unavailable.
- Even published serosurveys must be interpreted with caution in the absence of reliable sensitivity and specificity data.
- Sample size limitations
- Large sample sizes are difficult to achieve.
- Studies are often biased toward species that are relatively easy to capture, leaving voids in understanding of more elusive species.
- Study designs often utilize convenience sampling, and results may not be applicable to a broader population.
- Longitudinal studies are rarely feasible
- Most wildlife sampling is done on a cross-sectional basis.
- Individuals can be tracked over time using mark-recapture studies, but these are labor-intensive and associated with biases in trappability of animals.
- Permit requirements for wildlife research are daunting
- Institutional animal use and care committees may be unfamiliar with wildlife field studies (Sikes and Bryan 2016).
- A priori estimates of sample size or infection prevalence are often unknown.
- Protocols for anesthesia and sampling are often optimized for laboratory animals and not translatable to wildlife.
- Statuses of host exposure or infection in relation to disease are ill-defined
- Serosurveys demonstrate exposure and alone should not be used to evaluate reservoir potential.
- Wildlife pathology investigations can provide key information on pathogenesis and population impacts, but are rarely conducted across populations.
- Limited experimental infection data
- Wildlife species are not commonly used as animal models.
- Expectations for progression of disease and time course of infectiousness are typically extracted from domestic and laboratory species.
Ecological Framework for Defining and Characterizing Reservoirs
Definition of a Reservoir
The definition of a reservoir is much discussed and has been refined in recent years. Haydon et al. (2002) defined a reservoir as “one or more epidemiologically connected populations or environments in which the pathogen can be permanently maintained and from which infection is transmitted to the defined target population.” This definition is appropriate for the multihost transmission system of T. cruzi (Gürtler and Cardinal 2015). The target population is defined as the population or host species of interest or concern. Here, we discuss candidate wildlife reservoirs of T. cruzi in the context of the target populations of humans, dogs, and nonhuman primates, all of which are associated with increasing diagnoses of Chagas disease in the southern United States.
In previous definitions, a criterion for a reservoir host is that it does not develop disease as a result of infection with the pathogen (Keane and Miller 2003). However, it is clear from many systems that this is not a requirement for reservoir status. For example, rabies, Hendra, and Nipah viruses all have some pathogenicity to their reservoir host populations (Haydon et al. 2002). Degree and duration of disease can certainly influence the reservoir capacity of a host, however, by directly affecting the time during which it is available to pass the disease on to vectors or other hosts.
It is important to note that ability to be infected by the pathogen does not alone qualify a species as a reservoir. Thus, serological studies on their own are of limited use in determining reservoir potential, as they merely indicate exposure to the pathogen, and give little to no information about the ability of the host to infect vectors or other hosts. They can, however, be used in combination with other data to calculate reservoir competence. Because infection with T. cruzi is generally considered to be life-long, hosts that harbor anti-T. cruzi antibodies are also interpreted to be currently infected (Hall et al. 2007), though more research is needed in wildlife species. For this reason, in contrast to many other zoonotic pathogen systems, T. cruzi seroprevalence estimates can be considered interchangeable with infection prevalence estimates. Additionally, in vector-borne diseases, the presence of infected reservoirs alone does not pose a risk to the target host; the vector must be present and must come into contact with both the reservoirs and target hosts.
Importance of Identifying Reservoirs
The identification of reservoirs is imperative for guiding intervention strategies to reduce transmission in multihost pathogen systems. While it is unlikely that any intervention into the sylvatic cycle could block transmission completely, identifying which species are the most important reservoirs serving to infect those vectors most likely to contact humans or other target hosts (pet dogs, nonhuman primates) could help to guide strategies to reduce spillover. For example, field vaccination of white-footed mice (Peromyscus leucopus), the principle reservoir of Borrelia burgdorferi in the northeastern United States, reduced the infection prevalence of the tick vector in the study sites (Richer et al. 2014). Additionally, aerial distribution of oral baits laden with a rabies vaccine targeted to raccoons has created an immune barrier to halt the westward spread of raccoon strain rabies in the northeast, and a similar program targeted to coyotes helped eliminate the canine strain of rabies from Texas (Slate et al. 2005).
Heterogeneity in Pathogen Transmission
The rate of pathogen transmission (basic reproductive number, R0) is not homogeneous across individuals or host species. Study of heterogeneities in transmission of vector-borne diseases and human sexually transmitted diseases led to the empirical 20/80 rule, which states that in general, 20% of the host population contributes to 80% of the net transmission potential (Woolhouse et al. 1997). Thus, interventions that do not completely block transmission from the most important 20% of the population would be much less effective than predicted given homogeneity of transmission potential. In extreme cases of transmission heterogeneity, only a few key individuals, known as “superspreaders,” contribute disproportionately to the number of transmission events (Lloyd-Smith et al. 2005). This concept of superspreaders has been applied to whole species within multihost transmission systems; for example, American robins serve as a superspreader of West Nile virus (Kilpatrick et al. 2006). Within a reservoir species, heterogeneity in contribution to transmission has been noted for guinea pig (Cavia porcellus) reservoirs of T. cruzi in urban Peru, where most individuals quickly control parasitaemia, but a subset of animals remains highly infectious to vectors for many months (Levy et al. 2015). Conversely, certain host species may have a relatively lower transmission potential and act to dampen the spread of pathogens, termed supersuppressors or dilution hosts. Examples here include Virginia opossum (Didelphis virginiana), which consume the ticks that vector the Lyme disease pathogen Borrelia burgdorferi and therefore serve as an ecological trap, and Northern cardinals and Mimidae spp. that are fed upon by a disproportionate number of mosquitoes but are only moderately competent hosts for West Nile virus (Levine et al. 2016; Ostfeld and Keesing 2000). However, some of these incompetent hosts still contribute to the overall transmission system by serving as bloodmeal sources and amplifying vector populations, as has been shown for deer in the Lyme disease system (Dobson and Randolph 2011) and for chickens with T. cruzi (Gürtler and Cardinal 2015).
Measures of Reservoir Importance
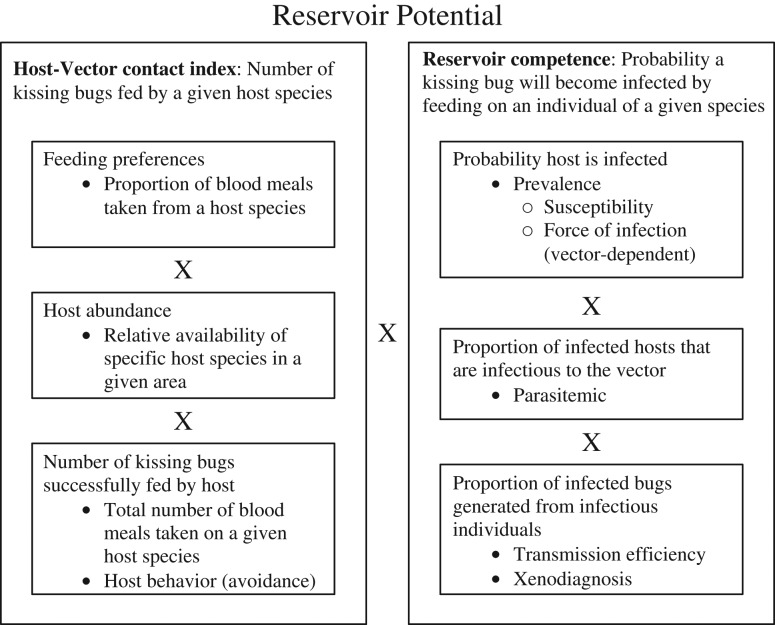
Measures of the relative importance of different host species as reservoirs of a pathogen have been refined over the years in various disease systems. These have been expressed in terms of reservoir potential, reservoir competence, and reservoir capacity. The concept of reservoir potential was first introduced in the Lyme disease system and defined as the relative contribution made by a host species to the horizontal infection of a vector population (Mather et al. 1989). Reservoir potential (Figure 2) is calculated as the product of the number of vectors fed by an individual of a given species and “realized reservoir competence,” the probability that a vector feeding on a host species becomes infected (Brunner et al. 2008). Reservoir competence is therefore the product of the prevalence of host infection and host infectiousness. Considering pathogens for which the reservoir is composed of a group of connected populations (metapopulation), reservoir capacity is defined as a weighted measure of the potential of a host metapopulation to support long-term persistence of a pathogen in the absence of external imports (Viana et al. 2014). Using these concepts, Gürtler and Cardinal (2015) explored the relative contribution of certain domestic and peridomestic reservoirs of T. cruzi in light of three parameters: (1) host susceptibility, infection, and survival; (2) host infectiousness; and (3) host-vector contact. Although the terminology and mathematics surrounding these concepts vary, it is clear that evaluating reservoirs of vector-borne pathogens necessitates quantitative measures of the vertebrate species and their interactions with vectors, and very few studies are designed to fill these knowledge gaps.
Figure 2.
A conceptual framework for evaluating wildlife reservoirs of T. cruzi by determining reservoir potential (Mather et al., 1989; Brunner et al., 2008), an index of the relative importance of a reservoir host as a source of infection to vectors.
T. cruzi Background
T. cruzi is a zoonotic vector-borne protozoan capable of infecting animals from virtually all mammalian orders. An estimated 6 million people are infected worldwide (WHO 2015), of which an estimated 240,000 to 300,000 reside in the United States, though the true burden of human disease in the United States is unknown due to a lack of recognition and reporting (Bern and Montgomery 2009; Manne-Goehler et al. 2016). The disease is enzootic in triatomine insect vectors, wild mammals, and dogs in the southern United States (Bern et al. 2011). Autochthonous transmission to humans was first reported in the United States in 1955 (N. C. Woody and H. B. Woody 1955) and is increasingly recognized as a public health threat (Cantey et al. 2012; Garcia et al. 2015). In addition to the human health burden, T. cruzi infection is also a significant veterinary health problem in the southern United States, with studies documenting 10–25% of dogs (Kjos et al. 2008; Tenney et al. 2014; S. A. Hamer, unpublished data) and a significant number of nonhuman primates (Bern et al. 2011; Dorn et al. 2012) being seropositive, the latter posing a threat to biomedical science initiatives that use nonhuman primate models.
There are 11 species of kissing bugs in the United States, and the highest species diversity of triatomines is found in Texas (Bern et al. 2011). The insect vector acquires the trypomastigote stage of the T. cruzi parasite during blood feeding on an infected host, and the parasite replicates as epimastigotes in the digestive tract of the bug, maturing to infective metacyclic trypomastigotes in the hindgut, which are passed in the feces. The parasite can be transmitted through the stercorarian route when the insect defecates the infectious stage of the parasite onto the host during or shortly after blood feeding, which is then rubbed into the bite wound, broken skin, or a mucous membrane. Oral transmission has been implicated in outbreaks of acute human Chagas disease following consumption of contaminated juices, and oral transmission through the consumption of vectors is likely very important in sylvatic cycles, especially for omnivorous or insectivorous wildlife. In an experimental infection study, four striped skunks (Mephitis mephitis) were infected with T. cruzi intravenously or per os (Davis et al. 1980). Ingestion of infected insects was shown to cause infection in opossums, raccoons, and woodrats and is a probable route of infection in dogs (Montenegro et al. 2002; Roellig, Ellis, and Yabsley 2009a; Ryckman et al. 1965; Yaeger 1971). Transmission may also occur via ingestion of a parasitemic animal (Thomas et al. 2007; Rocha et al. 2013). Additional alternative routes of transmission are transplacental and through blood transfusion or organ transplant.
T. cruzi is a genetically heterogeneous species and is comprised of seven strain types or discrete typing units, TcI-VI, and TcBat. TcI has been divided into TcIdom and TcIsyl, representing domestic and sylvatic isolates (Ramírez et al. 2013). These strain types are associated with different geographical locations, reservoir host species, and reportedly, clinical manifestations (Jansen et al. 2015; Ramírez et al. 2010). TcI and TcIV are the most commonly reported discrete typing units in the United States (Bern et al. 2011; Roellig et al. 2013), though TcII has been isolated from a small number of rodents (Herrera et al. 2015). More research is needed on the specific importance of these strain types in the United States and their relevance to outcome of infection.
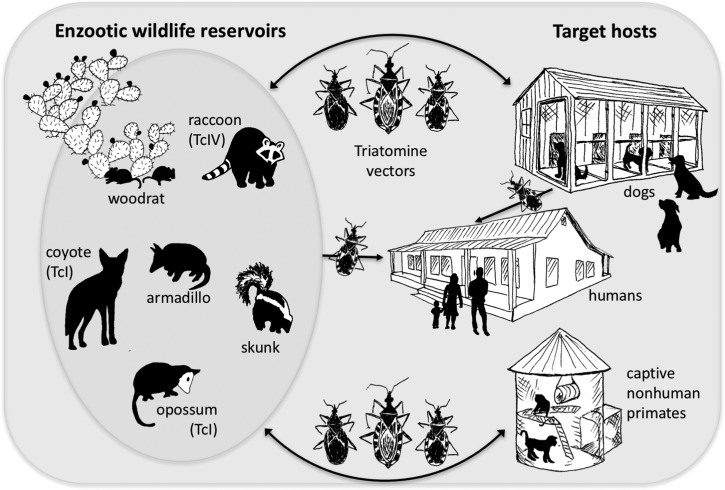
Across Latin America, T. cruzi is maintained in distinct transmission settings of domestic/peridomestic cycles—defined by vector species that are adapted to live predominantly in and around human dwellings and feed on inhabitants—and sylvatic cycles, with different vectors, reservoirs, and strain types associated with each (Zingales et al. 2012). Dogs, cats, commensal rodents, and domesticated guinea pigs serve as predominant reservoirs in the peridomestic and domestic settings, whereas opossums, armadillos, and rodents are major sylvatic reservoir hosts (Gürtler and Cardinal 2015; Jansen et al. 2015). In the United States, however, although there have been infrequent reports of both adult and nymphal kissing bugs found within homes in the United States (Curtis-Robles et al. 2015; Klotz et al. 2016; Navin et al. 1985; Wozniak et al. 2015), truly domestic transmission cycles are rare, owing in part to different standards of housing and different species of triatomines. Peridomestic and sylvatic bug activity is much more common, and transmission to humans and other target taxa results from spillover from the enzootic cycles (Figure 1). Wildlife are important in the maintenance of the parasite in these sylvatic cycles, and better characterizing their relative importance as reservoirs is important in understanding the transmission of T. cruzi in the United States.
Figure 1.
Current understanding of transmission cycles of T. cruzi in the southern United States with wildlife hosts and well-characterized strain-type associations. In contrast to transmission settings across South and Central America and Mexico, in the southern United States, exclusive domestic cycles appear less important in terms of risk to target hosts than does spillover from enzootic transmission. Original artwork by C. Hodo.
Characterizing Reservoirs of T. cruzi in the United States
Framework for Characterizing Reservoir Potential
Reservoir potential, introduced by Mather et al. (1989), is an index of the relative importance of a reservoir host as a source of infection to vectors, and provides a useful framework for evaluating host species in multihost pathogen transmission systems. There are numerous reports of T. cruzi infection in various wildlife species in the United States, but with little attention to the degree to which each species serves as a reservoir. Models of contact processes between triatomines and wildlife hosts concluded that the limiting factors of stercorarian transmission to hosts was dependent upon host species. In particular, the population density of vectors limited transmission to woodrats, whereas the population density of raccoons and opossums limited transmission to these hosts (Kribs-Zaleta 2010). However, the author acknowledged a severe lack of data underlying parameter estimates and did not attempt to quantify a reservoir potential for the hosts discussed. While there is indeed a significant paucity of data on some criteria necessary for calculating the reservoir potential of candidate species in the United States, we will discuss the available data to attempt to inform the following parameters, as outlined previously (Gürtler and Cardinal 2015): (1) host susceptibility (proportion of exposed hosts that get infected), (2) host infectiousness to triatomine vectors, and (3) vector-host contact (considering relative abundance of vectors and hosts and vector feeding preferences). The first two parameters can be combined to calculate a numerical index of reservoir competence. This, combined with measures of vector-host contact, informs reservoir potential (Figure 2). Further, we will discuss the additional consideration of host-strain type associations.
Candidate Species
We reviewed all published studies of T. cruzi-infected wildlife species in the United States to tabulate parameters to input into the reservoir potential conceptual framework to evaluate the relative importance of each species (Table 1). In total, we reviewed 77 published estimates of anti-T. cruzi antibodies or T. cruzi parasite infection in at least 26 wildlife species across 15 southern states, expanding upon those previously reviewed across the United States (Bern et al. 2011) and in Texas (Gunter et al. 2016). In Table 1, we combined reports of seroprevalence with direct parasite detection to calculate the overall prevalence (including seropositive animals and animals with evidence of parasite anywhere), because infection with T. cruzi is considered life-long such that hosts harboring anti-T. cruzi antibodies are also currently infected. Recognizing that not all infected hosts will be infectious to vectors at any given time, we then compiled reports that utilized PCR of blood, hemoculture, or microscopic methods (i.e., measures of parasitemia) to calculate an infectiousness index. For each wildlife host species, we then summarized the total number of positive animals over the total number of tested animals across all published reports to present species-specific aggregate overall prevalence and aggregate infectiousness indices for comparative purposes. It must be recognized, however, that each individual study is associated with its own biases, and so the aggregate measures we computed are not intended to be representative of all populations of a particular wildlife species across the southern United States. Further, some relatively understudied species may also have key ecological roles, but logistics of sampling have led to them being underrepresented. Below, we comment specifically on some of the key wildlife species most well represented in the literature in the context of the available data to address some of the key parameters in the reservoir potential equation.
Table 1.
Summary of Trypanosoma cruzi studies in wildlife in the United States, with results compiled as overall prevalence (including as positive animals harboring anti-T. cruzi antibodies and animals with evidence of parasite anywhere) and infectiousness index (including as positive animals with measures of parasitemia). For each wildlife species, an aggregate infection prevalence and aggregate infectiousness index was calculated for comparative purposes, although each individual study is associated with its own biases and so these metrics are not intended to represent all wildlife populations in the southern United States.
| Species | Statea | Overall prevalenceb | Infectiousness indexc | Method(s)a | References | ||||
|---|---|---|---|---|---|---|---|---|---|
| No. tested | No. positive | Prev. | No. tested | No. positive | % Infectious | ||||
| Raccoon (Procyon lotor) | |||||||||
| AL | 35 | 5 | 14.3% | 35 | 2 | 5.7% | Culture (heart and blood) | (Olsen et al. 1964) | |
| FL | 33 | 4 | 12.1% | 33 | 4 | 12.1% | Culture (blood) | (Schaffer et al. 1978) | |
| GA | 10 | 5 | 50.0% | 10 | 5 | 50.0% | Culture (blood) | (Schaffer et al. 1978) | |
| TX | 25 | 6 | 24.0% | 25 | 6 | 24.0% | Culture (blood) | (Schaffer et al. 1978) | |
| TX | 9 | 0 | 0.0% | Serology (IHA) | (Burkholder et al. 1980) | ||||
| OK | 8 | 5 | 62.5% | 8 | 5 | 62.5% | Culture (blood) | (John and Hoppe 1986) | |
| NC | 20 | 3 | 15.0% | 20 | 3 | 15.0% | Culture (blood) | (Karsten et al. 1992) | |
| GA | 54 | 12 | 22.2% | 54 | 12 | 22.2% | Culture (blood) | (Pung et al. 1995) | |
| GA | 30 | 13 | 43.3% | 30 | 13 | 43.3% | Culture (blood), blood smear | (Pietrzak and Pung 1998) | |
| TN | 3 | 2 | 66.7% | 3 | 2 | 66.7% | Culture (blood) | (Herwaldt et al. 2000) | |
| GA, SC | 221 | 104 | 47.1% | Serology (IFA) | (Yabsley and Noblet 2002) | ||||
| VA | 464 | 153 | 33.0% | Serology (IFA) | (Hancock et al. 2005) | ||||
| KY | 44 | 19 | 43.2% | 44 | 17 | 38.6% | Serology (IFA), culture (blood) | (Groce 2008) | |
| AZ | 5 | 1 | 20.0% | Serology (IFA) | (Brown et al. 2010) | ||||
| FL | 70 | 38 | 54.3% | Serology (IFA) | (Brown et al. 2010) | ||||
| GA | 510 | 167 | 32.7% | 168 | 50 | 29.8% | Serology (IFA), culture (blood) | (Brown et al. 2010) | |
| MO | 109 | 74 | 67.9% | Serology (IFA) | (Brown et al. 2010) | ||||
| TN | 706 | 206 | 29.2% | Serology (IFA) | (Maloney et al. 2010) | ||||
| TX | 20 | 18 | 90.0% | 20 | 12 | 60.0% | Culture (blood), PCR | (Charles et al. 2012) | |
| TX | 70 | 49 | 70.0% | 18 | 14 | 77.8% | PCR (heart, blood) | (Curtis-Robles et al. 2016) | |
| TX | 24 | 15 | 62.5% | 18 | 9 | 50.0% | PCR (heart, blood) | Hodo, unpublished data | |
| TX | 2 | 2 | 100% | 2 | 2 | 100% | PCR (heart, blood) | Hodo, unpublished data | |
| Raccoon aggregate | 2472 | 901 | 36.4% | 488 | 156 | 32.0% | |||
| Woodrat (Neotoma spp.) | |||||||||
| Neotoma micropus | TX | 100 | 32 | 32.0% | 100 | 31 | 31.0% | Culture (blood), xenodiagnosis | (Packchanian 1942) |
| Neotoma micropus | TX | 30 | 7 | 23.3% | 30 | 7 | 23.3% | Culture (blood), blood smear | (Burkholder et al. 1980) |
| Neotoma micropus | TX | 159 | 42 | 26.4% | PCR (liver) | (Pinto et al. 2009) | |||
| Neotoma micropus | TX | 104 | 50 | 48.1% | 104 | 35 | 33.7% | Serology (IFA, ICT), blood smear, culture (blood), PCR (blood) | (Charles et al. 2012) |
| Neotoma floridana | LA | 15 | 11 | 73.3% | PCR (heart, liver, skeletal muscle, spleen) | (Herrera et al. 2015) | |||
| Neotoma macrotis | CA | 49 | 7 | 14.3% | 49 | 7 | 14.3% | PCR (blood) | (Shender et al. 2016) |
| Neotoma floridana | TX | 1 | 0 | 0.0% | 1 | 0 | 0.0% | PCR (heart, blood) | Hodo, unpublished data |
| Woodrat aggregate | 458 | 149 | 32.5% | 284 | 80 | 28.2% | |||
| Opossum (Didelphis virginiana) | |||||||||
| TX | 8 | 8 | 100.0% | 8 | 8 | 100.0% | Culture (blood), xenodiagnosis | (Packchanian 1942) | |
| TX | 391 | 63 | 16.1% | 391 | 63 | 16.1% | Blood smear | (Eads et al. 1963) | |
| AL | 126 | 17 | 13.5% | 126 | 14 | 11.1% | Culture (heart and blood) | (Olsen et al. 1964) | |
| OK | 10 | 0 | 0.0% | 10 | 0 | 0.0% | Culture (blood) | (John and Hoppe 1986) | |
| LA | 48 | 18 | 37.5% | 48 | 16 | 33.3% | Culture (blood), histopathology | (Barr et al. 1991) | |
| NC | 12 | 1 | 8.3% | 12 | 1 | 8.3% | Culture (blood) | (Karsten et al. 1992) | |
| GA | 39 | 6 | 15.4% | 39 | 6 | 15.4% | Culture (blood) | (Pung et al. 1995) | |
| KY | 48 | 15 | 31.3% | 48 | 0 | 0.0% | Serology (IFA), culture (blood) | (Groce 2008) | |
| FL | 27 | 14 | 51.9% | Serology (IFA) | (Brown et al. 2010) | ||||
| GA | 421 | 118 | 28.0% | 83 | 11 | 13.3% | Serology (IFA), culture (blood) | (Brown et al. 2010) | |
| GA | 29 | 3 | 10.3% | PCR (heart) | (Parrish and Mead 2010) | ||||
| VA | 6 | 1 | 16.7% | Serology (IFA) | (Brown et al. 2010) | ||||
| TX | 5 | 4 | 80.0% | 5 | 4 | 80.0% | PCR (heart, blood) | Hodo, unpublished data | |
| Opossum aggregate | 1170 | 268 | 22.9% | 770 | 123 | 16.0% | |||
| Striped skunk (Mephitis mephitis) | |||||||||
| CA | 1 | 1 | 100.0% | Serology, histology | (Ryan et al. 1985) | ||||
| AZ | 34 | 3 | 8.8% | Serology (IFA) | (Brown et al. 2010) | ||||
| GA | 1 | 1 | 100.0% | Serology (IFA) | (Brown et al. 2010) | ||||
| TX | 4 | 4 | 100.0% | 4 | 3 | 75.0% | Culture (blood), PCR (blood) | (Charles et al. 2012) | |
| TX | 3 | 2 | 66.7% | 3 | 2 | 66.7% | PCR (heart, blood) | Hodo, unpublished data | |
| Striped skunk aggregate | 43 | 11 | 25.6% | 7 | 5 | 71.4% | |||
| Nine-banded armadillo | |||||||||
| (Dasypus novemcinctus) | TX | 15 | 1 | 6.7% | 15 | 1 | 6.7% | Culture (blood), xenodiagnosis | (Packchanian 1942) |
| LA | 80 | 30 | 37.5% | 80 | 23 | 28.8% | Culture (blood); Serology (direct agglutination) | (Yaeger 1988) | |
| LA | 98 | 1 | 1.0% | 98 | 1 | 1.0% | Culture (blood) | (Barr et al. 1991) | |
| Armadillo aggregate | 193 | 32 | 16.6% | 193 | 25 | 13.0% | |||
| Coyote (Canis latrans) | |||||||||
| TX | 156 | 20 | 12.8% | Serology (IHA) | (Burkholder et al. 1980) | ||||
| TX | 134 | 19 | 14.2% | Serology (IFA) | (Grögl et al. 1984) | ||||
| GA | 23 | 1 | 4.3% | Serology (IFA) | (Brown et al. 2010) | ||||
| VA | 26 | 1 | 3.8% | Serology (IFA) | (Brown et al. 2010) | ||||
| GA | 27 | 2 | 7.4% | Serology (IFA) | (Gates et al. 2014) | ||||
| TN | 21 | 2 | 9.5% | Serology (ICT) | (Rosypal et al. 2014) | ||||
| TX | 84 | 12 | 14.3% | 23 | 4 | 17.4% | PCR (heart, blood) | (Curtis-Robles et al. 2016) | |
| TX | 199 | 16 | 8.0% | Serology (ICT) | (Garcia et al. 2016) | ||||
| TX | 97 | 8 | 8.2% | 92 | 3 | 3.3% | PCR (heart, blood) | Hodo, unpublished data | |
| Coyote aggregate | 767 | 81 | 10.6% | 115 | 7 | 6.1% | |||
| Gray fox | |||||||||
| (Urocyon cinereoargenteus) | SC | 26 | 2 | 7.7% | Serology (IFA) | (Rosypal et al. 2007) | |||
| GA | 21 | 0 | 0.0% | Serology (IFA) | (Brown et al. 2010) | ||||
| NC | 43 | 4 | 9.3% | Serology (ICT) | (Rosypal et al. 2010) | ||||
| VA | 11 | 2 | 18.2% | Serology (ICT) | (Rosypal et al. 2010) | ||||
| TX | 58 | 8 | 13.8% | 11 | 1 | 9.1% | PCR (heart, blood) | (Curtis-Robles et al. 2016) | |
| Gray fox aggregate | 159 | 16 | 10.1% | 11 | 1 | 9.1% | |||
| Bobcat (Lynx rufus) | |||||||||
| GA | 62 | 2 | 3.2% | Serology (IFA) | (Brown et al. 2010) | ||||
| TX | 14 | 2 | 14.3% | 2 | 0 | 0.0% | PCR (heart, blood) | (Curtis-Robles et al. 2016) | |
| Bobcat aggregate | 76 | 4 | 5.3% | 2 | 0 | 0.0% | |||
| Feral swine (Sus scrofa) | |||||||||
| GA | 110 | 0 | 0.0% | Serology (IFA) | (Brown et al. 2010) | ||||
| TX | 64 | 3 | 4.7% | 64 | 0 | 0.0% | PCR (heart, blood) | (Comeaux et al. 2016) | |
| Feral swine aggregate | 174 | 3 | 1.7% | 64 | 0 | 0.0% | |||
| Other rodents | |||||||||
| Perognathus hispidus, Liomys irrorattus, Onychomys leucogaster | TX | 45 | 6 | 13.3% | 45 | 6 | 13.3% | Culture (blood), blood smear | (Burkholder et al. 1980) |
| Otospermophilus beecheyi, Peromyscus maniculatus | CA | 23 | 2 | 8.7% | 23 | 2 | 8.7% | Serology (CF, IIF), culture | (Navin et al. 1985) |
| Mus musculus, Peromyscus pectoralis laceianus, P. leucopus, Sigmodon hispidus, Rattus rattus, Ictidomys mexicanus, Otospermophilus variegatus | TX | 28 | 5 | 17.9% | 28 | 5 | 17.9% | PCR (blood), culture (blood) | (Charles et al. 2012) |
| Mus musculus, Peromyscus gossypinus | LA | 44 | 34 | 77.3% | PCR (heart, liver, skeletal muscle, spleen) | (Herrera et al. 2015) | |||
| Rattus rattus | TX | 145 | 0 | 0.0% | 61 | 0 | 0.0% | PCR (heart, blood) | (Hodo et al., 2017) |
| Sigmodon hispidus | TX | 27 | 0 | 0.0% | 27 | 0 | 0.0% | PCR (heart, blood) | Hodo, unpublished data |
| Other rodents aggregate | 312 | 47 | 15.1% | 184 | 13 | 7.1% | |||
| Other species | |||||||||
| Ringtail (Bassariscus astutus) | AZ | 1 | 1 | 100.0% | Serology (IFA) | (Brown et al. 2010) | |||
| Badger (Taxidea taxus) | TX | 8 | 2 | 25.0% | Serology (IHA) | (Burkholder et al. 1980) | |||
| Bats (various species) | TX | 593 | 1 | 0.2% | PCR (heart) | (Hodo, Goodwin et al. 2016) | |||
CF, complement fixation; ELISA, enzyme-linked immunosorbent assay; ICT, immunochromatographic test; IFA, indirect fluorescent antibody; IHA, indirect hemagglutination assay; IIF, indirect immunofluorescence.
aExcluding results from nonendemic states (e.g., Maryland, Pennsylvania), or from studies using samples considered nondiagnostic for T. cruzi (e.g., kidney culture). Data from negative populations are shown when the same study also reported positive data for different states or species, or when a large sample size of animals was involved.
bOverall prevalence includes all measures of T. cruzi detection: serology, whole parasite detection (blood smear or culture), and PCR. In T. cruzi, self cure is considered extremely rare, so seropositive animals are considered to be infected.
cMeasures that detect parasite in the blood (culture, blood smear, PCR of blood) are used to calculate the infectiousness index, acknowledging that PCR may not necessarily represent live intact parasite.
Host Susceptibility
The gold standard methodology for elucidating host susceptibility to infection is through experimental infection, but such studies have only been conducted with T. cruzi on a limited number of wildlife species with small sample sizes (Davis et al. 1980; Roellig, Ellis, and Yabsley 2009b), discussed below. Additionally, infection studies may be limited in generalizability because of the marked heterogeneity in both the pathogen and hosts. Relative susceptibility can be inferred from reports of seroprevalence, when considering infection prevalence of vectors as well as that of other mammalian hosts in the same environment. A major limitation, however, are the numerous different methods used to determine infection, many of which have not been properly validated for use in wildlife species, or even in domestic species, given the absence of a gold standard diagnostic test. Because sensitivity and specificity of different existing diagnostic tests may vary widely across tests and species, it is difficult to compare or combine data from different studies. Further, because dynamics of local transmission vary by geographic location and lower prevalence of infection is expected in northern regions where vectors are not abundant, the positive predictive value of diagnostic tests is not uniform across studies. Despite these challenges, the available literature can be used to draw some conclusions about relative susceptibility of the wildlife community, and below we comment on some of the most well-studied species in the United States.
Raccoons (Procyon lotor) are the most frequently studied candidate T. cruzi reservoir species in the United States and have been studied across at least 13 states. Raccoons across the southern United States are consistently highly infected, with an aggregate overall prevalence of 36.4% and many individual studies showing overall prevalence in excess of 60% (Table 1); variation within geographic areas is likely an artifact of diagnostic method (Bern et al. 2011; Curtis-Robles et al. 2016). Raccoons have been experimentally inoculated with T. cruzi intravenously, par os, or though ingestion of infected bugs, and in two studies all of the inoculated raccoons became infected (Roellig, Ellis, and Yabsley 2009a, 2009b). The next most frequently studied species in the United States, the Virginia opossum, Didelphis virginiana, is the only opossum species in the United States. Many other Didelphis spp. and Philander opossum are recognized as key T. cruzi reservoirs across South America, Central America, and Mexico (Jansen and Roque 2010). The aggregate overall prevalence from 11 studies of naturally infected opossums is 22.9% (Table 1). Experimental infections with strain type TcI have yielded infected opossums, but attempts to inoculate opossums with TcIV did not result in a patent infection (Roellig, Ellis, and Yabsley 2009b). In another study, 3/7 opossums became infected after eating infected triatomine bugs (Yaeger 1971). Woodrats (Neotoma spp.) are recognized as key hosts for triatomine vectors, especially in the western United States, where triatomines infest the nests of the rats (Kjos et al. 2013; Kofoid and McCulloch 1916; Packchanian 1942; Ryckman et al. 1965; Shender et al. 2016). The seven studies of T. cruzi in woodrats show an aggregate overall prevalence of 32.5% (Table 1). Among the other less-studied candidate wildlife reservoir species in the southern United States that have shown some level of infection are coyotes, striped skunk, nine-banded armadillo, and gray fox, with aggregate infection prevalences of 10.6%, 26%, 17%, and 10%, respectively (Table 1).
Host Infectiousness
Xenodiagnosis, or the feeding of pathogen-free vectors on hosts in order to quantify the incidence of vector infection, is a gold standard method for determining host infectiousness. Xenodiagnosis of naturally infected T. cruzi reservoirs has been performed only on a very limited basis in the United States, with 2/2 woodrats and 5/8 opossums infecting xenodiagnostic triatomines (Packchanian 1942). Less direct indicators of host infectiousness include the presence of parasite in the blood, which can be detected via microscopy, hemoculture, or PCR. While PCR results do not necessarily reflect the presence of viable parasite, PCR positivity has been correlated with parasitemia in experimental studies (Caldas et al. 2012).
Of the 77 estimates of wildlife T. cruzi infection that we reviewed, 49 (63%) used methods that can inform the potential infectivity of the host. The aggregate infectiousness index for raccoons and opossums is 32% and 16%, respectively (Table 1). Experimental infections showed short duration of parasitemia in opossums compared with raccoons (Roellig, Ellis, and Yabsley 2009b). Supporting this, surveys of wild raccoons and opossums in Georgia and Florida showed increased blood culture-based parasite detection in raccoons compared with opossums, despite similar seroprevalence rates between the two species (Brown et al. 2010). Woodrats have an aggregate infectiousness index of 28.2% (Table 1). Only two studies have assessed the presence of parasite in the blood of coyotes, and these both used PCR (Curtis-Robles et al. 2016; C. L. Hodo and S. A. Hamer, unpublished data) and were located in central Texas, with an aggregate infectiousness index of 6%. Both of these studies were conducted in the winter and may not reflect the parasitemia status of coyotes throughout the year. The two studies from which skunk infectiousness can be inferred both have a very small sample size (total n = 7) but have an aggregate infectiousness index of 71%. Finally, armadillos in three studies were associated with aggregate infectiousness index of 13%, while foxes had an infectiousness index of 9% in one study (Table 1).
Vector-Host Contact
Although a host species may be highly infected and infectious, it serves as an important reservoir only if triatomine vectors feed on it, become infected, and subsequently transmit the parasite to the target hosts. Assessment of vector-host interactions is limited by a number of factors (Box 2), including opportunistic rather than systematic sampling of triatomines in the United States, limited blood meal analysis studies, and lack of information on the relative population densities of the host community. The primary means for quantifying vector-host contact in arthropod-borne disease studies is through blood meal analysis of vectors, through which the residual traces of a host bloodmeal in a vector’s digestive tract are identified to the genus or species level using immunologic or molecular methods. Extreme flexibility in triatomine feeding behavior has been demonstrated, with insects feeding opportunistically based on host availability (Gürtler et al. 2009; Rabinovich et al. 2011). We generated a qualitative indication of the generalist feeding behavior of kissing bugs in the southern United States by reviewing the five published triatomine bloodmeal analysis studies from this region (Table 2), but we caution that these data alone cannot be interpreted as a measure of kissing bug feeding preferences due to the aforementioned biases (Box 2).
Box 2. Complexities of triatomine vectors and Trypanosoma cruzi transmission that limit the ability to define vector-host interactions.
- Generalist vector feeding behavior results in large pool of candidate hosts
- Determining feeding preferences necessitates large-scale biodiversity survey encompassing multiple classes (mammals, reptiles, amphibians, birds).
- Opportunistic vector collection leads to biases in the apparent host community
- Systematic collection of triatomines has proved more difficult relative to that of ticks, mosquitoes, or other vectors.
- Triatomines are most commonly collected opportunistically (e.g., dispersing adults seen in areas frequented by humans) or through manual searches of known harborage sites such as wildlife dens and dog kennels, where hosts are obvious.
- Vectors may feed on many different hosts during their life cycle, which limits ability to pinpoint infection source
- T. cruzi infection is maintained transstadially, complicating the ability to incriminate which host species was the source of infection.
- Stercorarian transmission of the parasite results in dissociation of the transmission event from the act of blood feeding
- Vectorial capacity is difficult to calculate when transmission pathway is unknown.
- Molecular bloodmeal analysis of triatomine hindguts is challenging
- Status quo methods based on PCR and Sanger sequencing likely reveal only the most recently utilized host species.
- Human contamination may be intractable.
- Freshly engorged insects have the highest chance of success for incriminating host species.
Table 2.
Host species detected in triatomine blood meal analysis studies in the United States
| Study location (reference) | TX (Gorchakov et al., 2016) | LA (Waleckx et al., 2014) | AZ (Klotz et al., 2014) | TX (Kjos et al., 2013) | CA, AZ (Stevens et al., 2012) | |
|---|---|---|---|---|---|---|
| Bug collection sites | ih, oh, ru | ih, oh | z | dk, ih, oh, wr | CA: sy; AZ: sy, z | |
| Species detected in blood meal | Number of bugs with blood meal from each species | Total | ||||
| Human (Homo sapiens) | 40 | 21 | 10 | 1 | 5 | 77 |
| Woodrat (Neotoma spp.) | 2 | 1 | 1 | 47 | 51 | |
| Dog/wolf/coyote (Canis spp.) | 20 | 3 | 3a | 19 | 4a | 49 |
| Green tree frog (Hyla cinerea) | 23 | 23 | ||||
| Raccoon (Procyon lotor) | 5 | 12 | 1 | 18 | ||
| Cricket (Gryllus texensis/rubens) | 15 | 15 | ||||
| Cow (Bos taurus) | 2 | 6 | 5 | 13 | ||
| Pig (Sus scrofa) | 2 | 6 | 1 | 2 | 11 | |
| Cat (Felis catus) | 2 | 1 | 6 | 9 | ||
| Squirrel (Sciurus spp.) | 4 | 2 | 6 | |||
| Cottontail (Sylvilagus spp.) | 4 | 4 | ||||
| Mouse (Mus musculus) | 1 | 2 | 3 | |||
| Opossum (Didelphis virginiana) | 3 | 3 | ||||
| Rat (Rattus spp.) | 1 | 1 | 2 | |||
| Gray fox (Urocyon cinereoargenteus) | 2 | 2 | ||||
| Armadillo (Dasypus novemcinctus) | 2 | 2 | ||||
| Bighorn sheep (Ovis canadensis) | 2a | 2 | ||||
| Chicken (Gallus gallus) | 1 | 1 | ||||
| Deer (Odocoileus virginianus) | 1 | 1 | ||||
| Black vulture (Coragyps atratus) | 1 | 1 | ||||
| Turkey vulture (Cathartes aura) | 1 | 1 | ||||
| Evening bat (Nyctceius humeralis) | 1 | 1 | ||||
| Mustelid | 1 | 1 | ||||
| Porcupine (Erythizon dorsatum) | 1 | 1 | ||||
| Total bugs with blood mealb | 62 | 43 | 11 | 96 | 10 | 222 |
dk, dog kennel; ih, inside home; oh, outside home; ru, rural; sy, sylvatic habitat; wr, woodrat nest; z, zoological park.
aBlood meal may be from captive zoo animal.
bIn some cases, multiple host blood meals were detected in single bugs, so the sum of individual blood meals is greater than the total number of bugs tested.
Raccoon blood has commonly been detected in the gut contents of triatomine bugs in the southern states. In one report of blood meals from triatomine bugs collected in rural peridomestic settings in Texas, raccoon blood was detected in 5/62 bugs (Gorchakov et al. 2016). Another study of bugs in residential settings in Texas also identified a raccoon blood meal in a single Triatoma gerstaeckeri (Kjos et al. 2013). In Louisiana, 12 of 49 Triatoma sanguisuga were found to contain a raccoon blood meal (Waleckx et al. 2014). Our own unpublished data include four raccoon blood meals in citizen-collected triatomines collected from central, south, and west Texas (S. A. Hamer, unpublished data). Additionally, there are three reports of raccoon blood being detected in the same bug that had also fed on a human (Gorchakov et al. 2016), creating a scenario of spillover risk. Canids are the second most common blood meal source detected in triatomines in the United States (Table 2), but unfortunately, most blood meal analysis studies do not use methods capable of differentiating between Canis species so distinguishing coyote from dog blood meals is not feasible. Opossum blood meals were detected in a Triatoma protracta and two Triatoma recurva in a zoological park in Arizona (Klotz et al. 2009) and in a Triatoma indictiva found within a bedroom in Texas (S. A. Hamer, unpublished data). Two of the opossum-fed bugs from Arizona also had evidence of human blood-feeding (Table 2). Blood from woodrats unsurprisingly comprised the majority of blood meals detected in triatomines collected in or around woodrat nests (Kjos et al. 2013), and woodrat blood was also detected in three other blood meal analysis studies (Klotz et al. 2014; Waleckx et al. 2014; Gorchakov et al. 2016). Woodrat blood co-occured with a human blood meal in a bug found inside a house in Texas (Gorchakov et al. 2016). Other wildlife species represented in triatomine blood meals include armadillo, cottontail rabbit, gray fox, porcupine, house mouse, roof rat, and skunk, as well as a number of species refractory to T. cruzi infection (e.g., insects, birds, reptiles, and amphibians) (Table 2; S. A. Hamer, unpublished data).
Host-Strain Type Associations
Growing evidence suggests that certain T. cruzi strain types are associated with particular host species as well as different clinical outcomes in humans (Gürtler and Cardinal 2015; Ramírez et al. 2010; Zingales et al. 2012). Experimental studies in dogs have demonstrated differing clinical, pathologic, and immunologic outcomes resulting from infection with different strains. For example, dogs infected with T. cruzi isolates from an armadillo and opossum developed acute and chronic myocarditis, while dogs infected with an isolate from a dog did not develop disease (Barr, Gossett, et al. 1991). Increased numbers of inflammatory cells were observed in the heart of dogs infected with TcI compared to TcII (Duz et al. 2014). Strain types TcI and TcIV are enzootic in the United States (Bern et al. 2011), and TcII has recently been detected in a small number of rodents in Louisiana (Herrera et al. 2015). While the sample size is admittedly small (n = 5), thus far the only locally infected humans in the United States that have been definitively strain typed have been infected with TcI (Roellig et al. 2008). Similarly, while domestic dogs are infected with both TcI and TcIV, preliminary evidence suggests the majority of dogs suffering from chronic heart disease are infected with TcI (C. L. Hodo and S. A. Hamer, unpublished data). Therefore, it is possible that reservoir hosts harboring TcI may be more important in the context of spillover risk to humans and dogs than those carrying TcIV. TcI and TcIV infections have been documented in nonhuman primates at facilities throughout the United States, but strain type has not yet been associated with disease status (Bern et al., 2011; C. L. Hodo and S. A. Hamer, unpublished data). Opossums throughout the Americas are predominantly infected with TcI (Bern et al. 2011; Zingales et al. 2012), while raccoons are almost exclusively infected with TcIV (Bern et al. 2011; Curtis-Robles et al. 2016; Roellig et al. 2008). Attempts to experimentally infect opossums with a TcIV isolate from a raccoon did not result in infection (Roellig, Ellis, and Yabsley 2009b). Both TcI and TcIV have been detected in skunks and armadillos (Charles et al. 2012; C. L. Hodo and S. A. Hamer, unpublished data; Roellig et al. 2008), while only TcI has been detected in coyotes (Curtis-Robles et al. 2016; C. L. Hodo and S. A. Hamer, unpublished data). Woodrats in Texas (Neotoma micropus) were infected with either TcI or TcIV, and two Neotoma floridana in Louisiana were infected with TcI, while a third was co-infected with TcI and TcII (Herrera et al. 2015).
Summary and Conclusion
Reservoir potential is heterogeneous across space, given changes in the composition of wildlife, vector, and parasite communities. Accordingly, the biological relevance of the reservoir potential framework depends upon the spatial scale of the empirical data. As a starting point, we have reviewed and aggregated the available data on candidate wildlife T. cruzi reservoirs from across 15 states that encompass vastly diverse ecosystems, and future studies at a finer spatial resolution will be useful in identifying key reservoirs in different epidemiological settings. Our review highlights three key knowledge gaps that remain before reservoir potential can more comprehensively be evaluated, and filling these gaps should form the framework for future study.
Knowledge Gap #1: Measuring Host Infectiousness and Infection Dynamics
Diagnostics for T. cruzi exposure or infection in wildlife rarely involve methods that directly inform infectiousness to kissing bug vectors—a key parameter for understanding reservoir potential. This knowledge gap could be addressed in the United States with more xenodiagnosis studies, which have routinely been done in Central and South America (L. Herrera and Urdaneta-Morales 1997; Gürtler et al. 2007; Carrasco et al. 2012). However, laboratory colonies of uninfected kissing bugs in the United States are rare and high maintenance, and Institutional Biosafety Committee approval of xenodiagnoses protocols is challenging. To resolve this, one approach would be to concurrently conduct xenodiagnoses along with quantitative PCR, which determines genome copies of T. cruzi relative to a house-keeping gene. This approach could determine a “threshold” of parasitemic infectiousness that, once determined, could be used in place of xenodiagnoses.
Infectiousness may not be constant over time, depending on host-level factors or infectious dose. Therefore, aside from measuring infectiousness of naturally infected animals in a cross-sectional fashion, important knowledge could be gained from studies designed to measure susceptibility, dynamics of infectiousness over time, and pathology in wildlife species. Some experimental infection studies have been performed in wildlife species such as raccoon (Roellig, Ellis, and Yabsley 2009b), opossum (Roellig, Ellis, and Yabsley 2009b; Yaeger 1971), and skunk (Davis et al. 1980), but sample sizes are so small that it is difficult to draw conclusions about susceptibility across the entire species. Longitudinal studies in naturally infected wildlife are logistically difficult and labor intensive (Box 1), but could provide invaluable data on dynamics of infectiousness over time. Pathology studies of T. cruzi-infected wildlife have been conducted on a limited basis (Barr et al. 1991; Charles et al. 2012; Curtis-Robles et al. 2016; Packchanian 1942; Pietrzak and Pung 1998; Ryan et al. 1985), but more thorough investigation could shed light on infection dynamics, tissue tropisms, and population-level effects of infection.
Knowledge Gap #2: Measuring Vector-Host Contact
Understanding triatomine feeding patterns, and thus host-vector contact, through the use of blood meal analysis presents several challenges (Box 2). Because each triatomine may feed dozens of times throughout the nymphal instars and in the adult life stage, future blood meal analysis studies should use methods that allow the detection of mixed species and historic bloodmeals and should incorporate estimates of the relative abundance of available vertebrate hosts in the area sampled. Additionally, bugs found within and directly around human housing with wildlife blood meals are of interest and can help to indicate the risk of spillover from these sylvatic transmission cycles. Finally, when vector infection data are combined with bloodmeal identification, the infective bloodmeal index (Zárate et al., 1980; Gürtler et al. 2007) can be calculated, although the infective host may not definitively be identified given transstadial passage of T. cruzi that could have been acquired from one or more hosts.
Knowledge Gap #3: Determining Epidemiological Relevance of T. cruzi Strains in Enzootic Transmission
Molecular epidemiological investigations to source-track transmission of the most pathogenic strains in target hosts could incriminate enzootic reservoirs that could be targeted in control interventions, and this field of study applied to T. cruzi transmission in the United States is not as advanced as that in South America (Fernández et al. 2014). While raccoons are associated with the highest aggregate overall T. cruzi prevalence (36.4%), the available studies reveal that they are disproportionately infected with TcIV. The significance of this strain for human health is unknown relative to TcI, which has been more frequently implicated in human and canine disease. For this reason, wildlife reservoirs that are infected with TcI such as opossums and coyotes, despite the lower aggregate overall prevalence in the latter (10%), may play a greater role as reservoirs of the strain that is pathogenic to target populations of humans and dogs. Further, from a wildlife health perspective, the pathogenic effects of T. cruzi in general, and specific T. cruzi strains in particular, on individual wildlife hosts are largely unknown. Future work should include studies designed to determine differences in clinical outcome between parasite strain types in target hosts as well as in infection dynamics in reservoirs.
This review has illuminated the significant gaps in knowledge that will need to be addressed in future research in order to better characterize the reservoir potential of wildlife species for T. cruzi and other vector-borne diseases. While raccoons, opossums, woodrats, and skunks appear to rise to the top in importance as reservoirs of T. cruzi in the United States, other understudied species may have similar or even greater importance. Additionally, more data are needed on the association of particular strain types with disease outcomes. In light of the increasing human and veterinary health burden of vector-borne zoonotic disease, a detailed understanding of wildlife reservoirs will provide necessary data for protecting human and animal health.
Acknowledgments
We thank Gabriel Hamer and Rachel Curtis-Robles for intellectual contributions. Carolyn Hodo is supported by National Institutes of Health fellowship 2T32OD011083-06.
References
- Barr SC, Brown CC, Dennis VA, Klei TR. 1991. The lesions and prevalence of Trypanosoma cruzi in opossums and armadillos from southern Louisiana. J Parasitol 77:624–627. [PubMed] [Google Scholar]
- Barr SC, Gossett KA, Klei TR. 1991. Clinical, clinicopathologic, and parasitologic observations of trypanosomiasis in dogs infected with North American Trypanosoma cruzi isolates. Am J Vet Res 52:954–960. [PubMed] [Google Scholar]
- Bern C, Kjos S, Yabsley MJ, Montgomery SP. 2011. Trypanosoma cruzi and Chagas’ disease in the United States. Clin Microbiol Rev 24:655–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bern C, Montgomery SP. 2009. An estimate of the burden of Chagas disease in the United States. Clin Infect Dis 49:e52–e54. [DOI] [PubMed] [Google Scholar]
- Brown EL, Roellig DM, Gompper ME, Monello RJ, Wenning KM, Gabriel MW, Yabsley MJ. 2010. Seroprevalence of Trypanosoma cruzi among eleven potential reservoir species from six states across the southern United States. Vector Borne Zoonotic Dis 10:757–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner JL, LoGiudice K, Ostfeld RS. 2008. Estimating reservoir competence of Borrelia burgdorferi hosts: prevalence and infectivity, sensitivity, and specificity. J Med Entomol 45:139–147. [DOI] [PubMed] [Google Scholar]
- Burkholder JE, Allison TC, Kelly VP. 1980. Trypanosoma cruzi (Chagas) (Protozoa: Kinetoplastida) in invertebrate, reservoir, and human hosts of the lower Rio Grande valley of Texas. J Parasitol 66:305–311. [PubMed] [Google Scholar]
- Caldas S, Caldas IS, de Figueiredo Diniz L, de Lima WG, de Paula Oliveira R, Cecílio AB, Ribeiro I, Talvani A, Bahia MT. 2012. Real-time PCR strategy for parasite quantification in blood and tissue samples of experimental Trypanosoma cruzi infection. Acta Trop 123:170–177. [DOI] [PubMed] [Google Scholar]
- Cantey PT, Stramer SL, Townsend RL, Kamel H, Ofafa K, Todd CW, Currier M, Hand S, Varnado W, Dotson E, Hall C, Jett PL, Montgomery SP. 2012. The United States Trypanosoma cruzi infection study: evidence for vector-borne transmission of the parasite that causes Chagas disease among United States blood donors. Transfusion 52:1922–1930. [DOI] [PubMed] [Google Scholar]
- Carrasco HJ, Segovia M, Llewellyn MS, Morocoima A, Urdaneta-Morales S, Martínez C, Martínez CE, Garcia C, Rodríguez M, Espinosa R, de Noya BA, Díaz-Bello Z, Herrera L, Fitzpatrick S, Yeo M, Miles MA, Feliciangeli MD. 2012. Geographical distribution of Trypanosoma cruzi genotypes in Venezuela. PLoS Negl Trop Dis 6:e1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles RA, Kjos S, Ellis AE, Barnes JC, Yabsley MJ. 2012. Southern plains woodrats (Neotoma micropus) from southern Texas are important reservoirs of two genotypes of Trypanosoma cruzi and host of a putative novel Trypanosoma Species. Vector Borne Zoonotic Dis 13:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeaux JM, Curtis-Robles R, Lewis BC, Cummings KJ, Mesenbrink BT, Leland BR, Bodenchuk MJ, Hamer SA. 2016. Survey of feral swine (Sus scrofa) infection with the agent of Chagas disease (Trypanosoma cruzi) in Texas, 2013–14. J Wildl Dis 52:627–630. [DOI] [PubMed] [Google Scholar]
- Curtis-Robles R, Lewis BC, Hamer SA. 2016. High Trypanosoma cruzi infection prevalence associated with rare cardiac pathology among wild carnivores in central Texas. Int J Parasitol Parasites Wildl 5:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis-Robles R, Wozniak EJ, Auckland LD, Hamer GL, Hamer SA. 2015. Combining public health education and disease ecology research: Using citizen science to assess Chagas disease entomological risk in Texas. PLoS Negl Trop Dis 9:e0004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis-Robles R*, Zecca IB*, Roman-Cruz V, Carbajal ES, Auckland LD, Flores I, Millard AV, Hamer SA. 2017. Trypanosoma cruzi (agent of Chagas disease) in sympatric human and dog populations in colonias of the Lower Rio Grande Valley of Texas, USA. Am J Trop Med Hyg 94:805–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DS, Russell LH, Adams LG, Yaeger RG. 1980. An experimental infection of Trypanosoma cruzi in striped skunks (Mephitis mephitis). J Wildl Dis 16:403–406. [DOI] [PubMed] [Google Scholar]
- Dobson A, Randolph SE. 2011. Modelling the effects of recent changes in climate, host density and acaricide treatments on population dynamics of Ixodes ricinus in the UK. J Appl Ecol 48:1029–1037. [Google Scholar]
- Dorn PL, Daigle ME, Combe CL, Tate AH, Stevens L, Phillippi-Falkenstein KM. 2012. Low prevalence of Chagas parasite infection in a nonhuman primate colony in Louisiana. J Am Assoc Lab Anim Sci 51:443–447. [PMC free article] [PubMed] [Google Scholar]
- Duz ALC, Vieira PM de A, Roatt BM, Aguiar-Soares RDO, Cardoso JM de O, Oliveira FCB de, Reis LES, Tafuri WL, Veloso VM, Reis AB, et al. . 2014. The TcI and TcII Trypanosoma cruzi experimental infections induce distinct immune responses and cardiac fibrosis in dogs. Mem Inst Oswaldo Cruz 109:1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eads RB, Trevino HA, Campos EG. 1963. Triatoma (Hemiptera: Reduviidae) infected with Trypanosoma cruzi in south Texas wood rat dens. Southwest Nat 8:38. [Google Scholar]
- Fernández MDP, Cecere MC, Lanati LA, Lauricella MA, Schijman AG, Gürtler RE, Cardinal MV. 2014. Geographic variation of Trypanosoma cruzi discrete typing units from Triatoma infestans at different spatial scales. Acta Trop 140:10–18. [DOI] [PubMed] [Google Scholar]
- Garcia MN, Aguilar D, Gorchakov R, Rossmann SN, Montgomery SP, Rivera H, Woc-Colburn L, Hotez PJ, Murray KO. 2015. Evidence of autochthonous Chagas disease in southeastern Texas. Am J Trop Med Hyg 92:325–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MN, O’Day S, Fisher-Hoch S, Gorchakov R, Patino R, Feria-Arroyo TP, Laing ST, Lopez JE, Ingber A, Jones KM, Murray KO. 2016. One health interactions of Chagas disease vectors, canid hosts, and human residents along the Texas-Mexico border. PLoS Negl Trop Dis 10:e0005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates M, Gerhold RW, Wilkes RP, Gulsby WD, Maestas L, Rosypal A, Miller KV, Miller DL. 2014. Parasitology, virology, and serology of free-ranging coyotes (Canis latrans) from Central Georgia, USA. J Wildl Dis 50:896–901. [DOI] [PubMed] [Google Scholar]
- Gorchakov R, Trosclair LP, Wozniak EJ, Feria-Arroyo PT, Garcia MN, Gunter SM, Murray KO. 2016. Trypanosoma cruzi infection prevalence and bloodmeal analysis in Triatomine vectors of Chagas disease from rural peridomestic locations in Texas, 2013–2014. J Med Entomol 53:911–918. [DOI] [PubMed] [Google Scholar]
- Groce BC. 2008. Trypanosoma cruzi in wild raccoons and opossums from Kentucky. PhD dissertation, Western Kentucky University. [Google Scholar]
- Grögl M, Kuhn RE, Davis DS, Green GE. 1984. Antibodies to Trypanosoma cruzi in coyotes in Texas. J Parasitol 70:189–191. [PubMed] [Google Scholar]
- Gunter SM, Brown EL, Gorchakov R, Murray KO, Garcia MN. 2016. Sylvatic transmission of Trypanosoma cruzi among domestic and wildlife reservoirs in Texas, USA: A review of the historical literature. Zoonoses Public Health epub. Available online (http://onlinelibrary.wiley.com/doi/10.1111/zph.12330/full), accessed on December 5, 2016. [DOI] [PubMed]
- Gürtler RE, Cardinal MV. 2015. Reservoir host competence and the role of domestic and commensal hosts in the transmission of Trypanosoma cruzi. Acta Trop 151:32–50. [DOI] [PubMed] [Google Scholar]
- Gürtler RE, Ceballos LA, Ordóñez-Krasnowski P, Lanati LA, Stariolo R, Kitron U. 2009. Strong host-feeding preferences of the vector Triatoma infestans modified by vector density: Implications for the epidemiology of Chagas disease. PLoS Negl Trop Dis 3:e447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gürtler RE, Cécere MC, Lauricella MA, Cardinal MV, Kitron U, Cohen JE. 2007. Domestic dogs and cats as sources of Trypanosoma cruzi infection in rural northwestern Argentina. Parasitology 134:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CA, Polizzi C, Yabsley MJ, Norton TM. 2007. Trypanosoma cruzi prevalence and epidemiologic trends in lemurs on St. Catherines Island, Georgia. J Parasitol 93:93–96. [DOI] [PubMed] [Google Scholar]
- Hancock K, Zajac AM, Pung OJ, Elvinger F, Rosypal AC, Lindsay DS. 2005. Prevalence of antibodies to Trypanosoma cruzi in raccoons (Procyon lotor) from an urban area of northern Virginia. J Parasitol 91:470–472. [DOI] [PubMed] [Google Scholar]
- Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. 2002. Identifying reservoirs of infection: A conceptual and practical challenge. Emerg Infect Dis 8:1468–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera CP, Licon MH, Nation CS, Jameson SB, Wesson DM. 2015. Genotype diversity of Trypanosoma cruzi in small rodents and Triatoma sanguisuga from a rural area in New Orleans, Louisiana. Parasit Vectors 8:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera L, Urdaneta-Morales S. 1997. Synanthropic rodent reservoirs of Trypanosoma (Schizotrypanum) cruzi in the valley of Caracas, Venezuela. Rev Inst Med Trop Sao Paulo 39:279–282. [DOI] [PubMed] [Google Scholar]
- Herwaldt BL, Grijalva MJ, Newsome AL, McGhee CR, Powell MR, Nemec DG, Steurer FJ, Eberhard ML. 2000. Use of polymerase chain reaction to diagnose the fifth reported US case of autochthonous transmission of Trypanosoma cruzi, in Tennessee, 1998. J Infect Dis 181:395–399. [DOI] [PubMed] [Google Scholar]
- Hodo CL, Bertolini NR, Bernal JC, VandeBerg JL, Hamer SA. 2017. Lack of Trypanosoma cruzi infection in urban roof rats (Rattus rattus) at a Texas facility housing naturally infected nonhuman primates. J Am Assoc Lab Anim Sci 56:1–6. [PMC free article] [PubMed] [Google Scholar]
- Hodo CL, Goodwin CC, Mayes BC, Mariscal JA, Waldrup KA, Hamer SA. 2016. Trypanosome species, including Trypanosoma cruzi, in sylvatic and peridomestic bats of Texas, USA. Acta Trop 164:259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen AM, Roque A. 2010. Domestic and wild mammalian reservoirs In: Tibayrenc M, Telleria J, eds. American trypanosomiasis: Chagas disease one hundred years of research. Boston: Elsevier; p 249–276. [Google Scholar]
- Jansen AM, Xavier SCC, Roque ALR. 2015. The multiple and complex and changeable scenarios of the Trypanosoma cruzi transmission cycle in the sylvatic environment. Acta Trop 151:1–15. [DOI] [PubMed] [Google Scholar]
- John DT, Hoppe KL. 1986. Trypanosoma cruzi from wild raccoons in Oklahoma. Am J Vet Res 47:1056–1059. [PubMed] [Google Scholar]
- Karsten V, Davis C, Kuhn R. 1992. Trypanosoma cruzi in wild raccoons and opossums in North Carolina. J Parasitol 78:547–549. [PubMed] [Google Scholar]
- Keane CB, Miller BF. 2003. Miller-Keane Encyclopedia and Dictionary of Medicine, Nursing and Allied Health. Philadelphia: Saunders by Elsevier, Inc. [Google Scholar]
- Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. 2006. Host heterogeneity dominates West Nile virus transmission. Proc Biol Sci 273:2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos SA, Marcet PL, Yabsley MJ, Kitron U, Snowden KF, Logan KS, Barnes JC, Dotson EM. 2013. Identification of bloodmeal sources and Trypanosoma cruzi infection in triatomine bugs (Hemiptera: Reduviidae) from residential settings in Texas, the United States. J Med Entomol 50:1126–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjos SA, Snowden KF, Craig TM, Lewis B, Ronald N, Olson JK. 2008. Distribution and characterization of canine Chagas disease in Texas. Vet Parasitol 152:249–256. [DOI] [PubMed] [Google Scholar]
- Klotz SA, Dorn PL, Klotz JH, Pinnas JL, Weirauch C, Kurtz JR, Schmidt J. 2009. Feeding behavior of triatomines from the southwestern United States: An update on potential risk for transmission of Chagas disease. Acta Trop 111:114–118. [DOI] [PubMed] [Google Scholar]
- Klotz SA, Schmidt JO, Dorn PL, Ivanyi C, Sullivan KR, Stevens L. 2014. Free-roaming kissing bugs, vectors of Chagas disease, feed often on humans in the Southwest. Am J Med 127:421–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz SA, Shirazi FM, Boesen K, Beatty NL, Dorn PL, Smith S, Schmidt JO. 2016. Kissing bug (Triatoma spp.) intrusion into homes: Troublesome bites and domiciliation. Environ Health Insights 10:45–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid CA, Donat F. 1933. Experimental Infection with Trypanosoma cruzi from intestine of cone-nose bug, Triatoma protracta. Proc Soc Exp Biol Med 30:489–491. [Google Scholar]
- Kofoid CA, McCulloch I. 1916. On Trypanosoma triatomae, a new flagellate from a Hemipteran bug from the nests of the wood rat Neotoma fuscipes. Univ Calif Pub Zool. 16:113–126. [Google Scholar]
- Kribs-Zaleta C. 2010. Estimating contact process saturation in sylvatic transmission of Trypanosoma cruzi in the United States. PLoS Negl Trop Dis 4:e656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RS, Mead DG, Hamer GL, Brosi BJ, Hedeen DL, Hedeen MW, McMillan JR, Bisanzio D, Kitron UD. 2016. Supersuppression: Reservoir competency and timing of mosquito host shifts combine to reduce spillover of West Nile virus. Am J Trop Med Hyg 95:1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Tustin A, Castillo-Neyra R, Mabud TS, Levy K, Barbu CM, Quispe-Machaca VR, Ancca-Juarez J, Borrini-Mayorí K, Naquira-Velarde C, Ostfeld RS; Chagas Disease Working Group in Arequipa Peru . 2015. Bottlenecks in domestic animal populations can facilitate the emergence of Trypanosoma cruzi, the aetiological agent of Chagas disease. Proc Biol Sci 282:20142807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Smith JO, Schreiber SJ, Kopp PE, Getz WM. 2005. Superspreading and the effect of individual variation on disease emergence. Nature 438:355–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney J, Newsome A, Huang J, Kirby J, Kranz M, Wateska A, Dunlap B, Yabsley MJ, Dunn JR, Jones TF, Moncayo AC. 2010. Seroprevalence of Trypanosoma cruzi in raccoons from Tennessee. J Parasitol 96:353–358. [DOI] [PubMed] [Google Scholar]
- Manne-Goehler J, Umeh CA, Montgomery SP, Wirtz VJ. 2016. Estimating the burden of Chagas disease in the United States. PLoS Negl Trop Dis 10:e0005033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather TN, Wilson ML, Moore SI, Ribeiro JM, Spielman A. 1989. Comparing the relative potential of rodents as reservoirs of the Lyme disease spirochete (Borrelia burgdorferi). Am J Epidemiol 130:143–150. [DOI] [PubMed] [Google Scholar]
- Montenegro VM, Jimenez M, Dias JCP, Zeledon R. 2002. Chagas disease in dogs from endemic areas of Costa Rica. Mem Inst Oswaldo Cruz 97:491–494. [DOI] [PubMed] [Google Scholar]
- Navin TR, Roberto RR, Juranek DD, Limpakarnjanarat K, Mortenson EW, Clover JR, Yescott RE, Taclindo C, Steurer F, Allain D. 1985. Human and sylvatic Trypanosoma cruzi infection in California. Am J Public Health 75:366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen PF, Shoemaker JP, Turner HF, Hays KL. 1964. Incidence of Trypanosoma cruzi (Chagas) in wild vectors and reservoirs in East-Central Alabama. J Parasitol 50:599–603. [PubMed] [Google Scholar]
- Ostfeld RS, Keesing F. 2000. Biodiversity and disease risk: The case of Lyme disease. Conserv Biol 14:722–728. [Google Scholar]
- Packchanian A. 1942. Reservoir hosts of Chagas’ disease in the State of Texas: Natural infection of nine-banded armadillo (Dasypus novemcinctus texanus), house mice (Mus musculus), opossum (Didelphis virginiana), and wood rats (Neotoma micropus micropus), with Trypanosoma cruzi in the state of Texas. Am J Trop Med Hyg s1-22:623–631. [Google Scholar]
- Parrish EA, Mead AJ. 2010. Determining the prevalence of Trypanosoma cruzi in road-killed opossums (Didelphis virginiana) from Baldwin County, Georgia, using polymerase chain reaction. Ga J Sci 68:132–139. [Google Scholar]
- Pietrzak SM, Pung OJ. 1998. Trypanosomiasis in raccoons from Georgia. J Wildl Dis 34:132–136. [DOI] [PubMed] [Google Scholar]
- Pinto CM, Baxter BD, Hanson JD, Méndez-Harclerode FM, Suchecki JR, Grijalva MJ, Fulhorst CF, Bradley RD. 2009. Using museum collections to detect pathogens. Emerg Infect Dis 16:356–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pung OJ, Banks CW, Jones DN, Krissinger MW. 1995. Trypanosoma cruzi in wild raccoons, opossums, and triatomine bugs in southeast Georgia, U.S.A. J Parasitol 81:324–326. [PubMed] [Google Scholar]
- Rabinovich JE, Kitron UD, Obed Y, Yoshioka M, Gottdenker N, Chaves LF. 2011. Ecological patterns of blood-feeding by kissing-bugs (Hemiptera: Reduviidae: Triatominae). Mem Inst Oswaldo Cruz 106:479–494. [DOI] [PubMed] [Google Scholar]
- Ramírez JD, Guhl F, Rendón LM, Rosas F, Marin-Neto JA, Morillo CA. 2010. Chagas cardiomyopathy manifestations and Trypanosoma cruzi genotypes circulating in chronic Chagasic patients. PLoS Negl Trop Dis 4:e899–e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez JD, Tapia-Calle G, Guhl F. 2013. Genetic structure of Trypanosoma cruzi in Colombia revealed by a high-throughput nuclear multilocus sequence typing (nMLST) approach. BMC Genet 14:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richer LM, Brisson D, Melo R, Ostfeld RS, Zeidner N, Gomes-Solecki M. 2014. Reservoir targeted vaccine against Borrelia burgdorferi: a new strategy to prevent Lyme disease transmission. J Infect Dis 209:1972–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha FL, Roque A, de Lima JS, Cheida CC. 2013. Trypanosoma cruzi infection in neotropical wild carnivores (Mammalia: Carnivora): At the top of the T. cruzi transmission chain. PLoS One 8:e67463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig DM, Brown EL, Barnabé C, Tibayrenc M, Steurer FJ, Yabsley MJ. 2008. Molecular typing of Trypanosoma cruzi isolates, United States. Emerg Infect Dis 14:1123–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig DM, Ellis AE, Yabsley MJ. 2009. a. Oral transmission of Trypanosoma cruzi with opposing evidence for the theory of carnivory. J Parasitol 95:360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig DM, Ellis AE, Yabsley MJ. 2009. b. Genetically different isolates of Trypanosoma cruzi elicit different infection dynamics in raccoons (Procyon lotor) and Virginia opossums (Didelphis virginiana). Int J Parasitol 39:1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roellig DM, Savage MY, Fujita AW, Barnabé C, Tibayrenc M, Steurer FJ, Yabsley MJ. 2013. Genetic variation and exchange in Trypanosoma cruzi isolates from the United States. PLoS ONE 8:e56198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosypal AC, Smith T, Alexander A, Weaver M, Stewart R, Houston A, Gerhold R, Van Why K, Dubey JP. 2014. Serologic survey of antibodies to Trypanosoma cruzi in coyotes and red foxes from Pennsylvania and Tennessee. J Zoo Wildl Med 45:991–993. [DOI] [PubMed] [Google Scholar]
- Rosypal AC, Tidwell RR, Lindsay DS. 2007. Prevalence of antibodies to Leishmania infantum and Trypanosoma cruzi in wild canids from South Carolina. J Parasitol 93:955–957. [DOI] [PubMed] [Google Scholar]
- Rosypal AC, Tripp S, Lewis S, Francis J, Stoskopf MK, Larsen RS, Lindsay DS. 2010. Survey of antibodies to Trypanosoma cruzi and Leishmania spp. in gray and red fox populations from North Carolina and Virginia. J Parasitol 96:1230–1231. [DOI] [PubMed] [Google Scholar]
- Ryan CP, Hughes PE, Howard EB. 1985. American trypanosomiasis (Chagas’ disease) in a striped skunk. J Wildl Dis 21:175–176. [DOI] [PubMed] [Google Scholar]
- Ryckman RE, Folkes DL, Olsen LE, Robb PL, Ryckman AE. 1965. Epizootiology of Trypanosoma cruzi in southwestern North America. J Med Entomol 2:87–108. [DOI] [PubMed] [Google Scholar]
- Schaffer GD, Hanson WL, Davidson WR, Nettles VF. 1978. Hematotropic parasites of translocated raccoons in the southeast. J Am Vet Med Assoc 173:1148–1151. [PubMed] [Google Scholar]
- Shender LA, Lewis MD, Rejmanek D, Mazet JAK. 2016. Molecular diversity of Trypanosoma cruzi detected in the vector Triatoma protracta from California, USA. PLoS Negl Trop Dis 10:e0004291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shender L, Niemela M, Conrad P, Goldstein T, Mazet J. 2016. Habitat management to reduce human exposure to Trypanosoma cruzi and western conenose bugs (Triatoma protracta). Ecohealth 13:525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikes RS, Bryan JA. 2016. Institutional animal care and use committee considerations for the use of wildlife in research and education. ILAR J 56:335–341. [DOI] [PubMed] [Google Scholar]
- Slate D, Rupprecht CE, Rooney JA, Donovan D, Lein DH, Chipman RB. 2005. Status of oral rabies vaccination in wild carnivores in the United States. Virus Res 111:68–76. [DOI] [PubMed] [Google Scholar]
- Stevens L, Dorn PL, Hobson J, la Rua de NM, Lucero DE, Klotz JH, Schmidt JO, Klotz SA. 2012. Vector blood meals and Chagas disease transmission potential, United States. Emerg Infect Dis 18:646–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenney TD, Curtis-Robles R, Snowden KF, Hamer SA. 2014. Shelter dogs as sentinels for Trypanosoma cruzi transmission across Texas. Emerg Infect Dis 20:1323–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas ME, Rasweiler Iv JJ, D’Alessandro A. 2007. Experimental transmission of the parasitic flagellates Trypanosoma cruzi and Trypanosoma rangeli between triatomine bugs or mice and captive neotropical bats. Mem Inst Oswaldo Cruz 102:559–565. [DOI] [PubMed] [Google Scholar]
- Viana M, Mancy R, Biek R, Cleaveland S, Cross PC, Lloyd-Smith JO, Haydon DT. 2014. Assembling evidence for identifying reservoirs of infection. Trends Ecol Evol 29:270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waleckx E, Suarez J, Richards B, Dorn PL. 2014. Triatoma sanguisuga blood meals and potential for Chagas disease, Louisiana, USA. Emerg Infect Dis 20:2141–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody NC, Woody HB. 1955. American trypanosomiasis (Chagas’ disease); first indigenous case in the United States. J Am Med Assoc 159:676–677. [DOI] [PubMed] [Google Scholar]
- Woolhouse ME, Dye C, Etard JF, Smith T, Charlwood JD, Garnett GP, Hagan P, Hii JL, Ndhlovu PD, Quinnell RJ, Watts CH, Chandiwana SK, Anderson RM. 1997. Heterogeneities in the transmission of infectious agents: Implications for the design of control programs. Proc Natl Acad Sci USA 94:338–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO [World Health Organization] 2015. Chagas disease in Latin America: An epidemiological update based on 2010 estimates. Wkly Epidemiol Rec. [PubMed]
- Wozniak EJ, Lawrence G, Gorchakov R, Alamgir H, Dotson E, Sissel B, Sarkar S, Murray KO. 2015. The biology of the triatomine bugs native to South Central Texas and assessment of the risk they pose for autochthonous Chagas disease exposure. J Parasitol 101:520–528. [DOI] [PubMed] [Google Scholar]
- Yabsley MJ, Noblet GP. 2002. Seroprevalence of Trypanosoma cruzi in raccoons from South Carolina and Georgia. J Wildl Dis 38:75–83. [DOI] [PubMed] [Google Scholar]
- Yaeger RG. 1971. Transmission of Trypanosoma cruzi infection to opossums via the oral route. J Parasitol 57:1375–1376. [PubMed] [Google Scholar]
- Yaeger RG. 1988. The prevalence of Trypanosoma cruzi infection in armadillos collected at a site near New Orleans, Louisiana. Am J Trop Med Hyg 38:323–326. [DOI] [PubMed] [Google Scholar]
- Zárate LG, Morales López G, Cabrera Ozuna M, García Santiago G, Zárate RJ. 1980. The biology and behavior of Triatoma barberi (Hemiptera: Reduviidae) in Mexico. IV. Feeding and defecation patterns. J Med Ent 21:548–560. [DOI] [PubMed] [Google Scholar]
- Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, Schijman AG, Llewellyn MS, Lages-Silva E, Machado CR, Andrade SG, Sturm NR. 2012. The revised Trypanosoma cruzi subspecific nomenclature: Rationale, epidemiological relevance and research applications. Infect Genet Evol 12:240–253. [DOI] [PubMed] [Google Scholar]