Abstract
Aims
Luma is a recently discovered, evolutionarily conserved protein expressed in mammalian heart, which is associated with the LInker of Nucleoskeleton and Cytoskeleton (LINC) complex. The LINC complex structurally integrates the nucleus and the cytoplasm and plays a critical role in mechanotransduction across the nuclear envelope. Mutations in several LINC components in both humans and mice result in various cardiomyopathies, implying they play essential, non-redundant roles. A single amino acid substitution of serine 358 to leucine (S358L) in Luma is the unequivocal cause of a distinct form of arrhythmogenic cardiomyopathy. However, the role of Luma in heart has remained obscure. In addition, it also remains to be determined how the S358L mutation in Luma leads to cardiomyopathy.
Methods and results
To determine the role of Luma in the heart, we first determined the expression pattern of Luma in mouse heart. Luma was sporadically expressed in cardiomyocytes throughout the heart, but was highly and uniformly expressed in cardiac fibroblasts and vascular smooth muscle cells. We also generated germline null Luma mice and discovered that germline null mutants were viable and exhibited normal cardiac function. Luma null mice also responded normally to pressure overload induced by transverse aortic constriction. In addition, localization and expression of other LINC complex components in both cardiac myocytes and fibroblasts was unaffected by global loss of Luma. Furthermore, we also generated and characterized Luma S358L knock-in mice, which displayed normal cardiac function and morphology.
Conclusion
Our data suggest that Luma is dispensable for murine cardiac development and function and that the Luma S358L mutation alone may not cause cardiomyopathy in mice.
Keywords: Arrhythmogenic right ventricular cardiomyopathy, Inner nuclear membrane, Linker of nucleoskeleton to cytoskeleton complex, Nuclear envelope, Outer nuclear membrane
1. Introduction
Luma (also known as TMEM43 or transmembrane protein 43) is highly evolutionarily conserved and is expressed in mammalian heart.1,2 Luma was first identified in a proteomics screen for nuclear envelope proteins in neuroblastoma cells3 and was subsequently shown to localize to the inner nuclear membrane (INM) where it interacts with the LInker of Nucleoskeleton and Cytoskeleton (LINC) complex, including LINC complex proteins SUN2, Emerin, Lamins A/C, and B1.1,3–6 Localization of Luma in mouse embryonic fibroblasts (MEFs) is dependent on A-type Lamins, as Luma is mislocalized to the endoplasmic reticulum in MEFs derived from Lamin A/C-deficient mice.1 Furthermore, depletion of Luma in HeLa cells results in mislocalization of Emerin,1 suggesting a potential role for Luma in LINC complex function.
The LINC complex connects the nuclear lamina to the cytoskeleton,7,8 provides structural support to the nucleus,7,9–12 and plays an essential role in regulating gene expression13–17 and mechanotransduction.5,13–15,18–21 Many mutations in genes that encode either core components or regulators/effectors of the LINC complex, including Emerin, Lamins A/C, B1, and B2, Nesprins, and SUN-domain proteins, have been identified as causing skeletal or cardiac myopathy.2,18,22–32 Human genetic linkage studies have revealed that a single amino acid substitution of serine 358 to leucine (S358L) in Luma is the unequivocal cause of fully penetrant arrhythmogenic right ventricular cardiomyopathy (ARVC), characterized by ventricular tachycardia, fibro-fatty replacement of cardiomyocytes, heart failure, and sudden cardiac death.2,33–36 It has also been reported that mutations in Luma are associated with Emery–Dreifuss Muscular Dystrophy (EDMD)-related myopathy.4
Although there are several in vitro studies in cultured cardiomyocytes and non-cardiomyocytes addressing potential roles of Luma and effects of the S358L mutation,35,37–39 no studies have addressed the role of Luma during cardiac development and function in vivo. There are also no in vivo studies to address molecular mechanisms by which the Luma S358L mutation leads to cardiomyopathy in animal models. To investigate the in vivo role of Luma in mammalian heart, we first determined the expression pattern of Luma in mouse heart. We found that Luma was sporadically expressed in cardiomyocytes, but was highly and uniformly expressed in cardiac fibroblasts and vascular smooth muscle cells (vSMCs). As Luma was expressed in most cell types in the heart, we took a global knockout (KO) approach to study effects of Luma loss on cardiac function. Echocardiography of Luma KO mice revealed no defects in cardiac function up to 20 months of age, and Luma KO mice responded normally to pressure overload induced by transverse aortic constriction (TAC). Furthermore, hemodynamic analysis revealed that Luma KOs had normal contractile function in response to treatment with the beta-1 agonist dobutamine. Given these data, we hypothesized that the S358L mutation was likely a gain-of-function mutation. Therefore, we generated S358L knock-in (KI) mice using a CRISPR/Cas9 approach to ensure that the gene encoding the S358L mutant Luma would be expressed under the control of the endogenous Luma locus. Luma KI mice displayed normal cardiac function at baseline. Taken together, our data suggest that Luma is dispensable for murine cardiac development and function, and that the Luma S358L mutation alone may not cause cardiomyopathy in mice.
2. Methods
2.1 Study approval
All animal procedures were approved by the UCSD Animal Care and Use Committee and were in full compliance with the guidelines for the care and use of laboratory animals from the National Institutes of Health. UCSD has an Animal Welfare Assurance (A3033-01) on file with the Office of Laboratory Animal Welfare and is fully accredited by AAALAC International.
For isolation of adult hearts, mice were sacrificed by cervical dislocation following anaesthesia by intraperitoneal injection of a mixture of ketamine (100 mg/kg) and xylazine (5 mg/kg), while the depth of anaesthesia was monitored by toe pinch. For isolation of neonatal cardiomyocytes, neonatal mice were sacrificed by decapitation.
2.2 Generation of Luma global KO and S358L KI mice
Embryonic stem cells containing the TMEM43tm1a(EUCOMM)Wtsi KO first construct were obtained from the IMPC Consortium, injected into blastocysts, and implanted in pseudo-pregnant female mice as previously described (see Supplementary material online, Figure S3A).40 Chimeric Luma floxed (Lumaf/+) mice were crossed with Sox2-Cre deleter mice to obtain Luma+/− mice.41
Luma KI mice were generated by using a CRISPR/Cas9 approach.42 Briefly, CRISPR RNA (crRNA), trans-activating crRNA, Cas9 protein were injected along with the donor oligonucleotide into C57/B6J mouse zygotes, which were designed to replace TCC (Serine) with TTA (Leucine) (see Supplementary material online, Figure S4A–D). Genotyping by polymerase chain reaction (PCR) analyses confirmed the presence of the mutant allele (see Supplementary material online, Figure S4C). Two founder heterozygous KI mice were crossed for three generations into C57/B6J to dilute any potential off-target effects.
2.3 Echocardiography, hemodynamics, and TAC
Mice were anesthetized with 0.5% isoflurane and underwent echocardiography using
VisualSonics, SonoSite FUJIFILM, Vevo 2100 ultrasound system with a linear transducer 32–55 MHz as described previously.40,43,44 Measurements of heart rate, left ventricular internal dimensions at end of diastole and systole (LVIDd, LVIDs, respectively), end-diastolic interventricular septal thickness (IVSd), and LV posterior wall thickness (LVPWd) were determined from the LV M-mode tracing. Percentage fractional shortening (%FS) was used as an indicator of systolic cardiac function. For hemodynamics, 10-week-old adult mice were anesthetized using 100 mg/kg ketamine and 10 mg/kg xylazine and subjected to hemodynamic measurements as described previously.40N = 8 mice were used for each genotype. For TAC, 10-week-old male mice weighing 20–28 g were anaesthetized with ketamine/xylazine and aortas banded with a 27.5 gauge (for <25 g mice) or 27 gauge (for mice ≥25 g) needle as previously described.45
2.4 Immunofluorescence
Hearts from Collagen1a1-GFP, Tbx18-H2B-GFP, and HCN4-tdTomato expressing mice were perfusion fixed with 4% Paraformaldehyde (PFA) or ice-cold acetone, respectively, and dehydrated using sucrose gradients followed by embedding in a sucrose/optical cutting temperature (OCT) mix as previously described.46 All other hearts were isolated and rinsed in phosphate buffered saline (PBS), immersed in cold isopentane, and embedded in OCT on dry ice. Unfixed sections (10 μm) were cut and fixed in acetone at −20 °C for 5 min. After permeabilization with washing buffer (PBS with 0.2% TX-100) sections were incubated with the indicated antibodies (see Supplementary material online, Table S1) overnight in blocking buffer (PBS with 2% normal donkey serum, 3% bovine serum albumin) in a humidified chamber at 4 °C. Sections were rinsed in wash buffer and incubated at room temperature with the indicated fluorescently conjugated secondary antibodies and DAPI (1 : 500) diluted in blocking buffer for 1 h. Slides were rinsed in wash buffer and mounted in mounting buffer (Dako, Santa Clara, CA). Sections were imaged as described previously.47–49 Briefly, using an oil-immersed ×100 objective, with 1.35 numerical aperture, on an inverted microscope controlled by a DeltaVision system (Applied Precision, Issaquah, WA).
2.5 Western blotting
Ventricles were lysed in 50 μL/mg tissue as described previously.48 Briefly, frozen heart tissue and lysis buffer was pulverized into a fine powder using a pestle and mortar, followed by sonication to shear DNA. Lysates were centrifuged at 13 900 g at 4 °C for 15 min, supernatants aspirated and loaded onto 4–12% NuPage Bis/Tris gels for separation before transfer onto a nitrocellulose membrane (Bio-Rad, Hercules, CA) at 4 °C at a constant voltage of 55 V in transfer buffer (25 mM tris, 190 mM glycine, 20% methanol, pH 8.3). After blocking for 1–2 h in tris-buffered saline containing 0.1% Tween 20 and 5% non-fat dry milk, membranes were incubated overnight at 4 °C with the indicated primary antibody (see Supplementary material online, Table S1) in blocking buffer. Blots were washed and incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. Immunoreactive protein bands were visualized using enhanced chemiluminescence reagent (Thermo, Carlsbad, CA).
2.6 Primary cell isolations
Neonatal cardiomyocytes were isolated from postnatal day one to two pups using a combination of trypsin and collagenase type II enzymes as described previously.50 Briefly, hearts were isolated, rinsed in PBS, and incubated overnight with trypsin at 4 °C. The next day hearts were digested with warm trypsin followed by two collagenase type II treatments. Cells were then pre-plated to remove fibroblasts and other cell types, and the floating cardiomyocytes aspirated, counted, and seeded onto Laminin-coated glass-bottomed MatTek dishes. Fibroblasts were trypsinized, counted, and plated on MatTek dishes.
2.7 Scratch wound assays
Isolated fibroblasts were counted and seeded onto 35 mm plastic dishes. A sterile scratch was made using a P200 tip, detached cells were rinsed with medium, and phase-contrast images were recorded every 3 h using an Olympus Analogue Video Camera System mounted on a Nikon TMS microscope. Image analysis was performed by tracing the edge of the wound using Image J and the remaining wound was calculated using Microsoft Excel.
2.8 Histology
Hearts were isolated from age- and sex-matched littermates and washed in PBS before being fixed overnight in formalin. Hearts were subsequently dehydrated in 70% ethanol and embedded in paraffin and cut into 10 μm sections. Sections were stained using Masson’s Trichrome, mounted and imaged using a Hamamatsu NanoZoomer 2.0HT Slide Scanning System (Hamamatsu, Bridgewater, NJ) as described previously.51
2.9 Real-time PCR
Total RNA was extracted from mouse hearts using Trizol reagent according to manufacturer’s recommendations (Life Technologies, Carlsbad, CA). Complementary DNA was synthesized using MMLV Reverse Transcriptase (Bio-Rad). Primers for RT–PCR are listed in Supplementary material online, Table S2. RT–PCR reactions were performed using Sso-Fast EvaGreen Real Time PCR (Bio-Rad) master mix in 96-well low-profile PCR plates in the CFX96 Bio-Rad Thermocycler.
2.10 Human tissues
Human specimens were obtained from the Sydney Heart Bank at the University of Sydney and signed patient consent was obtained for all samples in this tissue bank. Human Research Ethics Committee approval was obtained by St Vincent’s Hospital (#H03/118) and by the University of Sydney Human Research Ethics Committee (#12146, #159401) and conforms to the principles in the Declaration of Helsinki. Human tissue was used in accordance with the ethical guidelines of King’s College London (College Research Ethics Committee 04/05–74; REC reference 12/EM/0106), under current UK law. Samples from the left ventricular free walls of patients from non-failing donor hearts, which were cardiopleged, but not required for heart transplantation, were provided by the Australian Red Cross Blood Transfusion Service. Typically, these hearts were not transplanted because of tissue incompatibility.
2.11 Statistics
Data are presented as mean ± standard error of the mean unless indicated otherwise. We used two-tailed Student’s t-test or two-way analysis of variance (ANOVA), with Sadik’s post-hoc test for comparisons among groups as indicated. Analysis was performed using Microsoft Excel or Graphpad Prism software. P-values of <0.05 were considered significant.
3. Results
3.1 Luma is expressed in most cell types, predominantly in fibroblasts and coronary vSMCs in mouse and human myocardium
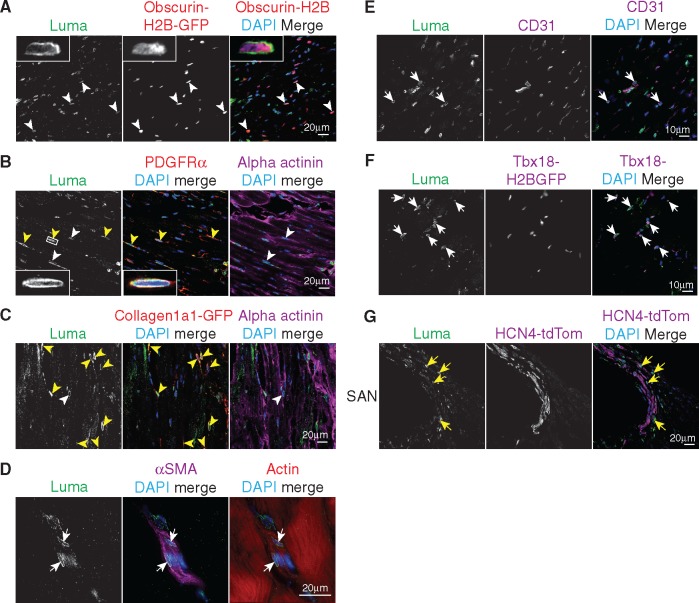
Luma messenger RNA and protein are expressed in human hearts.2,33 We first confirmed expression of Luma in mouse hearts, as well as skeletal muscle, and a number of other tissues by western blot analysis (see Supplementary material online, Figure S1A). Although widely expressed, Luma was abundantly expressed in heart (see Supplementary material online, Figure S1A). To investigate Luma subcellular localization and expression within various cell types of the heart, we next performed immunofluorescence microscopy utilizing a panel of antibodies and various green fluorescent protein (GFP) and tdTomato indicator mouse lines (Figure 1A–G). Although Luma was localized to the nuclear envelope in 71% of cardiomyocytes (Obscurin-H2B-GFP,52,53Figure 1A), it was expressed in all cardiac fibroblasts of all four chambers of heart [platelet-derived growth factor receptor alpha (PDGFRα), Figure 1B and Collagen1a1-GFP,52,53Figure 1C and see Supplementary material online, Figure S1B]. Immunofluorescence analysis of Luma in vSMCs [alpha smooth muscle actin (αSMA), Figure 1D], endothelial cells (ECs; CD31, Figure 1E), and pericytes (Tbx18-H2B-GFP,54Figure 1F) revealed high expression in vSMCs, whereas Luma was rarely observed in ECs and only in some pericytes. Because mutations in Luma lead to fatal arrhythmias in humans,2,33 we predicted that Luma might be enriched in regions of the heart responsible for cardiac conduction. To test this hypothesis, we used the HCN4CreERT2-tdTomato indicator line to mark the cardiac conduction system.55 However, Luma was not observed in the sinoatrial node, atrioventricular node, or Purkinje fibres (Figure 1G and data not shown).
Figure 1.
Luma is expressed in most cell types in the heart. Adult hearts from WT and various cell-type-specific indicator mouse lines were fixed and analysed under immunofluorescence microscopy utilizing a panel of antibodies and various indicator mouse lines for various cell-type-specific markers (labelled as magenta/red). (A) Obscurin-H2B-GFP (red) expressing mice were used to mark cardiomyocyte nuclei. Note that Luma was expressed in myocytes (white arrowheads). Inset, high magnification view of a cardiomyocyte nucleus expressing Luma. (B) PDGFRα to mark fibroblasts (red) and alpha-actinin (magenta). Inset, high-magnification fibroblast nucleus in white box. (C) Collagen1a1-GFP (red, fibroblast marker). Note that Luma was expressed in all fibroblasts (yellow arrowheads) and some cardiomyocytes (white arrowheads). (D) αSMA was used to mark smooth muscle cells (magenta) and phalloidin for actin (red). Note that Luma was also expressed in smooth muscle cells (white arrows). (E) CD31 was used to mark ECs (magenta). Note that Luma was found in some ECs (white arrow). (F) Tbx18-H2B-GFP expressing hearts were used as a pericyte marker. Note that Luma was found in some pericytes (white arrows). (G) HCN4-tdTomato was used as a conduction system marker and the sinoatrial node (SAN) region was imaged. Note that Luma was not found in these cells (yellow arrows). DNA is stained with DAPI (blue).
Consistent with the expression pattern of Luma in mouse myocardium, immunofluorescence staining of human myocardium obtained from the left ventricle of non-failing adults revealed that, as in mouse heart, Luma concentrated at the nuclear envelope and was predominantly found in vSMCs and cardiac interstitial cells (see Supplementary material online, Figure S2).
3.2 Luma KO mice are viable and have normal cardiac function and morphology
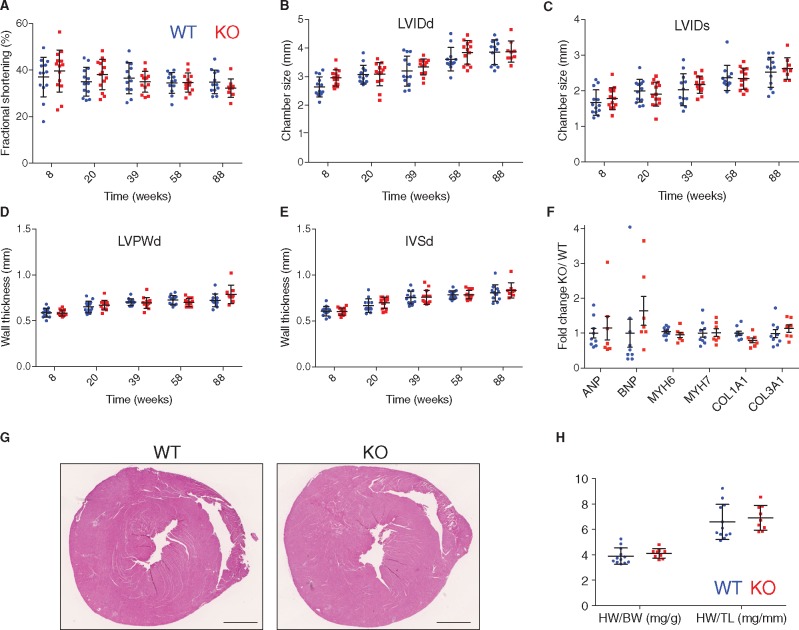
To investigate the in vivo role of Luma in cardiac function, we generated Luma KO mice by crossing targeted mice containing Luma floxed alleles with global deleter Sox2-cre mice41 (see Supplementary material online, Figure S3A and B). Luma KO mice were born at expected Mendelian ratios with 48 out of an expected 50 pups (from 202 pups total) observed from Luma heterozygous null crosses (see Supplementary material online, Figure S3C). Western blotting and immunofluorescence analyses confirmed loss of Luma expression in KO mice (see Supplementary material online, Figure S3D–F). Given this result, we next evaluated cardiac function of Luma KO and wild type (WT) mice over the course of 20 months by echocardiography (Figure 2A–E). Cardiac function of Luma KO mice was comparable to WT littermates (32% vs. 35% FS), as were LVIDd (both 3.9 mm) and LVIDs (2.6 vs. 2.5 mm), LVPWd (0.79 vs. 0.72 mm), and IVSd (0.83 vs. 0.81 mm) at 88 weeks (Figure 2A–E). We observed no changes in any of these parameters, irrespective of their gender. We also observed no change in expression levels of the foetal gene program (ANP, BNP, Myh6, Myh7) and pro-fibrotic markers (Collagen 1a1 and 3a1) by quantitative RT–PCR analysis (Figure 2F). Complementary to the echocardiography and gene expression analyses, hematoxylin and eosin staining revealed no evidence of morphological defects (Figure 2G). There were also no changes in heart weight to body weight and heart weight to tibia length ratios of Luma KO mice when compared to WT littermates (Figure 2H).
Figure 2.
Luma KO mice display no overt defects in cardiac function and morphology. Echocardiographic measurements of (A) FS, (B) LVIDd, (C) LVIDs, (D) LVPWd, and (E) IVSd over the course of 88 weeks in WT (n = 13; 7 females, 6 males) and Luma KO mice (n = 14; 5 females, 9 males). (F) Quantitative real-time PCR analysis of foetal genes, ANP, BNP, Myh6, Myh7, and pro-fibrotic markers, Collagen 1a1 and 3a1, in WT and Luma KO mouse hearts at 88 weeks (n = 7–9 per genotype). (G) Macroscopic cross-sectional views of hematoxylin and eosin stained hearts isolated from WT and Luma KO mice at 88 weeks. Scale bar: 1 mm. (H) HW/BW and HW/TL of WT vs. Luma KO mice at 88 weeks (n = 9–12 per genotype). P > 0.05 according to two-way ANOVA with Sidak’s post-hoc test (A–F, H).
3.3 Localization of most LINC complex proteins and nuclear Lamins is unaffected in Luma KO cardiomyocytes and fibroblasts
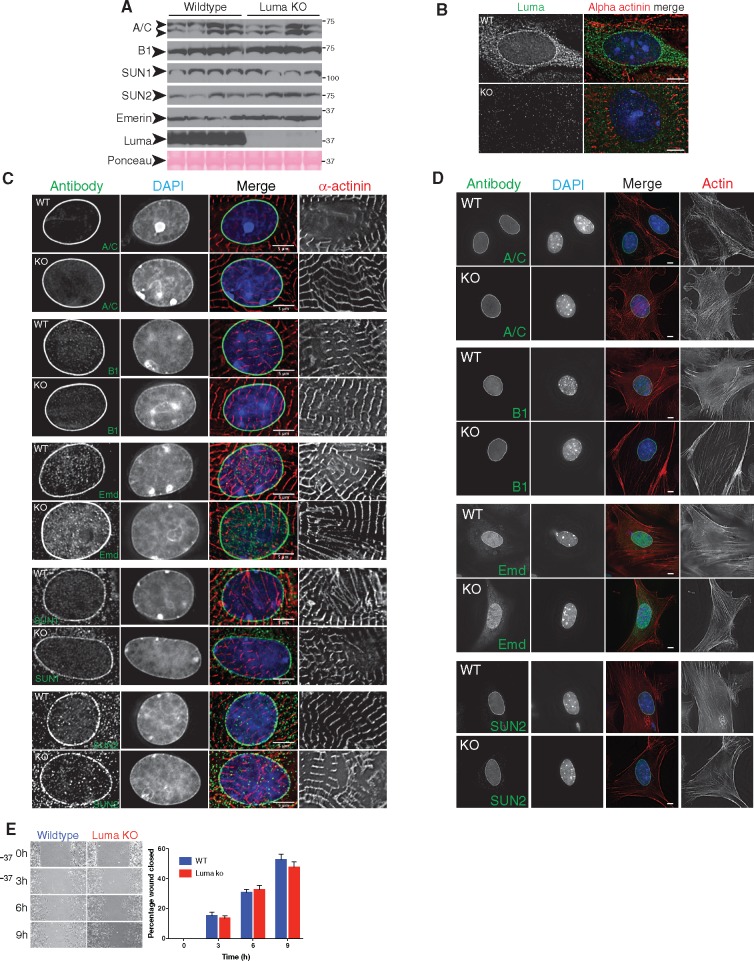
Since ablation of other LINC complex proteins has been shown to affect LINC complex integrity,18,48 we hypothesized that known interaction partners of Luma may be compensating for loss of Luma in Luma KO mice. Western blot analysis revealed that levels of Lamins A/C, B1, SUN2, and Emerin were unchanged in Luma KO vs. WT mice at 88 weeks (Figure 3A; see Supplementary material online, Figure S3G). Given that Luma binds to LINC complex-associated proteins Lamin A/C, B1, Emerin, and SUN2,1,4 and knockdown of Luma in HeLa cells led to mislocalization of Emerin,1 we isolated both cardiomyocytes and cardiac fibroblasts from WT and Luma KO neonatal hearts and performed immunostaining for various LINC complex and LINC complex-associated components. As expected, Luma localized to the nuclear envelope in cardiomyocytes (Figures 1A and 3B) and fibroblasts (Figure 1B). Localization of Lamins A/C, B1, Emerin, SUN1, and SUN2 were unchanged between WT and Luma KO in both cell types, where they localized to the nuclear envelope (Figure 3C and D). Isolation and staining of adult cardiomyocytes from WT and Luma KO mice showed similar localization of the LINC components (data not shown). Because Luma was expressed at high levels in fibroblasts, and the LINC complex has been implicated in cell migration,56,57 we hypothesized that Luma KO fibroblasts might display migration defects. To test this, we performed scratch-wound assays, but observed that Luma KO fibroblasts were able to migrate at the same rate as WT fibroblasts (Figure 3E).
Figure 3.
LINC complex protein levels and localizations are unaffected in Luma KO mice. (A) Western blot analysis of Lamins A/C (A/C), B1 (B1), SUN1, SUN2, Emerin, and Luma in WT and Luma KO hearts. Ponceau stain serves as loading control. Arrowheads show the predicted molecular weight (MW) of the indicated proteins. MW is denoted in kDa (n = 4 per genotype). (B) Immunofluorescence analysis of Luma (green) in WT and Luma KO neonatal cardiomyocytes. Cardiomyocytes were stained with alpha actinin (red) and DAPI (blue). Note that Luma localizes predominantly to the nuclear envelope, and this signal is absent in Luma KO cells. (C, D) Immunofluorescence analysis of (C) neonatal cardiomyocytes and (D) fibroblasts isolated from WT and Luma KO hearts. Cells were fixed and stained for nuclear lamina proteins Lamin A/C (A/C), Lamin B1 (B1); nuclear envelope protein Emerin (Emd); and LINC components SUN1 and SUN2. Cardiomyocytes were stained with alpha actinin (red); fibroblasts were stained with phalloidin (red) and DAPI (blue). Note that all proteins remained localized to the nuclear envelope in WT and KO cells. Scale bar: 5 μm. (B–D) n = 30–40 cells per genotype from three independent isolations. (E) Scratch wound assay of WT and Luma KO neonatal fibroblasts. Note that Luma KO cells were able to migrate at normal rates compared to WT cells (n = 3 biological replicates, n = 6–9 scratches/replicate). P > 0.05 according to the Student’s t-test.
3.4 Luma KO mice exhibit normal hypertrophic response to pressure overload
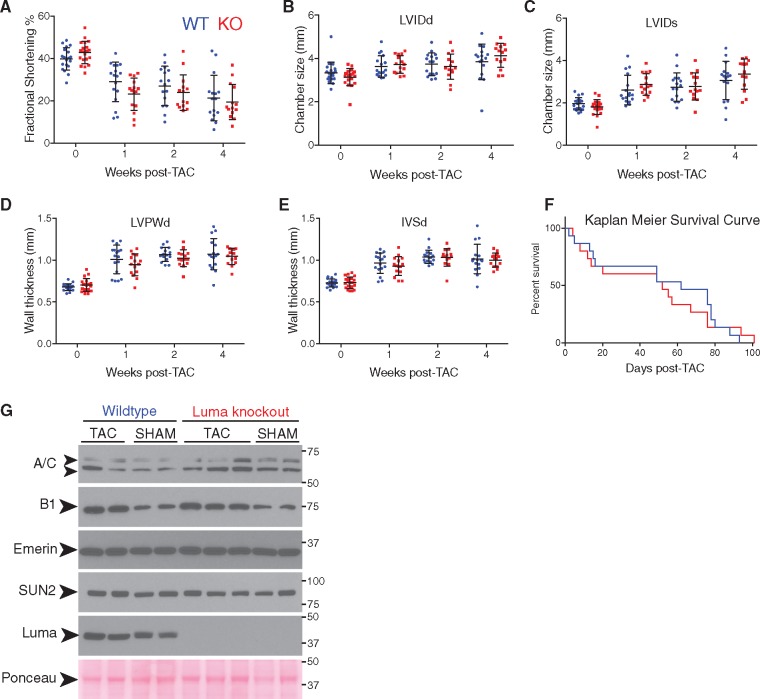
Lamin A/C heterozygous null mice display an attenuated hypertrophic response following pressure overload by TAC.58 Given that Luma interacts with Lamin A/C, and that LINC complex components have been shown to play both structural and mechanotransduction roles,18–21 we hypothesized that Luma KO mice may also display an attenuated hypertrophic response to pressure overload. To test this hypothesis, we performed TAC on 10-week-old mice followed by serial echocardiography measurements at 1 and 2 weeks, post-TAC. As expected, when compared to SHAM-operated animals, WT mice subjected to TAC underwent remodelling, as evidenced by a progressive decrease in cardiac function, and increasing dimensions of the left ventricular chambers and wall thickness (Figure 4A–E). However, there were no significant differences in any of these parameters between Luma KO and WT littermates (P > 0.05, two-way ANOVA), suggesting that Luma KO hearts exhibit a normal hypertrophic response to pressure overload. At 4 weeks post-TAC, both WT and Luma KO mice progressed to heart failure, with comparable FS (mean values 19.5% and 21.3%, respectively); chamber sizes, LVIDd (4.1 and 3.8 mm, respectively), LVIDs (3.4 and 3.1 mm, respectively); wall thicknesses, LVPWd (1.0 and 1.1 mm, respectively), IVSd (both 1.0 mm); with no significant differences observed (P > 0.05, two-way ANOVA) (Figure 4A–E). Furthermore, Kaplan–Meier survival analysis found no significant difference between Luma KO and WT littermates post-TAC (median survivals of 52 and 62 days, respectively, P = 0.87, Mantel–Cox χ2 test) (Figure 4F). Western blot analysis detected no difference in expression levels of binding partners of Luma (Figure 4G; see Supplementary material online, Figure S3H).
Figure 4.
Luma KO mice exhibit normal hypertrophic response to pressure overload. Echocardiographic measurements of (A) FS, (B) LVIDd, (C) LVIDs, (D) LVPWd, and (E) IVSd) at baseline (0) and 1, 2, and 4 weeks post-TAC surgery in WT and Luma KO mice. Note that as expected, cardiac function significantly deceased after TAC and chamber dimensions and wall thicknesses increased. However, there were no significant changes observed between WT and Luma KO mice. (n = 20 males per genotype). (F) Kaplan–Meier survival analysis showed no significant change in survival between WT and Luma KO mice post-TAC. (G) Western blot analysis of Lamins A/C (A/C), B1 (B1), Emerin, SUN2, and Luma in WT and Luma KO mice 2 weeks post-TAC and SHAM surgeries. Ponceau stain serves as a loading control (n = 4–5 per genotype). Arrowheads show the predicted MW of the indicated proteins. MW is denoted in kDa. P > 0.05 according to two-way ANOVA with Sidak’s post-hoc test (A–E). P = 0.87, Mantel–Cox χ2 test (F).
3.5 Contractile function of Luma KO mice is unaffected in response to beta-adrenergic stimulation
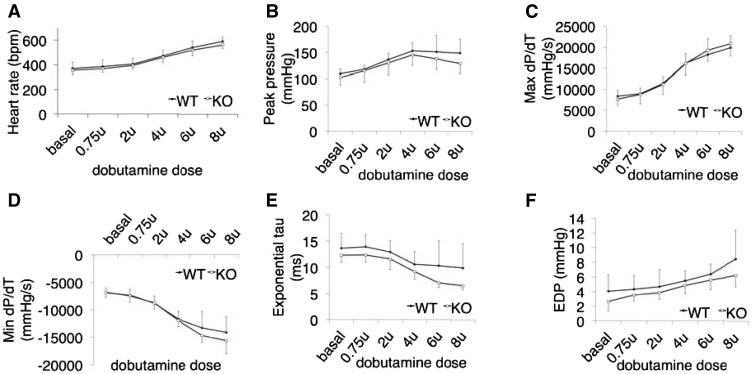
Given that Luma KO mice showed normal cardiac function at 88 weeks of age and had similar hypertrophic response and heart failure progression as their control littermates following TAC, we next investigated cardiac contractile function following beta-adrenergic stimulation. Ten-week-old mice were treated with increasing amounts of dobutamine and subjected to hemodynamic analyses. Results demonstrated that Luma KO mice responded similarly to WT littermates, with no impairment of contractile function in response to beta-adrenergic stimulation (Figure 5A–F). Taken together, Luma KO mice displayed no overt cardiac defects, responded normally to pressure overload, and had normal contractile function.
Figure 5.
Contractile function of Luma KO mice is unaffected following beta-adrenergic stimulation. Invasive hemodynamics measurements of (A) heart rate, (B) peak pressure, (C) maximum rate of pressure change in the left ventricle (dP/dT), (D) minimum rate of (dP/dT), (E) exponential tau (index of LV relaxation), and (F) end-diastolic pressure in WT and Luma KO mice in response to increasing levels of beta-adrenergic agonist dobutamine (n = 8 males per genotype). P > 0.05 according to the Student’s t-test (A–F).
3.6 Luma S358L KI mice have normal cardiac function
Given that no changes in cardiac function were observed following global loss of Luma, we reasoned that the S358L mutation in Luma that causes ARVC in humans might be a gain-of-function mutation. To test whether the Luma S358L mutation is a gain-of-function mutation, we generated KI mice using a CRISPR/Cas9 approach (see Supplementary material online, Figure S4A–D). Two founder heterozygous mutant mice were crossed for three generations into a C57B6J background to dilute potential off-target effects. Heterozygous mutants were then interbred to produce homozygous S358L mutants. From 154 pups analysed, 72 homozygous (expected 77) and 35 heterozygous (expected 38.5) mutants were present at weaning, suggesting that homozygous null mutants were present at expected Mendelian ratios (χ2 test, P = 0.28) (see Supplementary material online, Figure S4E).
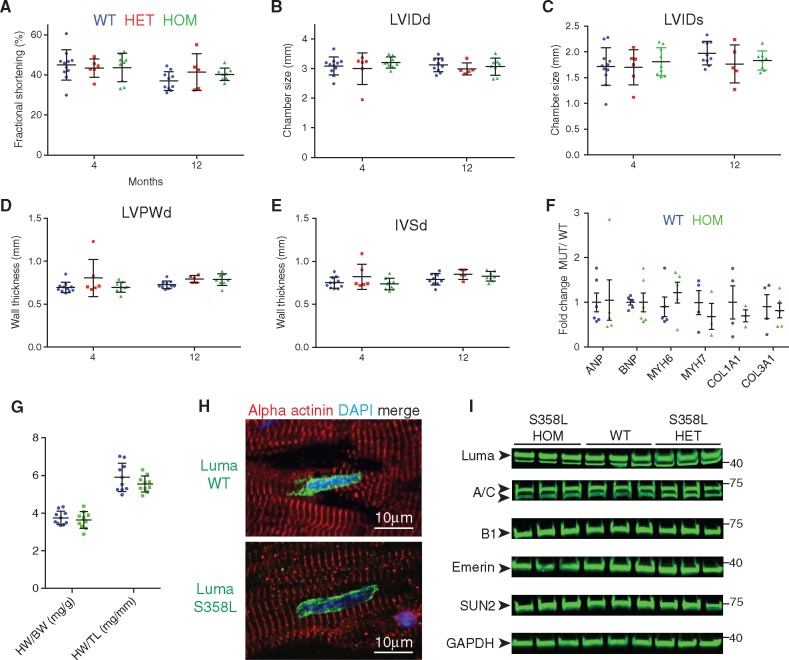
After generating S358L mice, we performed echocardiography over the course of 12 months on the S358L mutants. Surprisingly, we observed no changes between S358L heterozygous, homozygous, and WT littermates in cardiac function (FS, 41%, 40%, 37%, respectively); LVIDd (3.0, 3.1, and 3.1 mm, respectively); LVIDs (1.8, 1.8, and 2.0 mm, respectively); LVPWd (0.79, 0.79, and 0.72 mm, respectively); IVSd (0.85, 0.83, and 0.79 mm, respectively) at 1 year of age (Figure 6A–E). Next, we checked expression levels of foetal genes (ANP, BNP, Myh6, Myh7) and pro-fibrotic genes (Collagen-1a1, Collagen-3a1) in 2- to 3-month-old mice. Consistent with our echocardiographic analysis, we observed no changes in these factors, which suggested an absence of remodelling in mutants (Figure 6F). The indices of hypertrophy (heart weight to body weight ratio, HW : BW and heart weight to tibia length ratio, HW : TL) were unchanged between WT and S358L mutant mice (Figure 6G). Consistent with the lack of an ARVC-like phenotype, we observed no incidences of sudden death or premature lethality of S358L mutant mice compared to their WT littermates.
Figure 6.
Luma S358L KI mice display no overt defects in cardiac function and morphology. Echocardiographic measurements of (A) FS, (B) LVIDd, (C) LVIDs, (D) LVPWd, and (E) IVSd over the course of 12 months in WT (n = 11; 9 females, 2 males), Luma S358L heterozygous (HET) mice (n = 6; 3 females, 3 males), Luma S358L homozygous (HOM) (n = 9; 5 females, 4 males). (F) Quantitative real-time PCR analysis of foetal genes, ANP, BNP, Myh6, Myh7, and pro-fibrotic markers, Collagen 1a1 and 3a1, in WT and Luma S358L homozygous mouse hearts at 2–3 months (n = 3–6 per genotype). (G) HW/BW and HW/TL ratios of WT vs. Luma KO mice at 47 weeks (n = 10 per genotype). (H) Immunofluorescence analysis of adult hearts isolated from WT and S358L homozygous mice. Sections were stained with antibodies raised against Luma (green), alpha actinin (red), and DAPI (blue). Note that Luma S358L has a similar localization to WT Luma. (I) Western blot analysis of Lamins A/C (A/C), B1 (B1), Emerin, SUN2, and Luma in WT and Luma S358L KI hearts. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) serves as a loading control. Arrowheads show the predicted MW of the indicated proteins. MW is indicated in kDa (n = 3 per genotype). P > 0.05 according to two-way ANOVA with Sidak’s post-hoc test (A–G).
3.7 Luma S358L mutant localizes normally to the nuclear envelope in Luma S358L mutant myocardium and levels of LINC proteins are unchanged in Luma S358L mutant cardiac tissue
The mutant S358L Luma protein has been reported to have an altered subcellular localization in HL1 cells, where it localizes to microfilaments and structures resembling phagosomes.38 However, the subcellular localization of the Luma S358L mutant in heart has not been investigated when expressed under the control of the endogenous Luma locus. We found that the mutant S358L Luma protein localized to the nuclear envelope as was observed for WT Luma (Figure 6H). Previously, it was reported that Luma staining in human myocardium was reduced in patients with S358L mutations,33 however we found that Luma levels were similar in WT and both S358L heterozygous and homozygous mice (Figure 6i). In vivo analysis of S358L mutant mouse hearts suggested cardiac function was normal and that Luma localization was unaffected. Next we checked the levels of binding partners of Luma to investigate any potential alterations in expression levels of these proteins. As with Luma KO mice, levels of Lamins A/C, B1, Emerin, and SUN2 were unchanged in S358L mutant mice when compared to those in WT littermates (Figure 6J).
4. Discussion
Luma is an INM protein associated with the LINC complex1 and is highly expressed in heart2 (and this study). Previous human genetic linkage studies have revealed that an autosomal-dominant S358L mutation in Luma is the unequivocal cause of fully penetrant ARVC.2,33–35 To investigate the role of Luma in mammalian heart, we determined the expression pattern of Luma in mouse heart, using an antibody that had been validated by comparing western blot and immunostaining signals in WT and Luma null hearts. We found that Luma was sporadically expressed in cardiomyocytes, but was highly and uniformly expressed in cardiac fibroblasts and vSMCs in both mouse and human myocardium. We then confirmed that Luma was selectively expressed at the nuclear envelope in cardiomyocytes and cardiac fibroblasts. Localization and expression of other LINC complex components was unaffected in Luma KO hearts and cardiac myocytes. Furthermore, we found that Luma was expressed in the same cell types in human myocardium. Interestingly, others have reported that Luma localizes to the sarcolemma and intercalated disc in human and pig hearts.33,39 However, whether these antibodies recognize Luma or a protein with a similar epitope to Luma that resides at these structures remains to be seen.
We then generated and characterized Luma KO mice. Surprisingly, Luma null mice were viable and displayed no defects in cardiac function, even at 2 years of age. In addition, Luma KO mice responded normally to pressure overload induced by TAC. Explanations as to why no cardiac phenotype was observed in global Luma KO mice include: (i) Luma KO mice were generated in an inbred C57B6 background, which may lack certain genetic modifiers required to exacerbate phenotypes associated with alterations in Luma function. Mice of a different strain may more closely model the human phenotype. (ii) Function of Luma in humans or mice may not be equivalent. This was shown to be the case for the LINC-associated component Emerin. X-linked EDMD in humans, caused by ablation or reduction of Emerin expression by a mutation that leads to a premature stop codon, was not recapitulated in Emerin KO mice, which displayed relatively mild phenotypes compared to X-EDMD patients. Only when both Emerin and its interaction partner LAP1 were ablated was a significant phenotype observed in mice. In this regard, it is should be pointed out that there is no report to show that Luma nonsense mutations in human result in cardiomyopathy.
Given that no changes in cardiac function were observed following global loss of Luma, we reasoned that the S358L mutation in Luma that causes ARVC in human might be a gain-of-function mutation. To test whether the Luma S358L mutation is a gain-of-function mutation, we generated and characterized Luma KI mice. We found that Luma S358L KI mice had normal cardiac function, that Luma S358L mutant protein localized normally to the nuclear envelope, and that levels of LINC proteins were unchanged in Luma S358L mutant cardiac tissue. Our observation of no baseline cardiac phenotype in Luma KI mice was quite unexpected, given that human genetic linkage studies have unequivocally revealed that the S358L mutation in Luma results in cardiomyopathy.2,33 In addition, human and mouse orthologues of Luma share 93% identity and the serine 358 residue and adjacent region is 100% conserved between human and mouse.1 However, it should be pointed out that many patients with the S358L mutation develop late-onset cardiomyopathy.2 We only measured cardiac function of Luma KI mice, which were in a pure C57/B6 genetic background, at baseline conditions up to 12 months of age. Future studies are needed to determine whether Luma S358L KI mice in C67/B6 or other genetic backgrounds would eventually develop cardiomyopathy with greater age and to determine whether Luma S358L KI mice respond normally to cardiac stress, such as TAC-induced pressure overload.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgements
We would like to thank Cris dos Remedios (University of Sydney) and Elisabeth Ehler (King’s College London) for kindly providing the non-failing human heart samples.
Funding
M.J.S. and X.F. were supported by American Heart Association postdoctoral fellowships (13POST17060120, 16POST30960067). J.V. was supported by a CIRM postdoctoral fellowship (TG2-01154). J.C. and S.M.E. were funded by grants from the National Institutes of Health (NIH) HL123626, HL130295, HL106968, HL123626, HL119967, HL123747, HL128773-01. J.C. was also funded by Foundation Leducq (TNE-13CVD04) and is the American Heart Association Endowed Chair in Cardiovascular Research. Microscopy was supported by NIH grant P30 NS047101.
Conflict of interest: none declared.
References
- 1. Bengtsson L, Otto H.. Luma interacts with emerin and influences its distribution at the inner nuclear membrane. J Cell Sci 2008;121:536–548. [DOI] [PubMed] [Google Scholar]
- 2. Merner ND, Hodgkinson KA, Haywood AF, Connors S, French VM, Drenckhahn JD, Kupprion C, Ramadanova K, Thierfelder L, McKenna W, Gallagher B, Morris-Larkin L, Bassett AS, Parfrey PS, Young TL.. Arrhythmogenic right ventricular cardiomyopathy type 5 is a fully penetrant, lethal arrhythmic disorder caused by a missense mutation in the TMEM43 gene. Am J Hum Genet 2008;82:809–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dreger M, Bengtsson L, Schoneberg T, Otto H, Hucho F.. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc Natl Acad Sci U S A 2001;98:11943–11948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang WC, Mitsuhashi H, Keduka E, Nonaka I, Noguchi S, Nishino I, Hayashi YK.. TMEM43 mutations in Emery–Dreifuss muscular dystrophy-related myopathy. Ann Neurol 2011;69:1005–1013. [DOI] [PubMed] [Google Scholar]
- 5. Stroud MJ, Banerjee I, Veevers J, Chen J.. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure, function, and disease. Circ Res 2014;114:538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schirmer EC, Florens L, Guan T, Yates JR III, Gerace L.. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science 2003;301:1380–1382. [DOI] [PubMed] [Google Scholar]
- 7. Sosa BA, Rothballer A, Kutay U, Schwartz TU.. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 2012;149:1035–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D.. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 2006;172:41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S.. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol 2006;26:3738–3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Padmakumar VC, Libotte T, Lu W, Zaim H, Abraham S, Noegel AA, Gotzmann J, Foisner R, Karakesisoglou I.. The inner nuclear membrane protein Sun1 mediates the anchorage of Nesprin-2 to the nuclear envelope. J Cell Sci 2005;118:3419–3430. [DOI] [PubMed] [Google Scholar]
- 11. Starr DA, Han M.. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science 2002;298:406–409. [DOI] [PubMed] [Google Scholar]
- 12. Starr DA, Fridolfsson HN.. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol 2010;26:421–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaalouk DE, Lammerding J.. Mechanotransduction gone awry. Nat Rev Mol Cell Biol 2009;10:63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lombardi ML, Lammerding J.. Keeping the LINC: the importance of nucleocytoskeletal coupling in intracellular force transmission and cellular function. Biochem Soc Trans 2011;39:1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mammoto A, Mammoto T, Ingber DE.. Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci 2012;125:3061–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Puckelwartz MJ, Depreux FF, McNally EM.. Gene expression, chromosome position and lamin A/C mutations. Nucleus 2011;2:162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang N, Tytell JD, Ingber DE.. Mechanotransduction at a distance: mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol 2009;10:75–82. [DOI] [PubMed] [Google Scholar]
- 18. Banerjee I, Zhang J, Moore-Morris T, Pfeiffer E, Buchholz KS, Liu A, Ouyang K, Stroud MJ, Gerace L, Evans SM, McCulloch A, Chen J, Wang D-Z.. Targeted ablation of nesprin 1 and nesprin 2 from murine myocardium results in cardiomyopathy, altered nuclear morphology and inhibition of the biomechanical gene response. PLoS Genet 2014;10:e1004114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J.. Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 2013;497:507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT.. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J Cell Biol 2005;170:781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT.. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004;113:370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Puckelwartz MJ, Kessler EJ, Kim G, Dewitt MM, Zhang Y, Earley JU, Depreux FF, Holaska J, Mewborn SK, Pytel P, McNally EM.. Nesprin-1 mutations in human and murine cardiomyopathy. J Mol Cell Cardiol 2010;48:600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang J, Felder A, Liu Y, Guo LT, Lange S, Dalton ND, Gu Y, Peterson KL, Mizisin AP, Shelton GD, Lieber RL, Chen J.. Nesprin 1 is critical for nuclear positioning and anchorage. Hum Mol Genet 2010;19:329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bione S, Maestrini E, Rivella S, Mancini M, Regis S, Romeo G, Toniolo D.. Identification of a novel X-linked gene responsible for Emery–Dreifuss muscular dystrophy. Nat Genet 1994;8:323–327. [DOI] [PubMed] [Google Scholar]
- 25. Bonne G, Di Barletta MR, Varnous S, Becane HM, Hammouda EH, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea JA, Duboc D, Fardeau M, Toniolo D, Schwartz K.. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery–Dreifuss muscular dystrophy. Nat Genet 1999;21:285–288. [DOI] [PubMed] [Google Scholar]
- 26. Mounkes LC, Kozlov SV, Rottman JN, Stewart CL.. Expression of an LMNA-N195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum Mol Genet 2005;14:2167–2180. [DOI] [PubMed] [Google Scholar]
- 27. Arimura T, Helbling-Leclerc A, Massart C, Varnous S, Niel F, Lacene E, Fromes Y, Toussaint M, Mura AM, Keller DI, Amthor H, Isnard R, Malissen M, Schwartz K, Bonne G.. Mouse model carrying H222P-Lmna mutation develops muscular dystrophy and dilated cardiomyopathy similar to human striated muscle laminopathies. Hum Mol Genet 2005;14:155–169. [DOI] [PubMed] [Google Scholar]
- 28. Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ Jr, Spudich S, De Girolami U, Seidman JG, Seidman C, Muntoni F, Muehle G, Johnson W, McDonough B.. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med 1999;341:1715–1724. [DOI] [PubMed] [Google Scholar]
- 29. Taylor MR, Slavov D, Gajewski A, Vlcek S, Ku L, Fain PR, Carniel E, Di Lenarda A, Sinagra G, Boucek MM, Cavanaugh J, Graw SL, Ruegg P, Feiger J, Zhu X, Ferguson DA, Bristow MR, Gotzmann J, Foisner R, Mestroni L, Familial Cardiomyopathy Registry Research Group. Thymopoietin (lamina-associated polypeptide 2) gene mutation associated with dilated cardiomyopathy. Hum Mutat 2005;26:566–574. [DOI] [PubMed] [Google Scholar]
- 30. Zhang Q, Bethmann C, Worth NF, Davies JD, Wasner C, Feuer A, Ragnauth CD, Yi Q, Mellad JA, Warren DT, Wheeler MA, Ellis JA, Skepper JN, Vorgerd M, Schlotter-Weigel B, Weissberg PL, Roberts RG, Wehnert M, Shanahan CM.. Nesprin-1 and -2 are involved in the pathogenesis of Emery Dreifuss muscular dystrophy and are critical for nuclear envelope integrity. Hum Mol Genet 2007;16:2816–2833. [DOI] [PubMed] [Google Scholar]
- 31. Mejat A, Misteli T.. LINC complexes in health and disease. Nucleus 2010;1:40–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Puckelwartz MJ, Kessler E, Zhang Y, Hodzic D, Randles KN, Morris G, Earley JU, Hadhazy M, Holaska JM, Mewborn SK, Pytel P, McNally EM.. Disruption of nesprin-1 produces an Emery Dreifuss muscular dystrophy-like phenotype in mice. Hum Mol Genet 2009;18:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Christensen AH, Andersen CB, Tybjaerg-Hansen A, Haunso S, Svendsen JH.. Mutation analysis and evaluation of the cardiac localization of TMEM43 in arrhythmogenic right ventricular cardiomyopathy. Clin Genet 2011;80:256–264. [DOI] [PubMed] [Google Scholar]
- 34. Hodgkinson KA, Connors SP, Merner N, Haywood A, Young TL, McKenna WJ, Gallagher B, Curtis F, Bassett AS, Parfrey PS.. The natural history of a genetic subtype of arrhythmogenic right ventricular cardiomyopathy caused by a p.S358L mutation in TMEM43. Clin Genet 2013;83:321–331. [DOI] [PubMed] [Google Scholar]
- 35. Rajkumar R, Sembrat JC, McDonough B, Seidman CE, Ahmad F.. Functional effects of the TMEM43 Ser358Leu mutation in the pathogenesis of arrhythmogenic right ventricular cardiomyopathy. BMC Med Genet 2012;13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Milting H, Klauke B, Christensen AH, Musebeck J, Walhorn V, Grannemann S, Munnich T, Ari T, Rasmussen TB, Jensen HK, Mogensen J, Baecker C, Romaker E, Laser KT, zu Knyphausen E, Kassner A, Gummert J, Judge DP, Connors S, Hodgkinson K, Young T-L, van der Zwaag PA, van Tintelen JP, Anselmetti D.. The TMEM43 Newfoundland mutation p.S358L causing ARVC-5 was imported from Europe and increases the stiffness of the cell nucleus. Eur Heart J 2015;36:872–881. [DOI] [PubMed] [Google Scholar]
- 37. Jiang C, Zhu Y, Zhou Z, Gumin J, Bengtsson L, Wu W, Songyang Z, Lang FF, Lin X.. TMEM43/Luma is a key signaling component mediating EGFR-induced NF-kappaB activation and tumor progression. Oncogene 2017;36:2813–2823. [DOI] [PubMed] [Google Scholar]
- 38. Siragam V, Cui X, Masse S, Ackerley C, Aafaqi S, Strandberg L, Tropak M, Fridman MD, Nanthakumar K, Liu J, Sun Y, Su B, Wang C, Liu X, Yan Y, Mendlowitz A, Hamilton RM, Ai X.. TMEM43 mutation p.S358L alters intercalated disc protein expression and reduces conduction velocity in arrhythmogenic right ventricular cardiomyopathy. PLoS One 2014;9:e109128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Franke WW, Dorflinger Y, Kuhn C, Zimbelmann R, Winter-Simanowski S, Frey N, Heid H.. Protein LUMA is a cytoplasmic plaque constituent of various epithelial adherens junctions and composite junctions of myocardial intercalated disks: a unifying finding for cell biology and cardiology. Cell Tissue Res 2014;357:159–172. [DOI] [PubMed] [Google Scholar]
- 40. Zhang Z, Stroud MJ, Zhang J, Fang X, Ouyang K, Kimura K, Mu Y, Dalton ND, Gu Y, Bradford WH, Peterson KL, Cheng H, Zhou X, Chen J.. Normalization of Naxos plakoglobin levels restores cardiac function in mice. J Clin Invest 2015;125:1708–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hayashi S, Lewis P, Pevny L, McMahon AP.. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 2002;119(Suppl. 1):S97–S101. [DOI] [PubMed] [Google Scholar]
- 42. Ma X, Chen C, Veevers J, Zhou X, Ross RS, Feng W, Chen J.. CRISPR/Cas9-mediated gene manipulation to create single-amino-acid-substituted and floxed mice with a cloning-free method. Sci Rep 2017;7:42244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sheikh F, Ouyang K, Campbell SG, Lyon RC, Chuang J, Fitzsimons D, Tangney J, Hidalgo CG, Chung CS, Cheng H, Dalton ND, Gu Y, Kasahara H, Ghassemian M, Omens JH, Peterson KL, Granzier HL, Moss RL, McCulloch AD, Chen J.. Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease. J Clin Invest 2012;122:1209–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang X, Bogomolovas J, Wu T, Zhang W, Liu C, Veevers J, Stroud MJ, Zhang Z, Ma X, Mu Y, Lao DH, Dalton ND, Gu Y, Wang C, Wang M, Liang Y, Lange S, Ouyang K, Peterson KL, Evans SM, Chen J.. Loss-of-function mutations in co-chaperone BAG3 destabilize small HSPs and cause cardiomyopathy. J Clin Invest 2017;127:3189–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rockman HA, Ross RS, Harris AN, Knowlton KU, Steinhelper ME, Field LJ, Ross J Jr, Chien KR.. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A 1991;88:8277–8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Boogerd CJ, Aneas I, Sakabe N, Dirschinger RJ, Cheng QJ, Zhou B, Chen J, Nobrega MA, Evans SM.. Probing chromatin landscape reveals roles of endocardial TBX20 in septation. J Clin Invest 2016;126:3023–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stroud MJ, Nazgiewicz A, McKenzie EA, Wang Y, Kammerer RA, Ballestrem C.. GAS2-like proteins mediate communication between microtubules and actin through interactions with end-binding proteins. J Cell Sci 2014;127:2672–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stroud MJ, Feng W, Zhang J, Veevers J, Fang X, Gerace L, Chen J.. Nesprin 1alpha2 is essential for mouse postnatal viability and nuclear positioning in skeletal muscle. J Cell Biol 2017;216:1915–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stroud MJ, Kammerer RA, Ballestrem C.. Characterization of G2L3 (GAS2-like 3), a new microtubule- and actin-binding protein related to spectraplakins. J Biol Chem 2011;286:24987–24995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fang X, Stroud MJ, Ouyang K, Fang L, Zhang J, Dalton ND, Gu Y, Wu T, Peterson KL, Huang HD, Chen J, Wang N.. Adipocyte-specific loss of PPARgamma attenuates cardiac hypertrophy. JCI Insight 2016;1:e89908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirai M, Arita Y, McGlade CJ, Lee KF, Chen J, Evans SM.. Adaptor proteins NUMB and NUMBL promote cell cycle withdrawal by targeting ERBB2 for degradation. J Clin Invest 2017;127:569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Moore-Morris T, Guimaraes-Camboa N, Banerjee I, Zambon AC, Kisseleva T, Velayoudon A, Stallcup WB, Gu Y, Dalton ND, Cedenilla M, Gomez-Amaro R, Zhou B, Brenner DA, Peterson KL, Chen J, Evans SM.. Resident fibroblast lineages mediate pressure overload-induced cardiac fibrosis. J Clin Invest 2014;124:2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, Chen J.. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. J Cell Sci 2009;122:2640–2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guimaraes-Camboa N, Cattaneo P, Sun Y, Moore-Morris T, Gu Y, Dalton ND, Rockenstein E, Masliah E, Peterson KL, Stallcup WB, Chen J, Evans SM.. Pericytes of multiple organs do not behave as mesenchymal stem cells in vivo. Cell Stem Cell 2017;20:345–359.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mezzano V, Liang Y, Wright AT, Lyon RC, Pfeiffer E, Song MY, Gu Y, Dalton ND, Scheinman M, Peterson KL, Evans SM, Fowler S, Cerrone M, McCulloch AD, Sheikh F.. Desmosomal junctions are necessary for adult sinus node function. Cardiovasc Res 2016;111:274–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Luke Y, Zaim H, Karakesisoglou I, Jaeger VM, Sellin L, Lu W, Schneider M, Neumann S, Beijer A, Munck M, Padmakumar VC, Gloy J, Walz G, Noegel AA.. Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J Cell Sci 2008;121:1887–1898. [DOI] [PubMed] [Google Scholar]
- 57. Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D.. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J 2007;93:2542–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cupesi M, Yoshioka J, Gannon J, Kudinova A, Stewart CL, Lammerding J.. Attenuated hypertrophic response to pressure overload in a lamin A/C haploinsufficiency mouse. J Mol Cell Cardiol 2010;48:1290–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.