Abstract
Many multicellular organisms rely on symbiotic associations for support of metabolic activity, protection, or energy. Understanding the mechanisms involved in controlling such interactions remains a major challenge. In an unbiased approach we identified key players that control the symbiosis between Hydra viridissima and its photosynthetic symbiont Chlorella sp. A99. We discovered significant up-regulation of Hydra genes encoding a phosphate transporter and glutamine synthetase suggesting regulated nutrition supply between host and symbionts. Interestingly, supplementing the medium with glutamine temporarily supports in vitro growth of the otherwise obligate symbiotic Chlorella, indicating loss of autonomy and dependence on the host. Genome sequencing of Chlorella sp. A99 revealed a large number of amino acid transporters and a degenerated nitrate assimilation pathway, presumably as consequence of the adaptation to the host environment. Our observations portray ancient symbiotic interactions as a codependent partnership in which exchange of nutrients appears to be the primary driving force.
Research organism: Other
eLife digest
All animals host microorganisms; some of which form ‘symbiotic’ relationships with their host that are mutually beneficial. For instance, the human gut shelters tens of thousands of species of bacteria that break down our food for us, and corals, jellyfish or sea anemones can extract energy directly from sunlight thanks to the algae that live inside their cells.
Hydra, a small freshwater animal, lives in a symbiotic relationship with algae called Chlorella that it carries inside its cells. Once an independent organism, Chlorella has evolved in such a way that, in nature, it cannot exist without Hydra anymore. In turn, the algae produce sugars to fuel the animal when it cannot get food from the environment. Yet, despite over 30 years of research, it still remains unclear how exactly the relationship between Hydra and Chlorella works, and how it came to be. Understanding how these two organisms live together could help researchers to figure out the general principles that guide symbiotic interactions.
Nitrogen is an element that is essential for life, and organisms can extract it from various sources, such as nitrates or the amino acid glutamine. Here, Hamada, Schröder et al. sequenced the entire genome of Chlorella. This revealed that Chlorella has lost someof the genes required to obtain nitrates, and to process them into nitrogen. However, the genetic analysis showed that the algae express genes that allow them to import amino acids.
In turn, analysis of the genes expressed by Hydra when it lives in symbiosis with Chlorella showed that the animal turns on genetic information needed to make glutamine. It thus seems that Hydra creates glutamine which Chlorella can import; the algae then process this amino acid to obtain the nitrogen they need. Hamada, Schröder et al. also discovered that if the environment was artificially enriched in glutamine, Chlorella could live on their own outside of Hydra for a while.
The results suggest that symbiotic relationships, such as the one between Hydra and Chlorella, were established because the organisms became dependent on each other for essential nutrients. This co-dependency is strengthened if the organisms lose the ability to produce the nutrients on their own. However, this partnership may be altered when the environment changes too much, especially if the balance of nutrients available gets tipped. For example, if seas that are normally poor in nutrients become suddenly rich in these elements, this may disrupt the existence of symbiotic organisms such as corals.
Introduction
Symbiosis has been a prevailing force throughout the evolution of life, driving the diversification of organisms and facilitating rapid adaptation of species to divergent new niches (Moran, 2007; Joy, 2013; McFall-Ngai et al., 2013). In particular, symbiosis with photosynthetic symbionts is observed in many species of cnidarians such as corals, jellyfish, sea anemones and hydra, contributing to the ecological success of these sessile or planktonic animals (Douglas, 1994; Davy et al., 2012). Among the many animals dependent on algal symbionts, inter-species interactions between green hydra Hydra viridissima and endosymbiotic unicellular green algae of the genus Chlorella have been a subject of interest for decades (Muscatine and Lenhoff, 1963; Roffman and Lenhoff, 1969). Such studies not only provide insights into the basic ‘tool kit’ necessary to establish symbiotic interactions, but are also of relevance in understanding the resulting evolutionary selective processes (Muscatine and Lenhoff, 1965a; 1965b; Thorington and Margulis, 1981).
The symbionts are enclosed in the host endodermal epithelial cells within perialgal vacuoles called ‘symbiosomes’. The interactions at play here are clearly metabolic: the algae depend on nutrients that are derived from the host or from the environment surrounding the host, while in return the host receives a significant amount of photosynthetically fixed carbon from the algae. Previous studies have provided evidence that the photosynthetic symbionts provide their host with maltose, enabling H. viridissima to survive periods of starvation (Muscatine and Lenhoff, 1963; Muscatine, 1965; Roffman and Lenhoff, 1969; Cook and Kelty, 1982; Huss et al., 1994). Chlorella-to-Hydra translocation of photosynthates is critical for polyps to grow (Muscatine and Lenhoff, 1965b; Mews, 1980; Douglas and Smith, 1983; 1984). Presence of symbiotic algae also has a profound impact on hydra´s fitness by promoting oogenesis (Habetha et al., 2003; Habetha and Bosch, 2005).
Pioneering studies performed in the 1980 s (McAuley and Smith, 1982; Rahat and Reich, 1984) showed that there is a great deal of adaptation and specificity in this symbiotic relationship. All endosymbiotic algae found in a single host polyp are clonal and proliferation of symbiont and host is tightly correlated (Bossert and Dunn, 1986; McAuley, 1986a). Although it is not yet known how Hydra controls cell division in symbiotic Chlorella, Chlorella strain A99 is unable to grow outside its polyp host and is transmitted vertically to the next generation of Hydra, indicating loss of autonomy during establishment of its symbiotic relationship with this host (Muscatine and McAuley, 1982; Campbell, 1990; Habetha et al., 2003).
Molecular phylogenetic analyses suggest that H. viridissima is the most basal species in the genus Hydra and that symbiosis with Chlorella was established in the ancestral viridissima group after their divergence from non-symbiotic Hydra groups (Martínez et al., 2010; Schwentner and Bosch, 2015). A recent phylogenetic analysis of different strains of green hydra resulted in a phylogenetic tree that is topologically equivalent to that of their symbiotic algae (Kawaida et al., 2013), suggesting these species co-evolved as a result of their symbiotic relationship. Although our understanding of the factors that promote symbiotic relationships in cnidarians has increased (Shinzato et al., 2011; Davy et al., 2012; Lehnert et al., 2014; Baumgarten et al., 2015; Ishikawa et al., 2016), very little is known about the molecular mechanisms allowing this partnership to persist over millions of years.
Recent advances in transcriptome and genome analysis allowed us to identify the metabolic interactions and genomic evolution involved in achieving the Hydra-Chlorella symbiotic relationship. We present here the first characterization, to our knowledge, of genetic complementarity between green Hydra and Chlorella algae that explains the emergence and/or maintenance of a stable symbiosis. We also provide here the first report of the complete genome sequence from an obligate intracellular Chlorella symbiont. Together, our results show that exchange of nutrients is the primary driving force for the symbiosis between Chlorella and Hydra. Subsequently, reduction of metabolic pathways may have further strengthened their codependency. Our findings provide a framework for understanding the evolution of a highly codependent symbiotic partnership in an early emerging metazoan.
Results
Discovery of symbiosis-dependent Hydra genes
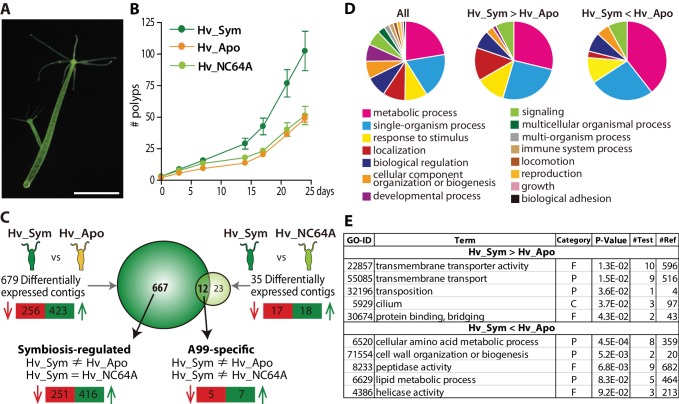
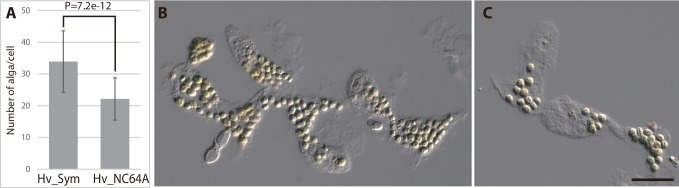
As tool for our study we used the green hydra H. viridissima (Figure 1A) colonized with symbiotic Chlorella sp. strain A99 (abbreviated here as Hv_Sym), aposymbiotic H. viridissima from which the symbiotic Chlorella were removed (Hv_Apo), as well as aposymbiotic H.viridissima, which have been artificially infected with Chlorella variabilis NC64A (Hv_NC64A). The latter is symbiotic to the single-cellular protist Paramecium (Karakashian and Karakashian, 1965). Although an association between H. viridissima and Chlorella NC64A can be maintained for some time, both their growth rate (Figure 1B) and the number of NC64A algae per Hydra cell (Figure 1—figure supplement 1) are significantly reduced compared to the symbiosis with native symbiotic Chlorella A99.
Figure 1. Hydra growth and differential expression of Hydra genes resulting from symbiosis.
(A) Hydra viridissima strain A99 used for this study. Scale bar, 2 mm. (B) Growth rates of polyps grown with native symbiotic Chlorella A99 (Hv_Sym, dark green), Aposymbiotic polyps from which Chlorella were removed (Hv_Apo, orange) and aposymbiotic polyps reinfected with Chlorella variabilis NC64A (Hv_NC64A, light green). Average of the number of hydra in each experimental group (n = 6) is represented. Error bars indicate standard deviation. (C) Graphic representation of differentially expressed genes identified by microarray. The transcriptome of Hv_Sym is compared with that of Hv_Apo and Hv_NC64A with the number of down-regulated contigs in Hv_Sym shown in red and those up-regulated in green. Genes differentially expressed in Hv_Sym compared to both Hv_Apo and Hv_NC64A are given as ‘A99-specific’, those differentially expressed between Hv_A99 and Hv_Apo but not Hv_NC64A as ‘Symbiosis-regulated’. (D) GO distribution of Biological Process at level two in all contigs (All), up-regulated contigs (Hv_Sym > Hv_Apo) and down-regulated contigs (Hv_Sym < Hv_Apo) in Hv_Sym. (E) Overrepresented GO terms in up-regulated contigs (Hv_Sym > Hv_Apo) and down-regulated contigs (Hv_Sym < Hv_Apo). Category, F: molecular function, C: cellular component, P: biological process. P-values, probability of Fisher’s exact test. #Test, number of corresponding contigs in differentially expressed contigs. #Ref, number of corresponding contigs in all contigs.
Figure 1—figure supplement 1. Chlorella sp. A99 and Chlorella variabilis NC64A in Hydra viridissima A99.
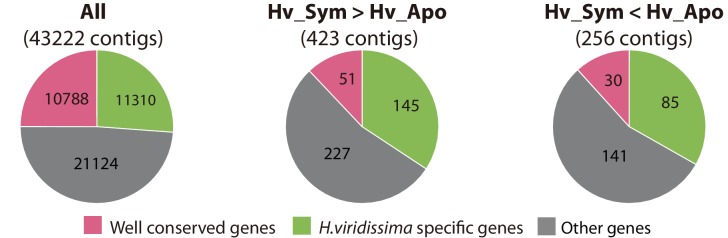
Figure 1—figure supplement 2. Conserved genes and species-specific genes differentially expressed in symbiotic Hydra.
Figure 1—figure supplement 3. Glutamine synthetase (GS) genes in Cnidarians.
H.H. viridissima genes involved in the symbiosis with Chlorella algae were identified by microarray based on the contigs of H. viridissima A99 transcriptome (NCBI GEO Platform ID: GPL23280). For the microarray analysis, total RNA was extracted from the polyps after light exposure for six hours. By comparing the transcriptomes of Hv_Sym and Hv_Apo, we identified 423 contigs that are up-regulated and 256 contigs that are down-regulated in presence of Chlorella A99 (Figure 1C). To exclude genes involved in oogenesis and embryogenesis, only contigs differently expressed with similar patterns in both sexual and asexual Hv_Sym were recorded. Interestingly, contigs whose predicted products had no discernible homologs in other organisms including other Hydra species were overrepresented in these differentially expressed contigs (Chi-squared test p<0.001) (Figure 1—figure supplement 2). Such taxonomically restricted genes (TRGs) are thought to play important roles in the development of evolutionary novelties and morphological diversity within a given taxonomic group (Khalturin et al., 2009; Tautz and Domazet-Lošo, 2011).
We further characterized functions of the differentially expressed Hydra genes by Gene Ontology (GO) terms (Ashburner et al., 2000) and found the GO term ‘localization’ overrepresented among up-regulated contigs (Hv_Sym > Hv_Apo), whereas the GO term ‘metabolic process’ was enriched among down-regulated contigs (Hv_Sym < Hv_Apo) (Figure 1D). More specifically, the up-regulated contigs included many genes related to ‘transmembrane transporter activity’, ‘transmembrane transport’, ‘transposition’, ‘cilium’ and ‘protein binding, bridging’ (Figure 1E). In the down-regulated contig set, the GO classes ‘cellular amino acid metabolic process’, ‘cell wall organization or biogenesis’ and ‘peptidase activity’ were overrepresented (Figure 1E). These results suggest that the Chlorella symbiont affects core metabolic processes and pathways in Hydra. Particularly, carrier proteins and active membrane transport appear to play a prominent role in the symbiosis.
As next step, we used GO terms, domain search and similarity search to further analyze the differentially expressed contigs between Hv_Sym and Hv_Apo (Supplementary file 1). As the genes with GO terms related to localization and transport, we identified 27 up-regulated contigs in Hv_Sym (Table 1). Interestingly, this gene set included a contig showing sequence similarity to the glucose transporter GLUT8 gene, which was previously reported to be up-regulated in the symbiotic state of the sea anemone Aiptasia (Lehnert et al., 2014; Sproles et al., 2018). Thus, a conserved mechanism may be responsible for photosynthate transport from the symbiont into the host cytoplasm across the symbiosome membrane. Further, a contig encoding a carbonic anhydrase (CA) enzyme was up-regulated in Hv_Sym (Table 1). CA catalyzes the interconversion of HCO3 and CO2. Similar to the GLUT8 gene, carbonic anhydrase also appears to be up-regulated in symbiotic corals and anemones (Weis et al., 1989; Grasso et al., 2008; Ganot et al., 2011; Lehnert et al., 2014). It appears plausible that for efficient photosynthesis in symbiotic algae, the host may need to convert CO2 to the less freely diffusing inorganic carbon (HCO3) to maintain it in the symbiosome (Lucas and Berry, 1985; Weis et al., 1989; Barott et al., 2015). We also observed up-regulation of contigs encoding proteins involved in vesicular and endosomal trafficking, such as spe-39 protein, otoferlin, protein fam194b and V-type proton ATPase 21 kda proteolipid, which may have a function in nutrition exchange between host and symbiont and maintenance of proper condition in the symbiosome. Upregulated genes also include genes encoding rhamnospondin and fibrillin, known to be involved in cell adhesion and extracellular matrix, and retention of the symbiont at the proper site in the Hydra cells.
Table 1. List of differentially expressed genes between Hv_Sym and Hv_Apo, which are likely to be involved in symbiotic relationship.
| Probename | Fold change | Human_BestHit | blast2GO_Description | |||
|---|---|---|---|---|---|---|
| Hv_Sym /Hv_Apo |
Hv_Sym_sexy /Hv_Apo |
Hv_NC64A /Hv_Sym |
||||
| Localization and Transport | ||||||
| Hv_Sym > Hv_Apo | ||||||
| rc_6788 | 9.87 | 8.00 | 1.01 | helicase conserved c-terminal domain containing protein |
||
| rc_10246 | 8.26 | 5.15 | 1.82 | protein | ||
| rc_6298 | 7.10 | 4.73 | 0.99 | hypothetical protein LOC220081 | protein fam194b | |
| 2268 | 6.96 | 3.58 | 1.26 | protein Daple | viral a-type inclusion protein | |
| 10548 | 6.74 | 6.89 | 0.73 | transient receptor potential cation channel subfamily M member three isoform d |
transient receptor potential cation channel subfamily m member 3-like |
|
| rc_1290 | 6.44 | 7.18 | 0.99 | tetratricopeptide repeat protein eight isoform B |
tetratricopeptide repeat protein 8 | |
| 18736 | 6.04 | 6.34 | 1.03 | BTB/POZ domain-containing protein KCTD9 |
btb poz domain-containing protein kctd9-like; unnamed protein product |
|
| rc_9270 | 5.96 | 10.03 | 1.37 | PREDICTED: hypothetical protein LOC100131693 |
eukaryotic translation initiation factor 4e | |
| NPNHRC_15697 | 3.85 | 2.74 | 0.62 | major facilitator superfamily domain- containing protein 1 |
||
| 290 | 3.68 | 3.73 | 1.32 | splicing factor, arginine/ serine-rich 6 |
splicing arginine serine-rich 4 | |
| rc_9596 | 3.56 | 4.19 | 1.62 | BTB/POZ domain-containing protein KCTD10 |
btb poz domain-containing adapter for cul3-mediated degradation protein 3 |
|
| rc_6774 | 3.34 | 3.32 | 1.31 | solute carrier family 43, member 2 |
large neutral amino acids transporter small subunit 4 |
|
| rc_26218 | 3.29 | 2.91 | 0.41 | sodium-dependent phosphate transport protein 2A isoform 1 |
sodium-dependent phosphate transport protein 2b |
|
| NPNHRC_26094 | 3.20 | 3.98 | 1.31 | SPE-39 proteinid="T5" | spe-39 protein | |
| 9096 | 3.10 | 2.20 | 0.69 | otoferlin isoform d | otoferlin | |
| rc_21349 | 2.89 | 4.25 | 0.78 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 |
5 -amp-activated protein kinase catalytic subunit alpha-2 |
|
| npRC_14488 | 2.88 | 2.65 | 0.71 | solute carrier family 2, facilitated glucose transporter member 8 |
solute carrier family facilitated glucose transporter member 8-like |
|
| 8863 | 2.75 | 2.70 | 0.81 | ATP-binding cassette, sub-family B, member 10 precursor |
abc transporter b family protein | |
| rc_11896 | 2.49 | 2.56 | 1.52 | ATP-binding cassette, sub-family B, member 10 precursor |
abc transporter b family member 25-like | |
| rc_6842 | 2.41 | 3.35 | 1.59 | hypothetical protein LOC112752 isoform 2 |
intraflagellar transport protein 43 homolog | |
| 5242 | 2.36 | 3.35 | 1.22 | growth arrest-specific protein 8 | growth arrest-specific protein 8 | |
| 5815 | 2.23 | 2.47 | 0.78 | plasma membrane calcium- transporting ATPase 4 isoform 4a |
plasma membrane calcium atpase | |
| 8765 | 2.22 | 3.25 | 0.91 | growth arrest-specific protein 8 | growth arrest-specific protein 8 | |
| NPNH_14052 | 2.19 | 2.17 | 0.79 | V-type proton ATPase 21 kDa proteolipid subunit isoform 2 |
v-type proton atpase 21 kda proteolipid subunit-like |
|
| rc_2499 | 2.18 | 2.03 | 1.47 | endoplasmic reticulum-Golgi intermediate compartment protein three isoform a |
endoplasmic reticulum-golgi intermediate compartment protein 3 isoform 2 |
|
| rc_13969 | 2.08 | 3.09 | 0.97 | major facilitator superfamily | ||
| (IPR023561) Carbonic anhydrase, alpha-class | ||||||
| rc_24825 | 2.49 | 2.38 | 0.83 | protein tyrosine phosphatase, receptor type, G precursor |
receptor-type tyrosine-protein phosphatase gamma |
|
| Cell Adhesion and extracelluar matrix | ||||||
| Hv_Sym > Hv_Apo | ||||||
| 7915 | 4.01 | 5.09 | 0.94 | fibrillin-2 precursor | fibrillin-1- partial | |
| npRC_24163 | glutamate3.69 | 3.59 | 1.32 | semaphorin 5A precursor | rhamnospondin 1 | |
| Immunity, apoptosis and recognition | ||||||
| Hv_Sym > Hv_Apo | ||||||
| (IPR000157) Toll/interleukin-1 receptor homology (TIR) domain | ||||||
| 5168 | 9.28 | 4.92 | 0.61 | protein; PREDICTED: uncharacterized protein LOC100893943 |
||
| 12749 | 5.13 | 3.35 | 1.26 | PREDICTED: uncharacterized protein LOC100893943 [Strongylocentrotus purpuratus] |
||
| (IPR011029) DEATH-like | ||||||
| 6508 | 6.70 | 5.10 | 0.64 | PREDICTED: hypothetical protein [Hydra magnipapillata] |
||
| rc_2417 | 5.39 | 2.70 | 1.01 | nod3 partial; PREDICTED: uncharacterized protein LOC100206003 |
||
| (IPR002398) Peptidase C14, caspase precursor p45 | ||||||
| NPNH_21275 | 2.36 | 3.53 | 1.18 | caspase seven isoform alpha precursor |
caspase d | |
| (IPR016187) C-type lectin fold | ||||||
| 11411 | 2.93 | 2.98 | 0.75 | C-type mannose receptor 2 | PREDICTED: similar to predicted protein, partial [Hydra magnipapillata] |
|
| Hv_Sym < Hv_Apo | ||||||
| (IPR000488) Death | ||||||
| 7319 | 0.45 | 0.31 | 1.10 | probable ubiquitin carboxyl- terminal hydrolase CYLD isoform 2 |
ubiquitin carboxyl-terminal hydrolase cyld | |
| (IPR001875) Death effector domain | ||||||
| RC_FV81RT001CSTY | 0.31 | 0.39 | 0.93 | astrocytic phosphoprotein PEA-15 | fadd | |
| Chitinase | ||||||
| Hv_Sym > Hv_Apo | ||||||
| (IPR001223) Glycoside hydrolase, family 18, catalytic domain | ||||||
| rc_4450 | 2.78 | 3.83 | 0.66 | chitinase 2 | ||
| Hv_Sym < Hv_Apo | ||||||
| (IPR000726) Glycoside hydrolase, family 19, catalytic | ||||||
| FPVQZVL01EAWBY | 0.21 | 0.16 | 1.78 | endochitinase 1-like | ||
| 1028 | 0.23 | 0.18 | 1.47 | endochitinase 1-like | ||
| Oxidative Stress Response | ||||||
| Hv_Sym > Hv_Apo | ||||||
| np_1276 | 5.99 | 7.16 | 0.78 | glutaredoxin-2, mitochondrial isoform 2 |
cpyc type | |
| 10926 | 3.9 | 2.3 | 0.8 | hydroxysteroid dehydrogenase- like protein 2 |
hydroxysteroid dehydrogenase-like protein 2 |
|
| 469 | 2.97 | 3.53 | 0.76 | cytochrome P450 3A7 | cytochrome p450 | |
| FV81RT001DCTAQ | 2.69 | 2.50 | 0.75 | oxidoreductase NAD-binding domain-containing protein one precursor |
oxidoreductase nad-binding domain- containing protein 1 |
|
| 696 | 2.30 | 3.24 | 0.69 | methionine-R-sulfoxide reductase B1 |
selenoprotein 1; methionine-r-sulfoxide reductase b1-a-like |
|
| 6572 | 2.23 | 2.15 | 1.06 | L-xylulose reductase | l-xylulose reductase | |
| 13298 | 2.10 | 3.49 | 0.64 | eosinophil peroxidase preproprotein |
peroxidase | |
| npRC_6975 | 2.04 | 2.77 | 1.42 | methionine-R-sulfoxide reductase B1 |
selenoprotein 1; methionine-r-sulfoxide reductase b1-a-like |
|
| (IPR024079) Metallopeptidase, catalytic domain | ||||||
| Hv_array_4952 | 4.77 | 13.31 | 0.72 | meprin A subunit beta precursor |
zinc metalloproteinase nas-4-like | |
| Hv_array_rc_3992 | 2.66 | 2.23 | 1.27 | matrix metalloproteinase seven preproprotein |
matrix metalloproteinase-24-like | |
| Hv_Sym < Hv_Apo | ||||||
| RC_FWZAEML02HKSC | 0.255 | 0.153 | 1.444 | ascorbate peroxidase | ||
| np_14962 | 0.293 | 0.455 | 1.390 | tryptophan 5-hydroxylase 2 | phenylalanine hydroxylase | |
| rc_4151 | 0.318 | 0.463 | 1.693 | phenylalanine-4-hydroxylase | phenylalanine hydroxylase | |
| 2835 | 0.384 | 0.344 | 1.787 | u1 small nuclear ribonucleoprotein 70 kda | ||
| rc_11426 | 0.413 | 0.458 | 1.591 | short-chain dehydrogenase/ reductase family 9C member 7 |
uncharacterized oxidoreductase -like | |
| FWZAEML02IC34R | 0.427 | 0.448 | 1.159 | aldehyde dehydrogenase 5A1 isoform two precursor |
succinate-semialdehyde mitochondrial-like | |
| FWZAEML02HKSCO | 0.454 | 0.307 | 0.833 | ascorbate peroxidase | ||
| (IPR004045) Glutathione S-transferase, N-terminal | ||||||
| RC_FWZAEML02GGHN | 0.09 | 0.07 | 1.81 | hematopoietic prostaglandin D synthase |
glutathione s-transferase family member (gst-7) |
|
| (IPR024079) Metallopeptidase, catalytic domain | ||||||
| rc_11270 | 0.14 | 0.20 | 1.33 | meprin A subunit beta precursor | protein; zinc metalloproteinase nas-4-like | |
| rc_RSASM_15059 | 0.22 | 0.29 | 1.42 | ---NA--- | ||
| 2111 | 0.37 | 0.43 | 1.74 | meprin A subunit beta precursor | zinc metalloproteinase nas-4-like | |
| 12451 | 0.50 | 0.39 | 0.78 | meprin A subunit alpha precursor | zinc metalloproteinase nas-13- partial | |
| (IPR013122) Polycystin cation channel, PKD1/PKD2 | ||||||
| 28854 | 0.37 | 0.28 | 0.94 | polycystin-2 | receptor for egg jelly partial | |
| 15774 | 0.40 | 0.26 | 0.76 | polycystic kidney disease protein 1-like two isoform a |
protein | |
Photosynthesis by symbiotic algae imposes Reactive Oxygen Species (ROS) that can damage lipids, proteins and DNA in the host cells (Lesser, 2006). Therefore, in symbiosis with photosynthetic organisms an appropriate oxidative stress response of the host is required for tolerance of the symbiont. Indeed, an increase of antioxidant activities in symbiotic states of cnidarians has been reported previously (Richier et al., 2005) and it has been suggested that ROS produced by stress could be the major trigger of symbiosis breakdown during coral bleaching (Lesser, 2006; Weis, 2008). To understand the oxidative stress response in green hydra, we searched the differentially expressed genes with the GO terms ‘response to oxidative stress’, ‘oxidation-reduction process’ and ‘oxidoreductase activity’. In Hv_Sym, contigs for peroxidase, methionine-r-sulfoxide reductase/selenoprotein and glutaredoxin, which are known to be related to oxidative stress response were up-regulated (Table 1). On the other hand, some contigs encoding glutathione S-transferase and ascorbate peroxidase were down-regulated in Hv_Sym. In addition, two contigs encoding polycystin were down-regulated in Hv_Sym. Polycystin is an intracellular calcium release channel that is inhibited by ROS (Montalbetti et al., 2008) and is also down-regulated in a different strain of symbiotic green hydra (Ishikawa et al., 2016). In addition, six contigs encoding metalloproteinases showed differential expression between Hv_Sym and Hv_Apo. Although metalloproteinases have many functions such as cleavage of cell surface proteins and remodeling of the extracellular matrix, in a previous study they also were found to play a role in the oxidative stress response (Császár et al., 2009). A key antioxidant in the oxidative stress response in symbiotic cnidarians turns out to be glutathione (Sunagawa et al., 2009; Meyer and Weis, 2012). The gene encoding glutathione S-transferase was previously observed to be downregulated in corals, sea anemones, different strains of green hydra and Paramecium (Kodama et al., 2014; Lehnert et al., 2014; Ishikawa et al., 2016; Mohamed et al., 2016). Our study supports this view (Table 1) and may point to a conserved feature of oxidative stress response in algal-animal symbiosis.
Previous studies have suggested that during establishment of coral–algal symbiosis the host immune response may be partially suppressed (Weis et al., 2008; Mohamed et al., 2016). Our observations in Hydra together with previous findings in corals indicate that regulation of symbiosis by innate immunity pathways indeed may be a general feature of cnidarian symbiosis. Among the differentially expressed contigs we identified a number of genes involved in innate immunity and apoptosis. Pattern recognition receptors (PRRs) and the downstream innate immunity and apoptosis pathways are thought to play important roles in various symbiotic interactions including cnidarian-dinoflagellate symbiosis (Davy et al., 2012). In Hv_Sym we found two up-regulated contigs that contain a Toll/interleukin-1 receptor (TIR) domain (Table 1). TIR is a known PRR that is inserted in the host cell membrane and plays an important role in the innate immune system by specifically recognizing microbial-associated molecular patterns, such as flagellin, lipopolysaccharide (LPS) and peptidoglycan (Hoving et al., 2014). Furthermore, we found one up-regulated contig with similarity to a mannose receptor gene with C-type lectin domain (Table 1). This is worth noting since C-type lectin receptors bind carbohydrates and some of them are known to function as PRRs. Host lectin-algal glycan interactions have been proposed to be involved in infection and recognition of symbionts in some cnidarians including green hydra, sea anemones and corals (Meints and Pardy, 1980; Lin et al., 2000; Wood-Charlson et al., 2006). Interestingly, up-regulation of C-type lectin genes was also observed during onset of cnidarian–dinoflagellate symbiosis (Grasso et al., 2008; Schwarz et al., 2008; Sunagawa et al., 2009; Mohamed et al., 2016).
Furthermore, contigs encoding chitinase enzymes also were differentially expressed between Hv_Sym and Hv_Apo (Table 1). Chitinases are involved in degradation of chitin, which is a component of the exoskeleton of arthropods and the cell wall of fungi, bacteria and some Chlorella algae (Kapaun and Reisser, 1995), and also might play a role in host-defense systems for pathogens which have chitinous cell wall. Chitinases are classified into two glycoside hydrolase families, GH18 and GH19, with different structures and catalytic mechanisms. In Hv_Sym two contigs encoding GH18 chitinases were up-regulated, while one contig encoding a GH19 chitinase was down-regulated, suggesting that the enzymes involved in chitin degradation are sensitive to the presence or absence of symbiotic Chlorella.
To narrow down the number of genes specifically affected by the presence of the native symbiont Chlorella A99, we identified 12 contigs that are differentially expressed in symbiosis with Chlorella A99, but not in presence of foreign Chlorella NC64A (Figure 1C A99-specific). Independent qPCR confirmed the differential expression pattern for 10 of these genes (Table 2). The genes up-regulated by the presence of the symbiont encode a Spot_14 protein, a glutamine synthetase (GS) and a sodium-dependent phosphate (Na/Pi) transport protein in addition to a H. viridissima specific gene (rc_12891: Sym-1) and a Hydra genus specific gene (rc_13570: Sym-2) (Table 2). Hydra genes down-regulated by the presence of Chlorella A99 were two H. viridissima-specific genes and three metabolic genes encoding histidine ammonia-lyase, acetoacetyl-CoA synthetase and 2-isopropylmalate synthase (Table 2). Of the up-regulated genes, Spot_14 is described as thyroid hormone-responsive spot 14 protein reported to be induced by dietary carbohydrates and glucose in mammals (Tao and Towle, 1986; Brown et al., 1997). Na/Pi transport protein is a membrane transporter actively transporting phosphate into cells (Murer and Biber, 1996). GS plays an essential role in the metabolism of nitrogen by catalyzing the reaction between glutamate and ammonia to form glutamine (Liaw et al., 1995). Interestingly, out of the three GS genes H. viridissima contains only GS-1 was found to be up-regulated by the presence of the symbiont (Figure 1—figure supplement 3). The discovery of these transcriptional responses points to an intimate metabolic exchange between the partners in a species-specific manner.
Table 2. List of genes differentially expressed in Hv_Sym compared to both Hv_Apo and Hv_NC64A (‘A99-specific’).
Fold change of expression level determined by microarray analysis and qPCR analysis
| Hv_Sym > Hv_Apo, Hv_NC64A | ||||||
|---|---|---|---|---|---|---|
| Probe name (gene ID) | Microarray | qPCR | Gene annotation | InterProScan | ||
| Sym/Apo | Sym/NC64A | Sym/Apo | Sym/NC64A | |||
| rc_13579 | 12.8 | 4.0 | 11.2 | 4.0 | (Hydra specific) | |
| rc_12891 | 9.0 | 2.9 | 14.6 | 6.9 | (Hydra viridis specific) | |
| 27417 | 4.5 | 4.8 | 3.0 | 3.0 | IPR009786 Spot_14 | |
| rc_26218 | 3.3 | 2.4 | 2.5 | 2.3 | sodium-dependent phosphate transport protein |
PTHR10010 Sodium-dependent phosphate transport protein 2C |
| 1046 | 3.1 | 2.1 | 2.2 | 1.6 | glutamine synthetase | |
| Hv_Sym < Hv_Apo, Hv_NC64A | ||||||
| Probe name (gene ID) | Microarray | qPCR | Gene Annotation | InterProScan | ||
| Apo/Sym | NC64A/Sym | Apo/Sym | NC64A/Sym | |||
| NPNHRC_26859 | 83.2 | 9.7 | ∞ | ∞ | (Hydra viridis specific) | |
| RC_FVQRUGK01AXSJ | 13.7 | 2.6 | 2.1 | 1.5 | acetoacetyl-CoA synthetase | |
| rc_14793 | 7.2 | 4.1 | 9.4 | 4.8 | 2-isopropylmalate synthase | IPR013785 Aldolase_TIM, |
| FV81RT002HT2FL | 2.8 | 2.0 | 3.1 | 1.8 | histidine ammonia-lyase | IPR001106 Aromatic_Lyase IPR008948 L-Aspartase-like |
| NPNHRC_12201 | 2.7glutamate | 2.3 | 2.6 | 2.5 | (Hydra viridis specific) | |
To better understand the specificity of Hydra´s response to the presence of the foreign symbiont, we also identified the genes differentially expressed in Hydra polyps hosting a non-native Chlorella NC64A (Hv_NC64A) compared to both polyps hosting the obligate symbiont Chlorella A99 (Hv_A99) and aposymbiotic Hydra (Hv_Apo). We found 19 contigs that were up-regulated and 45 contigs that were down-regulated in presence of NC64A, which strikingly did not include any genes related to immunity or oxidative stress response (Supplementary file 1). Instead, the differentially expressed contigs showed similarity to methylase genes involved in ubiquinone menaquinone biosynthesis and secondary metabolite synthesis such as n-(5-amino-5-carboxypentanoyl)-l-cysteinyl-d-valine synthase and non-ribosomal peptide synthase. Four differentially expressed contigs specifically up-regulated in Hv_NC64A encoded ubiquitin carboxyl-terminal hydrolases, (Table 3).
Table 3. List of annotated genes up-regulated in Hv_NC64A compared to Hv_Sym.
| Probename | Hv_NC64A/ Hv_Sym |
Hv_Apo/ Hv_Sym |
Hv_Sym_sexy/ Hv_Sym |
Blast2GO description |
|---|---|---|---|---|
| rc_1623 | 4.57 | 1.64 | 5.98 | methylase involved in ubiquinone menaquinone biosynthesis |
| 28947 | 3.52 | 1.59 | 0.63 | non-ribosomal peptide synthetase |
| 1353 | 3.13 | 1.63 | 0.10 | nuclear protein set |
| 14347 | 2.69 | 2.40 | 0.54 | n-(5-amino-5-carboxypentanoyl)-l -cysteinyl-d-valine synthase |
| SSH_397 | 2.67 | 2.39 | 0.50 | n-(5-amino-5-carboxypentanoyl)-l -cysteinyl-d-valine synthase |
| RC_FWZAEML01C7BP | 2.28 | 0.82 | 0.41 | ubiquitin carboxyl-terminal hydrolase family protein |
| RC_FVQRUGK01EOXS | 2.25 | 1.52 | 0.53 | ubiquitin carboxyl-terminal hydrolase family protein |
| rc_11710 | 2.15 | 1.26 | 0.31 | ubiquitin carboxyl-terminal hydrolase family protein |
| 1677 | 2.10 | 1.19 | 0.38 | ubiquitin carboxyl-terminal hydrolase family protein |
| rc_363 | 2.21 | 1.04 | 0.76 | gcc2 and gcc3 family protein |
Symbiont-dependent Hydra genes are up-regulated by photosynthetic activity of Chlorella A99
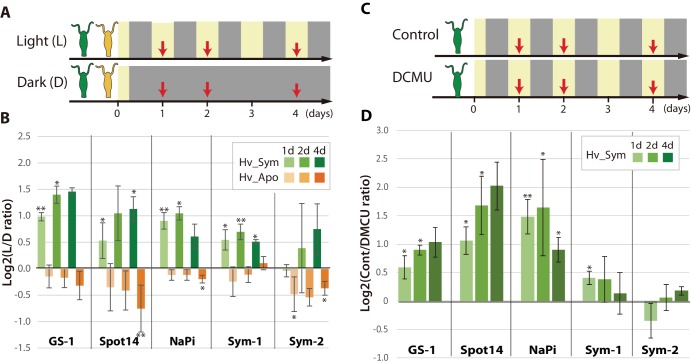
To test whether photosynthetic activity of the symbiont is required for up-regulation of gene expression, Hv_Sym was either cultured under a standard 12 hr light/dark alternating regime or continuously in the dark for 1 to 4 days prior to RNA extraction (Figure 2A). Interestingly, four (GS1, Spot14, Na/Pi and Sym-1) of five genes specifically activated by the presence of Chlorella A99 showed significant up-regulation when exposed to light (Figure 2B), indicating the relevance of photosynthetic activity of Chlorella. This up-regulation was strictly dependent on presence of the algae, as in aposymbiotic Hv_Apo the response was absent (Figure 2B). On the other hand, symbiosis-regulated Hydra genes not specific for Chlorella A99 (Figure 1C Symbiosis-regulated, Table 4) appear to be not up-regulated in a light-dependent manner (Figure 2—figure supplement 1). These genes are involved in Hydra´s innate immune system (e.g. proteins containing Toll/interleukin-1 receptor domain or Death domain) or in signal transduction (C-type mannose receptor, ephrin receptor, proline-rich transmembrane protein 1, ‘protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (p58 repressor)’). That particular transcriptional changes observed in Hydra rely solely on the photosynthetic activity of Chlorella A99 was confirmed by substituting the dark incubation with selective chemical photosynthesis inhibitor DCMU (Dichorophenyl-dimethylurea) (Vandermeulen et al., 1972), which resulted in a similar effect (Figure 2C,D).
Figure 2. Differential expression of Hydra genes under influence of Chlorella photosynthesis.
(A) Sampling scheme. Hv_Sym (green) and Hv_Apo (orange) were cultured under a standard light-dark regime (Light: L) and in continuous darkness (Dark: D), and RNA was extracted from the polyps at the days indicated by red arrows. (B) Expression difference of five A99-specific genes in Hv_Sym (green bars) and Hv_Apo (orange bars) between the light-dark condition and darkness. The vertical axis shows log scale (log2) fold changes of relative expression level in Light over Dark. (C) Sampling scheme of inhibiting photosynthesis. (D) Differential expression of the five A99-specific genes under conditions allowing (Control) or inhibiting photosynthesis (DCMU). The vertical axis shows log scale (log2) fold changes of relative expression level in Control over DCMU treated. T-tests were performed between Light and Dark (B), and DCMU and Control (D). For each biological replicate (n = 3) 50 hydra polyps were used for total RNA extraction. Error bars indicate standard deviation. P-value of t-test, *<0.05, **<0.01.
Figure 2—figure supplement 1. Differential expression of symbiosis-dependent Hydra genes grown under light/dark condition and in darkness.
Table 4. List of the genes differentially expressed between Hv_Sym and Hv_Apo.
Fold change of expression level determined by microarray analysis and qPCR
| Hv_Sym > Hv_Apo | ||||
|---|---|---|---|---|
| Probe name (gene ID) |
Microarray | qPCR | Gene annotation | InterProScan |
| Sym/Apo | Sym/Apo | |||
| 5168 | 9.3 | 7.4 | IPR000157 TIR_dom PTHR23097 Tumor necrosis factor receptor superfamily member |
|
| 6508 | 6.7 | 2.9 | IPR011029:DEATH-like_dom | |
| 11411 | 2.9 | 2.0 | C-type mannose receptor 2 | IPR000742 EG-like_dom IPR001304 C-type_lectin |
| 26108 | 7.2 | 7.2 | ephrin type-A receptor six isoform a | |
| rc_2417 | 5.4 | 3.5 | IPR000488 Death_domain | |
| rc_24563 | 6.1 | 6.7 | Proline-rich transmembrane protein 1 | IPR007593 CD225/Dispanin_fam PTHR14948 NG5 |
| rc_9398 | 6.2 | 5.4 | protein-kinase, interferon-inducible double stranded RNA dependent inhibitor, repressor of (P58 repressor) |
PTHR11697 general transcription factor 2-related zinc finger protein |
| Hv_Sym < Hv_Apo | ||||
|
Probe name
(gene ID) |
Microarray | qPCR | Gene Annotation | InterProScan |
| Apo/Sym | Apo/Sym | |||
| rc_10789 | 2.5 | 3.7 | endoribonuclease Dicer | IPR000999 RNase_III_dom PTHR1495 helicase-related |
| rc_12826 | 3.0 | 2.3 | interferon regulatory factor 1 | IPR001346 Interferon_reg_fact_DNA-bd_dom; IPR011991 WHTH_DNA-bd_dom PTHR11949 interferon regulatory factor |
| rc_8898 | 6.1 | 4.1 | leucine-rich repeat-containing protein 15 isoform b |
IPR001611 Leu-rich_rp PTHR24373 Toll-like receptor 9 |
| FV81RT001CSTY | 3.2 | 2.0 | astrocytic phosphoprotein PEA-15 | IPR001875 DED, IPR011029 DEATH-like_dom |
| RSASM_17752 | 4.0 | 2.1 | CD97 antigen isoform two precursor | IPR000832 GPCR_2_secretin-like PTHR12011 vasoactive intestinal polypeptide receptor 2 |
Symbiont-dependent Hydra genes are expressed in endodermal epithelial cells and up-regulated by sugars
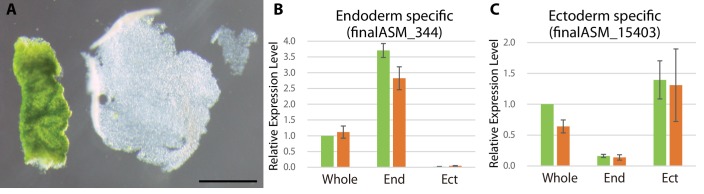
To further characterize the symbiont induced Hydra genes, we performed whole mount in situ hybridization (Figure 3A–F) and quantified transcripts by qPCR using templates from isolated endoderm and ectoderm (Figure 3—figure supplement 1), again comparing symbiotic and aposymbiotic polyps (Figure 3G–I). The GS-1 gene and the Spot14 gene are expressed both in ectoderm and in endoderm (Figure 3A,B) and both genes are strongly up-regulated in the presence of the symbiont (Figure 3G,H). In contrast, the Na/Pi gene was expressed only in the endoderm (Figure 3C) and there it was strongly up-regulated by the symbiont (Figure 3I). Since Chlorella sp. A99 colonizes endodermal epithelial cells only, the impact of algae on symbiosis-dependent genes in both the ectodermal and the endodermal layer indicates that photosynthetic products can be transported across these two tissue layers or some signals can be transduced by cell-cell communication.
Figure 3. Spatial expression patterns of genes coding for glutamine synthetase, Spot 14 and Na/Pi-transporter.
(A-F); Whole mount in situ hybridization using antisense (A–C) and sense probes (D-F; negative controls) for glutamine synthetase-1 (GS-1; left), Spot 14 (center) and Na/Pi-transporter (NaPi; right). Inserts show cross sections of the polyp’s body. (G–I) Relative expression levels of whole animal (whole), isolated endoderm (End) and isolated ectoderm (Ect) tissue of Hv_Sym (green bars) and Hv_Apo (orange bars). For each biological replicate (n = 3) 10–20 hydra polyps were used for total RNA extraction of endodermal and ectodermal tissue. T-test was performed between Hv_Sym and Hv_apo. Pvalue, *<0.05, **<0.01. (J) Expression change of genes GS-1, Spot14, NaPi, Sym-1 and Sym-2 following exposure to 25, 50 and 100 mM maltose in Hv_Apo. For each biological replicate (n = 3) 50 hydra polyps were used for total RNA extraction The vertical axis shows log scale (log2) fold changes of relative expression level of maltose-treated over the untreated Hv_Apo control. T-test was performed between maltose-treated in each concentration and control (*: p value <0.05) and Kruskal-Wallis test (†: p value <0.05) in the series of 48 hr treatment were performed. Error bars indicate standard deviation.
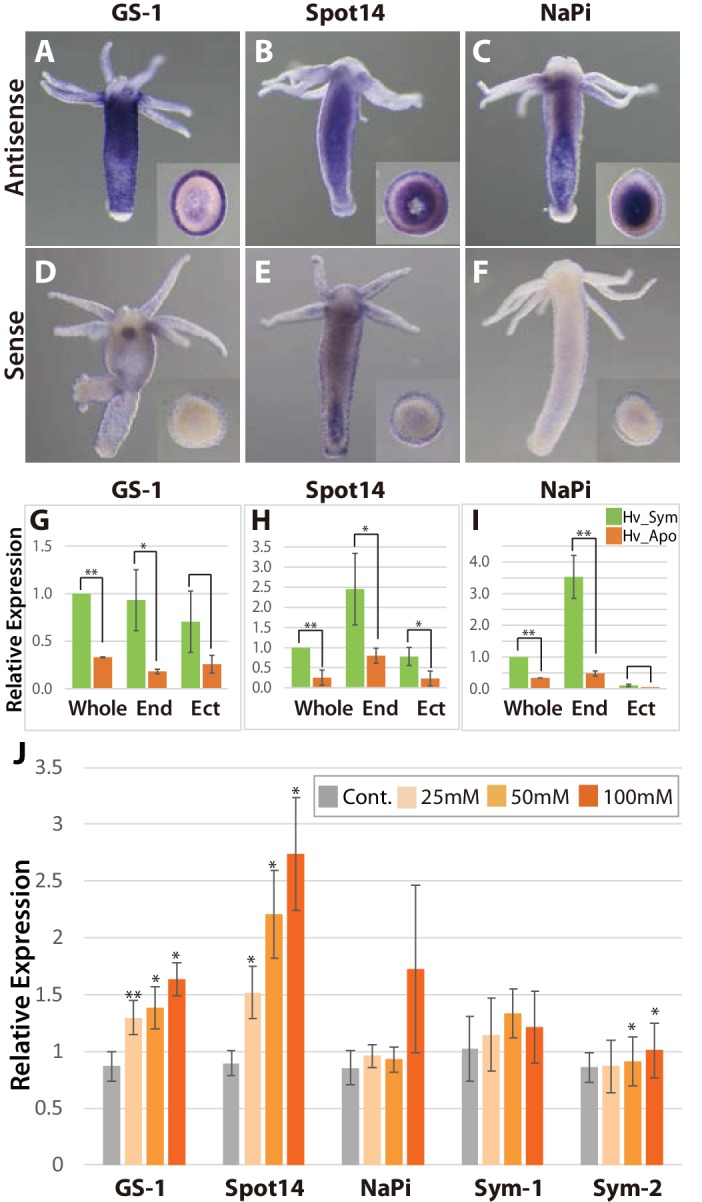
Figure 3—figure supplement 1. Tissue isolation of green hydra.
Figure 3—figure supplement 2. Effects of sugars on Hydra growth.
To more closely dissect the nature of the functional interaction between Hydra and Chlorella and to explore the possibility that maltose released from the algae is involved in A99-specific gene regulation, we cultured aposymbiotic polyps (Hv_Apo) for 2 days in medium containing various concentrations of maltose (Figure 3J). Of the five A99 specific genes, GS-1 and the Spot14 gene were up-regulated by maltose in a dose-dependent manner; the Na/Pi gene was only up-regulated in 100 mM maltose and the Hydra specific genes Sym-1 and Sym-2 did not show significant changes in expression by exposure to maltose (Figure 3J). This provides strong support for previous views that maltose excretion by symbiotic algae contributes to the stabilization of this symbiotic association (Cernichiari et al., 1969). When polyps were exposed to glucose instead of maltose, the genes of interest were also transcriptionally activated in a dose-dependent manner, while sucrose had no effect (Figure 3—figure supplement 2A–D). Exposure to low concentrations of galactose increased transcriptional activity but at high concentration it did not, indicating a substrate inhibitor effect for this sugar. That the response to glucose is similar or even higher compared to maltose after 6 hr of treatment (Figure 3—figure supplement 2E), suggests that Hydra cells transform maltose to glucose as a source of energy. In animals including cnidarians, several glucose transporters have been identified (Sproles et al., 2018), but yet no maltose transporters. This is consistent with the view that maltose produced by the symbiont is digested to glucose in the symbiosome and translocated to the host cytoplasm through glucose transporters.
The Chlorella A99 genome records a symbiotic life style
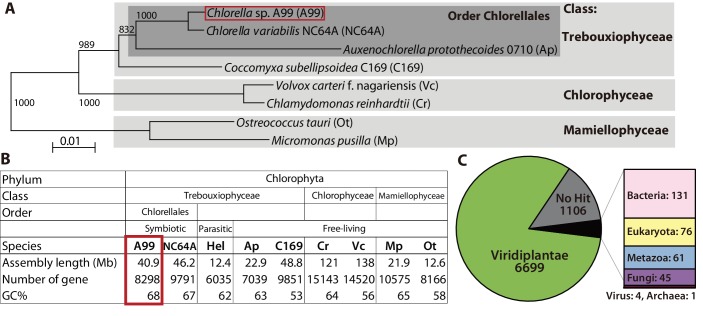
To better understand the symbiosis between H. viridissima and Chlorella and to refine our knowledge of the functions that are required in this symbiosis, we sequenced the genome of Chlorella sp. strain A99 and compared it to the genomes of other green algae. The genome of Chlorella sp. A99 was sequenced to approximately 211-fold coverage, enabling the generation of an assembly comprising a total of 40.9 Mbp (82 scaffolds, N50 = 1.7 Mbp) (Table 5). Chlorella sp. A99 belongs to the family Chlorellaceae (Figure 4A) and of the green algae whose genomes have been sequenced it is most closely related to Chlorella variabilis NC64A (NC64A) (Merchant et al., 2007; Palenik et al., 2007; Worden et al., 2009; Blanc et al., 2010; Prochnik et al., 2010; Blanc et al., 2012; Gao et al., 2014; Pombert et al., 2014). The genome size of the total assembly in strain A99 was similar to that of strain NC64A (46.2 Mb) (Figure 4B). By k-mer analysis (k-mer = 19), the genome size of A99 was estimated to be 61 Mbp (Marçais and Kingsford, 2011). Its GC content of 68%, is the highest among the green algae species recorded (Figure 4B). In the A99 genome, 8298 gene models were predicted. As shown in Figure 4C, about 80% of these predicted genes have extensive sequence similarity to plant genes, while 13% so far have no similarity to genes of any other organisms (Figure 4C). It is also noteworthy that 7% of the A99 genes are similar to genes of other kingdoms but not to Hydra, indicating the absence of gene transfer from Hydra to the symbiont genome (Figure 4C).
Table 5. Summary of sequence data for assembling Chlorella sp. A99 genome sequences.
| Number of reads | 85469010 | |
| Number of reads assembled | 61838513 | |
| Number of bases | 17398635102 | |
| Scaffolds | Contigs | |
| Total length of sequence | 40934037 | 40687875 |
| Total number of sequences | 82 | 7455 |
| Maximum length of sequence | 4003385 | 171868 |
| N50 | 1727419 | 12747 |
| GC contents (%) | 68.07% | 69.95% |
Figure 4. Comparison of key features deduced from the Chlorella A99 genome with other green algae.
(A) Phylogenetic tree of eight genome sequenced chlorophyte green algae including Chlorella sp. A99. The NJ tree is based on sequences of the 18S rRNA gene, ITS1, 5.8S rRNA gene, ITS2 and 28S rRNA gene. (B) Genomic features and taxonomy of the sequenced chlorophyte green algae. Hel: Helicosporidium sp. ATCC50920. (C) The proportion of similarity of Chlorella A99 gene models to those of other organisms.
The Chlorella A99 genome provides evidences for extensive nitrogenous amino acid import and an incomplete nitrate assimilation pathway
Several independent lines of evidence demonstrate that nitrogen limitation and amino-acid metabolism have a key role in the Chlorella–Hydra symbiosis and that symbiotic Chlorella A99 depends on glutamine provided by its host (Rees, 1986; McAuley, 1987a; 1987b; McAuley, 1991; Rees, 1991;1989). To identify Chlorella candidate factors for the development and maintenance of the symbiotic life style, we therefore used the available genome information to assess genes potentially involved in amino acid transport and the nitrogen metabolic pathway.
When performing a search for the Pfam domain ‘Aa_trans’ or ‘AA_permease’ to find amino acid transporter genes in the A99 genome, we discovered numerous genes containing the Aa_trans domain (Table 6A). In particular, A99 contains many orthologous genes of amino acid permease 2 and of transmembrane amino acid transporter family protein (solute carrier family 38, sodium-coupled neutral amino acid transporter), as well as NC64A (Table 6B, Supplementary file 2). Both of these gene products are known to transport neutral amino acids including glutamine. This observation is supporting the view that import of amino acids is an essential feature for the symbiotic way of life of Chlorella.
Table 6. Amino acid transporter genes in Chlorella sp. A99 (A99), Chlorella variabilis NC64A (NC64A), Coccomyxa subellipsoidea C-169 (C169), Volvox carteri (Vc), Micromonas pusilla (Mp) and Ostreococcus tauri (Ot) and Chlamydomonas reinhardtii (Cr).
| A. The number of Pfam domains related to amino acids transport | |||||||
| Pfam domain name | A99 | NC64A | c169 | Cr | Vc | Mp | Ot |
| Aa_trans | 30 | 38 | 21 | 9 | 7 | 9 | 8 |
| AA_permease | 4 | 6 | 15 | 5 | 6 | 1 | 1 |
| B. Ortholog groups including Aa_trans domain containing genes overrepresented in symbiotic Chlorella | |||||||
| Ortholog group ID: Gene annotation | A99 | NC64A | c169 | Cr | Vc | Mp | Ot |
| OG0000040: amino acid permease 2 | 12 | 12 | 6 | 3 | 1 | 0 | 0 |
| OG0000324: transmembrane amino acid transporter family protein (solute carrier family 38, sodium-coupled neutral amino acid transporter) |
6 | 7 | 1 | 2 | 1 | 0 | 0 |
In symbiotic organisms, loss of genes often occurs due to the strictly interdependent relationship (Ochman and Moran, 2001; Wernegreen, 2012), raising the possibility that Chlorella A99 might have lost some essential genes. To test this hypothesis, we searched the Chlorella A99 genome for genes conserved across free-living green algae Coccomyxa subellipsoidea C169 (C169), Chlamydomonas reinhardtii (Cr) and Volvox carteri (Vc). In a total of 9851 C169 genes, we found 5701 genes to be conserved in Cr and Vc (Supplementary file 3). Of these, 238 genes did not match to any gene models and genomic regions in Chlorella A99 and thus were considered as gene loss candidates. Interestingly, within these 238 candidates, genes with the GO terms ‘transport’ in biological process and ‘transporter activity’ in molecular function were overrepresented (Figure 5). In particular, the 28 genes annotated to these GO terms encoded nitrate transporter, urea transporter and molybdate transporter, which are known to be involved in nitrogen metabolism (Table 7). Beside ammonium, nitrate and urea are major nitrogen sources for plants, whereas molybdate is a co-factor of the nitrate reductase, an important enzyme in the nitrate assimilation pathway. These transporter genes are conserved across green algae including Chlorella NC64A (Sanz-Luque et al., 2015; Gao et al., 2014) and appear to be lost in the Chlorella A99 genome.
Figure 5. Genes missing in the genome of Chlorella A99.
Functional categorization of genes present in Coccomyxa subellipsoidea C169 (A, C) and genes missing in Chlorella A99 (B, D) by GO terms using Bast2GO. Multilevel pie charts show enrichment of GO’ Biological Process’ terms (A, B) and GO ‘Molecular Function’ terms (C, D) on the lowest level, which cover at least 10% of the total amount of annotated sequences.
Table 7. List of Coccomyxa subellipsoidea C169 (C169) genes, which are present in Chlamydomonas reinhardtii and Volvox carteri, but missing in the genome of Chlorella A99.
| UniProt ID in C169 | Description |
|---|---|
| F1DPL8_9CHLO | ATP synthase F0 subunit 6 (mitochondrion) |
| F1DPL7_9CHLO | cytochrome c oxidase subunit 3 (mitochondrion) |
| I0YZU4_9CHLO | equilibrative nucleoside transporter 1 |
| I0Z311_9CHLO | equilibrative nucleoside transporter family |
| I0YZC9_9CHLO | high affinity nitrate transporter |
| I0Z2L2_9CHLO | hypothetical protein COCSUDRAFT_28432 |
| I0YJ99_9CHLO | hypothetical protein COCSUDRAFT_34498 |
| I0YKQ1_9CHLO | hypothetical protein COCSUDRAFT_45098 |
| I0YYD3_9CHLO | hypothetical protein COCSUDRAFT_65897 |
| I0YYP5_9CHLO | importin-4 isoform X1 |
| I0YQQ1_9CHLO | low-CO2-inducible membrane |
| I0YJD4_9CHLO | MFS transporter |
| I0YTY0_9CHLO | molybdate transporter 2 |
| F1DPM0_9CHLO | NADH dehydrogenase subunit 3 (mitochondrion) |
| F1DPM4_9CHLO | NADH dehydrogenase subunit 6 (mitochondrion) |
| F1DPM8_9CHLO | NADH dehydrogenase subunit 9 (mitochondrion) |
| I0Z357_9CHLO | plasma membrane phosphate transporter Pho87 |
| I0Z9Y1_9CHLO | pre translocase subunit |
| I0YPT2_9CHLO | transcription and mRNA export factor ENY2-like |
| I0Z976_9CHLO | transport SEC23 |
| I0Z3Q6_9CHLO | tyrosine-specific transport -like isoform X1 |
| I0YXU9_9CHLO | urea active transporter |
| I0YRT0_9CHLO | urea active transporter |
| I0YRL4_9CHLO | urea-proton symporter DUR3 |
| I0YUF9_9CHLO | urea-proton symporter DUR3 |
| I0YJS6_9CHLO | urea-proton symporter DUR3 |
| I0YQ78_9CHLO | urea-proton symporter DUR3-like |
| I0YIH7_9CHLO | Zip-domain-containing protein |
In nitrogen assimilation processes, plants usually take up nitrogen in the form of nitrate (NO3-) via nitrate transporters (NRTs) or as ammonium (NH4+) via ammonium transporters (AMT) (Figure 6A). In higher plants, two types of nitrate transporters, NRT1 and NRT2, have been identified (Krapp et al., 2014). Some NRT2 require nitrate assimilation-related component 2 (NAR2) to be functional (Quesada et al., 1994). NO3- is reduced to nitrite by nitrate reductase (NR), NO2- is transported to the chloroplast by nitrate assimilation-related component1 (NAR1), and NO2- is reduced to NH4+ by nitrite reductase (NiR). NH4+ is incorporated into glutamine (Gln) by glutamine synthetase (GS), and Gln is incorporated into glutamate (Glu) by NADH-dependent glutamine amide-2-oxoglutarate aminotransferase (GOGAT), also known as glutamate synthase. This pathway is highly conserved among plants and all of its major components, including NRT1 and NRT2, NAR1 and NAR2, NR, NiR, AMT, GOGAT and GS, are present in the 10 green algae species that have been genome-sequenced so far (with the exception of NRT1, which is absent in Micromonas pusilla) (Sanz-Luque et al., 2015). In Symbiodinium, the photosynthetic symbiont of marine invertebrates, all these components of the nitrogen assimilation pathway were also observed (Supplementary file 4) (Shoguchi et al., 2013; Lin et al., 2015; Aranda et al., 2016; Sproles et al., 2018).
Figure 6. Nitrogen assimilation pathways in Chlorella A99.
(A) Schematic diagram of the nitrogen assimilation pathway in plants showing the function of nitrate transporters NRT1 (peptides/nitrate transporter) and NRT2 (nitrate/nitrite transporter), nitrate assimilation-related components NAR1 and NAR2, nitrate reductase NR, nitrite reductase NiR, ammonium transporter AMT, glutamate synthetase GOGAT and glutamine synthetase GS. Genes shown in red boxes (NRT2, NAR2 and NiR) were not found in the Chlorella sp. A99 genome. (B) Table showing the number of nitrogen assimilation genes in Chlorella sp. A99 (A99), Chlorella variabilis NC64A (NC64A), Coccomyxa subellipsoidea C169 (C169), Volvox carteri f. nagariensis (Vc), Chlamydomonas reinhardtii (Cr), Ostreococcus tauri (Ot) and Micromonas pusilla (Mp). (C) Gene clusters of nitrate assimilation genes around the shared NR genes (blue) in the genomes of NC64A, C169 and A99. Red boxes show nitrate assimilation genes absent in A99 and gray boxes depict other genes. Numbers below the boxes are JGI protein IDs of NC64A and C169. Numbers below the genes of A99 are JGI protein IDs of the best hit genes in NC64A and C169 and their gene name.
Figure 6—figure supplement 1. PCR of nitrate assimilation genes.
Based on the annotation by Sanz-Luque et al. (Sanz-Luque et al., 2015), we searched these nitrogen assimilation genes in the Chlorella A99 genome, using ortholog grouping and a reciprocal BLAST search using the protein sequences from other green algae (Figure 6B, Supplementary file 5). As expected, the Chlorella A99 genome contains many homologues of the genes involved in nitrogen assimilation in plants including genes encoding NRT1, NAR1, NR, AMT, GS and GOGAT (Figure 6B). Intriguingly, our systematic searches failed to identify representative genes for NRT2, NAR2 and NiR in the Chlorella A99 genome (Figure 6B). We confirmed the absence of the NRT2 and NiR genes by PCR using primers designed for the conserved regions of these genes and which failed to produce a product with genomic DNA as a template (Figure 6—figure supplement 1). Due to the weak sequence conservation of the NAR2 gene in the three algae genomes, PCR of that gene was not performed. Taken together, our observations indicate that Chlorella A99 algae appears to lack NRT2, NAR2 and NiR.
Since in many fungi, cyanobacteria and algae species, nitrate assimilation genes are known to act in concert and a gene cluster of NR and NiR genes is conserved between different green algae (Sanz-Luque et al., 2015), we next investigated the level of genomic clustering of the nitrate assimilation pathway genes in the Chlorella genome. Comparing the genomes of NC64A and C169 revealed the presence of a cluster of NR and NiR genes (Figure 6C). In NC64A, two NRT2 genes, together with genes for NAR2, NR and NiR are clustered on scaffold 21. In C169, one of the NR genes and NiR are clustered together, whereas the second NR gene is separate. Interestingly, analysis of the sequences around the NR gene in the Chlorella A99 genome provided no evidence for the presence of a co-localized NiR gene or any other nitrate assimilation genes, nor any conserved gene synteny to NC64A and C169 (Figure 6C). Therefore, our comparative genomic analyses points to an incomplete and scattered nitrogen metabolic pathway in symbiotic Chlorella A99, which lacks essential transporters and enzymes for nitrate assimilation as well as the clustered structure of nitrate assimilation genes.
Supplementing the medium with glutamine allows temporary in vitro growth of symbiotic Chlorella A99
The absence of genes essential for nitrate assimilation in the Chlorella A99 genome (Figure 6) is consistent with its inability to grow outside the Hydra host cell (Habetha and Bosch, 2005) and indicates that Chlorella symbionts are dependent on metabolites provided by their host. We hypothesized that Chlorella is unable to use nitrite and ammonium as a nitrogen source, and that it relies on Hydra assimilating ammonium to glutamine to serve as the nitrogen source. To test this hypothesis and to examine utilization of nitrogen compounds of A99, we isolated Chlorella A99 from Hv_Sym and cultivated it in vitro using modified bold basal medium (BBM) (Nichols and Bold, 1965) containing the same amount of nitrogen in the form of NO3-, NH4+, Gln or casamino acids (Figure 7). As controls, Chlorella variabilis NC64A (NC64A) isolated from Hv_NC64A and free-living C169 were used. To confirm that the cultured A99 is not contamination, we amplified and sequenced the genomic region of the 18S rRNA gene by PCR (Figure 7—figure supplement 1) and checked this against the genomic sequence of A99. Kamako et al. reported that free-living alga Chlorella vulgaris Beijerinck var. vulgaris grow in media containing only inorganic nitrogen compounds as well as in media containing casamino acids as a nitrogen source, while NC64A required amino acids for growth (Kamako et al., 2005). Consistent with these observations, C169 grew in all tested media and NC64A grew in media containing casamino acids and Gln, although its growth rate was quite low in presence of NH4+ and NO3- (Figure 7). Remarkably, Chlorella A99 increased in cell number for up to 8 days in media containing casamino acids and Gln (Figure 7). Similar to NC64A, A99 did not grow in presence of NH4+ and NO3-. The growth rates of both A99 and NC64A were higher in medium containing a mixture of amino acids (casamino acids) than the single amino acid Gln. In contrast to NC64A, A99 could not be cultivated permanently in casamino acids or glutamine supplemented medium, indicating that additional growth factors are necessary to maintain in vitro growth of this obligate symbiont. Thus, although in vitro growth of A99 can be promoted by adding Glu and amino acids to the medium, A99 cannot be cultured permanently in this enriched medium, indicating that other host derived factors remain to be uncovered.
Figure 7. Growth of green algae in presence of various nitrogen sources.
The growth rate of Chlorella A99 (A99), Chlorella variabilis NC64A (NC64A) and Coccomyxa subellipsoidea C-169 (C169) by in vitro culture was assessed for different nitrogen sources with casamino acids (blue), glutamine (orange), ammonium (gray) and nitrate (yellow). Mean number of algae per ml were determined at 4, 8, 12 days after inoculation with 106 cell/ml. Error bars indicate standard deviation.
Figure 7—figure supplement 1. PCR of 18S rRNA genes in cultured algae.
Discussion
Metabolic co-dependence in Hydra-Chlorella symbiosis
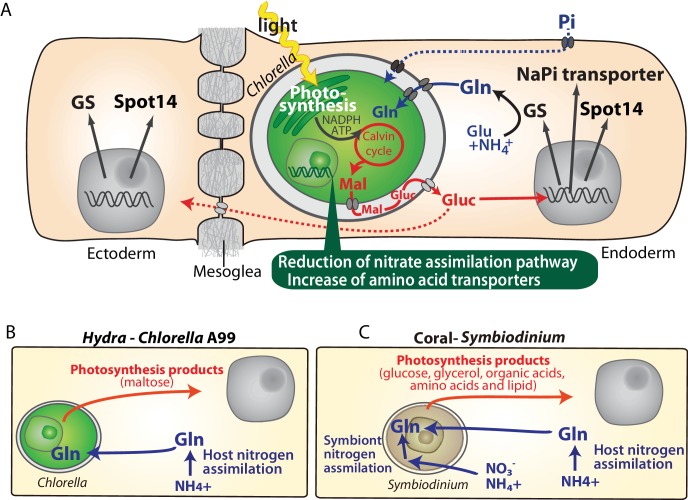
Sequencing of the Chlorella A99 genome in combination with the transcriptome analyses of symbiotic, aposymbiotic and NC64A-infected H. viridissima polyps has enabled the identification of genes with specific functions in this symbiotic partnership. The Hydra-Chlorella symbiosis links carbohydrate supply from the photosynthetic symbiont to glutamine synthesis by the host. Characteristics of the symbiont genome obviously reflect its adaptation to this way of life, including an increase in amino acid transporters and degeneration of the nitrate assimilation pathway. This conclusion is based on six observations: (i) Expression of some genes including GS-1, Spot 14 and NaPi is specifically up-regulated in the presence of Chlorella A99 (Figure 1C, Table 2), and (ii) they are induced by both, photosynthetic activity of Chlorella and by supplying exogenous maltose or glucose (Figures 2 and 3J, Figure 3—figure supplement 2). Maltose produced by the symbiont is likely to be digested to glucose in symbiosome and translocated to the host cytoplasm through glucose transporters (Figure 8A). Upregulation of a GLUT8 gene in the symbiotic state of green hydra may reflect activation of sugar transport (Table 1). These results indicate that maltose release by photosynthesis of the symbiont enhances nutrition supply including glutamine by the host (Figure 8A). (iii) Symbiotic Chlorella A99 cannot be cultivated in vitro in medium containing a single inorganic nitrogen source (Figure 7). Since medium containing glutamine supports in vitro growth of A99, this organism appears to depend on glutamine provided by the Hydra host. (iv) The genome of Chlorella A99 contains multiple amino acid transporter genes (Table 6), but lacks genes involved in nitrate assimilation (Figure 6), pointing to amino acids as main source of nitrogen and a degenerated nitrate assimilation pathway. As for ammonium, which is one of the main nitrogen sources in plants, previous studies have reported the inability of symbiotic algae to take up ammonium because of the low peri-algal pH (pH 4–5) that stimulates maltose release (Douglas and Smith, 1984; Rees, 1989; McAuley, 1991; Dorling et al., 1997). Since Chlorella apparently cannot use nitrite and ammonium as a nitrogen source, it seems that Hydra has to assimilate ammonium to glutamine and provides it to Chlorella A99 (Figure 8A).
Figure 8. Molecular interactions in the symbiosis of cnidarians.
(A) Summary of symbiotic interactions between Hydra and Chlorella A99. During light conditions, Chlorella A99 performs photosynthesis and produces maltose (Mal), which is secreted into the Hydra symbiosome where it is possibly digested to glucose (Gluc), shown in red. The sugar induces expression of Hydra genes encoding glutamine synthetase (GS), Na/Pi transporter (NaPi) and Spot14. GS catalyzes the condensation of glutamate (Glu) and ammonium (NH4+) to form glutamine (Gln), which is used by Chlorella as a nitrogen source. Since the sugar also up-regulates the NaPi gene, which controls intracellular phosphate levels, it might be involved in the supply of phosphorus to Chlorella as well (blue broken line). The sugar is transported to the ectoderm (red broken line) and there induces the expression of GS and Spot14. In the Chlorella A99 genome, degeneration of the nitrate assimilation system and an increase of amino acid transporters was observed (green balloon). (B, C) Comparison between Hydra-Chlorella symbiosis and coral-Symbiodinium symbiosis. Red indicates transfer of photosynthesis products from the symbiont to the host, and blue indicates transfer of nitrogen sources from the host to the symbiont. While the host organisms Hydra and coral can assimilate NH4+ to Gln (B, C), assimilation of inorganic nitrogen by Symbiodinidium plays an important role for the symbiotic system in coral (C).
(v) While polyps with native symbiont Chlorella A99 grew faster than aposymbiotic ones, symbiosis with foreign algae NC64A had no effect on the growth of polyps at all (Figure 1B). (vi) Hydra endodermal epithelial cells host significantly fewer NC64A algae than A99 (Figure 1—figure supplement 1) providing additional support for the view of a tightly regulated codependent partnership in which exchange of nutrients appears to be the primary driving force. Previous studies have reported that symbiotic Chlorella in green hydra releases significantly larger amounts of maltose than NC64A (Mews and Smith, 1982; Rees, 1989). In addition, Rees reported that Hydra polyps containing high maltose releasing algae had a high GS activity, whereas aposymbiotic Hydra or Hydra with a low maltose releasing algae had lower GS activity (Rees, 1986). Although the underlying mechanism of how maltose secretion and transportation from Chlorella is regulated is still unclear, the amount of maltose released by the symbiont could be an important symbiont-derived driver or stabilizer of the Hydra–Chlorella symbiosis.
More general lessons for animal-algal symbiosis
Transcriptome comparison between symbiotic and aposymbiotic H. viridissima demonstrated that symbiosis-regulated genes are involved in oxidative stress response and innate immunity. The fact that PRRs and apoptosis-related genes, are also differentially expressed in a number of other symbiotic cnidarians (Table 1), suggests innate immunity as conserved mechanism involved in controlling the development and maintenance of stable symbiotic interactions. Furthermore, the exchange of nitrogenous compounds and photosynthetic products between host and symbiont observed here in the Hydra-Chlorella symbiosis is also observed in marine invertebrates such as corals, sea anemones and giant clams associated with Symbiodinium algae (Figure 8B,C). Despite these similarities, however, there are also conspicuous differences among symbiotic cnidarians in particular with respect to the nutrients provided by the symbiont to the host. For example, symbiotic Chlorella algae in green hydra, Paramecium and fresh water sponges provide their photosynthetic products in form of maltose and glucose (Figure 8B) (Brown and Nielsen, 1974; Wilkinson, 1980; Kamako and Imamura, 2006). In contrast, Symbiodinium provides glucose, glycerol, organic acids, amino acids as well as lipids to its marine hosts (Figure 8C) (Muscatine and Cernichiari, 1969; Lewis and Smith, 1971; Trench, 1971; Kellogg and Patton, 1983). A former transcriptome analysis of amino acid biosynthetic pathways suggested that Symbiodinium can synthesize almost all amino acids (Shinzato et al., 2014). Gene loss in cysteine synthesis pathway in the coral host Acropora digitifera seems to reflect the dependency on the amino acids provided by the Symbiodinium symbiont (Shinzato et al., 2011). In contrast to Symbiodinium which can assimilate inorganic nitrogen such as nitrate and ammonium (Lipschultz and Cook, 2002; Grover et al., 2003; Tanaka et al., 2006; Yellowlees et al., 2008), the symbiotic Chlorella algae in Hydra and Paramecium can only use amino acids as a nitrogen source (Figure 6) (Kamako et al., 2005).
In efforts to explain the metabolic efficiency of nitrogen use in symbiotic organisms, two models have been proposed: the ‘nitrogen conservation’ and the ‘nitrogen recycling’ hypothesis. The nitrogen conservation hypothesis suggests that photosynthetic carbon compounds from the symbiont are used preferentially by the host respiration, which reduces catabolism of nitrogenous compounds (Rees and Ellard, 1989; Szmant et al., 1990; Wang and Douglas, 1998). The ‘nitrogen recycling’ hypothesis suggests that symbionts assimilate nitrogenous waste (ammonium) of the host into valuable, organic compounds, which then are translocated back to the host (Figure 8C Symbiont nitrogen assimilation) (Lewis and Smith, 1971; Muscatine and Porter, 1977; Falkowski et al., 1993; Wang and Douglas, 1998). Our observation that in symbiotic green hydra many genes involved in amino acid metabolism are down-regulated (Figure 1E) is consistent with the assumption of reduction of amino acid consumption by respiration.
In addition to the nitrogen recycling hypothesis, it has been proposed that also corals, sea anemones, Paramecium and green hydra hosts can assimilate ammonium into amino acids (Figure 8B,C Host nitrogen assimilation) (Miller and Yellowlees, 1989; Rees, 1989; Szmant et al., 1990; Rees, 1991; Wang and Douglas, 1998; Lipschultz and Cook, 2002). Ammonia assimilation by the host implies that the host controls the nitrogen status to regulate metabolism of the symbionts, which may be involved in controlling the number of symbionts within the host cell. For organisms such as corals living in oligotrophic sea, inorganic nitrogen assimilation and recycling may be necessary to manage the nitrogen sources efficiently. In contrast, for Hydra and Paramecium living in a relatively nutrient-rich environment may be advantageous in terms of metabolic efficiency that the symbiont abandons its ability to assimilate inorganic nitrogen and specializes in the supply of photosynthetic carbohydrate to the host.
Genome evolution in symbiotic Chlorella sp. A99
Metabolic dependence of symbionts on host supply occasionally results in genome reduction and gene loss. For example, symbiotic Buchnera bacteria in insects are missing particular genes in essential amino acid pathways (Shigenobu et al., 2000; Hansen and Moran, 2011). The fact that the corresponding genes of the host are up-regulated in the bacteriocyte, indicates complementarity and syntrophy between host and symbiont. Similarly, in Chlorella A99 the nitrogen assimilation system could have been lost as a result of continuous supply of nitrogenous amino acids provided by Hydra.
Compared to Chlorella NC64A, the closest relative to Chlorella A99 among the genome-sequenced algae, genome size and total number of genes in Chlorella A99 were found to be smaller (Figure 4B). Although both A99 and NC64A cannot be cultivated using inorganic nitrogen sources (Figure 7) (Kamako et al., 2005), NC64A, unlike A99, obtains all major nitrogen assimilation genes and their cluster structure on the chromosome (Figure 6) (Sanz-Luque et al., 2015). NR and NiR activities were found to be induced by nitrate in free-living Chlorella, but not in Chlorella NC64A, indicating mutations in the regulatory region (Kamako et al., 2005). Considering the phylogenetic position of NC64A and the symbiotic Chlorella of green hydra (Kawaida et al., 2013), the disability of nitrate assimilation in A99 and NC64A seems to have evolved independently, suggesting convergent evolution in a similar symbiotic environment.
Although our findings indicate that genome reduction in Chlorella A99 is more advanced than in Chlorella NC64A, genome size and total number of genes do not differ much between the Trebouxiophyceae (A99, NC64A and C169) (Figure 4B). By contrast, the parasitic algae Helicosporidium and Auxochlorella have significantly smaller genome sizes and number of genes indicating extensive genome reduction (Gao et al., 2014; Pombert et al., 2014). The apparently unchanged complexity of the Chlorella A99 genome suggests a relatively early stage of this symbiotic partnership. Thus, gene loss in metabolic pathways could occur as a first step of genome reduction in symbionts caused by the adaptation to continuous nutrient supply from the host. Taken together, our study suggests metabolic-codependency as the primary driving force in the evolution of symbiosis between Hydra and Chlorella.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Hydra viridissima A99) |
Hydra viridissima A99 | PMID: 16351895 | ||
| Strain, strain background (Chlorella sp. A99) |
Chlorella sp. A99 | PMID: 16351895 | NCBI BioProject ID: PRJNA412448 |
|
| Strain, strain background (Chlorella variabilis NC64A) |
Chlorella variabilis NC64A | Microbial Culture Collection at the National Institute for Environmental Studies |
NIES-2541 | |
| Strain, strain background (Coccomyxa subellipsoidea C-169) |
Coccomyxa subellipsoidea
C-169 |
Microbial Culture Collection at the National Institute for Environmental Studies |
NIES-2166 | |
| Strain, strain background (Chlamydomonas reinhardtii) |
Chlamydomonas reinhardtii | Microbial Culture Collection at the National Institute for Environmental Studies |
NIES-2235 | |
| Commercial assay or kit | TruSeq DNA LT Sample Prep Kit |
Illumina | FC-121–2001 | |
| Commercial assay or kit | Nextera Mate Pair Sample Preparation Kit |
Illumina | FC-132–1001 | |
| Commercial assay or kit | Miseq reagent kit v3 | Illumina | MS-102–3003 | |
| Commercial assay or kit | HiSeq SBS kit v4 | Illumina | FC-401–4003 | |
| Commercial assay or kit | BigDye Terminator v3.1 Cycle Sequencing Kit |
Thermo Fisher Scientific | 4337454 | |
| Commercial assay or kit | 4 × 44K Hydra viridissima A99 Custom-Made Microarray |
Agilent Technologies | NCBI GEO Platform ID: GPL23280 |
|
| Commercial assay or kit | GE Hybridization Kit and GE Wash Pack |
Agilent Technologies | 5188–5242, 5188–5327 | |
| Commercial assay or kit | High Sensitivity DNA Kit | Agilent Technologies | 5067–4626 | |
| Commercial assay or kit | RNA6000 nano kit | Agilent Technologies | 5067–1511 | |
| Commercial assay or kit | Low Input Quick Amp Labeling Kit |
Agilent Technologies | 5190–2305 | |
| Commercial assay or kit | PureLink RNA Mini Kit | Thermo Fisher Scientific | 12183018A | |
| Commercial assay or kit | Fermentas First Strand cDNA Synthesis Kit |
Thermo Fisher Scientific | K1621 | |
| Chemical compound, drug |
Trizol reagent | Thermo Fisher Scientific | 15596026 | |
| Chemical compound, drug |
AmpliTaq Gold 360 Master Mix |
Thermo Fisher Scientific | 4398901 | |
| Chemical compound, drug |
ISOPLANT II | Nippon Gene | 316–04153 | |
| Chemical compound, drug |
GoTaq qPCR Master Mix | Promega | A6002 | |
| Chemical compound, drug |
KOD FX Neo | TOYOBO | KFX-201 | |
| Software, algorithm | Feature Extraction Software |
Agilent Technologies | RRID:SCR_014963 | |
| Software, algorithm | Newbler | 454 Life Sciences, Roche Diagnostics |
RRID:SCR_011916 | |
| Software, algorithm | SSPACE | PMID: 21149342 | RRID:SCR_005056 | |
| Software, algorithm | GapCloser | PMID: 23587118 | RRID:SCR_015026 | |
| Software, algorithm | NCBI BLAST | PMID: 2231712 | RRID:SCR_004870 | |
| Software, algorithm | CEGMA | PMID: 17332020 | RRID:SCR_015055 | |
| Software, algorithm | Augustus: Gene Prediction | PMID: 16845043 | RRID:SCR_008417 | |
| Software, algorithm | Blast2GO | PMID: 16081474 | RRID:SCR_005828 | |
| Software, algorithm | Hmmer | PMID: 9918945 | RRID:SCR_005305 | |
| Software, algorithm | CLUSTALX2 | PMID: 17846036 | RRID:SCR_002909 | |
| Software, algorithm | BioEdit | Nucleic Acid Symposium Series 41, 95–98 |
RRID:SCR_007361 | |
| Software, algorithm | Njplot | Biochimie 78, 364–369 | NA | |
| Software, algorithm | OrthoFinder | PMID: 26243257 | NA |
Biological materials and procedures
Experiments were carried out with the Australian Hydra viridissima strain A99, which was obtained from Dr. Richard Campbell, Irvine. Polyps were maintained at 18°C on a 12 hr light/dark cycle and fed with Artemia two or three times a week. Aposymbiotic (algae free) polyps were obtained by photobleaching using 5 μM DCMU (3-(3,4-dichlorophenyl)−1,1-dimethylurea) as described before (Pardy, 1976; Habetha et al., 2003). Experiments were carried out with polyps starved for 3–6 days. Isolation of endodermal layer and ectodermal layer was performed as described by Kishimoto et al. (Kishimoto et al., 1996). Symbiotic Chlorella were isolated as described before by Muscatine and McAuley (Muscatine, 1983; McAuley, 1986). Chlorella variabilis NC64A (NIES-2541), Coccomyxa subellipsoidea C-169 (NIES-2166) and Chlamydomonas reinhardtii (NIES-2235) were obtained from the Microbial Culture Collection at the National Institute for Environmental Studies (Tsukuba, Japan).
Nucleic acid preparation
Total RNA of Hydra was extracted by use of the Trizol reagent and PureLink RNA Mini Kit (Thermo Fisher Scientific) after lysis and removal of algae by centrifugation. The genomic DNA of green algae was extracted using ISOPLANT II (Nippon Gene, Tokyo, Japan) following DNase I treatment to degrade contaminant DNA. Quantity and quality of DNA and RNA were checked by NanoDrop (Thermo Scientific Inc., Madison, USA) and BioAnalyzer (Agilent Technologies, Santa Clara, USA).
Microarray analysis
Total RNA for synthesis of cRNA targets was extracted from about 100 green hydra for each experimental group. Experiments were carried out using three biological replicates. cRNA labeled with cyanine-3 were synthesized from 400 ng total Hydra RNA using a Low Input Quick Amp Labeling Kit for one color detection (Agilent Technologies). A set of fluorescently labeled cRNA targets was employed in a hybridization reaction with 4 × 44K Custom-Made Hydra viridissima Microarray (Agilent Technologies) contributing a total of 43,222 transcripts that was built by mRNA-seq data (NCBI GEO Platform ID: GPL23280) (Bosch et al., 2009). Hybridization and washing were performed using the GE Hybridization Kit and GE Wash Pack (Agilent Technologies) after which the arrays were scanned on an Agilent Technologies G2565BA microarray scanner system with SureScan technology following protocols according to the manufacturer's instructions. The intensity of probes was extracted from scanned microarray images using Feature Extraction 10.7 software (Agilent Technologies). All algorithms and parameters used in this analysis were used with default conditions. Background-subtracted signal-intensity values (gProcessedSignal) generated by the Feature Extraction software were normalized using the 75th percentile signal intensity among the microarray. Those genes differentially expressed between two samples were determined by average of fold change (cut of >2.0) and Student's t-test (p<0.1). The data series are accessible at NCBI GEO under accession number GSE97633.
Quantitative real time RT-PCR
Total RNA was extracted from 50 green hydra polyps for each biological replicate independently. For reverse transcription of total RNA First Strand cDNA Synthesis Kit (Fermentas, Ontario, Canada) was used. Real-time PCR was performed using GoTaq qPCR Master Mix (Promega, Madison, USA) and ABI Prism 7300 (Applied Biosystems, Foster City, USA). All qPCR experiments were performed in duplicate with three biological replicates each. Values were normalized using the expression of the tubulin alpha gene. Primers used for these experiments are listed in Supplementary file 6A.
Whole mount in situ hybridization
Expression patterns of specific Hydra genes were detected by whole mount in situ hybridization with digoxigenin (DIG)-labelled RNA probes. Specimens were fixed in 4% paraformaldehyde. Hybridization signal was visualized using anti-DIG antibodies conjugated to alkaline phosphatase and NBT/BCIP staining solution (Roche). DIG-labeled sense probes (targeting the same sequences as the antisense probes) were used as a control. Primers used for these experiments are listed in Supplementary file 6B.
Genome sequencing and gene prediction
For genome sequencing of Chlorella sp. A99, Chlorella sp. A99 was isolated from H. viridissima A99 and genomic DNA was extracted. Paired-end library (insert size: 740 bp) and mate-pair libraries (insert size: 2.2 and 15.2 kb) were made using Illumina TruSeq DNA LT Sample Prep Kit and Nextera Mate Pair Sample Preparation Kit respectively (Illumina Inc., San Diego, USA), following the manufacturer's protocols. Genome sequencing was performed using Illumina Miseq and Hiseq 2000 platforms. Sequence reads were assembled using Newbler Assembler version 2.8 (Roche, Penzberg, Germany) and subsequent scaffolding was performed by SSPACE (Boetzer et al., 2011). Gaps inside the scaffolds were closed with the paired-end and mate-pair data using GapCloser of Short Oligonucleotide Analysis Package (Luo et al., 2012). To overcome potential assembly errors arising from tandem repeats, sequences that aligned to another sequence by more than 50% of the length using blastn (1e-50) were removed from the assembly. The completeness of the genome was evaluated using CEGMA v2.4 (Core Eukaryotic Genes Mapping Approach) based on mapping of the 248 most highly conserved core eukaryotic genes (CEGs) on the assembled genome (Parra et al., 2007). The completeness of complete and partial CEGs in the A99 scaffolds was 80 and 88%, respectively. The fraction of repetitive sequences was 12%. Gene model was predicted by AUGUSTUS 3.0.1 using model parameters for NC64A (Stanke et al., 2006). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession PCFQ00000000 (BioProject ID: PRJNA412448). Genome sequences and gene models are also accessible at the website of OIST Marine Genomics Unit Genome Project (http://marinegenomics.oist.jp/chlorellaA99/viewer/info?project_id=65).
Analysis of genes in Hydra viridissima and Chlorella
Annotation of transcriptome contigs and prediction of gene models was performed by use of BLAST, Gene Ontology (Ashburner et al., 2000) and blast2go (Conesa et al., 2005). To examine the conservation of H. viridissima contigs among metazoans, homology searches by blastx (evalue 1E-5) were performed using protein databases obtained from NCBI for Drosophila melanogaster and Homo sapiens, from the JGI genome portal (http://genome.jgi.doe.gov/) for Branchiostoma floridae, Nematostella vectensis, from Echinobase (http://www.echinobase.org/EchinoBase/) for Strongylocentrotus pupuratus, from Compagen for Hydra magnipapillata, and from the OIST marine genomics Genome browser ver.1.1 (http://marinegenomics.oist.jp/coral/viewer/info?project_id=3) for Acropora digitifera.
For comparative analysis of gene models of Chlorella sp. A99 and other algae, domain searches against the Pfam database (Pfam-A.hmm) were performed using HMMER (Eddy, 1998; Finn et al., 2016), and ortholog gene grouping was done using OrthoFinder (Emms and Kelly, 2015). The sequences of the reference genes and genomes were obtained from the database of the JGI genome portal for Chlorella variabilis NC64A, Coccomyxa subellipsoidea C-169, Volvox carteri, Micromonas pusilla, and Ostreococcus tauri, from NCBI for Auxenochlorella protothecoides 0710, from Phytozome (http://phytozome.jgi.doe.gov/pz/portal.html) for Chlamydomonas reinhardtii, from OIST Marine Genomics (http://marinegenomics.oist.jp/symb/viewer/info?project_id=21) for Symbiodinium minutum, Symbiodinium kawagutti genome, from Dinoflagellate Resources (http://web.malab.cn/symka_new/) for Symbiodinium kawagutti and Reefgenomics (http://reefgenomics.org/) for Symbiodinium microadriaticum) (Merchant et al., 2007; Palenik et al., 2007; Worden et al., 2009; Blanc et al., 2010; Prochnik et al., 2010; Blanc et al., 2012)
Nitrogen assimilation genes in Chlorella A99 were identified by orthologous gene groups and reciprocal blast searches. The number of genes for nitrate assimilation genes, glutamine synthetase and glutamate synthetase, and clustering of such genes were systematically reported by (Sanz-Luque et al., 2015). We used these data as reference for searches of nitrogen assimilation genes, and further nitrogen assimilation genes were searched by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (Kanehisa and Goto, 2000). JGI genome browsers of Chlorella variabilis NC64A and Coccomyxa subellipsoidea C-169 were also used for retrieving genes and checking gene order on the scaffolds.
Phylogenetic analysis
For a phylogenetic tree of chlorophyte green algae, the sequences of 18S rRNA gene, ITS1, 5.8S rRNA gene, ITS2 and 28S rRNA gene were obtained from scaffold20 of Chlorella A99 genome sequence, and from NCBI nucleotide database entries for Chlorella variabilis NC64A (FM205849.1), Auxenochlorella protothecoides 0710 (NW_011934479.1), Coccomyxa subellipsoidea C169 (AGSI01000011.1), Volvox carteri f. nagariensis (NW_003307662.1), Chlamydomonas reinhardtii (FR865576.1), Ostreococcus tauri (GQ426340.1) and Micromonas pusilla (FN562452.1). Multiple alignments were produced with CLUSTALX (2.1) with gap trimming (Larkin et al., 2007). Sequences of poor quality that did not well align were deleted using BioEdit (Hall, 1999). Phylogenetic analyses were performed using the Neighbor-Joining method by CLUSTALX with the default parameters (1000 bootstrap tests and 111 seeds). Representative phylogenetic trees were drawn by using NJ plot (Perrière and Gouy, 1996).
PCR amplification of nitrate assimilation genes in green algae
Primers were designed based on the conserved region of the NRT2 gene, NiR and NR genes (positive control) identified by comparison of genes from Chlorella variabilis NC64A (NC64A), Coccomyxa subellipsoidea C169 (C169), and Chlamydomonas reinhardtii (Cr) which belongs to Chlorophyceae class of green algae. Primers for NAR2 could not be designed because of insufficient conservation. As positive controls, amplicons were produced for NR of all the green algae examined and of NRT2 and NiR from NC64A, C169 and Cr, after which their sequences were checked. KOD FX Neo (TOYOBO, Tokyo, Japan) was used under the following conditions: an initial denaturation phase (94°C for 120 s) followed by 36 cycles of (98°C for 30 s, 69°C for 100 s) for NiR, (98°C for 30 s, 58°C for 30 s and 68°C for 210 s) for NRT2 and (98°C for 30 s, 59°C for 30 s and 68°C for 60 s) for NR. In each case, 10 ng gDNA was used as a template. The primers used are described in Supplementary file 6C. PCR products were sequenced to confirm amplification of the target genes using ABI PRISM 3100 Genetic Analyzer (Thermo Fisher Scientific Inc., Madison, USA) using BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific).
In vitro culture of algae
To isolate symbiotic algae, polyps were quickly homogenized in 0.25% sodium dodecyl sulfate (SDS) solution and centrifuged at 3000 g for 1 min. The pellet was resuspended in 0.05% SDS and centrifuged at 500 g for 5 min. Isolated A99, NC64A and C169 were washed by sterilized Bold Basal Medium (Bischoff and Bold, 1963) modified by the addition of 0.5% glucose, 1.2 mg/L vitamine B1 (Thiaminhydrochloride), 0.01 mg/L vitamine B12 (Cyanocobalamin) (Supplementary file 7) and incubated for two days in modified Bold Basal Medium with 50 mg/l ampicillin and streptomycin. The algae were cultivated in 5 ml of modified Bold Basal Medium (BBM) with the same amount of nitrogen (2.9 mM NaNO3, NH4Cl, glutamine or 426 mg/l casamino acids) and 5 mg/l Carbendazim (anti-fungal) with fluorescent illumination (12 hr light, 12 hr dark) at 20˚C. Mean numbers of algae per ml were calculated from three tubes enumerated at 4, 8, and 12 days after inoculation with 106 cell/sml using a hemocytometer. After cultivation, gDNA was isolated from the A99 cultured in Gln-containing BBM and casamino acid-containing BBM and A99 was isolated from green hydra directly. A partial genomic region of the 18S rRNA gene was amplified by PCR and sequenced to confirm absence of contamination by other algae. PCR was performed using AmpliTaq Gold (Thermo Fisher Scientific). Sequencing was performed as described above. The primers used are described in Supplementary file 6D.
Acknowledgements
We thank Trudy Wassenaar for critical reading the text and for discussion. We also thank the DNA Sequencing Section, IT Section and Kanako Hisata in the Okinawa Institute of Science and Technology (OIST) for excellent technical support. The computations for this work were partially performed on the NIG supercomputer at the ROIS National Institute of Genetics. We are thankful to Angela Douglas for sustained exchanges and discussions on symbiosis in hydra and two anonymous referees for their constructive comments on a previous version of this manuscript. This work was supported in part by JSPS KAKENHI Grant-in-Aid for Young Scientists (B) 25840132 and Scientific Research (C) 15K07173 to MH. and by the Deutsche Forschungsgemeinschaft (DFG) (CRC1182 ‘Origin and Function of Metaorganisms’). TCGB. gratefully appreciates support from the Canadian Institute for Advanced Research (CIFAR).
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Thomas CG Bosch, Email: tbosch@zoologie.uni-kiel.de.
Paul G Falkowski, Rutgers University, United States.
Funding Information
This paper was supported by the following grants:
Japan Society for the Promotion of Science Young Scientists (B) 25840132 to Mayuko Hamada.
Japan Society for the Promotion of Science Scientific Research (C) 15K07173 to Mayuko Hamada.
Deutsche Forschungsgemeinschaft CRC1182 to Thomas CG Bosch.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Funding acquisition, Investigation, Visualization, Writing—original draft.
Formal analysis, Investigation, Visualization, Writing—review and editing.
Formal analysis, Investigation, Writing—review and editing.
Formal analysis, Investigation.
Resources, Writing—review and editing.
Investigation, Writing—review and editing, Library preparation and genome sequencing.
Formal analysis, Writing—review and editing.
Formal analysis, Writing—review and editing.
Supervision, Project administration, Writing—review and editing.
Conceptualization, Supervision, Funding acquisition, Writing—original draft, Writing—review and editing.
Additional files
Gene expression of green hydra with native symbiotic Chlorella A99 (Hv_Sym), that in sexual phase (Hv_Sym_sexy), aposymbiotic polyps from which symbiotic Chlorella were removed (Hv_Apo) and aposymbiotic polyps reinfected with Chlorella variabilis NC64A (Hv_NC64A) were compared.
(B) Blast best hit genes of Arabidopsis thaliana in Chlorella sp. A99 genes belonging to OG0000040 and OG0000324.
Data availability
Microarray information and the data series are accessible at NCBI GEO under accession number GPL23280 and GSE97633 respectively. All the results of microarray analysis are included in Supplementary Table 1. The Whole Genome Shotgun project of Chlorella sp. A99 has been deposited at DDBJ/ENA/GenBank under the accession PCFQ00000000 (BioProject ID: PRJNA412448). Genome sequences and gene models are also accessible at the website of OIST Marine Genomics Unit Genome Project (http://marinegenomics.oist.jp/chlorellaA99/viewer/info?project_id=65). All data generated by qPCR are included in Source Data: Figure2, Figure2 - Figure supplement 1, Source Data: Figure3, Source Data: Figure3 - Figure Supplement 2 and Source Data: Table 2, Table 4
The following datasets were generated:
Mayuko Hamada, author. Chlorella sp. A99 genome sequence and gene models. 2018 http://marinegenomics.oist.jp/chlorellaA99/viewer/info?project_id=65 Publicly available at OIST Marine Genomics Unit (Chlorella sp. A99)
Mayuko Hamada, author. Chlorella sp. A99 genome sequence. 2018 http://www.ncbi.nlm.nih.gov/bioproject/412448 Publicly available at NCBI BioProject (Accession no: PCFQ00000000)
Fraune S, author; Bosch TC, author. Agilent-029560 Hydra viridissima transcriptome-based custom microarray. 2017 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL23280 Publicly available at the NCBI Gene Expression Omnibus (accession no: GPL23280)
Fraune S, author; Kürn U, author; Bosch TC, author. Identification of genes involved in symbiosis of green hydra and Chlorella. 2017 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97633 Publicly available at the NCBI Gene Expression Omnibus (accession no: GSE97633)
The following previously published datasets were used:
Blanc G, author; Duncan G, author; Agarkova I, author; Borodovsky M, author; Gurnon J, author; Kuo A, author; Lindquist E, author; Lucas S, author; Pangilinan J, author; Polle J, author; Salamov A, author; Terry A, author; Yamada T, author; Dunigan DD, author; Grigoriev IV, author; Claverie JM, author; Van Etten JL, author. The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. 2010 https://genome.jgi.doe.gov/ChlNC64A_1/ChlNC64A_1.home.html Publicly available at JGI MycoCosm (Chlorella variabilis NC64A)
Blanc G, author; Agarkova I, author; Grimwood J, author; Kuo A, author; Brueggeman A, author; Dunigan DD, author; Gurnon J, author; Ladunga I, author; Lindquist E, author; Lucas S, author; Pangilinan J, author; Proschold T, author; Salamov A, author; Schmutz J, author; Weeks D, author; Yamada T, author; Lomsadze A, author; Borodovsky M, author; Claverie JM, author; Grigoriev IV, author; Van Etten JL, author. The genome of the polar eukaryotic microalga Coccomyxa subellipsoidea reveals traits of cold adaptation. 2012 https://genome.jgi.doe.gov/Coc_C169_1/Coc_C169_1.home.html Publicly available at JGI MycoCosm (Coccomyxa subellipsoidea C-169 v2.0)
Prochnik SE, author; Umen J, author; Nedelcu AM, author; Hallmann A, author; Miller SM, author; Nishii I, author; Ferris P, author; Kuo A, author; Mitros T, author; Fritz-Laylin LK, author; Hellsten U, author; Chapman J, author; Simakov O, author; Rensing SA, author; Terry A, author; Pangilinan J, author; Kapitonov V, author; Jurka J, author; Salamov A, author; Shapiro H, author; Schmutz J, author; Grimwood J, author; Lindquist E, author; Lucas S, author; Grigoriev IV, author; Schmitt R, author; Kirk D, author; Rokhsar DS, author. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. 2010 https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias=Org_Vcarteri Publicly available at JGI Phytozome (Volvox carteri v2.1)
Merchant SS, author; Prochnik SE, author; Vallon O, author; Harris EH, author; Karpowicz SJ, author; Witman, author; GB, author; Terry A, author; Salamov, author; A, author; Fritz-Laylin LK, author; Marechal-Drouard L, author; Marshall WF, author; Qu LH, author; Nelson DR, author; Sanderfoot AA, author; Spalding MH, author; Kapitonov VV, author; Ren Q, author; Ferris P, author; Lindquist E, author; Shapiro H, author; Lucas SM, author; Grimwood J, author; Schmutz J, author; Cardol P, author; Cerutti H, author; Chanfreau G, author; Chen CL, author; Cognat V, author; Croft MT, author; Dent R, author; Dutcher S, author; Fernandez E, author; Fukuzawa H, author; Gonzalez-Ballester D, author; Gonzalez-Halphen D, author; Hallmann A, author; Hanikenne M, author; Hippler M, author; Inwood W, author; Jabbari K, author; Kalanon M, author; Kuras R, author; Lefebvre PA, author; Lemaire SD, author; Lobanov AV, author; Lohr M, author; Manuell A, author; Meier I, author; Mets L, author; Mittag M, author; Mittelmeier T, author; Moroney JV, author; Moseley J, author; Napoli C, author; Nedelcu AM, author; Niyogi K, author; Novoselov SV, author; Paulsen IT, author; Pazour G, author; Purton S, author; Ral JP, author; Riano-Pachon DM, author; Riekhof W, author; Rymarquis L, author; Schroda M, author; Stern D, author; Umen J, author; Willows R, author; Wilson N, author; Zimmer SL, author; Allmer J, author; Balk J, author; Bisova K, author; Chen CJ, author; Elias M, author; Gendler K, author; Hauser C, author; Lamb MR, author; Ledford H, author; Long JC, author; Minagawa J, author; Page MD, author; Pan J, author; Pootakham W, author; Roje S, author; Rose A, author; Stahlberg E, author; Terauchi AM, author; Yang P, author; Ball S, author; Bowler C, author; Dieckmann CL, author; Gladyshev VN, author; Green P, author; Jorgensen R, author; Mayfield S, author; Mueller-Roeber B, author; Rajamani S, author; Sayre RT, author; Brokstein P, author; Dubchak I, author; Goodstein D, author; Hornick L, author; Huang YW, author; Jhaveri J, author; Luo Y, author; Martinez D, author; Ngau WC, author; Otillar B, author; Poliakov A, author; Porter A, author; Szajkowski L, author; Werner G, author; Zhou, author; K, author; Grigoriev IV, author; Rokhsar DS, author; Grossman AR, author. The Chlamydomonas genome reveals the evolution of key animal and plant functions. 2007 http://phytozome.jgi.doe.gov/pz/portal.html Publicly available at JGI Phytozome (Chlamydomonas reinhardtii v5.5)
Worden AZ, author; Lee JH, author; Mock T, author; Rouze P, author; Simmons MP, author; Aerts AL, author; Allen AE, author; Cuvelier ML, author; Derelle E, author; Everett MV, author; Foulon E, author; Grimwood J, author; Gundlach H, author; Henrissat B, author; Napoli C, author; McDonald SM, author; Parker MS, author; Rombauts S, author; Salamov A, author; Von Dassow P, author; Badger JH, author; Coutinho PM, author; Demir E, author; Dubchak I, author; Gentemann C, author; Eikrem W, author; Gready JE, author; John U, author; Lanier W, author; Lindquist EA, author; Lucas S, author; Mayer KF, author; Moreau H, author; Not F, author; Otillar R, author; Panaud O, author; Pangilinan J, author; Paulsen I, author; Piegu B, author; Poliakov A, author; Robbens S, author; Schmutz J, author; Toulza E, author; Wyss T, author; Zelensky A, author; Zhou K, author; Armbrust EV, author; Bhattacharya D, author; Goodenough UW, author; Van de Peer Y, author; Grigoriev IV, author. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. 2009 https://genome.jgi.doe.gov/MicpuC2/MicpuC2.home.html Publicly available at JGI MycoCosm (Micromonas pusilla CCMP1545)
Palenik B, author; Grimwood J, author; Aerts A, author; Rouze P, author; Salamov A, author; Putnam N, author; Dupont C, author; Jorgensen R, author; Derelle E, author; Rombauts S, author; Zhou K, author; Otillar R, author; Merchant SS, author; Podell S, author; Gaasterland T, author; Napoli C, author; Gendler K, author; Manuell A, author; Tai V, author; Vallon O, author; Piganeau G, author; Jancek S, author; Heijde M, author; Jabbari K, author; Bowler C, author; Lohr M, author; Robbens S, author; Werner G, author; Dubchak I, author; Pazour GJ, author; Ren Q, author; Paulsen I, author; Delwiche C, author; Schmutz J, author; Rokhsar D, author; Van de Peer Y, author; Moreau H, author; Grigoriev IV, author. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. 2007 https://genome.jgi.doe.gov/Ostta4221_3/Ostta4221_3.home.html Publicly available at JGI MycoCosm (Ostreococcus tauri RCC4221 v3.0)
Finn RD, author; Coggill P, author; Eberhardt RY, author; Eddy SR, author; Mistry J, author; Mitchell AL, author; Potter SC, author; Punta M, author; Qureshi M, author; Sangrador-Vegas A, author; Salazar GA, author; Tate J, author; Bateman A, author. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279-285. 10.1093/nar/gkv1344. 2016 ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam31.0/ Available to download at the ftp site. (dataset name: Pfam-A.hmm)
References
- Aranda M, Li Y, Liew YJ, Baumgarten S, Simakov O, Wilson MC, Piel J, Ashoor H, Bougouffa S, Bajic VB, Ryu T, Ravasi T, Bayer T, Micklem G, Kim H, Bhak J, LaJeunesse TC, Voolstra CR. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Scientific Reports. 2016;6:39734. doi: 10.1038/srep39734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barott KL, Venn AA, Perez SO, Tambutté S, Tresguerres M. Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. PNAS. 2015;112:607–612. doi: 10.1073/pnas.1413483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, Simakov O, Esherick LY, Liew YJ, Lehnert EM, Michell CT, Li Y, Hambleton EA, Guse A, Oates ME, Gough J, Weis VM, Aranda M, Pringle JR, Voolstra CR. The genome of Aiptasia, a sea anemone model for coral symbiosis. PNAS. 2015;112:11893–11898. doi: 10.1073/pnas.1513318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff HW, Bold HC. Some Soil Algae From Enchanted Rock and Related Algal Species. Austin: University of Texas; 1963. [Google Scholar]
- Blanc G, Agarkova I, Grimwood J, Kuo A, Brueggeman A, Dunigan DD, Gurnon J, Ladunga I, Lindquist E, Lucas S, Pangilinan J, Pröschold T, Salamov A, Schmutz J, Weeks D, Yamada T, Lomsadze A, Borodovsky M, Claverie JM, Grigoriev IV, Van Etten JL. The genome of the polar eukaryotic microalga coccomyxa subellipsoidea reveals traits of cold adaptation. Genome Biology. 2012;13:R39. doi: 10.1186/gb-2012-13-5-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Duncan G, Agarkova I, Borodovsky M, Gurnon J, Kuo A, Lindquist E, Lucas S, Pangilinan J, Polle J, Salamov A, Terry A, Yamada T, Dunigan DD, Grigoriev IV, Claverie JM, Van Etten JL. The chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. The Plant Cell. 2010;22:2943–2955. doi: 10.1105/tpc.110.076406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- Bosch TC, Augustin R, Anton-Erxleben F, Fraune S, Hemmrich G, Zill H, Rosenstiel P, Jacobs G, Schreiber S, Leippe M, Stanisak M, Grötzinger J, Jung S, Podschun R, Bartels J, Harder J, Schröder JM. Uncovering the evolutionary history of innate immunity: the simple metazoan Hydra uses epithelial cells for host defence. Developmental & Comparative Immunology. 2009;33:559–569. doi: 10.1016/j.dci.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Bossert P, Dunn KW. Regulation of intracellular algae by various strains of the symbiotic Hydra viridissima. Journal of Cell Science. 1986;85:187–195. doi: 10.1242/jcs.85.1.187. [DOI] [PubMed] [Google Scholar]
- Brown JA, Nielsen PJ. Transfer of photosynthetically produced carbohydrate from endosymbiotic chlorellae to Paramecium bursaria. The Journal of Protozoology. 1974;21:569–570. doi: 10.1111/j.1550-7408.1974.tb03702.x. [DOI] [PubMed] [Google Scholar]
- Brown SB, Maloney M, Kinlaw WB. "Spot 14" protein functions at the pretranslational level in the regulation of hepatic metabolism by thyroid hormone and glucose. Journal of Biological Chemistry. 1997;272:2163–2166. doi: 10.1074/jbc.272.4.2163. [DOI] [PubMed] [Google Scholar]
- Campbell RD. Transmission of symbiotic algae through sexual reproduction in hydra: movement of algae into the oocyte. Tissue and Cell. 1990;22:137–147. doi: 10.1016/0040-8166(90)90017-4. [DOI] [PubMed] [Google Scholar]
- Cernichiari E, Muscatine L, Smith DC. Maltose excretion by the symbiotic algae of Hydra viridis. Proceedings of the Royal Society B: Biological Sciences. 1969;173:557–576. doi: 10.1098/rspb.1969.0077. [DOI] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Cook CB, Kelty MO. Glycogen, protein, and lipid content of green, aposymbiotic, and nonsymbiotic hydra during starvation. Journal of Experimental Zoology. 1982;222:1–9. doi: 10.1002/jez.1402220102. [DOI] [Google Scholar]
- Császár NBM, Seneca FO, van Oppen MJH. Variation in antioxidant gene expression in the scleractinian coral Acropora millepora under laboratory thermal stress. Marine Ecology Progress Series. 2009;392:93–102. doi: 10.3354/meps08194. [DOI] [Google Scholar]
- Davy SK, Allemand D, Weis VM. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiology and Molecular Biology Reviews. 2012;76:229–261. doi: 10.1128/MMBR.05014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorling M, McAuley PJ, Hodge H. Effect of pH on growth and carbon metabolism of maltose-releasing Chlorella (Chlorophyta) European Journal of Phycology. 1997;32:19–24. doi: 10.1080/09541449710001719335. [DOI] [Google Scholar]
- Douglas A, Smith DC. The green hydra symbiosis. VIII. mechanisms in symbiont regulation. Proceedings of the Royal Society B: Biological Sciences. 1984;221:291–319. doi: 10.1098/rspb.1984.0035. [DOI] [Google Scholar]
- Douglas AE, Smith DC. The Cost of Symbionts to the Host in the Green Hydra Symbiosis. Berlin: W. DeGriiyter and Co; 1983. [Google Scholar]
- Douglas AE. Symbiotic Interactions. Oxford; New York: Oxford University Press; 1994. [Google Scholar]
- Eddy SR. Profile hidden markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowski PG, Dubinsky Z, Muscatine L, McCloskey L. Population control in symbiotic corals. BioScience. 1993;43:606–611. doi: 10.2307/1312147. [DOI] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. The pfam protein families database: towards a more sustainable future. Nucleic Acids Research. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Moya A, Magnone V, Allemand D, Furla P, Sabourault C. Adaptations to endosymbiosis in a cnidarian-dinoflagellate association: differential gene expression and specific gene duplications. PLoS Genetics. 2011;7:e1002187. doi: 10.1371/journal.pgen.1002187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Wang Y, Shen Y, Yan D, He X, Dai J, Wu Q. Oil accumulation mechanisms of the oleaginous microalga chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics. 2014;15:582. doi: 10.1186/1471-2164-15-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso LC, Maindonald J, Rudd S, Hayward DC, Saint R, Miller DJ, Ball EE. Microarray analysis identifies candidate genes for key roles in coral development. BMC Genomics. 2008;9:540. doi: 10.1186/1471-2164-9-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover R, Maguer J-F, Allemand D, Ferrier-Pagés C. Nitrate uptake in the scleractinian coral Stylophora pistillata. Limnology and Oceanography. 2003;48:2266–2274. doi: 10.4319/lo.2003.48.6.2266. [DOI] [Google Scholar]
- Habetha M, Anton-Erxleben F, Neumann K, Bosch TC. The Hydra viridis/Chlorella symbiosis. growth and sexual differentiation in polyps without symbionts. Zoology. 2003;106:101–108. doi: 10.1078/0944-2006-00104. [DOI] [PubMed] [Google Scholar]
- Habetha M, Bosch TC. Symbiotic Hydra express a plant-like peroxidase gene during oogenesis. Journal of Experimental Biology. 2005;208:2157–2165. doi: 10.1242/jeb.01571. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acid Symposium Series. 1999;41:95–98. [Google Scholar]
- Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proceedings of the National Academy of Sciences. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors, microbial recognition and immunity. Cellular Microbiology. 2014;16:185–194. doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huss VAR, Holweg C, Seidel B, Reich V, Rahat M, Kessler E. There is an ecological basis for host/symbiont specificity in chlorella/Hydra symbioses. Endocytobiosis and Cell Research. 1994;10:35–46. [Google Scholar]
- Ishikawa M, Yuyama I, Shimizu H, Nozawa M, Ikeo K, Gojobori T. Different endosymbiotic interactions in two Hydra species reflect the evolutionary history of endosymbiosis. Genome Biology and Evolution. 2016;8:2155–2163. doi: 10.1093/gbe/evw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joy JB. Symbiosis catalyses niche expansion and diversification. Proceedings of the Royal Society B: Biological Sciences. 2013;280:20122820. doi: 10.1098/rspb.2012.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamako S, Imamura N. Effect of Japanese paramecium bursaria extract on photosynthetic carbon fixation of symbiotic algae. The Journal of Eukaryotic Microbiology. 2006;53:136–141. doi: 10.1111/j.1550-7408.2005.00084.x. [DOI] [PubMed] [Google Scholar]
- Kamako S-ichiro, Hoshina R, Ueno S, Imamura N. Establishment of axenic endosymbiotic strains of Japanese Paramecium bursaria and the utilization of carbohydrate and nitrogen compounds by the isolated algae. European Journal of Protistology. 2005;41:193–202. doi: 10.1016/j.ejop.2005.04.001. [DOI] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapaun E, Reisser W. A chitin-like glycan in the cell wall of a chlorella sp. (Chlorococcales, chlorophyceae) Planta. 1995;197:577–582. doi: 10.1007/BF00191563. [DOI] [Google Scholar]
- Karakashian SJ, Karakashian MW. Evolution and symbiosis in the genus Chlorella and related algae. Evolution. 1965;19:368–377. doi: 10.1111/j.1558-5646.1965.tb01728.x. [DOI] [Google Scholar]
- Kawaida H, Ohba K, Koutake Y, Shimizu H, Tachida H, Kobayakawa Y. Symbiosis between Hydra and chlorella: molecular phylogenetic analysis and experimental study provide insight into its origin and evolution. Molecular Phylogenetics and Evolution. 2013;66:906–914. doi: 10.1016/j.ympev.2012.11.018. [DOI] [PubMed] [Google Scholar]
- Kellogg RB, Patton JS. Lipid droplets, medium of energy exchange in the symbiotic Anemone condylactis gigantea: a model coral polyp. Marine Biology. 1983;75:137–149. doi: 10.1007/BF00405996. [DOI] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. More than just orphans: are taxonomically-restricted genes important in evolution? Trends in Genetics. 2009;25:404–413. doi: 10.1016/j.tig.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Kishimoto Y, Murate M, Sugiyama T. Hydra regeneration from recombined ectodermal and endodermal tissue. I. Epibolic ectodermal spreading is driven by cell intercalation. Journal of Cell Science. 1996;109:763–772. doi: 10.1242/jcs.109.4.763. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Suzuki H, Dohra H, Sugii M, Kitazume T, Yamaguchi K, Shigenobu S, Fujishima M. Comparison of gene expression of Paramecium bursaria with and without chlorella variabilis symbionts. BMC Genomics. 2014;15:183. doi: 10.1186/1471-2164-15-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince AS, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F. Nitrate transport and signalling in Arabidopsis. Journal of Experimental Botany. 2014;65:789–798. doi: 10.1093/jxb/eru001. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lehnert EM, Mouchka ME, Burriesci MS, Gallo ND, Schwarz JA, Pringle JR. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3: Genes|Genomes|Genetics. 2014;4:277–295. doi: 10.1534/g3.113.009084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Annual Review of Physiology. 2006;68:253–278. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- Lewis DH, Smith DC. The autotrophic nutrition of symbiotic marine coelenterates with special reference to hermatypic corals. I. Movement of photosynthetic products between the symbionts. Proceedings of the Royal Society B: Biological Sciences. 1971;178:111–129. doi: 10.1098/rspb.1971.0055. [DOI] [Google Scholar]
- Liaw SH, Kuo I, Eisenberg D. Discovery of the ammonium substrate site on glutamine synthetase, a third cation binding site. Protein Science. 1995;4:2358–2365. doi: 10.1002/pro.5560041114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KL, Wang JT, Fang LS. Participation of glycoproteins on zooxanthellal cell walls in the establishment of a symbiotic relationship with the Sea Anemone, aiptasia pulchella. Zoological Studies. 2000;39:172–178. [Google Scholar]
- Lin S, Cheng S, Song B, Zhong X, Lin X, Li W, Li L, Zhang Y, Zhang H, Ji Z, Cai M, Zhuang Y, Shi X, Lin L, Wang L, Wang Z, Liu X, Yu S, Zeng P, Hao H, Zou Q, Chen C, Li Y, Wang Y, Xu C, Meng S, Xu X, Wang J, Yang H, Campbell DA, Sturm NR, Dagenais-Bellefeuille S, Morse D. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science. 2015;350:691–694. doi: 10.1126/science.aad0408. [DOI] [PubMed] [Google Scholar]
- Lipschultz F, Cook CB. Uptake and assimilation of 15N-ammonium by the symbiotic sea anemones Bartholomea annulata and Aiptasia pallida: conservation versus recycling of nitrogen. Marine Biology. 2002;140:489–502. doi: 10.1007/s00227-001-0717-1. [DOI] [Google Scholar]
- Lucas WJ, Berry JA. Inorganic carbon transport in aquatic photosynthetic organisms. Physiologia Plantarum. 1985;65:539–543. doi: 10.1111/j.1399-3054.1985.tb08687.x. [DOI] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y, Tang J, Wu G, Zhang H, Shi Y, Liu Y, Yu C, Wang B, Lu Y, Han C, Cheung DW, Yiu S-M, Peng S, Xiaoqian Z, Liu G, Liao X, Li Y, Yang H, Wang J, Lam T-W, Wang J. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez DE, Iñiguez AR, Percell KM, Willner JB, Signorovitch J, Campbell RD. Phylogeny and biogeography of Hydra (Cnidaria: Hydridae) using mitochondrial and nuclear DNA sequences. Molecular Phylogenetics and Evolution. 2010;57:403–410. doi: 10.1016/j.ympev.2010.06.016. [DOI] [PubMed] [Google Scholar]
- Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley PJ, Smith DC. The green Hydra symbiosis. V. Stages in the intracellular recognition of algal symbionts by digestive cells. Proceedings of the Royal Society B: Biological Sciences. 1982;216:7–23. doi: 10.1098/rspb.1982.0058. [DOI] [Google Scholar]
- McAuley PJ. Isolation of viable uncontaminated chlorella from green hydra1. Limnology and Oceanography. 1986;31:222–224. doi: 10.4319/lo.1986.31.1.0222. [DOI] [Google Scholar]
- McAuley PJ. The cell cycle of symbiotic Chlorella. III. Numbers of algae in green hydra digestive cells are regulated at digestive cell division. Journal of cell science. 1986a;85:63–71. doi: 10.1242/jcs.85.1.63. [DOI] [PubMed] [Google Scholar]
- McAuley PJ. Nitrogen limitation and amino-acid metabolism of Chlorella symbiotic with green hydra. Planta. 1987a;171:532–538. doi: 10.1007/BF00392303. [DOI] [PubMed] [Google Scholar]
- McAuley PJ. Quantitative estimation of movement of an amino acid from host to Chlorella symbionts in green hydra. The Biological Bulletin. 1987b;173:504–512. doi: 10.2307/1541696. [DOI] [PubMed] [Google Scholar]
- McAuley PJ. Amino acids as a nitrogen source for Chlorella symbiotic with green hydra. In: Williams R. B, Cornelius P. F. S, Hughes R. G, Robson E. A, editors. Coelenterate Biology: Recent Research on Cnidaria and Ctenophora: Proceedings of the Fifth International Conference on Coelenterate Biology, 1989. Dordrecht: Springer Netherlands; 1991. pp. 369–376. [DOI] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. Animals in a bacterial world, a new imperative for the life sciences. PNAS. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meints RH, Pardy RL. Quantitative demonstration of cell surface involvement in a plant-animal symbiosis: lectin inhibition of reassociation. Journal of Cell Science. 1980;43:239–251. doi: 10.1242/jcs.43.1.239. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernández E, Fukuzawa H, González-Ballester D, González-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riaño-Pachón DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martínez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mews LK, Smith DC. The green Hydra symbiosis. VI. What is the role of maltose transfer from alga to animal? proceedings of the royal society of London. Series B: Biological Sciences. 1982;216:397–413. doi: 10.1098/rspb.1982.0083. [DOI] [Google Scholar]
- Mews LK. The green hydra symbiosis. III. The biotrophic transport of carbohydrate from alga to animal. Proceedings of the Royal Society B: Biological Sciences. 1980;209:377–401. doi: 10.1098/rspb.1980.0101. [DOI] [Google Scholar]
- Meyer E, Weis VM. Study of cnidarian-algal symbiosis in the "omics" age. The Biological Bulletin. 2012;223:44–65. doi: 10.1086/BBLv223n1p44. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Yellowlees D. Inorganic nitrogen uptake by symbiotic marine cnidarians: a critical review. Proceedings of the Royal Society B: Biological Sciences. 1989;237:109–125. doi: 10.1098/rspb.1989.0040. [DOI] [Google Scholar]
- Mohamed AR, Cumbo V, Harii S, Shinzato C, Chan CX, Ragan MA, Bourne DG, Willis BL, Ball EE, Satoh N, Miller DJ. The transcriptomic response of the coral Acropora digitifera to a competent Symbiodinium strain: the symbiosome as an arrested early phagosome. Molecular Ecology. 2016;25:3127–3141. doi: 10.1111/mec.13659. [DOI] [PubMed] [Google Scholar]
- Montalbetti N, Cantero MR, Dalghi MG, Cantiello HF. Reactive oxygen species inhibit polycystin-2 (TRPP2) cation channel activity in term human syncytiotrophoblast. Placenta. 2008;29:510–518. doi: 10.1016/j.placenta.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Moran NA. Symbiosis as an adaptive process and source of phenotypic complexity. PNAS. 2007;104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murer H, Biber J. Molecular mechanisms of renal apical na/phosphate cotransport. Annual Review of Physiology. 1996;58:607–618. doi: 10.1146/annurev.ph.58.030196.003135. [DOI] [PubMed] [Google Scholar]
- Muscatine L, Cernichiari E. Assimilation of photosynthetic products of zooxanthellae by a reef coral. The Biological Bulletin. 1969;137:506–523. doi: 10.2307/1540172. [DOI] [PubMed] [Google Scholar]
- Muscatine L, Lenhoff HM. Symbiosis: on the role of algae symbiotic with Hydra. Science. 1963;142:956–958. doi: 10.1126/science.142.3594.956. [DOI] [PubMed] [Google Scholar]
- Muscatine L, Lenhoff HM. Symbiosis of Hydra and algae. I. Effects of some environmental cations on growth of symbiotic and aposymbiotic Hydra. The Biological Bulletin. 1965a;128:415–424. doi: 10.2307/1539903. [DOI] [Google Scholar]
- Muscatine L, Lenhoff HM. Symbiosis of Hydra and algae. II. Effects of limited food and starvation on growth of symbiotic and aposymbiotic Hydra. The Biological Bulletin. 1965b;129:316–328. doi: 10.2307/1539848. [DOI] [Google Scholar]
- Muscatine L, McAuley PJ. Transmission of symbiotic algae to eggs of green Hydra. Cytobios. 1982;33:111–124. [PubMed] [Google Scholar]
- Muscatine L, Porter JW. Reef corals: mutualistic symbioses adapted to Nutrient-Poor environments. BioScience. 1977;27:454–460. doi: 10.2307/1297526. [DOI] [Google Scholar]
- Muscatine L. Symbiosis of Hydra and algae—III. Extracellular products of the algae. Comparative Biochemistry and Physiology. 1965;16:77–92. doi: 10.1016/0010-406X(65)90165-9. [DOI] [PubMed] [Google Scholar]
- Muscatine L. Hydra: Research Methods. New York: Plenum Press; 1983. [Google Scholar]
- Nichols HW, Bold HC. Trichosarcina polymorpha Gen. et sp. nov. Journal of Phycology. 1965;1:34–38. doi: 10.1111/j.1529-8817.1965.tb04552.x. [DOI] [Google Scholar]
- Ochman H, Moran NA. Genes lost and genes found: evolution of bacterial pathogenesis and symbiosis. Science. 2001;292:1096–1099. doi: 10.1126/science.1058543. [DOI] [PubMed] [Google Scholar]
- Palenik B, Grimwood J, Aerts A, Rouzé P, Salamov A, Putnam N, Dupont C, Jorgensen R, Derelle E, Rombauts S, Zhou K, Otillar R, Merchant SS, Podell S, Gaasterland T, Napoli C, Gendler K, Manuell A, Tai V, Vallon O, Piganeau G, Jancek S, Heijde M, Jabbari K, Bowler C, Lohr M, Robbens S, Werner G, Dubchak I, Pazour GJ, Ren Q, Paulsen I, Delwiche C, Schmutz J, Rokhsar D, Van de Peer Y, Moreau H, Grigoriev IV. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. PNAS. 2007;104:7705–7710. doi: 10.1073/pnas.0611046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardy RL. The morphology of green Hydra endosymbionts as influenced by host strain and host environment. Journal of Cell Science. 1976;20:655–669. doi: 10.1242/jcs.20.3.655. [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- Perrière G, Gouy M. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie. 1996;78:364–369. doi: 10.1016/0300-9084(96)84768-7. [DOI] [PubMed] [Google Scholar]
- Pombert JF, Blouin NA, Lane C, Boucias D, Keeling PJ. A lack of parasitic reduction in the obligate parasitic green alga Helicosporidium. PLoS Genetics. 2014;10:e1004355. doi: 10.1371/journal.pgen.1004355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, Hellsten U, Chapman J, Simakov O, Rensing SA, Terry A, Pangilinan J, Kapitonov V, Jurka J, Salamov A, Shapiro H, Schmutz J, Grimwood J, Lindquist E, Lucas S, Grigoriev IV, Schmitt R, Kirk D, Rokhsar DS. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science. 2010;329:223–226. doi: 10.1126/science.1188800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada A, Galván A, Fernández E. Identification of nitrate transporter genes in Chlamydomonas reinhardtii. The Plant Journal. 1994;5:407–419. doi: 10.1111/j.1365-313X.1994.00407.x. [DOI] [PubMed] [Google Scholar]
- Rahat M, Reich V. Intracellular infection of aposymbiotic Hydra viridis by a foreign free-living chlorella sp.: initiation of a stable symbiosis. Journal of Cell Science. 1984;65:265–277. doi: 10.1242/jcs.65.1.265. [DOI] [PubMed] [Google Scholar]
- Rees TAV, Ellard FM. Nitrogen conservation and the green Hydra symbiosis. Proceedings of the Royal Society B: Biological Sciences. 1989;236:203–212. doi: 10.1098/rspb.1989.0021. [DOI] [Google Scholar]
- Rees TAV. The green Hydra symbiosis and ammonium I. the role of the host in ammonium assimilation and its possible regulatory significance. Proceedings of the Royal Society B: Biological Sciences. 1986;229:299–314. doi: 10.1098/rspb.1986.0087. [DOI] [Google Scholar]
- Rees TAV. The green Hydra symbiosis and Ammonium II. ammonium assimilation and release by freshly isolated symbionts and cultured algae. Proceedings of the Royal Society B: Biological Sciences. 1989;235:365–382. doi: 10.1098/rspb.1989.0005. [DOI] [Google Scholar]
- Rees TAV. Are symbiotic algae nutrient deficient? Proceedings of the Royal Society B: Biological Sciences. 1991;243:227–233. doi: 10.1098/rspb.1991.0036. [DOI] [Google Scholar]
- Richier S, Furla P, Plantivaux A, Merle PL, Allemand D. Symbiosis-induced adaptation to oxidative stress. Journal of Experimental Biology. 2005;208:277–285. doi: 10.1242/jeb.01368. [DOI] [PubMed] [Google Scholar]
- Roffman B, Lenhoff HM. Formation of polysaccharides by Hydra from substrates produced by their endosymbiotic algae. Nature. 1969;221:381–382. doi: 10.1038/221381a0. [DOI] [PubMed] [Google Scholar]
- Sanz-Luque E, Chamizo-Ampudia A, Llamas A, Galvan A, Fernandez E. Understanding nitrate assimilation and its regulation in microalgae. Frontiers in Plant Science. 2015;6:899. doi: 10.3389/fpls.2015.00899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz JA, Brokstein PB, Voolstra C, Terry AY, Manohar CF, Miller DJ, Szmant AM, Coffroth MA, Medina M. Coral life history and symbiosis: functional genomic resources for two reef building caribbean corals, Acropora palmata and Montastraea faveolata. BMC Genomics. 2008;9:97. doi: 10.1186/1471-2164-9-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwentner M, Bosch TC. Revisiting the age, evolutionary history and species level diversity of the genus Hydra (Cnidaria: hydrozoa) Molecular Phylogenetics and Evolution. 2015;91:41–55. doi: 10.1016/j.ympev.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–87. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Shinzato C, Inoue M, Kusakabe M. A snapshot of a coral "holobiont": a transcriptome assembly of the scleractinian coral, porites, captures a wide variety of genes from both the host and symbiotic zooxanthellae. PLoS ONE. 2014;9:e85182. doi: 10.1371/journal.pone.0085182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzato C, Shoguchi E, Kawashima T, Hamada M, Hisata K, Tanaka M, Fujie M, Fujiwara M, Koyanagi R, Ikuta T, Fujiyama A, Miller DJ, Satoh N. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature. 2011;476:320–323. doi: 10.1038/nature10249. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, Takeuchi T, Hisata K, Tanaka M, Fujiwara M, Hamada M, Seidi A, Fujie M, Usami T, Goto H, Yamasaki S, Arakaki N, Suzuki Y, Sugano S, Toyoda A, Kuroki Y, Fujiyama A, Medina M, Coffroth MA, Bhattacharya D, Satoh N. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Current Biology. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- Sproles AE, Kirk NL, Kitchen SA, Oakley CA, Grossman AR, Weis VM, Davy SK. Phylogenetic characterization of transporter proteins in the cnidarian-dinoflagellate symbiosis. Molecular Phylogenetics and Evolution. 2018;120:307–320. doi: 10.1016/j.ympev.2017.12.007. [DOI] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Research. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, Wilson EC, Thaler M, Smith ML, Caruso C, Pringle JR, Weis VM, Medina M, Schwarz JA. Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics. 2009;10:258. doi: 10.1186/1471-2164-10-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmant AM, Ferrer LM, FitzGerald LM. Nitrogen excretion and O:n ratios in reef corals: evidence for conservation of nitrogen. Marine Biology. 1990;104:119–127. doi: 10.1007/BF01313165. [DOI] [Google Scholar]
- Tanaka Y, Miyajima T, Koike I, Hayashibara T, Ogawa H. Translocation and conservation of organic nitrogen within the coral-zooxanthella symbiotic system of Acropora pulchra, as demonstrated by dual isotope-labeling techniques. Journal of Experimental Marine Biology and Ecology. 2006;336:110–119. doi: 10.1016/j.jembe.2006.04.011. [DOI] [Google Scholar]
- Tao TY, Towle HC. Coordinate regulation of rat liver genes by thyroid hormone and dietary carbohydrate. Annals of the New York Academy of Sciences. 1986;478:20–30. doi: 10.1111/j.1749-6632.1986.tb15518.x. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Lošo T. The evolutionary origin of orphan genes. Nature Reviews Genetics. 2011;12:692–702. doi: 10.1038/nrg3053. [DOI] [PubMed] [Google Scholar]
- Thorington G, Margulis L. Hydra viridis: transfer of metabolites between Hydra and symbiotic algae. The Biological Bulletin. 1981;160:175–188. doi: 10.2307/1540911. [DOI] [PubMed] [Google Scholar]
- Trench RK. The physiology and biochemistry of zooxanthellae symbiotic with marine coelenterates. I. The assimilation of photosynthetic products of zooxanthellae by two marine coelenterates. Proceedings of the Royal Society B: Biological Sciences. 1971;177:225–235. doi: 10.1098/rspb.1971.0024. [DOI] [Google Scholar]
- Vandermeulen JH, Davis ND, Muscatine L. The effect of inhibitors of photosynthesis on zooxanthellae in corals and other marine invertebrates. Marine Biology. 1972;16:185–191. doi: 10.1007/BF00346940. [DOI] [Google Scholar]
- Wang J, Douglas AE. Nitrogen recycling or nitrogen conservation in an alga-invertebrate symbiosis? The Journal of Experimental Biology. 1998;201:2445–2453. doi: 10.1242/jeb.201.16.2445. [DOI] [PubMed] [Google Scholar]
- Weis VM, Smith GJ, Muscatine L. A ?CO2 supply? mechanism in Zooxanthellate cnidarians: role of carbonic anhydrase. Marine Biology. 1989;100:195–202. doi: 10.1007/BF00391958. [DOI] [Google Scholar]
- Weis VM. Cellular mechanisms of cnidarian bleaching: stress causes the collapse of symbiosis. Journal of Experimental Biology. 2008;211:3059–3066. doi: 10.1242/jeb.009597. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. Endosymbiosis. Current Biology. 2012;22:R555–R561. doi: 10.1016/j.cub.2012.06.010. [DOI] [PubMed] [Google Scholar]
- Wilkinson CR. Nutrient translocation from green algal symbionts to the freshwater sponge Ephydatia fluviatilis. Hydrobiologia. 1980;75:241–250. doi: 10.1007/BF00006488. [DOI] [Google Scholar]
- Wood-Charlson EM, Hollingsworth LL, Krupp DA, Weis VM. Lectin/glycan interactions play a role in recognition in a coral/dinoflagellate symbiosis. Cellular Microbiology. 2006;8:1985–1993. doi: 10.1111/j.1462-5822.2006.00765.x. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, Allen AE, Cuvelier ML, Derelle E, Everett MV, Foulon E, Grimwood J, Gundlach H, Henrissat B, Napoli C, McDonald SM, Parker MS, Rombauts S, Salamov A, Von Dassow P, Badger JH, Coutinho PM, Demir E, Dubchak I, Gentemann C, Eikrem W, Gready JE, John U, Lanier W, Lindquist EA, Lucas S, Mayer KF, Moreau H, Not F, Otillar R, Panaud O, Pangilinan J, Paulsen I, Piegu B, Poliakov A, Robbens S, Schmutz J, Toulza E, Wyss T, Zelensky A, Zhou K, Armbrust EV, Bhattacharya D, Goodenough UW, Van de Peer Y, Grigoriev IV. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
- Yellowlees D, Rees TA, Leggat W. Metabolic interactions between algal symbionts and invertebrate hosts. Plant, Cell & Environment. 2008;31:679–694. doi: 10.1111/j.1365-3040.2008.01802.x. [DOI] [PubMed] [Google Scholar]