Abstract
DNA photolyases catalyze the blue light-dependent repair of UV light-induced damage in DNA. DNA photolyases are specific for either cyclobutane-type pyrimidine dimers or (6–4) photoproducts. PHR2 is a gene that in Chlamydomonas reinhardtii encodes a class II DNA photolyase which catalyzes the photorepair of cyclobutane-type pyrimidine dimers. Based on amino acid sequence analysis of PHR2, which indicates the presence of a chloroplast targeting sequence, PHR2 was predicted to encode the chloroplast photolyase of Chlamydomonas. Using a sensitive gene-specific in vivo repair assay, we found that overexpression of PHR2 in Chlamydomonas results in targeting of the protein to not only the chloroplast, but also to the nucleus. Overexpression of PHR2 photolyase in a photoreactivation-deficient mutant, phr1, results in a largely inactive product. The phr1 mutant was found to be deficient in both photorepair of a chloroplast gene, rbcL, and a nuclear gene, rDNA. These results suggest that PHR2 is the structural gene for the photolyase targeted to both the chloroplast and the nucleus, and that the PHR1 gene product is necessary for full activity of PHR2 protein. To our knowledge, the requirement for a second gene for full activity of a DNA photolyase is novel.
INTRODUCTION
Cyclobutane pyrimidine dimers (CPDs) and (6–4) photoproducts are the two most prevalent forms of DNA damage caused by UV light. Cells have developed a line of defense against these UV-induced lesions, named photoreactivation. During photoreactivation, enzymes known as DNA photolyases use blue light energy to reverse CPDs or (6–4) photoproducts directly (1–4). The DNA photolyases have been divided into two classes based on amino acid sequence. The class I photolyases were discovered first and have thus been characterized in more detail (3,5). Some class I photolyases are specific for (6–4) photoproducts, whereas others are specific for CPDs (6). In contrast, all class II DNA photolyases analyzed to date are CPD specific (5). The amino acid sequence of class II DNA photolyases is distantly related to the class I photolyases (7,8), and both have been shown to bind two chromophores. All photolyases bind FAD, which serves as the catalytic chromophore during photoreactivation. The second chromophore is responsible for harvesting energy from photoreactivating light and can either be 8-hydroxy-5-deazaflavin (8-HDF) or 5,10-methenyltetrahydrofolate (MTHF) (5).
The unicellular alga Chlamydomonas reinhardtii has been shown to have photolyase activity in both the chloroplast and the nucleus (9). This is in contrast to another model plant, Arabidopsis thaliana, which has been shown to photorepair only nuclear DNA (10). We have previously described the isolation of a photoreactivation-deficient mutant of Chlamydomonas, phr1, which was reported to be severely deficient in nuclear photoreactivation and apparently normal in photoreactivation of chloroplast DNA (11). More recently, we have reported the cloning and characterization of PHR2, a gene encoding a class II DNA photolyase in Chlamydomonas (12). Unexpectedly, the phr1 mutation did not map to the PHR2 locus. We also showed that PHR2 mRNA levels were approximately equal between wild-type and phr1 cells, ruling out a role of the PHR1 gene product in transcription of PHR2. Since phr1 appeared to be deficient in only nuclear photoreactivation and did not map to the PHR2 locus, we proposed that there were two photolyase genes in Chlamydomonas, one for repair of nuclear lesions and one for repair of chloroplast lesions. Amino acid sequence analysis of the PHR2 protein suggested the presence of a chloroplast targeting sequence. This information led us to propose that PHR2 encodes the chloroplast photolyase of Chlamydomonas (12).
Here we report that overexpression of PHR2 in Chlamydomonas results in an increased ability to photoreactivate DNA, not only in the chloroplast, but also in the nucleus. Utilizing a sensitive gene-specific in vivo repair assay we also report that phr1 appears to be photoreactivation deficient, not only in the nucleus, but also in the chloroplast. Finally, we show that overexpression of PHR2 in a phr1 background results in only partially active PHR2. That full activity of PHR2 is dependent on the function of a second gene product is a novel discovery and, to date, unprecedented for DNA photolyases.
MATERIALS AND METHODS
Chlamydomonas strains and culture conditions
The phr1 strain of Chlamydomonas was isolated in our laboratory following N-methyl-N′-nitro-N-nitrosoguanidine mutagenesis of strain 137C (mt+) (11). Strains G7 (PHR1), A3 (phr1) and D7 (PHR1) each contain the PHR2 overexpression construct HSP PHR2 Myc-His. G7 (PHR1) and D7 (PHR1) each overexpress PHR2 in a wild-type photoreactivation background, which is annotated here as (PHR1). A3 (phr1) overexpresses PHR2 in the photoreactivation-deficient background of phr1, annotated as (phr1). Strain G7 (PHR1) was generated by transformation of a cw15arg7 strain of Chlamydomonas with the plasmid HSP PHR Myc-His. The A3 (phr1) strain is the progeny of a cross between phr1 and G7 (PHR1). The D7 (PHR1) strain is the progeny of a cross between A3 (phr1) and the wild-type cc-124 (mt–). Strains were grown on Tris–acetate–phosphate (TAP) medium (13) supplemented with 80 mg/l arginine where required.
Construction of the HSP PHR2 Myc-His overexpression construct
In order to overexpress PHR2 in Chlamydomonas, the PHR2 gene (GenBank accession no. AF129458) was placed under control of the HSP70A–RBCS2 chimeric promoter. The plasmid pCB 745 (a generous gift of M. Schroda and C. F. Beck, Institute for Biology III, University of Freiburg, Freiburg, Germany) contains the HSP70A–RBCS2 promoter, which allows heat shock-inducible expression of genes under its control (14). To simplify engineering the PHR2 overexpression construct, the ∼0.5 kb XbaI–HindIII fragment of pCB 745 containing the HPS70A–RBCS2 fusion promoter was moved into pGEM-7Zf+ (Promega) and then transferred into pBluescript SK+ (Stratagene) using XbaI and BamHI, forming pBS HSP XB.
Additionally, the PHR2 gene was modified by first inserting a Myc-His tag in-frame at the C-terminus of the PHR2 coding region. This was accomplished using two oligonucleotides, MHSEN (ACGAGGAGCAGAAGCTGATCTCGGAGGAGGACCTGAACAGCGCCGTGGACCACCACCACCACCACCACTAGTAGAC) and MH-NON (CGGTCTACTAGTGGTGGTGGTGGTGGTGGTCCACGGCGCTGTTCAGGTCCTCCTCCGAGATCAGCTTCTGCTCCTCGTCA), which when hybridized together encoded the Myc-His tag utilizing Chlamydomonas codon usage preference. The double-stranded Myc-His oligonucleotide was then inserted into PHR2 utilizing a BseRI site located 7 nt upstream of the PHR2 termination codon and an AccI site located within the PHR2 termination codon. This construct was sequenced to verify that the Myc-His tag was accurate. To facilitate the cloning of PHR2 Myc-His into pBS HSP XB, PCR was performed at the 5′-end of PHR2 to engineer a BamHI site immediately upstream of the PHR2 translation start codon. The PCR product was generated using primers PL49 (CGGGATCCATGTCCAGTAAGCGCAAAGCC) and PL46 (AGTTGTCCGCCAGGCGCTGGT). Following sequencing of the PCR product, it was ligated into PHR2 Myc-His using the engineered BamHI site and a native BglII site, yielding an ∼10 kb BamHI–SalI fragment containing the entire PHR2 Myc-His gene. This ∼10 kb fragment was ligated into pBS HSP XB, forming HSP PHR2 Myc-His.
Transformation of Chlamydomonas
The cw15arg7 strain of Chlamydomonas was transformed with 5 µg HSP PHR2 Myc-His and 1 µg co-transforming DNA, pUC ARG7.8, following standard methods (15). The transformants were grown on TAP plates and screened by PCR to determine those that contained the PHR2 overexpression construct. PCR was performed on transformant genomic DNA using the PHR2-specific primers PL40 (ACACCTCACCACAACCCACCGCCAA) and PL51 (GCGTACGTGGCCTACGTCAGCAA) under the following conditions: the DNA was denatured at 95°C for 3 min, followed by 30 cycles of 95°C for 45 s, 58°C for 45 s and 72°C for 1 min. The reactions were then maintained at 72°C for 7 min. Transformants containing the PHR2 overexpression construct could be identified from those without because the PCR product generated with PL40 and PL51 includes the DNA encoding the Myc-His tag of the overexpression construct, which leads to a PCR product that is 66 bp larger than the product from the endogenous PHR2 gene. Transformants containing the HSP PHR2 Myc-His construct were then subjected to heat shock and anti-Myc western blot analysis to determine those that showed heat shock-inducible overexpression of PHR2 Myc-His. Southern blot analysis was also performed on the genomic DNA of transformants overexpressing PHR2 Myc-His to determine the number of transforming inserts.
Western blot analysis
Western blots were performed following standard protocols (16). Polyclonal rabbit antibodies against Myc and goat anti-rabbit IgG antibodies were purchased from Santa Cruz Biotechnologies. Anti-Myc was diluted 1:2500 and goat anti-rabbit antibodies were diluted 1:2000. Enhanced chemiluminescence (Amersham Pharmacia Biotech) was used for detection.
Southern blot analysis
Southern blots were performed as described previously (12). The HSP PHR2 Myc-His transformant Southern blot was probed with PHR2 5′ PE, a 0.7 kb PstI–BglII fragment from the 5′-end of PHR2.
Chlamydomonas cellular extracts
Cultures of Chlamydomonas were grown to mid log phase. When heat shock was desired the cells were placed at 39°C for 1 h and pelleted immediately afterward by centrifugation. Cells were resuspended at 4 ml/g wet weight in sonication buffer [50 mM Tris–HCl, pH 8.0, 2 mM dithiothreitol, 1 mM EDTA, 300 mM (NH4)2SO4]. After resuspension, the cells were sonicated for 4 × 15 s on ice. The sonicate was centrifuged in a microcentrifuge for 10 min at 4°C. The supernatant was removed and ultracentrifuged at 120 000 g for 1 h at 4°C. After ultracentrifugation, 0.45 g/ml (NH4)2SO4 was added to the supernatant and the mixture was placed on ice for 30 min. The sample was then centrifuged at 9800 g for 15 min at 4°C. The supernatant was discarded and the pellet was resuspended in dialysis buffer (25 mM HEPES, pH 8.5, 40 mM KCl, 1 mM EDTA, 2 mM dithiothreitol, 17% glycerol) and dialyzed for 12–16 h at 4°C. Following dialysis the total protein concentration of the extract was measured by Bio-Rad protein assay.
Gel mobility shift assays
The UV-irradiated DNA probe was created and prepared as described (17). Briefly, two complementary oligonucleotides were synthesized containing a region of eight repeating pyrimidines (CTAGTTTTTTTT and CTAGAAAAAAAA). The oligonucleotides were phosphorylated, annealed, ligated into XbaI-digested pUC18 and transformed into Escherichia coli. Plasmid DNA was isolated from individual transformants and sequenced. The plasmid DNA used throughout this paper, pGDST8-3, contained three strings of the eight consecutive pyrimidines. A HindIII–EcoRI 87 bp fragment was isolated and end-labeled with [α-32P]dATP using Klenow DNA polymerase. The probe was then irradiated with 4 kJ/m2 UV light.
The mobility shift reactions were prepared in the following manner. An aliquot of 7500 c.p.m. UV-irradiated DNA probe was incubated with 10 µg total protein from Chlamydomonas ammonium sulfate cellular extracts in the dark at room temperature for 30 min with 10 mM HEPES, pH 7.4, 200 mM NaCl, 16 mM KCl, 0.8 mM dithiothreitol, 2 µg poly(dI–dC)·poly(dI–dC) and 1.5 µg sonicated salmon sperm DNA. The mixture was then separated on a 5% polyacrylamide gel (19:1) containing 0.5× Tris–borate–EDTA (TBE). The gel was run at 220 V in 0.5× TBE running buffer, dried and exposed to X-ray film.
In vivo repair assays
In vivo repair assays were performed essentially as described (10,18). Cell cultures of Chlamydomonas were grown to late log phase and diluted to an A700 of 0.85. One aliquot of cells was reserved as the no UV control. The cells were placed in glass Petri dishes. In a darkened room, the Petri dishes were placed uncovered under a germicidal UV light (254 nm) with constant agitation for varying lengths of time depending on the assay being performed. Aliquots of the UV-irradiated cells were then dispensed into glass Petri dishes for varying photoreactivation treatments at 27 cm below two cool white fluorescent tubes. DNA was harvested from each cellular aliquot as described earlier. An equivalent amount of DNA from each aliquot, based on ethidium bromide staining, was digested with BamHI and SmaI for chloroplast assays or with BglII for nuclear assays. Following these digestions the buffer of the samples was adjusted to 50 mM Tris–HCl, pH 8.0, and 5 mM EDTA. The samples were divided into two equal volumes. One half was treated with T4 endonuclease (Epicentre) at 37°C for 1 h while the other half was mock-digested.
Immediately following these digestions, alkaline loading buffer was added to each sample to final concentrations of 50 mM NaOH, 1 mM EDTA, 2.5% Ficoll and 0.25% bromocresol purple. The samples were run on 0.4% alkaline agarose gels in 30 mM NaOH and 1 mM EDTA at 22 V at 4°C with constant recirculation of buffer. After electrophoresis the gel was treated for 2 × 15 min in neutralization buffer (1 M Tris–HCl, pH 8.0, and 1.5 M NaCl). The DNA was depurinated, denatured and neutralized using the standard methodology (16). Following these treatments, the DNA was transferred to a nylon membrane and Southern blot analysis was performed following the same procedures as described above. A 3.6 kb BamHI rDNA fragment from plasmid P-92 was used as probe to detect the repair of an 8.7 kb BglII fragment of Chlamydomonas nuclear DNA. Chloroplast DNA repair was measured using a 1.0 kb EcoRV–PstI rbcL fragment from plasmid P-67, which hybridizes to an ∼12 kb BamHI–SmaI fragment of chloroplast DNA or a 1.7 kb EcoRI–PstI atpB fragment from plasmid P-130, which hybridizes to an ∼8 kb BamHI fragment of chloroplast DNA. P-67, P-92 and P-130 were all acquired from the Chlamydomonas Genetics Center (http://www.biology.duke.edu/chlamy/). The number of CPDs per DNA fragment length was calculated as described (10).
Densitometry
All densitometry was performed using a ChemiImager 4000 low light imaging system (Alpha Innotech).
RESULTS
Overexpression of PHR2 in Chlamydomonas
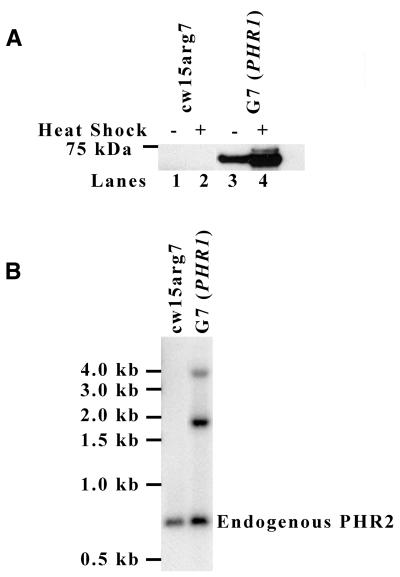
To overexpress PHR2 in Chlamydomonas we generated HSP PHR2 Myc-His, a construct that placed the PHR2 gene, modified to encode a Myc-His tag at the C-terminus of the PHR2 protein, under the control of a heat shock-inducible HSP70A–RBCS2 fusion promoter (14). As shown in Figure 1A, G7 (PHR1), a cell line transformed with the overexpression construct, has a heat shock-inducible protein that is recognized by an anti-Myc antibody and that migrates at ∼67 kDa, the predicted molecular mass of PHR2 Myc-His (Fig. 1A, lanes 3 and 4). A similar protein is not detected in untransformed cells (Fig. 1A, lanes 1 and 2). G7 (PHR1) cells show a significant level of PHR2 Myc-His expression without heat shock (Fig. 1A, lane 3). After heat shock in the G7 (PHR1) strain, a second band slightly larger than the expected 67 kDa appears (Fig. 1A, lane 4). An earlier report indicated that the HSP70A–RBCS2 fusion promoter produced two messages of different sizes following heat shock, indicating an alternative location for transcription initiation within the HSP70A region of the promoter (19). Based on this report, we analyzed the sequence of our PHR2 overexpression construct and located an ATG within the RBCS2 promoter that was in-frame with the PHR2 Myc-His gene. Translation from this AUG would add 58 amino acids to the N-terminus of the PHR2 protein, which is predicted to increase the molecular mass of the protein from 67 to 74 kDa. The migration of the heat shock-inducible second band during SDS–PAGE is consistent with a protein of this molecular mass (Fig. 1A, lane 4). This information leads us to believe that the larger heat shock-inducible protein detected by western blot is the product of an additional translation initiation site.
Figure 1.
Overexpression of PHR2 in Chlamydomonas. (A) Western blot analysis of whole cell lysates from cw15arg7 and G7 (PHR1) cells before and after heat shock. The G7 (PHR1) strain was derived by transformation of cw15arg7 with the overexpression construct HSP PHR2 Myc-His. Equal amounts of protein were separated by SDS–PAGE, transferred to PVDF and blotted with anti-Myc antibody. The position of the 75 kDa marker is indicated on the left. (B) Southern blot analysis of genomic DNA from cw15arg7 and G7 (PHR1) cells. DNA was digested with BglII and PstI and hybridized with radiolabeled PHR2 5′ PE probe. The endogenous PHR2 gene is indicated on the right.
Southern blot analysis of genomic DNA from G7 (PHR1) was performed to determine the number of HSP PHR2 Myc-His inserts. The PHR2-specific probe hybridized with what appeared to be three bands, endogenous PHR2 (∼0.7 kb), a band near 3.8 kb and a more intense band near 1.8 kb (Fig. 1B). Further Southern blot analysis revealed the presence of three HSP PHR2 Myc-His inserts (data not shown), explaining the increased intensity of the band near 1.8 kb.
Overexpressed PHR2 binds UV-damaged DNA in vitro
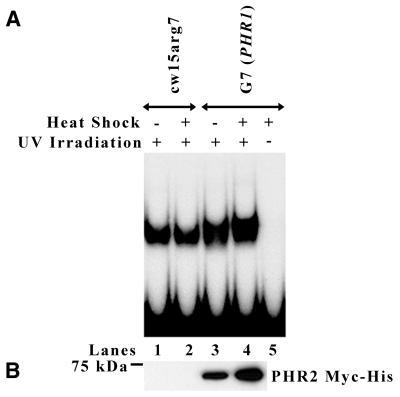
UV-irradiated DNA was used as a probe in gel mobility shift assays to test whether the overexpressed Myc-tagged PHR2 protein from Chlamydomonas could bind UV-irradiated DNA. For this assay it was necessary to use ammonium sulfate extracts of Chlamydomonas because some component of total cell extract inhibits binding of the photolyase to UV-irradiated DNA in vitro. Equivalent amounts of protein from extracts of heat shocked and non-heat shocked wild-type (cw15arg7) and G7 (PHR1) cells were mixed with a UV-irradiated DNA probe and run on a polyacrylamide gel in the dark. Under these conditions, DNA photolyase molecules will bind to the UV-damaged probe and remain bound throughout the experiment, since they cannot catalyze repair of the damage while in the dark. Extracts from both heat shocked and non-heat shocked wild-type cells showed nearly equivalent binding to the UV-irradiated probe (± 15%) (Fig. 2A, lanes 1 and 2). As expected, anti-Myc western blots of extracts from both heat shocked and non-heat shocked wild-type cells did not detect the presence of PHR2 Myc-His, indicating that an endogenous photolyase is responsible for binding UV-irradiated probe in these extracts (Fig. 2B, lanes 1 and 2). In contrast, the extract from non-heat shocked and heat shocked G7 (PHR1) cells bound 35 and 55% more probe, respectively, than extract from heat shocked wild-type cells (Fig. 2A, lanes 2–4). This increased ability to bind UV-irradiated probe by both types of G7 (PHR1) extract corresponds to the presence of PHR2 Myc-His, as determined by anti-Myc western blots (Fig. 2B, lanes 3 and 4). Direct comparison of lanes 1 and 2 with lanes 3 and 4 in Figure 2A shows that there is a slight decrease in mobility of some of the complexes formed using the G7 (PHR1) extracts. This is expected as the PHR2 protein overexpressed in G7 (PHR1) contains a Myc-His tag, leading to a larger protein than endogenous PHR2. When only the complexes with reduced mobility are compared between extracts from non-heat shocked and heat shocked G7 (PHR1) cells, the heat shocked extract binds 50% more probe than the extract from non-heat shocked cells. This difference corresponds to a 90% increase in the level of PHR2 Myc-His in the extract from heat shocked G7 (PHR1) cells versus the extract from non-heat shocked G7 (PHR1) cells, as determined by anti-Myc western blots (Fig. 2B, lanes 3 and 4). Significantly, the ability of overexpressed PHR2 Myc-His to bind DNA required that the DNA contained UV damage (Fig. 2A, lane 5).
Figure 2.
Overexpressed PHR2 Myc-His binds UV-irradiated DNA. (A) Aliquots of 10 µg ammonium sulfate protein extract from heat shocked and non-heat shocked cw15arg7 and G7 (PHR1) cells were incubated in the dark at 25°C for 30 min with radioactively labeled DNA probe that had or had not been UV-irradiated as indicated. The mixtures were then run on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. (B) Western blot analysis of ammonium sulfate protein extracts from cw15arg7 and G7 (PHR1) cells before (lanes 1 and 3) and after (lanes 2 and 4) heat shock. Equal amounts of protein were separated by SDS–PAGE, transferred to PVDF and blotted with anti-Myc antibody. The position of the 75 kDa marker is indicated on the left.
Overexpressed PHR2 repairs pyrimidine dimers in chloroplast and nuclear DNA of Chlamydomonas in vivo
To determine the intracellular localization of overexpressed PHR2, we utilized a gene-specific Southern blot-based in vivo repair assay to measure photoreactivation of both nuclear and chloroplast DNA sequences. Using this assay we were able to show that overexpressed PHR2 does catalyze the light-dependent repair of pyrimidine dimers in chloroplast DNA of Chlamydomonas. Unexpectedly, we also showed that overexpressed PHR2 photoreactivates pyrimidine dimers in the nucleus.
Since the PHR2 Myc-His overexpressing strain, G7 (PHR1), showed a significant level of PHR2 Myc-His expression without heat shock (Fig. 1A, lane 3), it was not necessary to heat shock the cells for any of the following in vivo repair assays. In these assays genomic DNA was isolated from cells following UV exposure and varying amounts of photoreactivation. The DNA was digested with restriction enzymes, divided equally and treated or mock-treated with T4 endonuclease, which cleaves DNA only at pyrimidine dimers. Following alkaline agarose gel electrophoresis, Southern blot analysis was performed using probes specific for either the nucleus (rDNA) or the chloroplast (rbcL). These probes were selected because they are multi-copy genes that provide extra sensitivity for detection by Southern blotting (20,21). Under these conditions DNA samples containing CPDs will appear as a smear on the Southern blot due to frequent T4 endonuclease cleavage, whereas repaired samples will not be cleaved by T4 endonuclease and will appear as a single band when probed. Comparison of the band intensities between samples treated with and without T4 endonuclease after similar light treatments reveals the level of repair. Although the results of these assays are specific for the repair of single genes within an organelle, we consider them representative of the repair occurring within the entire genome of that organelle.
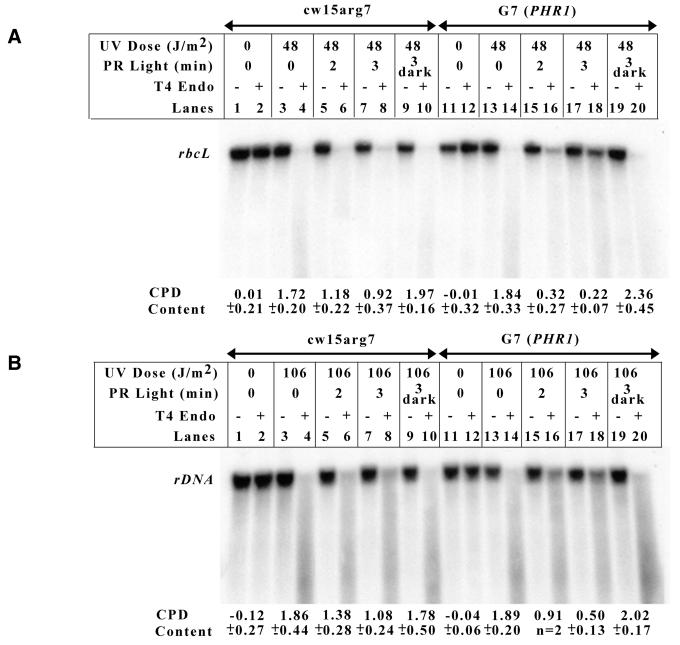
In vivo repair assays were performed comparing photoreactivation in the chloroplasts of G7 (PHR1) cells, which overexpress PHR2 Myc-His, and wild-type cells. Specifically, the assay measured the repair of CPDs in an ∼12 kb fragment of chloroplast DNA containing the Chlamydomonas rbcL gene. Figure 3A shows the Southern blot results from one chloroplast in vivo repair assay and beneath the Southern blot reports the number of pyrimidine dimers remaining in the DNA fragment following each treatment. A direct comparison of wild-type and G7 (PHR1) samples that were UV irradiated and then allowed to photoreactivate for 3 min indicates that the PHR2 overexpressing strain is able to repair more chloroplast DNA damage than the wild-type strain (Fig. 3A, compare lanes 7 and 8 with 17 and 18). Following 3 min photoreactivation the overexpressing strain has reduced the number of pyrimidine dimers by 88%, while the wild-type strain reduced the number by only 47%. Importantly, the number of dimers induced in the DNA of both strains by UV exposure was nearly equivalent (Fig. 3A, compare lanes 3 and 4 with 13 and 14). Also, the number of dimers remaining in the DNA of the dark control for each strain was similar to the number of dimers in the DNA of the strain immediately following UV exposure, which indicates that the repair seen in the photoreactivated samples is a light-dependent process (Fig. 3A, compare lanes 9 and 10 with 3 and 4, and lanes 19 and 20 with 13 and 14).
Figure 3.
Overexpressed PHR2 Myc-His photoreactivates both nuclear and chloroplast DNA in vivo. G7 (PHR1) and cw15arg7 cells were UV-irradiated and then immediately placed in the dark or under cool white fluorescent tubes. Genomic DNA was harvested from samples at the indicated times. (A) Repair in the chloroplast was monitored by digesting genomic DNA with BamHI and SmaI, followed by incubation in the presence or absence of T4 endonuclease. Following alkaline agarose gel electrophoresis, the samples were transferred and Southern blot analysis was performed using a 1.0 kb rbcL DNA fragment. The number of pyrimidine dimers on an ∼12 kb DNA fragment for each treatment is reported beneath the Southern blot as the average (± SD) of three separate experiments. (B) Photoreactivation in the nucleus was measured by digesting genomic DNA with BglII, followed by incubation in the presence or absence of T4 endonuclease. Alkaline agarose gel electrophoresis was performed, followed by Southern blot analysis with a 3.6 kb rDNA probe. The number of pyrimidine dimers on an 8.7 kb DNA fragment for each treatment is reported beneath the Southern blot as the average (± SD) of three separate experiments.
Similar assays were performed to compare the rate of photoreactivation in the nucleus of wild-type cells and the PHR2-overexpressing strain. For these assays, photoreactivation was measured using an 8.7 kb DNA fragment containing the Chlamydomonas rDNA gene. Figure 3B shows the Southern blot results from one experiment and beneath the blot reports the number of pyrimidine dimers in the DNA fragment following each treatment. Again, after 3 min photoreactivation the PHR2-overexpressing strain has repaired more of the damage to nuclear DNA than the wild-type strain (Fig. 3B, compare lanes 7 and 8 with 17 and 18). Following 3 min photoreactivation the overexpressing strain has reduced the number of pyrimidine dimers by 74%, while the wild-type strain has reduced the number by only 42%. As in the chloroplast, the number of dimers induced in the DNA of each strain is nearly the same (Fig. 3B, compare lanes 3 and 4 with lanes 13 and 14). The repair of nuclear DNA is also a light-dependent process, as shown by the lack of repair in samples maintained in the dark (Fig. 3B, lanes 9 and 10, and lanes 19 and 20).
Overexpressed PHR2 Myc-His does not complement the phr1 mutation
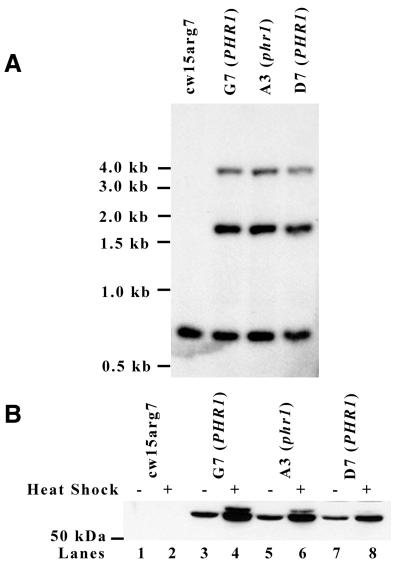
Once it was known that overexpressing PHR2 could increase nuclear photoreactivation in Chlamydomonas, it was of interest whether overexpressing PHR2 could complement the nuclear photoreactivation deficiency of the phr1 mutant. G7 (PHR1) was mated with phr1 to ensure that the context of the PHR2 Myc-His overexpression inserts in the phr1 genome would be the same as in the wild-type genome of G7 (PHR1). Tetrads of the cross were separated and strains derived from individual progeny of the cross were tested for sensitivity to 400 J/m2 UV light under photoreactivating conditions, which is a phenotype of the phr1 mutation. In theory, if PHR2 overexpression complements the phr1 mutation none of the strains from UV-sensitive progeny should overexpress PHR2. PCR analysis indicated that several of the UV-sensitive strains contained HSP PHR2 Myc-His inserts (data not shown). Southern blot analysis of a strain from one such progeny, A3 (phr1), suggested that it contained the same PHR2 overexpression inserts as G7 (PHR1) (Fig. 4A). Anti-Myc western blot analysis indicated that, similar to G7 (PHR1), a protein near 67 kDa was expressed at high levels in A3 (phr1) before and after heat shock (Fig. 4B). These data indicate that PHR2 overexpression does not complement the phr1 photoreactivation deficiency.
Figure 4.
Expression of PHR2 Myc-His in phr1 and wild-type backgrounds. (A) Southern blot analysis of genomic DNA from cw15arg7, G7 (PHR1), A3 (phr1) and D7 (PHR1) cells. The A3 (phr1) strain was generated by mating phr1 with G7 (PHR1). D7 (PHR1) was generated by crossing A3 (phr1) and the wild-type strain, cc-124. DNA was digested with BglII and PstI and hybridized with radiolabeled PHR2 5′ PE probe. The position of DNA markers is indicated on the left. (B) Western blot analysis of whole cell lysates from cw15arg7, G7 (PHR1), A3 (phr1) and D7 (PHR1) cells before and after heat shock as indicated. Equivalent protein amounts were separated by SDS–PAGE, transferred to PVDF and blotted with anti-Myc antibody. The position of the 50 kDa marker is shown on the left.
PHR2 Myc-His overexpressed in the phr1 mutant is largely inactive
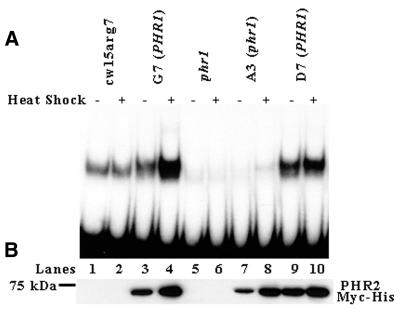
The inability of PHR2 overexpression to complement the phr1 mutation could be because the PHR2 protein is inactive or is not properly targeted to the nucleus and chloroplast. To determine whether PHR2 protein overexpressed in the phr1 background is active, ammonium sulfate extracts were prepared from heat shocked and non-heat shocked cultures of A3 (phr1). These extracts were then used in gel mobility shift assays using UV-irradiated DNA as probe. Anti-Myc western blot analysis indicates that extracts from both G7 (PHR1) and A3 (phr1) heat shocked cells have an increased level of PHR2 Myc-His in comparison with extracts from their non-heat shocked counterparts (Fig. 5B, compare lanes 3 with 4 and 7 with 8). Figure 5B also shows that extracts from heat shocked A3 (phr1) cells have relatively high levels of PHR2 Myc-His [∼80% of the PHR2 Myc-His found in extracts from heat shocked G7 (PHR1) cells] (lanes 4 and 8). In spite of this high level of PHR2 Myc-His, the amount of UV-irradiated probe bound by the extract from heat shocked A3 (phr1) cells (Fig. 5A, lane 8) is markedly less (∼2%) than probe bound by the extract from heat shocked G7 (PHR1) cells (Fig. 5A, lane 4). Remarkably, the amount of probe bound by extract from heat shocked A3 (phr1) cells (Fig. 5A, lane 8) is less than the amount bound by extract from heat shocked wild-type cells (Fig. 5A, lane 2). Also, the reduced ability of extract from heat shocked A3 (phr1) cells to bind UV-irradiated probe (Fig. 5A, lane 8) is similar to extract from heat shocked phr1 cells (Fig. 5A, lane 6), with the exception that the extract from heat shocked A3 (phr1) forms complexes with reduced mobility due to the Myc-His tag on the overexpressed PHR2 protein. This indicates that the overexpressed PHR2 protein in A3 (phr1) is only partially active.
Figure 5.
PHR2 Myc-His overexpressed in a phr1 background is largely inactive. (A) Equivalent amounts of protein from ammonium sulfate extracts of heat shocked and non-heat shocked cw15arg7, G7 (PHR1), phr1, A3 (phr1) and D7 (PHR1) cells were incubated in the dark at 25°C for 30 min with radioactively labeled DNA probe that had been UV irradiated. The mixtures were then run on a 5% non-denaturing polyacrylamide gel and visualized by autoradiography. (B) Western blot analysis of ammonium sulfate protein extracts from cw15arg7, G7 (PHR1), phr1, A3 (phr1) and D7 (PHR1) cells before and after heat shock. Equal amounts of protein was separated by SDS–PAGE, transferred to PVDF and blotted with anti-Myc antibody. The position of the 75 kDa marker is shown on the left.
As a control to determine whether the inactivity of the overexpressed PHR2 protein in A3 (phr1) was due to the phr1 mutation, we backcrossed the overexpression constructs out of the phr1 background into a wild-type strain of Chlamydomonas, cc-124. UV-resistant progeny from this cross containing the PHR2 overexpression construct were isolated. Southern blot analysis of D7 (PHR1), a strain derived from a product of the cross, indicated that it had the same inserts as G7 (PHR1) and A3 (phr1) (Fig. 4A). As in G7 (PHR1) and A3 (phr1), western blot analysis of D7 (PHR1) cells before and after heat shock showed significant levels of PHR2 Myc-His expression (Fig. 4B, lanes 7 and 8). Western blot analysis also indicated that extract from heat shocked D7 (PHR1) cells had high levels of PHR2 Myc-His [∼90% of the PHR2 Myc-His found in extract from heat shocked G7 (PHR1) cells] (Fig. 5B, lanes 4 and 10). Unlike extract from heat shocked A3 (phr1) cells (Fig. 5A, lane 8), extract from heat shocked D7 (PHR1) cells bound large amounts of UV-irradiated probe [70% of the amount bound by extract from heat shocked G7 (PHR1) cells] (Fig. 5A, lanes 4 and 10). These data support the hypothesis that the partial inactivation of the overexpressed PHR2 Myc-His in the phr1 background is due to the phr1 mutation.
The Chlamydomonas mutant phr1 is deficient in pyrimidine dimer repair in both the nucleus and the chloroplast
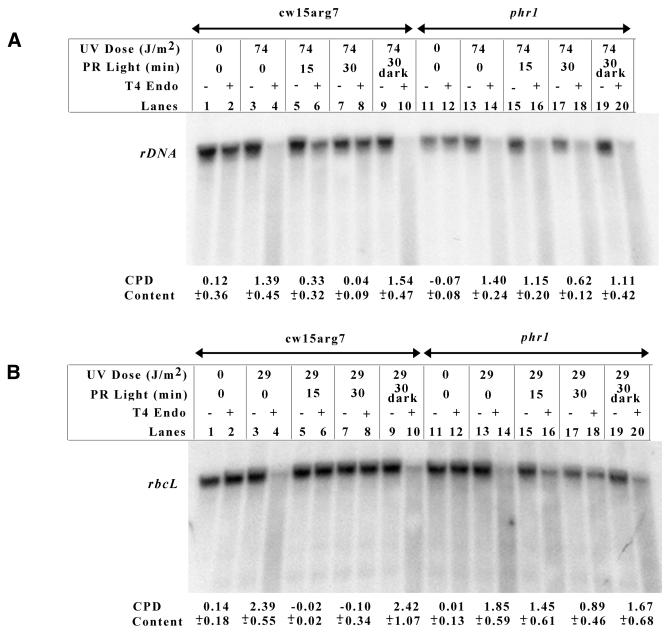
Initial characterization of phr1 indicated that it was severely deficient in photoreactivation of nuclear DNA, but apparently normal in the photorepair of chloroplast DNA (11). Since in vivo repair assays had shown that overexpression of PHR2 Myc-His resulted in an increased ability to photoreactivate in both the chloroplast and the nucleus and once it was shown that PHR2 overexpression could not complement the phr1 photoreactivation deficiency, we decided to re-examine the photoreactivation capabilities of phr1 using the in vivo repair assay. Figure 6A shows one nuclear in vivo repair assay performed with phr1 cells. As before, nuclear photoreactivation was monitored using an 8.7 kb DNA fragment containing the rDNA gene. The number of pyrimidine dimers on the DNA fragment for each treatment is reported beneath the Southern blot. The results from the assay are in agreement with our previous findings that phr1 is severely deficient in the photoreactivation of nuclear DNA when compared to wild-type cells treated under the same conditions (Fig. 6A, compare lanes 7 and 8 with 17 and 18). Following 30 min photoreactivation wild-type cells have repaired 97% of the dimers, while phr1 cells have only repaired 56%. Again, an equivalent amount of DNA damage was induced in each cell type (Fig. 6A, compare lanes 3 and 4 with lanes 13 and 14) and the repair is light dependent (Fig. 6A, lanes 9 and 10, and lanes 19 and 20).
Figure 6.
phr1 cells are deficient in photoreactivating chloroplast and nuclear DNA. Nuclear (A) and chloroplast (B) in vivo repair assays were performed with cw15arg7 and phr1 cells as described in Figure 3.
Figure 6B shows a chloroplast in vivo repair assay conducted on phr1 cells using the rbcL-specific probe and reports the number of dimers on the DNA fragment for each treatment. Unexpectedly, the results from the assays indicated that, when compared to wild-type cells, phr1 is severely deficient in chloroplast photoreactivation. This is most clearly seen in the samples treated with UV light and then allowed to photoreactivate for 30 min (Fig. 6B, compare lanes 7 and 8 with 17 and 18). Following 30 min photoreactivation, wild-type cells had repaired 100% of the pyrimidine dimers, while phr1 had repaired only 52%. As before, nearly the same amount of UV damage was induced in each of the cell types (Fig. 6B, compare lanes 3 and 4 with lanes 13 and 14). Also, the repair was light dependent (Fig. 6B, lanes 9 and 10, and lanes 19 and 20). This assay was repeated using a probe specific for a second chloroplast gene, atpB, and similar results were obtained (data not shown).
DISCUSSION
Through the use of a sensitive gene-specific in vivo repair assay, we have shown that Chlamydomonas cells that overexpress the class II DNA photolyase PHR2 have an increased ability to perform photoreactivation in both the chloroplast and the nucleus. Through the use of this same assay, we were also able to show that phr1, a photoreactivation-deficient mutant of Chlamydomonas, is deficient not only in nuclear photorepair, but also in chloroplast photoreactivation. Finally, we have shown that PHR2 requires functional phr1 for full activity.
Given that amino acid sequence analysis of PHR2 suggests that it contains a chloroplast targeting sequence, we anticipated that overexpression of PHR2 in Chlamydomonas would result in an increased ability to photoreactivate in the chloroplast. However, the finding that overexpressing PHR2 also increased photoreactivation in the nucleus was more surprising. It is possible that the localization of PHR2 to one of these organelles is a side-effect of overexpression. This seems unlikely, however, since we were able to show that the phr1 mutation affects both cellular compartments and is required for full activity of PHR2 protein. Although the class I DNA photolyase from Saccharomyces cerevisiae has been reported to localize to both the mitochondria and the nucleus (22), this is the first report of a class II DNA photolyase being targeted to two organelles, and the only photolyase that has been localized to the chloroplast and the nucleus. The signals responsible for directing PHR2 to these subcellular locations still remain to be determined. There are several examples of gene products from the human base excision repair pathway that are localized to two distinct cellular compartments, specifically the mitochondria and the nucleus. hMYH and hOGG1, which encode the human mutY homolog and 8-oxoguanine DNA glycosylase, respectively, are alternatively spliced to generate gene products specific for these two compartments (23–25). The human uracil-DNA glycosylase gene, UNG, uses alternative transcription starts and splicing to produce two proteins, UNG1 and UNG2, which are targeted to the mitochondria and the nucleus, respectively (26). It has also been reported that the human homolog of E.coli endonuclease III, hNTH1, contains dual transport signals that direct it to both the mitochondria and the nucleus (25,27). Northern blot analysis of Chlamydomonas poly(A)+ RNA with a PHR2-specific probe did not indicate the presence of more than one size of message, although the presence of two mRNAs with similar sizes encoding different protein products with different targeting sequences may not have been detectable (12). Additionally, anti-Myc western blot analysis of cells overexpressing the PHR2 Myc-His gene reveals one protein of the predicted molecular mass of PHR2 Myc-His (Fig. 1A, lane 3). Based on these data, we are inclined to believe that the PHR2 gene encodes a single protein that contains transport signals for both the nucleus and the chloroplast.
Our results indicating that phr1 is severely deficient in photoreactivating chloroplast DNA was problematical since data generated previously in our laboratory did not indicate such a severe deficiency (11). The assay originally used to monitor chloroplast photoreactivation in phr1 was based on the repair of DNA radiolabeled with thymidine, which is presumably specific for chloroplast DNA. Conversely, the assay used in this paper measured the repair of individual chloroplast genes, rbcL and atpB. Since these genes are known to reside in the chloroplast genome, we are led to believe that the assay used in this paper measures chloroplast photoreactivation more specifically than the radiolabeled thymidine assay.
The most intriguing result from our study was that PHR2 requires a functional phr1 gene for full activity. Traditionally, photoreactivation has been characterized as only requiring a single enzyme, DNA photolyase. However, an isolated report in the literature from 1981 described the isolation of phr2, a photoreactivation mutant of S.cerevisiae that complemented a strain carrying a mutation in PHR1, the structural photolyase gene (28). However, this report has never been confirmed or further characterized.
A possible explanation for the inactivity of PHR2 in the phr1 background is that phr1 is actually overproducing an inhibitor of DNA photolyase. However, we have previously reported that diploid strains heterozygous for the phr1 mutation have DNA photolyase activity similar to wild-type diploid strains (29). This evidence indicates that the mutation in phr1 is recessive and, as such, does not involve an inhibitor of DNA photolyase.
Reconstitution studies with the class I CPD photolyase from E.coli indicated that binding of photolyase to pyrimidine dimers in DNA requires FAD, but not MTHF (30). Additionally, it was shown that the ability of reconstituted photolyase to bind UV-damaged DNA is not affected by the redox state of FAD (30,31). More recently, the Arabidopsis thaliana class II DNA photolyase was expressed in E.coli as a 6×His fusion protein. The purified protein was shown to be catalytically active with FAD as the only detectable cofactor (32). This study suggests that, as with class I CPD photolyases, class II DNA photolyases require only FAD to bind pyrimidine dimers. It follows then that the reduced ability of phr1 extracts to bind UV-irradiated DNA could be caused by the presence of PHR2 molecules lacking FAD. The best method to test this hypothesis would be through a direct comparison of the chromophore composition of PHR2 molecules isolated from both wild-type and phr1 cells.
The exact nature of the phr1 activation step still awaits further definition. We have considered the possibility that PHR1 is involved in the biosynthesis of FAD. However, the photoreactivation deficiency of phr1 is quite severe, which would suggest that any change in FAD levels would have to be extensive. Since FAD is an essential cofactor in several metabolic processes, any significant change in FAD levels would be expected to have an impact on growth of the mutant. No such phenotype has been observed in phr1 cells. Alternatively, PHR1 could facilitate the insertion of FAD into PHR2. Although this is unprecedented for photolyases, mutation in a gene coding for a protein with this activity would not impact on other enzymes requiring FAD and would thus allow the apparently normal growth rate of the phr1 strain. It is also possible that PHR1 assists in the proper folding of PHR2. This could occur alone or in conjunction with insertion of FAD into the photolyase.
Based on the data presented in this paper, we propose that PHR2 is the photolyase responsible for photoreactivating both chloroplast and nuclear DNA in C.reinhardtii and that PHR2 cannot become fully active in the absence of functional PHR1. A second, somewhat less likely, model that can accommodate the data presented here is that PHR2 is only localized to one cellular compartment, probably the chloroplast, and that the increased repair observed in the nucleus of cells overexpressing PHR2 is simply an artifact. In this case, there would have to be an alternative gene encoding a nuclear photolyase and the product of this gene would also require functional PHR1 for full activity. We have previously cloned a gene, CPH1, predicted to encode a protein with sequence similarity to class I photolyases (33). However, CPH1 is now believed to encode a blue light photoreceptor, since recombinant protein expressed in E.coli shows no detectable photolyase activity. It seems unlikely that another photolyase-related gene exists in Chlamydomonas since extensive Southern blotting using a class I sequence probe from CPH1 and a class II probe from PHR2 does not indicate the presence of any closely related genes. Clearly, generating a PHR2 knockout is the ultimate way to determine whether the PHR2 protein is the sole photolyase responsible for photoreactivating nuclear and chloroplast DNA.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Cristoph Beck and Michael Schroda for providing the HSP70A–RBCS2 fusion promoter. We also thank Robin Miskimins for a critical review of and suggestions for the manuscript. This research was supported by grants from the National Institutes of Health (GM59857) and the US Department of Agriculture (National Research Initiative Competitive Grants Program no. 96-35304-3860).
References
- 1.Hearst J.E. (1995) The structure of photolyase: using photon energy for DNA repair. Science, 268, 1858–1859. [DOI] [PubMed] [Google Scholar]
- 2.Sancar A. (1996) No “end of history” for photolyases. Science, 272, 48–49. [DOI] [PubMed] [Google Scholar]
- 3.Sancar G.B. (2000) Enzymatic photoreactivation: 50 years and counting. Mutat. Res., 451, 25–37. [DOI] [PubMed] [Google Scholar]
- 4.Taylor C.B. (1997) Damage control. Plant Cell, 9, 111–114. [Google Scholar]
- 5.Kanai S., Kikuno,R., Toh,H., Ryo,H. and Todo,T. (1997) Molecular evolution of the photolyase-blue-light photoreceptor family. J. Mol. Evol., 45, 535–548. [DOI] [PubMed] [Google Scholar]
- 6.Todo T., Ryo,H., Yamamoto,K., Toh,H., Inui,T., Ayaki,H., Nomura,T. and Ikenaga,M. (1996) Similarity among Drosophila (6–4) photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science, 272, 109–112. [DOI] [PubMed] [Google Scholar]
- 7.Yasuhira S. and Yasui,A. (1992) Visible light-inducible photolyase gene from the goldfish Carassius auratus. J. Biol. Chem., 267, 25644–25647. [PubMed] [Google Scholar]
- 8.Yasui A., Eker,A.P.M., Yasuhira,S., Yajima,H., Kobayashi,T., Takao,M. and Oikawa,A. (1994) A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J., 13, 6143–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Small G.D. and Greimann,C.S. (1977) Photoreactivation and dark repair of ultraviolet light-induced pyrimidine dimers in chloroplast DNA. Nucleic Acids Res., 4, 2893–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen J.-J., Jiang,C.-Z. and Britt,A.B. (1996) Little or no repair of cyclobutyl pyrimidine dimers is observed in the organellar genomes of the young Arabidopsis seedling. Plant Physiol., 111, 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox J.L. and Small,G.D. (1985) Isolation of a photoreactivation-deficient mutant of Chlamydomonas. Mutat. Res., 146, 249–255. [DOI] [PubMed] [Google Scholar]
- 12.Petersen J.L., Lang,D.W. and Small,G.D. (1999) Cloning and characterization of a class II DNA photolyase from Chlamydomonas. Plant Mol. Biol., 40, 1063–1071. [DOI] [PubMed] [Google Scholar]
- 13.Gorman D.S. and Levine,R.P. (1965) Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA, 54, 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schroda M., Vallon,O., Wollman,F.-A. and Beck,C.F. (1999) A chloroplast-targeted heat shock protein 70 (HSP70) contributes to the photoprotection and repair of photosystem II during and after photoinhibition. Plant Cell, 11, 1165–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kindle K.L. (1990) High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc. Natl Acad. Sci. USA, 87, 1228–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 17.Todo T. and Ryo,H. (1992) Identification of cellular factors that recognize UV-damaged DNA in Drosophila melanogaster. Mutat. Res., 273, 85–93. [DOI] [PubMed] [Google Scholar]
- 18.Bohr V.A., Okumoto,D.S. and Hanawalt,P.C. (1986) Survival of UV-irradiated mammalian cells correlates with efficient DNA repair in an essential gene. Proc. Natl Acad. Sci. USA, 83, 3830–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroda M., Blöcker,D. and Beck,C.F. (2000) The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J., 21, 121–131. [DOI] [PubMed] [Google Scholar]
- 20.Marco Y. and Rochaix,J.-D. (1980) Organization of the nuclear ribosomal DNA of Chlamydomonas reinhardii. Mol. Gen. Genet., 177, 715–723. [DOI] [PubMed] [Google Scholar]
- 21.Harris E.H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 22.Yasui A., Yajima,H., Kobayashi,T., Eker,A.P.M. and Oikawa,A. (1992) Mitochondrial DNA repair by photolyase. Mutat. Res., 273, 231–236. [DOI] [PubMed] [Google Scholar]
- 23.Ohtsubo T., Nishioka,K., Imaiso,Y., Iwai,S., Shimokawa,H., Oda,H., Fujiwara,T. and Nakabeppu,Y. (2000) Identification of human MutY homolog (hMYH) as a repair enzyme for 2-hydroxyadenine in DNA and detection of multiple forms of hMYH located in nuclei and mitochondria. Nucleic Acids Res., 28, 1355–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takao M., Zhang,Q.-M., Yonei,S. and Yasui,A. (1999) Differential subcellular localization of human MutY homolog (hMYH) and the functional activity of adenine:8-oxoguanine DNA glycosylase. Nucleic Acids Res., 27, 3638–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takao M., Aburatani,H., Kobayashi,K. and Yasui,A. (1998) Mitochondrial targeting of human DNA glycosylases for repair of oxidative DNA damage. Nucleic Acids Res., 26, 2917–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nilsen H., Otterlei,M., Haug,T., Solum,K., Nagelhus,T.A., Skorpen,F. and Krokan,H.E. (1997) Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res., 25, 750–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda S., Biswalt,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]
- 28.MacQuillan A.M., Herman,A., Coberly,J.S. and Green,G. (1981) A second photoreactivation-deficient mutation in Saccharomyces cerevisiae. Photochem. Photobiol., 34, 673–677. [PubMed] [Google Scholar]
- 29.Munce D.B., Cox,J.L., Small,G.D., Vlcek,D., Podstavková,S. and Miadoková,E. (1993) Genetic and biochemical analysis of photolyase mutants of Chlamydomonas reinhardtii. Folia Microbiol., 38, 435–440. [Google Scholar]
- 30.Payne G., Wills,M., Walsh,C. and Sancar,A. (1990) Reconstitution of Escherichia coli photolyase with flavins and flavin analogues. Biochemistry, 29, 5706–5711. [DOI] [PubMed] [Google Scholar]
- 31.Jorns M.S., Wang,B., Jordan,S.P. and Chanderkar,L.P. (1990) Chromophore function and interaction in Escherichia coli DNA photolyase: reconstitution of the apoenzyme with pterin and/or flavin derivatives. Biochemistry, 29, 552–561. [DOI] [PubMed] [Google Scholar]
- 32.Kleiner O., Butenandt,J., Carell,T. and Batschauer,A. (1999) Class II DNA photolyase from Arabidopsis thaliana contains FAD as a cofactor. Eur. J. Biochem., 264, 161–167. [DOI] [PubMed] [Google Scholar]
- 33.Small G.D., Min,B. and Lefebvre,P.A. (1995) Characterization of a Chlamydomonas reinhardtii gene encoding a protein of the DNA photolyase/blue light photoreceptor family. Plant Mol. Biol., 28, 443–454. [DOI] [PubMed] [Google Scholar]