Abstract
Host specialization has important consequences for the diversification and ecological interactions of obligate pathogens. The anther-smut disease of natural plant populations, caused by Microbotryum fungi, has been characterized by specialized host-pathogen affinities, which contribute in part to the isolation among these numerous fungal species. This study investigated the molecular variation of Microbotryum pathogens within the geographic and host-specific distributions on wild Dianthus species in southern European Alps. With particular contrast to prior studies on this pathogen genus, a range of overlapping host specificities was observed for four delineated Microbotryum lineages on Dianthus hosts, and their frequent co-occurrence within single-host populations was quantified at local and regional scales. In addition to potential consequences for direct pathogen competition, the sympatry of Microbotryum lineages led to hybridization between them in many populations, and these admixed genotypes were shown to suffer significant meiotic sterility. Therefore, this investigation of the anther-smut fungi reveals how variation in the degrees of host specificity can have major implications for ecological interactions and genetic integrity of differentiated pathogen lineages.
Keywords: pathogen sympatry, Microbotryum violaceum, introgression, host range, host shift, secondary contact, generalist
Introduction
Infectious diseases have important impacts on all natural ecosystems, from effects on host individuals to determining community composition (Benitez et al. 2013; Bagchi et al. 2014; Bever et al. 2015), and pathogens themselves represent a major component of biodiversity (Dobson et al. 2008). The evolution of differentiated pathogen lineages is associated with barriers to gene flow that arise through geographic separation and through specialization onto different hosts (Rice 1987; de Vienne 2013; Kohn et al. 2005; Giraud et al. 2006, 2008a, 2010). However, where ecological specialization is not strong, the processes of natural migration or anthropogenic movement can lead to direct interactions between previously isolated pathogen lineages by their co-occurring in the same host population. There is great concern over such pathogen secondary contact because hybridization and competition may then favor the emergence of new disease properties, possibly more virulent or with new host affinities (Kuldau et al. 1999; Brasier 2000; Newcombe et al. 2000; Lin et al. 2007; Giraud et al. 2008a; Gladieux et al. 2011; Lemaire et al. 2016; Leroy et al. 2016; Stukenbrock 2013, 2016).
Examples of altered disease traits resulting from pathogen secondary contact and hybridization are often found in the agricultural and human medical literature (Schardl & Craven 2003; King et al. 2015). Hybrids between serotypes of the human pathogen Cryptococcus neoformans can yield more virulent genotypes (Lin et al. 2007). Secondary contact between differentiated pathogen populations of Venturia inaequalis on apples resulted in new virulent hybrids (Lemaire et al. 2016; Leroy et al. 2016). The hybrid rust pathogen Melampsora x columbiana emerged on poplars bred for resistance to the two parental rust species (Newcombe et al. 2000), with a similar emergence of hybrid powdery mildews on cereals (Menardo et al. 2016). Even in the absence of hybridization, competition between co-occurring pathogens can be a driver of pathogen virulence or diversification. A large body of theoretical work describes how competition might, positively or negatively, impact virulence, and there is support for these effects from experimental inoculation studies (reviewed by Alizon et al. 2013; Bull & Lauring 2014). In contrast, secondary contact between previously isolated pathogen lineages may lead to decrease disease severity, because the generated hybrids are less fit and/or because competition between lineages lower disease prevalence (Le Gac et al. 2007b, de Vienne et al. 2009a, Sloan et al 2008; Gold et al 2009). Given the current rates of global change, species redistributions are increasingly frequent, fostering ever more secondary contacts between pathogens (Desprez-Loustau et al. 2007; Gladieux et al. 2011, 2015; Lemaire et al. 2016). However, our ecological understanding of their interactions is limited by a lack of studies on natural disease systems where multiple related pathogen species co-occur.
The goal of this study was to determine the geographic and host-specific distributions of anther-smut pathogens on wild Dianthus hosts, and to assess the consequences of host-pathogen affinities for pathogen sympatry and hybridization. Anther-smut disease, caused by obligately pathogenic fungi in the genus Microbotryum, has served as an important model for disease ecology (Antonovics et al. 2002; Bernasconi et al. 2009), including the conditions that promote pathogen invasion (Fontaine et al. 2013, Gladieux et al. 2015; Feurtey et al. 2016), speciation (Le Gac et al. 2007a, 2007b; Refregier et al. 2010) and hybridization (de Vienne et al. 2009a; Gladieux et al. 2011). Most of this previous work has been with anther-smut diseases of a related host genus, Silene, where numerous pathogen species are highly specialized to different hosts and there is little hybridization (Le Gac et al. 2007b; Gladieux et al. 2011; Badouin et al. 2017). The monophyletic clade of anther-smut fungi on the host genus Dianthus is less well studied, but recent work suggests there is generally less host specificity compared to the pathogens on Silene hosts (Kemler et al. 2013). Some Microbotryum pathogens on Dianthus can infect overlapping sets of host species, and thus multiple fungal species can be found on a given Dianthus host species (Refregier et al. 2008; Kemler et al. 2013). In such systems where host specificity is low, there should be greater potential for different pathogen lineages to meet and interact directly in the same host population, and possibly to hybridize.
Here, we first address genetic structure and reproductive barriers among anther-smut fungi infecting Dianthus hosts in the southern European Alps using microsatellite variation and in vitro assays, and we show that there are several differentiated Microbotryum lineages. We then describe the geographic and host-specific distributions of these pathogens. Specifically, we aimed to: 1) identify genetically differentiated lineages of Microbotryum genotypes, and in particular previously unsampled lineages, 2) assess sterility of genotypes resulting from hybridization that may indicate reproductive isolation among the pathogen lineages, 3) determine pathogen distributions at different spatial scales in terms of host specialization and sympatry, and 4) assess the occurrence and distribution of pathogen genotypes resulting from hybridization, and in particular whether they are rare or frequent, which may also inform on their fitness relative to parental lineages. This study reveals important components of previously undetected pathogen diversity, and provides insights into the dynamics of multi-host multi-pathogen systems at the regional scale.
Methods
Study system
Microbotryum is a genus of basidiomycete fungi, in the subphylum Pucciniomycotina, that causes anther-smut disease on plants mostly in the Caryophyllaceae. In this disease, host pollen is replaced by fungal spores which are then spread primarily by insect pollinators (Fig. 1). Host plants are completely sterilized by infection because the anthers of all flowers are diseased and the ovary also fails to fully develop. The fungus persists only on perennial host species (Hood et al. 2010), forming systemic and persistent infections that have minimal effect on plant mortality (Thrall & Jarosz 1994). To complete the life cycle, the diploid smut spores from diseased anthers are deposition on a new host, after which they germinate, undergo meiosis and produce a haploid yeast stage with two mating types; mating among haploid cells must occur prior to infection of the new host plant (Giraud et al. 2008a). Prior studies on Silene hosts indicate that a single plant may sometimes harbor multiple genotypes of the pathogen, but there is frequently competitive exclusion, which is stronger between genetically more distant lineages (Hood 2003; Lopez-Villavicencio et al. 2007; Lopez-Villavicencio et al. 2011; Gold et al. 2009). The encounters inside diseased hosts are not a route to genetic exchange between pathogen genotypes because mating occurs on the host surface prior to infection.
Fig 1.
Dianthus pavonius illustrating symptoms of the anther-smut disease (left) and a healthy flower (right). Microbotryum species replace pollen of diseased plants with dark, powdery fungal spores, which can be carried by pollinators to healthy hosts.
Recent studies have aimed to identify species within the Microbotryum complex (Kemler et al. 2006; Denchev, 2007; Lutz et al. 2008; Denchev et al. 2009; Piątek et al. 2012, 2013). While concordance between multiple gene genealogies has been useful in these efforts, this criterion may not be optimal for young species with either incomplete gene coalescence or ongoing hybridization (reviewed in Cai et al. 2011). Nevertheless, several of the pathogens occurring on Dianthus hosts in Europe have been named as distinct species: Microbotryum dianthorum, Microbotryum carthusianorum, Microbotryum shykoffianum, and Microbotryum superbum (Denchev et al. 2009). In these cases it has been noted that pathogen sampling, and therefore taxonomic revision is incomplete and tentative (Le Gac et al. 2007b; Kemler et al. 2013).
It has been long recognized that Microbotryum from different host species are relatively specialized (Zillig 1921, Goldschmidt 1928). On the host genus Silene, Microbotryum species are generally host-specific, although there are exceptions as transient host-shifts (Antonovics et al. 2002; Gladieux et al. 2011). In contrast, on Dianthus multiple Microbotryum species have been reported from the same host species, and reciprocally some of the pathogen lineages can infect multiple host species with some apparent degree of overlapping host ranges (Le Gac et al. 2007b; Refregier et al. 2008; Kemler et al. 2013).
The host genus Dianthus represents a group of broadly distributed, mostly perennial herbs found commonly in habitats that include subalpine and alpine regions of Eurasia and Africa. Diversification in Europe of over 100 Dianthus species is believed to have coincided with regional climate changes in the Pleistocene and the onset of dry Mediterranean summer conditions (Valente et al. 2013).
Field collections
Surveys for anther-smut disease on Dianthus hosts were carried out in the southern European Alps from 2008 to 2011, in a focal region spanning latitudes 44.0 – 44.5N and longitudes 7.0 – 8.2E (Fig. 2, S1). A total of 443 pathogen samples were collected and genotyped from 81 populations across six Dianthus species, D. deltoides, D. furcatus, D. pavonius, D. seguieri, D. superbus, and D. sylvestris (Table S1). Flowering period largely overlaps for these species during the sampling period of early-July to mid-August, with D. deltoides and D. sylvestris flowering marginally earlier in the season. Sampling structure consisted of 19 valleys, within which populations/communities of a Dianthus species were defined as collections separated by at least 100m; a community was a collection site with more than one Dianthus species present and diseased. The Dianthus species were readily identified from morphological differences; in only one location were there a few plants of intermediate form, and no pathogen samples from those plants were included. The average number of samples per population was 5 (std. dev. = 4.3). To avoid potential contamination from pollinator-dispersed spores, Microbotryum samples consisted of mature unopened flower buds, one sample per diseased plant. These were stored at 4°C until being used for DNA extraction from excised individual anthers or for use in culturing in vitro.
Fig 2.
Distributions across southern European Alps of the Microbotryum pathogens and Dianthus host populations. Locations in the map are approximate, adjusted to allow non-overlapping representation of all pie graphs. Populations are represented by pie graphs, scaled by sample size (ranging from 1 to 28 samples). A) Population structure of 443 pathogen samples, inferred with the Bayesian clustering algorithm implemented in STRUCTURE, where four genetic clusters are indicated by colored wedges. Grey wedges indicate samples per site that did not meet the 0.9 confidence threshold for cluster assignment. B) Map of the populations of Dianthus hosts sampled for Microbotryum pathogens, indicating different host species compositions by sized and shaded pie graphs.
In 2012, additional samples were collected to determine if co-occurrence of sympatric Microbotryum lineages might exist at the level of the multiple-infections of individual host plants. From valleys labeled STA, PRA, and SBM (Table S1) multiple flowering stems per plant were sampled as separate flower buds. For the STA valley the numbers of plants and average number of stems per plant sampled were 22 plants averaging 3.1 stems, for PRA valley 27 plants averaging 2.8 stems, and for SBM valley 53 plants averaging 2.5 stems. Prior studies have shown that co-infection of the same host is unlikely to extend to the level of occupying the same flower (Day 1980; Hood 2003).
Microsatellite (Simple Sequence Repeat) genotyping
Eleven microsatellite markers were used by the methods described in Giraud (2004) with the isolation of diploid DNA of Microbotryum samples from Dianthus. Seven markers were from existing libraries (Giraud et al. 2002, 2008b), and four new markers were developed: mvD3, mvD5, mvD11, and mvD23 (Table S2). The new markers were found using the Repeat Finder program (http://www.proweb.org/proweb/Tools/selfblast.html) to search low coverage genome sequence data of Microbotryum isolated from Dianthus (454 sequencing at ca. 3x genome coverage; mean DNA fragments length of ca. 250 bp). PCR primers for 24 loci were designed using Primer3 (Rozen & Skaletsky 2000) to flank di- or trinucleotide repeat motifs of at least six iterations. Preliminary assays selected four marker loci showing polymorphism and amplification across all the Microbotryum samples.
Amplification of microsatellite loci was conducted using Chelex-extracted DNA (Bucheli et al. 2001) and fluorescent-labeled PCR primers. PCR was performed in a final volume of 10 μL containing 1× Phusion HF Buffer (New Englands Biolabs), 0.2 mmol/L deoxynucleoside-5′-triphosphate (Promega), 0.25 μmol/L forward and reverse primer, 1 U/μL Phusion HF DNA polymerase (New England Biolabs), and 0.8 μL of template DNA. Separate PCR reactions were started with an initialization step for 30 seconds at 98° C, 50 cycles of 10 seconds at 98°C, 30 seconds at the annealing temperature (Table S2), 30 seconds at 72°C, and 1 final extension cycle of 5 minutes at 72°C. PCR reactions with different primer dyes (Table S2) were multiplexed and a 600 LIZ dye size standard was added. Fragment analysis were run at the UMass Genomics Resources Lab on Applied Biosystems 3130 and 3130xl Genetic Analyzers run with Applied Biosystems five-dye chemistry. Alleles were scored with GENEMAPPER 4.0, and samples scored for the majority of loci (i.e. ≥6) were retained for the following analyses.
Population genetic subdivision and assessment of hybridization
Individual-based Bayesian clustering methods, implemented in STRUCTURE 2.3.3 (Pritchard et al. 2000), were used to investigate genetic subdivision of the 2008–20011 pathogen sample collection. This method is based on the use of Markov Chain Monte Carlo (MCMC) simulations to infer the assignment of genotypes into different numbers (K) of proposed clusters. The underlying algorithms minimize deviations from Hardy–Weinberg structure and linkage disequilibria within each cluster. Ten independently replicated runs of the analysis were carried out for successive K values (2≤K≤9), with 1,000,000 MCMC iterations after a burn-in of 100,000 steps. Outputs were processed with CLUMPP 1.1.2 (Jakobsson & Rosenberg 2007), for the identification of potentially distinct clusters among replicated runs for each K value. A G′-statistic greater than 80% was used to assign replicates to a common STRUCTURE mode. When multiple STRUCTURE modes were found (K > 3), we compared alternative modes. At each K, the different modes were not substantially different (H′ values > 0.70). We therefore only kept the major mode. To help assess the most relevant value of K, we determined the amount of additional information explained by increasing K using the rate of change in the log probability of data between successive K values (ΔK statistic) (Evanno et al. 2005). Individuals with a membership coefficient > 0.90 to a given cluster were assigned to that cluster, while samples with lower values were considered admixed genotypes as the result of hybridization between clusters. In addition to ΔK statistic and the proportion of individual samples assigned to clusters across K-values, we used methods assessing genetic subdivision independent of a population genetic model: factorial correspondence analysis (FCA), centered with respect to clusters, was performed with GENETIX v4.05 (Belkhir et al. 1996), and discriminant analysis of principal component (DAPC) was performed using the ADEGENET R package with the R software (dapc function)..
Patterns of isolation by distance were determined by assessing the correlation between matrices of geographic distances and genetic distances within groups of the cluster-assigned pathogens. Mantel test with 1,000 random permutations was performed between the individual coefficient of relatedness Fij (Loiselle et al. 1995) and a matrix of the natural logarithm of geographic distance between collection sites. These analyses were performed with SPAGeDI 1.3 (Hardy & Vekemans 2002).
Also, the spatial aggregation of pathogen clusters or host species was assessed using Mantel tests as correlations between geographic distances matrix between populations or valleys and the matrix of shared presence/absence of the cluster-assigned pathogens or host species among those populations or valleys. Using the most relevant K, as assessed based on STRUCTURE, FCA and sterility analyses, and cluster-assigned samples (i.e. membership coefficient > 0.90 to a given cluster), the presence of pathogen clusters was scored as a binary variable using GenAIEx 6.5 (Peakall & Smouse 2012).
We then tested whether the variation of the pathogen cluster distribution was similar among host species at the scales of individual plants, populations, and valley; at the latter two spatial scales the 3 of 81 populations or 4 of 19 valleys with multiple host species were excluded. For this analysis the SPSS 24 (SPSS Inc.) statistical software was used, conducting Fisher’s exact test with the Monte Carlo simulation (10,000 iterations) and the GLMz (Poisson probability distribution, log-linear model) for taking into account the confounded effects of host and location. Comparison of the proportions (arcsine transformed) of samples that were admixed genotypes from populations with or without combinations of cluster-assigned pathogens was performed using a univariate general linear model in SPSS with numbers of samples as a covariate.
Descriptive statistics
Genetic polymorphism of the clusters as described above (i.e. clusters inferred by STRUCTURE, FCA and sterility analyses) was quantified as allelic richness (AR) and private allele richness (AP) calculated with ADZE (Szpiech et al. 2008). Allelic richness and private allele richness were computed using sample sizes of N=10 (five individuals x two chromosomes) as the minimum common sample size across clusters for which all microsatellite loci had been successfully genotyped, and clusters were compared using Wilcoxon signed rank tests. Heterozygosity, Weir and Cockerham’s F-statistics and deviations from Hardy-Weinberg expectations were calculated with GENEPOP 4.0 (Raymond and Rousset 1995; Rousset 2008). Population-specific computations excluded admixed genotypes (i.e. retaining only samples with a membership coefficient > 0.9 to any cluster), leading to a sample size of N=318 individuals.
Sterility assays of admixed genotypes
Hybrid sterility, as a measure of reproductive isolation between clusters that were identified with the STRUCTURE and FCA analyses, was investigated by germination assays of Microbotryum spores from the same field-collected flowers as were genotyped. The diploid spores undergo meiosis to produce the tetrad of four haploid yeast-like sporidia (basidiospores) extending from the basidium (as illustrated in Hood & Antonovics 2000). Spores were germinated on potato dextrose agar and incubated at 22°C for 24 to 30 hours. The four post-meiotic haploid sporidia were separated by micromanipulation to areas on the agar surface away from other growing cultures, and the proportion of the sporidia that exhibited haploid viability (colony formation) was determined after 48 hours at 22°C.
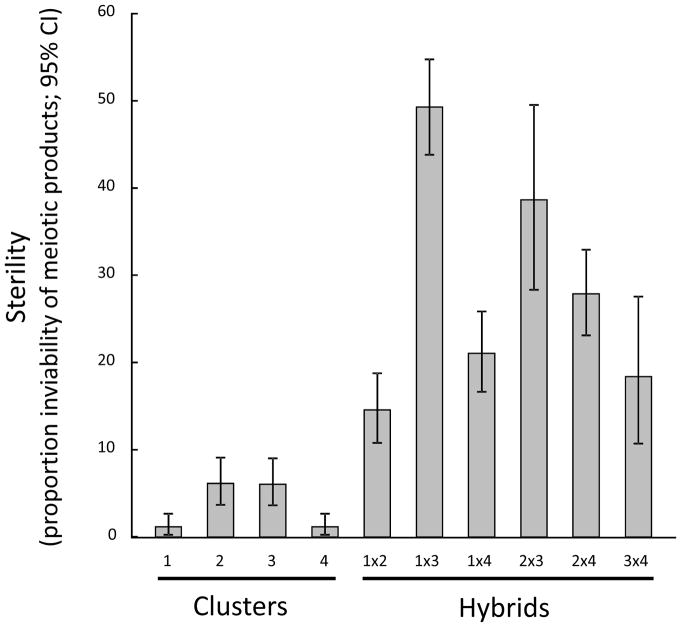
The sterility assays were carried out for five meiotic tetrads for up to 15 samples per each of four clusters and per each of the six possible types of admixed individuals between them. The putative parental clusters of the admixed genotypes were determined as the two clusters to which STRUCTURE analysis membership coefficients were greatest. Arcsine transformed data were used to determine 95% confidence intervals for the proportions of inviable meiotic products, which were back-transformed for plotting (Bolton & Bon 2009). The ability of yeast-like gametes to grow from the isolated post-meiotic cells was compared to non-admixed individuals using the t-test for independence on arcsine transformed proportions. Previous studies have shown that hybrids between different Microbotryum species may be viable but often suffer from sterility, due to the different karyotypes between species (de Vienne et al. 2009a).
DNA sequence analyses
For comparison with prior studies on Microbotryum from Dianthus, which used DNA sequence-based phylogenetics, two types of nuclear loci were investigated: 1) the internal transcribed spacer (ITS) region of the multi-copy ribosomal RNA gene as used by Kemler et al. (2013), and 2) elongation factor 1α (Ef1α) as used in Le Gac et al. (2007b), which were the two markers providing the best resolution among Microbotryum samples from Dianthus. We conducted bidirectional DNA sequencing for Ef1α and ITS for approximately 100 samples for which STRUCTURE analysis cluster membership coefficients were > 0.90 for the most likely K value and which spanned collection localities and hosts-of-origin (Table S3). DNA sequences were also retrieved from these prior studies for comparison.
In addition, ITS sequencing was performed on the Microbotryum samples collected as multiple stems from individual diseased host plants. Identities of the pathogen were assigned by their ITS sequence variants, and due to the multi-copy nature of ITS loci, admixed pathogens between lineages were identified by intra-genomic sequence polymorphisms at particular nucleotide positions in DNA sequencing electrophoregrams (James et al. 2009).
Results
Characterization of Microbotryum lineages
Analysis of microsatellite data for the 443 samples of Microbotryum infecting Dianthus hosts indicated the existence of four main pathogen clusters. Summary statistics on the microsatellite loci are presented in Table S4. Conclusion on the most relevant number of clusters was based upon a combination of metrics. The peak of ΔK value was found at K = 3, and it remained high for K = 4 and 5 (Fig. S2). For K-values of 2, 3 and 4, the proportion of samples assigned to clusters by membership coefficients > 0.90 was maximized in the range of 0.74 to 0.72, but then dropped markedly to 0.59 and 0.51 for K = 5 to 7 (Figs. S2 and S3). K = 4 was thus the highest K value defining well-delimited clusters, representing the finest strong genetic subdivision in the dataset, which may correspond to lineages isolated from extensive genetic exchange. This was also supported by the factorial correspondence analyses (FCA), which indicated strong separation of clusters at K = 4 (Fig. S5), while at K = 5 the additional cluster resulted in an FCA where the fifth clusters was poorly resolved. Discriminant analyses of principal component (DAPC) similarly showed that clusters at K = 4 were well separated by confidence inertia ellipses (Fig. S6). Hybrid sterility analyses below also indicated that the four clusters each contributed to the observed structure through partial meiotic failure of admixed genotypes. Analyses at K = 4 clusters are therefore presented below.
In the cluster assignment at K = 4, 318 of the 443 pathogen samples (72%) had membership coefficients > 0.90 assignment threshold, and 125 were considered admixed genotypes (Fig. S3). Among the 318 cluster-assigned Microbotryum samples, 143 samples were assigned with membership coefficients > 0.90 to Cluster 1 (Fig 2A, red color), 69 to Cluster 2 (Fig 2A, blue), and 53 to each of Cluster 3 (Fig 2A, green) and Cluster 4 (Fig 2A, yellow). The four clusters displayed similar significant deficiencies in heterozygotes as is characteristic Microbotryum fungi due to frequent selfing (Fontaine et al. 2013, Bueker et al. 2016) (Table 1). There was also similarity among clusters in patterns of allelic diversity (allele richness and private allele richness), with significant differences for these metrics present only between Clusters 1 and 3 (Tables 1 and S8). Levels of divergence in allelic frequencies between the clusters (FST) were strong for all pairwise comparisons, ranging from 0.268 to 0.399 (Table 2).
Table 1.
Genetic polymorphism of differentiation within cluster (K = 4) in Microbotryum fungi from Dianthus based on microsatellite variation.
| Clusters
|
||||
|---|---|---|---|---|
| 1 (red) | 2 (blue) | 3 (green) | 4 (yellow) | |
| N | 143 | 69 | 53 | 53 |
| Ho | 0.18 | 0.17 | 0.17 | 0.19 |
| HE | 0.51 | 0.63 | 0.70 | 0.51 |
| FIS* | 0.64 | 0.74 | 0.76 | 0.63 |
| AR (n=10) | 3.59 | 5.17 | 5.21 | 3.34 |
| AP (n=10) | 0.30 | 1.09 | 1.27 | 0.28 |
Admixed genotypes were not included in this analysis and thus analyses were conducted on a total of N = 318 individuals. N: Number of samples, Ho and HE : observed and expected heterozygosities, FIS : inbreeding coefficient, AR and AP: allelic richness and private allele richness, respectively, computed using sample sizes of n=10; color descriptors correspond to their use to indicate clusters in the figures.
All FIS values were significant at P < 0.05.
Table 2.
Genetic differentiation (FST) values between Microbotryum genetic clusters (K = 4) based on microsatellite variation.
| Clusters
|
||||
|---|---|---|---|---|
| 1 (red) | 2 (blue) | 3 (green) | 4 (yellow) | |
| 1 | ||||
| 2 | 0.276 | |||
| 3 | 0.399 | 0.262 | ||
| 4 | 0.325 | 0.268 | 0.362 | |
Admixed genotypes were not included in this analysis and thus analyses were conducted on a total of N = 318 individuals; color descriptors correspond to their use to indicate clusters in the figures.
Differentiation of the four identified pathogen clusters was supported by significant sterility of the field-collected admixed genotypes. Cluster membership coefficients less than the 0.90 threshold significantly predicted the inability of yeast-like gametes to grow from the isolated post-meiotic cells compared to non-admixed individuals (t-test for independence on transformed proportions, t(8) = −4.57, P = 0.002). Compared to samples from the four clusters, each of the six pairwise admixed types between them showed more haploid sterility for the immediate products of meiosis (Fig. 3, Fig. S7).
Fig. 3.
Sterility of admixed Microbotryum samples (i.e. with <0.9 confidence score for cluster assignment) compared to samples assigned to each of four genetic clusters. The putative parental clusters of the admixed genotypes were determined as the two clusters to which membership coefficients were greatest.
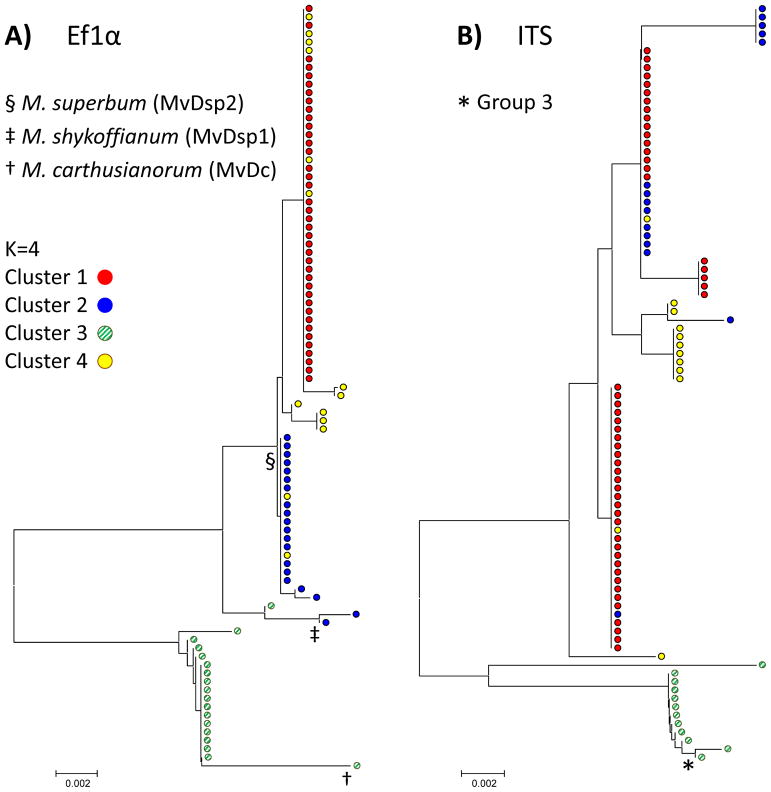
Analysis of DNA sequence data from two nuclear loci (i.e. Ef1α and ITS) allowed comparison of our population-based microsatellite study with prior delimitation among Microbotryum pathogens on Dianthus hosts. The three species of Microbotryum on Dianthus that were identified by Le Gac et al. (2007b) were represented in the current study (Fig. 4A), identified by having Ef1α sequences identical to the sequences used to delimit the species in that prior study. However, two of these three Ef1α sequence variants, for M. shykoffianum and M. carthusianum, did not encapsulate large numbers of samples in the current study, and Clusters 1 and 4 of the current study seemed to correspond to distinct lineages from those described previously based on multiple gene genealogies (Le Gac et al. 2007b). Among four main clades identified based on ITS sequence data in Kemler et al. (2013), only a single one, called Group 3, was represented in our dataset (Fig. 4B).
Fig. 4.
DNA sequence-based phylogenies of Microbotryum samples from Dianthus hosts for integration with prior studies. Four genetic clusters of Microbotryum samples are indicated by colored bars Approximately 100, cluster-assigned samples were included in both phylogenies. A) Phylogeny of elongation factor 1 α (Ef1α), with the placement of symbols immediately below the taxa with DNA sequences identical to Microbotryum species previously identified in Le Gac et al. (2007); M. carthusianorum NCBI accession DQ074540, M. shykoffianum accession DQ074549, and M. superbum accession DQ074547. B) Phylogeny of internal transcribed spacer (ITS) region of the multi-copy ribosomal RNA gene, with the symbol referring to a sequence identical to Microbotryum clade previously identified in Kemler et al. (2013); Group 3 accession JQ307901. The numbers of samples between the two trees differ due to a few failures of PCR or sequencing reactions.
The consistency with which DNA sequence-based variation discriminated microsatellite-based cluster assignments varied between Ef1α and ITS loci. In both the Ef1α and ITS phylogenies, Cluster 3 was most distant from the remaining samples (Fig. 4A,B). Ef1α tended to produce identical sequences for all samples of Cluster 1 and identical sequences for the majority of Cluster 2, and to also delimit these two clusters well. Cluster 4 was more dispersed among Ef1α variants, while it was well distinguished by ITS. In contrast, Clusters 1 and 2 were less well delimited by ITS than Ef1α.
Distributions and host affinities of Microbotryum lineages
The four pathogen lineages, identified as differentiated clusters above, were non-randomly distributed with respect to both location and host species (GMLz Wald-X2 for collection valley = 43.4, d.f. = 16, P < 0.001, and for host species = 27.5, d.f. = 4, P < 0.001). Indeed, geographic proximity was a significant predictor overall of sites sharing the same Microbotryum lineage (Mantel test, r = 0.216, P < 0.001 for 81 populations, and r = 0.164, P = 0.048 for 19 valleys). The lineage identified as Cluster 4 contributed most strongly to this spatial pattern being largely restricted to the southeastern portion of the sampling area (Fig. 2A); this pattern was not due to the restricted distribution of a particular host lineage to this region (Fig. 2B). Thus, the probability of Cluster 4’s presence in any two valleys as a function of the geographical distance between them approached significance (Mantel test, r = 0.260, P < 0.053 for 19 valleys) but this measure was not significant for the other pathogen lineages (smallest P-value among other clusters = 0.167). Within each of the four pathogen lineages there was no significant isolation by distance (Mantel tests with p-values within Cluster 1 = 0.44, Cluster 2 = 0.42, Cluster 3 = 0.46, Cluster 4 = 0.18).
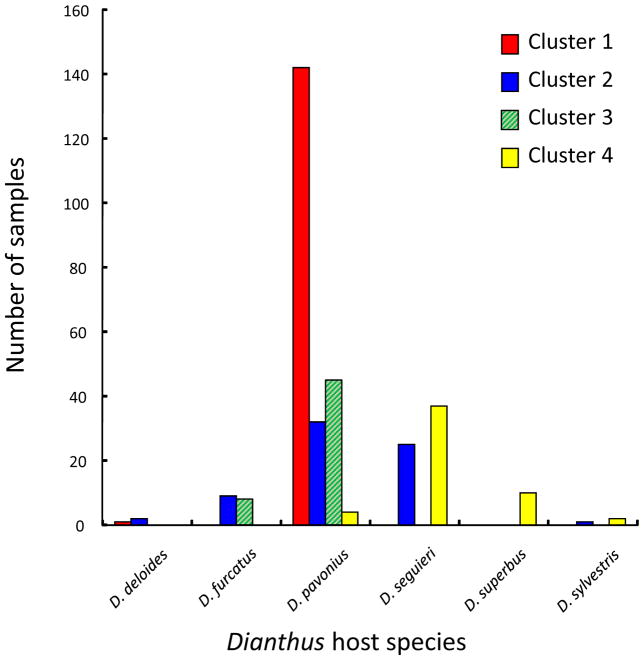
A non-random distribution of the four pathogen lineages among host species was also evident across hierarchical sampling scales, whether analyzing individual samples (Fig. 5, Fisher’s exact test, Monte-Carlo 10,000 iterations, P < 0.001) or treating populations or valleys as units of observation (Fig. S8; Fisher’s Exact Test, Monte-Carlo 10,000 iterations, P < 0.001 for populations and P = 0.009 for valleys). In particular, Cluster 1 was almost exclusively associated with Dianthus pavonius; only one out of 143 samples was isolated from another host, D. deltoides (Fig. 5). Cluster 3 also was most often associated with D. pavonius, though also with D. furcatus. The pathogen of Cluster 4 was most frequently associated with D. seguieri although also with three other hosts. The pathogen belonging to Cluster 2 was the most widespread, often on D. pavonius while also on four other hosts. The distribution of admixed pathogen samples among hosts is shown in Figure S9A.
Fig. 5.
Distribution of cluster-assigned samples of Microbotryum among six host species of Dianthus. Four genetic clusters of Microbotryum samples are indicated by colored bars.
Dianthus host species were thus associated with multiple Microbotryum lineages (Fig. 5). Within sampling locations, different pathogen lineages co-occurred, most often on the same host species (Fig. 2). Among the 56 populations where more than one cluster-assigned Microbotryum sample was collected, 17 (30%) contained multiple pathogen lineages. At the level of valleys, 12 of 19 (63%) contained multiple pathogen lineages. Pathogen co-occurrence, however, did not frequently extend down to the level of co-infecting individual host plants. Within three valleys where pathogen sympatry occurred (STA, SBM, and PRA; Table S1) and where multiple flowering stems were sampled from individual plants, there was only one instance of co-infection by different cluster-assigned pathogen genotypes among 102 plants sampled as multiple stems, averaging three stems per plant.
Sympatry of different diseased host species was markedly less common than sympatry of pathogen lineages; compared to the numbers of sites containing multiple pathogen lineages above, only three populations and four valleys contained diseased samples from more than one host species. Noting that the sample sizes were small, sympatry of diseased host species was not a significant predictor of sympatry for pathogen lineages; only one of three host-sympatric populations was a location of pathogen sympatry (binomial probability of at least one of three populations having pathogen sympatry, P = 0.657), and while each of four valleys with diseased host sympatries was also a location of pathogen sympatry this was not unexpected (binomial probability, P = 0.158). Regarding the distribution of just the host species among the sampled populations, geographic proximity was a significant predictor of populations sharing the same diseased Dianthus host species (Mantel test, r = 0.167, P = 0.010 for 81 populations), while this host spatial pattern was not significant at the among-valley scale (r = 0.217, P = 0.120 for 19 valleys) (Fig. 2).
Individuals characterized as admixed genotypes between pathogen lineages (cluster membership coefficients < 0.90) were found throughout the sampling region (Fig. 2), in 50 of 81 populations (62%) and in 15 of 19 valleys (79%). Admixed genotypes were found in 90% of populations with more than five pathogen samples. Controlling for the number of samples collected, the proportions of pathogens that were admixed did not depend significantly upon whether the population had a mixture of assigned pathogens lineages (arcsine transformed data, F = 0.972, P = 0.329). However, there was statistical bias among the different Dianthus host species for the ratio of admixed pathogens to cluster-assigned pathogens (Fisher’s Exact Test, Monte-Carlo 10,000 iterations, P = 0.020; Fig. S9A), driven by somewhat of an excess of admixed genotypes on D. furcatus. However, admixed pathogens were not derived from a common ancestral admixture of two other clusters, and the abundances of the six types of admixed pathogens (shown in Fig. S9B) appeared to occur by chance; the types did not differ significantly from the products of relative overall abundances of the four parental pathogen lineages (Fisher’s Exact Test, Monte-Carlo 10,000 iterations, P = 0.269).
Discussion
Our understanding of pathogen ecology has been greatly influenced by the recognition of cryptic genetic variation, helping to explain disease distributions through species-specific competition, specialization, and genetic exchange (Cai et al. 2011, de Leon & Nadler 2010). There are also significant implications for the choices of disease control strategies (Bickford et al. 2007). Previously subsumed under a single species name, revisions to the Microbotryum genus are ongoing (Le Gac et al. 2007b; Lutz et al. 2008; Denchev et al. 2009; Piątek et al. 2012, 2013), and studies show that anther-smut fungi on new host groups or geographic areas are likely to reveal substantial hidden diversity (Kemper et al. 2009). The current study on molecular variation, combined with host-of-origin and geographic distributions, contrasts strongly with prior characterizations of anther-smut lineages as highly specialized on a single host and with few hybrids in nature (Le Gac et al. 2007b; Bernasconi et al. 2009; Gladieux et al. 2011; Kemler et al. 2013). Although there was some degree of host specialization, host ranges can extend to several Dianthus species. As an important consequence of broader host ranges, sympatry of the differentiated pathogen lineages within host populations is remarkably frequent, providing opportunities for the direct interactions of pathogen competition and hybridization.
The level of differentiation among the Microbotryum lineages was informed by several lines of molecular and developmental or ecological evidence. The resolution of four lineages based on microsatellite data was supported by the high ΔK statistic, high proportion of samples assigned to clusters, and cluster separation in the FCA and DAPC analyses. Furthermore, DNA sequence variation at loci previously used for Microbotryum species-level phylogenetics (Le Gac et al. 2007; Kemler et al. 2013) tended to confirm the clustering of samples as based on these multi-locus markers. The phylogenetic resolution was most consistent with the separation of Cluster 3, and further phylogenetic analyses based on genome-wide multi-locus sequence data could aid in clarifying differences among the other lineages and whether additional subdivisions are present. Considering non-molecular criteria, the distinction of at least these four identified lineages is supported by their different distributions among host species and the significant sterility of the admixed genotypes, demonstrated by the failure of haploid meiotic products to grow in the yeast stage. Such sterility suggests substantial divergence in genic or chromosomal structures that prevent normal meiotic segregation (e.g. Dobzhansky 1933). A range of sterility values was observed within sample types, and while some such variation might reflect admixed genotypes not being F1 hybrids, the sterility of a few cluster-assigned samples at the 50% level may be due to a phenomenon of mating-type bias that is known to be an infrequent polymorphism causing such haploid viability in Microbotryum species (Thomas et al. 2003). Previous studies have shown that experimental hybrids between Microbotryum species from Silene do suffer particularly from the type of post-zygotic isolating mechanism observed here (de Vienne et al. 2009a; Le Gac et al. 2007a; Gibson et al. 2012; Buker et al. 2013). Microbotryum species often have different chromosome sizes and gametes of F1 hybrids display abnormal DNA content, higher than either parental species (de Vienne et al. 2009a), indicating that meiotic failure may arise from karyotype differences. Although the sampling in this study was more thorough at the population level than in previous ones, the sampling region was restricted and the number of host species was small relative to the known Dianthus distribution, even within Europe (Valente et al. 2010). Because only some of the DNA sequence variation for Microbotryum from Dianthus was overlapping with that reported by Le Gac et al. (2007b) and Kemler et al. (2013) the concordance of these datasets remains difficult to fully ascertain. It is highly probable that a substantially larger number of Microbotryum lineages may exist on Dianthus hosts elsewhere, and more studies that integrate molecular and organismal characterizations seem essential in advance of taxonomic delimitation to resolve species-level identities of Microbotryum on Dianthus.
The broader host ranges of anther-smut pathogens on Dianthus compared to prior generalizations on the Silene host genus is a particular contrast, as noted previously (Le Gac et al. 2007b; Kemler et al. 2013) and quantified here. Dianthus-Silene comparisons are quite old, indicated by Darwin (1875) in the following statement, “In the same family there may be a genus, as Dianthus, in which very many species can most readily be crossed; and another genus, as Silene, in which the most persevering efforts have failed to produce between extremely close species a single hybrid.” While we now know that a few closely related Silene species will cross (Kruckeberg 1961), the statement reflects a real difference between the genera in the relative timing of radiation, with morphologically diverse European Dianthus species radiating much more recently than most Silene species (Valente et al. 2010; see also Kemler et al. 2013). This more recent divergence may contribute to the ability of Microbotryum pathogens to shift between Dianthus host species if there are insufficient physiological differences to favor pathogen specialization. Consistent with this hypothesis, very recently diverged Silene host species, e.g. S. caroliniana and S. virginica, or S. vulgaris and S. uniflora, also tend to share the same Microbotryum species (Freeman et al. 2002; Piątek et al. 2013). However, two important considerations should also be noted. First, de Vienne et al. (2009b) showed that D. carthusianorum was unusually susceptible to cross-inoculation of Microbotryum pathogens even when they originated from Silene or Saponaria species. Therefore, we cannot exclude the possibility that the lack of specialized affinities of the Microbotryum-Dianthus system may be due to broad susceptibility as a Dianthus ‘host trait’ separate from the recent radiation. Second, among the four Microbotryum lineages studied here, there was one almost entirely restricted to D. pavonius suggesting that it has strong host specialization. Clearly, broader sampling across host species and DNA based phylogenetic studies should be complemented by experimental inoculation to help resolve the causes of variation in host-pathogen affinities.
A major consequence of lacking one-to-one host-pathogen affinities is that Dianthus species were infected by multiple Microbotryum lineages, very often in sympatry at the local population scale. However, this pathogen sympatry was unlikely to have been due to transient host-shifts because the co-occurrence of different diseased host species was rare. Instead, the pathogen distributions suggest that Dianthus hosts may harbor endemic populations of quite distinct anther-smut fungi; in a few cases a local host population even contained three Microbotryum lineages. One outcome of this sympatry is expected to be direct competition among these pathogen lineages. Microbotryum on Silene has been well studied as a model of multiple infections of individual hosts (Koskella et al. 2006; Lopez-Villavicencio et al. 2007; Tollenaere et al. 2016), and it has been shown that resident infections tend to prevent colonization by subsequent pathogens. On Silene hosts this within-host exclusion was stronger between Microbotryum species than among genotypes within species (Gold et al. 2009), which is consistent with the rarity of co-infection on Dianthus observed here even if sympatric pathogen lineages might be deposited together on the host surface prior to infection. The principle of competitive exclusion (e.g. Levin 1970) indicates that such sympatry in an obligate pathogen like Microbotryum would be an unstable condition, if it was the only factor affecting coexistence. Therefore, further research should be directed toward the nature of the interactions between co-occurring Microbotryum lineages, including the products of hybridization between them, and other factors such as disease transmission and resistance dynamics that might impact co-existence or recent secondary contact within host populations. The issue is not completely unique to Microbotryum on Dianthus, as co-occurrence of distinct anther-smut species on the same host species, while very rare in Silene, does have a counterpart in S. vulgaris, where three distinct Microbotryum species are endemic and specialized to this host (Le Gac et al. 2007b, Piątek et al. 2012).
The second important outcome of co-occurring Microbotryum lineages on Dianthus hosts was the very high prevalence of admixed pathogen genotypes, assuming the cluster-assignment threshold of 0.9. This group of fungi from Silene hosts is known to have a mating system that is predominantly selfing, via automixis (Giraud et al. 2008a), but hybridization can occur and has played an historic role in the evolution of Microbotryum species (Devier et al. 2010). In extant populations, Gladieux et al. (2011) showed that genetic exchange between Microbotryum species was predicated on contact sites between two Silene host species, each carrying their own specialized pathogen species. In general, divergence in the niches occupied by two host species can contribute to the allopatry of their respective pathogens. The lack of ecological specialization may also be especially important to the disease on Dianthus, where the lack of strict affinities of Microbotryum lineages to different host species contributes to direct contact, in sympatry on the same host, and the resulting frequent hybridization. Although failure to detect both parental lineages in some populations may be due to small sample sizes, the lack of observing populations that consist of only admixed genotypes, without any cluster-assigned samples, is consistent with our finding that hybridization leads to reduced fitness in the form of significant sterility. Because mating occurs on the plant surface prior to infection, and thus prior to competitive exclusion, the mixing of spores during pollinator movement between plants might frequently give rise hybridization even if fertility of the resulting genotypes is low. The extent to which crosses between lineages might persist through early generations or that they serve as a route for introgression between the pathogen lineages remains unclear and could contribute to the observed range of cluster-assignment scores or incongruence among datasets based on different loci.
In summary, this study provides insights into the complex distribution of Microbotryum pathogens specialized to natural populations of Dianthus hosts. Importantly, several Microbotryum lineages on Dianthus appear to lack narrow and strict host affinities, and sympatric contact between them drives the probabilities of both direct competition and hybridization. The full consequences of these processes for pathogen dynamics and evolution remain to be investigated, beyond the apparent reduced fertility of admixed genotypes. However, similar contact between closely related pathogens is becoming recognized in a range of systems, such as for the avian malaria parasites (Fallon et al. 2005; Ricklefs et al. 2014), suggesting Microbotryum might be a tractable model for a broadly occurring phenomenon in disease ecology. Furthermore, the common occurrence of admixed genotypes, contrasting sharply with the general patterns in observed Microbotryum on Silene hosts (Gladieux et al. 2011), adds to recent calls for investigating the circumstances that promote hybridization and the potential evolutionary consequences on pathogen populations (King et al. 2015). Indeed, although secondary contacts have been extensively studied in animals and plants, they have been little explored in pathogens, where the few recent examples show that they can have especially dramatic consequences on disease emergence (Lemaire et al. 2016, Leroy et al. 2016; Stukenbrock 2016).
Supplementary Material
Acknowledgments
This research was supported by NSF awards DEB-1115765, DEB-0747222. We thank Valentina Carasso, Ivan Pace, and Bruno Gallino at the Parco naturale del Marguareis for help with field work, and Felix Horns, Daniel Peterson, Molly Scott, and Paul Tyler for research assistance.
Footnotes
Author contributions
MEH, EP and CS conceived and designed the project with contributions of JA and TG. CS, SY, MEH and JA performed field collections. CS, EP, LR, AC, PG, EB, SY, TG and MEH contributed to molecular analyses and interpretations. MEH, EP and CS drafted the manuscript with contributions by all authors.
Data accessibility
Online Supporting Information: Sampling locations. Dryad: Microsatellite data, Structure input files. GenBank: DNA sequences accessions.
References
- Alizon S, de Roode JC, Michalakis Y. Multiple infections and the evolution of virulence. Ecology Letters. 2013;16:556–567. doi: 10.1111/ele.12076. [DOI] [PubMed] [Google Scholar]
- Antonovics J, Hood M, Partain J. The ecology and genetics of a host shift: Microbotryum as a model system. The American Naturalist. 2002;160:S40–S53. doi: 10.1086/342143. [DOI] [PubMed] [Google Scholar]
- Bagchi R, Gallery RE, Gripenberg S, Gurr SJ, Narayan L, Addis CE, Freckleton RP, Lewis OT. Pathogens and insect herbivores drive rainforest plant diversity and composition. Nature. 2014;506:85–88. doi: 10.1038/nature12911. [DOI] [PubMed] [Google Scholar]
- Badouin H, Gladieux P, Gouzy J, Siguenza S, Aguileta G, Snirc A, Le Prieur S, Jeziorski C, Branca A, Giraud T. Widespread selective sweeps throughout the genome of model plant pathogenic fungi and identification of effector candidates. Molecular Ecology. 2017 doi: 10.1111/mec.13976. In press. [DOI] [PubMed] [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. GENETIX 4.05, logiciel sous Windows TM pour la genetique des populations. Laboratoire genome, populations, interactions, CNRS UMR, 5000. 1996:1996–2004. [Google Scholar]
- Benitez MS, Hersh MH, Vilgalys R, Clark JS. Pathogen regulation of plant diversity via effective specialization. Trends in Ecology & Evolution. 2013;28:705–711. doi: 10.1016/j.tree.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Bernasconi G, Antonovics J, Biere A, Charlesworth D, Delph LF, Filatov D, Giraud T, Hood ME, Marais GAB, McCauley D, Pannell J, Shykoff JA, Vyskot B, Wolfe L, Widmer A. Silene as a model system in ecology and evolution. Heredity. 2009;103:5–14. doi: 10.1038/hdy.2009.34. [DOI] [PubMed] [Google Scholar]
- Bever JD, Mangan SA, Alexander HM. Maintenance of plant species diversity by pathogens. Annual Review of Ecology, Evolution, and Systematics. 2015;46:305–325. [Google Scholar]
- Bickford D, Lohman DJ, Sodhi NS, Ng PK, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends in Ecology and Evolution. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Bolton S, Bon C. Pharmaceutical statistics: practical and clinical applications. CRC Press; 2009. [Google Scholar]
- Brasier C. The rise of the hybrid fungi. Nature. 2000;405:134–135. doi: 10.1038/35012193. [DOI] [PubMed] [Google Scholar]
- Bucheli E, Gautschi B, Shykoff JA. Host-specific differentiation in the anther smut fungus Microbotryum violaceum as revealed by microsatellites. Journal of Evolutionary Biology. 2000;13:188–198. [Google Scholar]
- Bucheli E, Gautschi B, Shykoff J. Differences in population structure of the anther smut fungus Microbotryum violaceum on two closely related host species, Silene latifolia and S. dioica. Molecular Ecology. 2001;10:285–294. doi: 10.1046/j.1365-294x.2001.01146.x. [DOI] [PubMed] [Google Scholar]
- Bueker B, Eberlein C, Gladieux P, Schaefer A, Snirc A, Bennett DJ, Begerow D, Hood ME, Giraud T. Distribution and population structure of the anther smut Microbotryum silenes-acaulis parasitizing an arctic–alpine plant. Molecular Ecology. 2016;25:811–24. doi: 10.1111/mec.13512. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Lauring AS. Theory and empiricism in virulence evolution. PLoS Pathogen. 2014;10:e1004387. doi: 10.1371/journal.ppat.1004387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Giraud T, Zhang N, Begerow D, Cai G, Shivas RG. The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Diversity. 2011;50:121–133. [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or the preservation of favored races in the struggle for life. Murray; London, England, UK: 1859. [Google Scholar]
- Day AW. Competition and distribution studies of genetically marked strains of Ustilago violacea in the same host plant. Botanical Gazette. 1980;141:313–320. [Google Scholar]
- de Leon GPP, Nadler SA. What we don’t recognize can hurt us: a plea for awareness about cryptic species. Journal of Parasitology. 2010;96:453–464. doi: 10.1645/GE-2260.1. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Refregier G, Hood ME, Guigue A, Devier B, Vercken E, Smadja C, Deseille A, Giraud T. Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. Journal of Evolutionary Biology. 2009a;22:683–698. doi: 10.1111/j.1420-9101.2009.01702.x. [DOI] [PubMed] [Google Scholar]
- de Vienne DM, Hood ME, Giraud T. Phylogenetic determinants of potential host shifts in fungal pathogens. Journal of Evolutionary Biology. 2009b;22:2532–2541. doi: 10.1111/j.1420-9101.2009.01878.x. [DOI] [PubMed] [Google Scholar]
- Denchev CM, Giraud T, Hood ME. Three new species of anthericolous smut fungi on Caryophyllaceae. Mycologica Balcanica. 2009;6:79–84. [Google Scholar]
- Desprez-Loustau ML, Robin C, Buee M, Courtecuisse R, Garbaye J, Suffert F, Sache I, Rizzo DM. The fungal dimension of biological invasions. Trends in Ecology and Evolution. 2007;30:472–80. doi: 10.1016/j.tree.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Devier B, Aguileta G, Hood ME, Giraud T. Using phylogenies of pheromone receptor genes in the Microbotryum violaceum species complex to investigate possible events of speciation by hybridization. Mycologia. 2010;102:689–696. doi: 10.3852/09-192. [DOI] [PubMed] [Google Scholar]
- Dobzhansky T. Role of the autosomes in the Drosophila pseudoobscura hybrids. Proceedings of the National Academy of Sciences. 1933;19:950–953. doi: 10.1073/pnas.19.11.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson A, Lafferty KD, Kuris AM, Hechinger RF, Jetz W. Homage to Linnaeus: How many parasites? How many hosts? Proceedings of the National Academy of Sciences. 2008;105:S11482–S11489. doi: 10.1073/pnas.0803232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon SM, Bermingham E, Ricklefs RE. Host specialization and geographic localization of avian malaria parasites: A regional analysis in the Lesser Antilles. The American Naturalist. 2005;165:466–480. doi: 10.1086/428430. [DOI] [PubMed] [Google Scholar]
- Feurtey A, Gladieux P, Snirc A, Hood ME, Cornille A, Giraud T. Strong phylogeographic costructure between the anther smut fungus and its white campion host. New Phytologist. 2016 doi: 10.1111/nph.14191. in press. [DOI] [PubMed] [Google Scholar]
- Fontaine MC, Gladieux P, Hood ME, Giraud T. History of the invasion of the anther smut pathogen on Silene latifolia in North America. New Phytologist. 2013;198:946–56. doi: 10.1111/nph.12177. [DOI] [PubMed] [Google Scholar]
- Freeman AB, Duong KK, Shi TL, Hughes CF, Perlin MH. Isolates of Microbotryum violaceum from North American host species are phylogenetically distinct from their European host-derived counterparts. Molecular Phylogenetics and Evolution. 2002;23:158–170. doi: 10.1016/S1055-7903(02)00003-9. [DOI] [PubMed] [Google Scholar]
- Giraud T. Patterns of within population dispersal and mating of the fungus Microbotryum violaceum parasitizing the plant Silene latifolia. Heredity. 2004;93:559–65. doi: 10.1038/sj.hdy.6800554. [DOI] [PubMed] [Google Scholar]
- Giraud T, Fournier E, Vautrin D, Solignac M, Shykoff JA. Isolation of 44 polymorphic microsatellite loci in three host races of the phytopathogenic fungus Microbotryum violaceum. Molecular Ecology Notes. 2002;2:142–146. [Google Scholar]
- Giraud T, Villareal L, Austerlitz F, Le Gac M, Lavigne C. Importance of the life cycle in sympatric host race formation and speciation of pathogens. Phytopathology. 2006;96:280–287. doi: 10.1094/PHYTO-96-0280. [DOI] [PubMed] [Google Scholar]
- Giraud T, Yockteng R, Lopez-Villavicencio M, Refregier G, Hood M. Mating system of the anther smut fungus Microbotryum violaceum: selfing under heterothallism. Eukaryotic Cell. 2008a;7:765–775. doi: 10.1128/EC.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud T, Tockteng R, Marthey S, Chiapello H, Jonot O, Lopez-Villavicencio M, De Vienne DM, Hood ME, Refregier G, Gendrault-Jacquemard A, Wincker P. Permanent genetic resources: isolation of 60 polymorphic microsatellite loci in EST libraries of four sibling species of the phytopathogenic fungus complex Microbotryum. Molecular Ecology Resources. 2008b;8:387–392. doi: 10.1111/j.1471-8286.2007.01967.x. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Vercken E, Fontaine MC, Hood ME, Jonot O, Couloux A, Giraud T. Maintenance of fungal pathogen species that are specialized to different hosts: allopatric divergence and introgression through secondary contact. Molecular Biology and Evolution. 2011;28:459–471. doi: 10.1093/molbev/msq235. [DOI] [PubMed] [Google Scholar]
- Gladieux P, Feurtey A, Hood ME, Snirc A, Clavel J, Dutech C, Roy M, Giraud T. The population biology of fungal invasions. Molecular Ecology. 2015;24:1969–1986. doi: 10.1111/mec.13028. [DOI] [PubMed] [Google Scholar]
- Gold A, Giraud T, Hood ME. Within-host competitive exclusion among species of the anther-smut pathogen. BMC Ecology. 2009;9:11. doi: 10.1186/1472-6785-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschmidt V. Vererbungsversuche mit den biologischen Arten des Antherenbrandes (Ustilago violacea Pers.) Zeitschrift fuer Botanik. 1928;21:1–90. [Google Scholar]
- Hood ME, Antonovics J. Intratetrad mating, heterozygosity, and the maintenance of deleterious alleles in Microbotryum violaceum (=Ustilago violacea) Heredity. 2000;85:231–241. doi: 10.1046/j.1365-2540.2000.00748.x. [DOI] [PubMed] [Google Scholar]
- Hood ME. Dynamics of multiple infection and within-host competition by the anther-smut pathogen. The American Naturalist. 2003;162:122–133. doi: 10.1086/375539. [DOI] [PubMed] [Google Scholar]
- Hood ME, Mena-Ali JI, Gibson AK, Oxelman B, Giraud T, Yockyeng R, Arroyo MTK, Conti F, Pedersen A, Gladieux P, Antonovics J. Distribution of the anther-smut pathogen Microbotryum on species of the Caryophyllaceae. New Phytologist. 2010;187:217–229. doi: 10.1111/j.1469-8137.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SA, O’Kelly MJ, Carter DM, Davey RP, van Oudenaarden A, Roberts IN. Repetitive sequence variation and dynamics in the ribosomal DNA array of Saccharomyces cerevisiae as revealed by whole-genome resequencing. Genome Research. 2009;19:626–635. doi: 10.1101/gr.084517.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemler M, Lutz M, Goker M, Oberwinkler F, Begerow D. Hidden diversity in the non-caryophyllaceous plant-parasitic members of Microbotryum (Pucciniomycotina: Microbotryales) Systematics and Biodiversity. 2009;7:297–306. [Google Scholar]
- Kemler M, Martin MP, Telleria MT, Schafer AM, Yurkov A, Begerow D. Contrasting phylogenetic patterns of anther smuts (Pucciniomycotina: Microbotryum) reflect phylogenetic patterns of their caryophyllaceous hosts. Organisms Diversity and Evolution. 2013;13:111–126. [Google Scholar]
- King KC, Stelkens RB, Webster JP, Smith DF, Brockhurst MA. Hybridization in parasites: consequences for adaptive evolution, pathogenesis, and public health in a changing world. PLoS Pathogen. 2015;11:e1005098. doi: 10.1371/journal.ppat.1005098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B, Giraud T, Hood ME. Pathogen relatedness affects the prevalence of within-host competition. The American Naturalist. 2006;168:121–126. doi: 10.1086/505770. [DOI] [PubMed] [Google Scholar]
- Kruckeberg AR. Artificial crosses of Western North American Silenes. Brittonia. 1961;13:305–333. [Google Scholar]
- Le Gac M, Hood ME, Giraud T. Evolution of reproductive isolation within a parasitic fungal species complex. Evolution. 2007a;61:1781–1787. doi: 10.1111/j.1558-5646.2007.00144.x. [DOI] [PubMed] [Google Scholar]
- Le Gac M, Hood M, Fournier E, Giraud T. Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution. 2007b;61:15–26. doi: 10.1111/j.1558-5646.2007.00002.x. [DOI] [PubMed] [Google Scholar]
- Lemaire C, De Gracia M, Leroy T, Michalecka M, Lindhard-Pedersen H, Guerin F, Gladieux P, Le Cam B. Emergence of new virulent populations of apple scab from nonagricultural disease reservoirs. New Phytologist. 2016;209:1220–1229. doi: 10.1111/nph.13658. [DOI] [PubMed] [Google Scholar]
- Leroy T, Caffier V, Celton JM, Anger N, Durel CE, Lemaire C, Le Cam B. When virulence originates from nonagricultural hosts: evolutionary and epidemiological consequences of introgressions following secondary contacts in Venturia inaequalis. New Phytologist. 2016;210:1443–1452. doi: 10.1111/nph.13873. [DOI] [PubMed] [Google Scholar]
- Levin SA. Community equilibria and stability, and an extension of the competitive exclusion principle. The American Naturalist. 1970;104:413–423. [Google Scholar]
- Lopez-Villavicencio M, Jonot O, Coantic A, Hood ME, Enjalbert J, Giraud T. Multiple infections by the anther smut pathogen are frequent and involve related strains. PLoS Pathogen. 2007;3:e176. doi: 10.1371/journal.ppat.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Villavicencio M, Courjol F, Gibson AK, Hood ME, Jonot O, Shykoff JA, Giraud T. Competition, cooperation among kin, and virulence in multiple infections. Evolution. 2011;65:1357–66. doi: 10.1111/j.1558-5646.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- Lutz M, Piątek M, Kemler M, Chlebicki A, Oberwinkler F. Anther smuts of Caryophyllaceae: molecular analyses reveal further new species. Mycological Research. 2008;112:1280–96. doi: 10.1016/j.mycres.2008.04.010. [DOI] [PubMed] [Google Scholar]
- Menardo F, Praz CR, Wyder S, Ben-David R, Bourras S, Matsumae H, McNally KE, Parlange F, Riba A, Roffler S, Schaefer LK. Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nature Genetics. 2016;48:201–5. doi: 10.1038/ng.3485. [DOI] [PubMed] [Google Scholar]
- Newcombe G, Stirling B, McDonald S, Bradshaw HD. Melampsora× columbiana, a natural hybrid of M. medusae and M. occidentalis. Mycological Research. 2000;104:261–74. [Google Scholar]
- Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012;28:2537–2539. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piątek M, Lutz M, Ronikier A, Kemler M, Świderska-Burek U. Microbotryum heliospermae, a new anther smut fungus parasitic on Heliosperma pusillum in the mountains of the European Alpine System. Fungal Biology. 2012;116:185–195. doi: 10.1016/j.funbio.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Piątek M, Lutz M, Kemler M. Microbotryum silenes-saxifragae sp. nov. sporulating in the anthers of Silene saxifraga in southern European mountains. IMA fungus. 2013;4:29–40. doi: 10.5598/imafungus.2013.04.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refregier G, Le Gac M, Jabbour F, Widmer A, Shykoff JA, Yockteng R, Hood ME, Giraud T. Cophylogeny of the anther smut fungi and their caryophyllaceous hosts: Prevalence of host shifts and importance of delimiting parasite species for inferring cospeciation. BMC Evolutionary Biology. 2008;8:100. doi: 10.1186/1471-2148-8-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refregier G, Hood M, Giraud T. No Evidence of Reproductive Character displacement between two sister fungal species causing anther smut disease in Silene. International Journal of Plant Sciences. 2010;171:847–859. [Google Scholar]
- Rice W. Speciation via habitat specialization: the evolution of reproductive isolation as a correlated character. Evolutionary Ecology. 1987;1:301–314. [Google Scholar]
- Ricklefs RE, Outlaw DC, Svensson-Coelho M, Medeiros MC, Ellis VA, Latta S. Species formation by host shifting in avian malaria parasites. Proceedings of the National Academy of Sciences. 2014;111:14816–14821. doi: 10.1073/pnas.1416356111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH. Evolution, selection and isolation: a genomic view of speciation in fungal plant pathogens. New Phytologist. 2013;199:895–907. doi: 10.1111/nph.12374. [DOI] [PubMed] [Google Scholar]
- Stukenbrock EH. The role of hybridization in the evolution and emergence of new fungal plant pathogens. Phytopathology. 2016;106:104–12. doi: 10.1094/PHYTO-08-15-0184-RVW. [DOI] [PubMed] [Google Scholar]
- Schardl CL, Craven KD. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Molecular Ecology. 2003;12:2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x. [DOI] [PubMed] [Google Scholar]
- Thomas A, Shykoff J, Jonot O, Giraud T. Sex-ratio bias in populations of the phytopathogenic fungus Microbotryum violaceum from several host species. International Journal of Plant Sciences. 2003;164:641–647. [Google Scholar]
- Thrall PH, Jarosz AM. Host-pathogen dynamics in experimental populations of Silene alba and Ustilago violacea. I. Ecological and genetic determinants of disease spread. Journal of Ecology. 1994;82:549–559. [Google Scholar]
- Tollenaere C, Susi H, Laine AL. Evolutionary and epidemiological implications of multiple infection in plants. Trends in Plant Science. 2016;21:80–90. doi: 10.1016/j.tplants.2015.10.014. [DOI] [PubMed] [Google Scholar]
- Valente L, Savolainen V, Vargas P. Unparalleled rates of species diversification in Europe. Proceedings of the Royal Society, B. 2010;277:1489–1496. doi: 10.1098/rspb.2009.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vienne DM, Refregier G, Hood ME, Guigue A, Devier B, Vercken E, Smadja C, Deseille A, Giraud T. Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. Journal of Evolutionary Biology. 2009;22:683–698. doi: 10.1111/j.1420-9101.2009.01702.x. [DOI] [PubMed] [Google Scholar]
- Zillig H. Ueber spezialisierte Formen beim Antherenbrand, Ustilago violacea (Pers.) Fuckel. Zentralblatt fuer Bakteriologie und Parasitenkunde. 1921;53:33–73. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.