Abstract
The role of host epigenetic mechanisms in the natural history of low-grade cervical intraepithelial neoplasia (CIN1) is not well characterized. We explored differential methylation of imprinted gene regulatory regions as predictors of the risk of CIN1 regression.
A total of 164 patients with CIN1 were recruited from 10 Duke University clinics for the CIN Cohort Study. Participants had colposcopies at enrollment and up to five follow-up visits over three years. DNA was extracted from exfoliated cervical cells for methylation quantitation at CpG (cytosine-phosphate-guanine) sites and human papillomavirus (HPV) genotyping. Hazard ratios (HR) and 95% confidence intervals (CI) were estimated using Cox regression to quantify the effect of methylation on CIN1 regression over two consecutive visits, compared to non-regression (persistent CIN1; progression to CIN2+; or CIN1 regression at a single time-point), adjusting for age, race, high-risk HPV (hrHPV), parity, oral contraceptive and smoking status.
Median participant age was 26.6 years (range: 21–64.4 years), 39.0% were African-American, and 11% were current smokers. Most participants were hrHPV-positive at enrollment (80.5%). Over one-third of cases regressed (n=53, 35.1%). Median time-to-regression was 12.6 months (range: 4.5–24.0 months). Probability of CIN1 regression was negatively correlated to methylation at IGF2AS CpG 5 (HR=0.41; 95% CI=0.23–0.77) and PEG10 DMR (HR=0.80; 95% CI=0.65–0.98).
Altered methylation of imprinted IGF2AS and PEG10 DMRs may play a role in the natural history of CIN1. If confirmed in larger studies, further research on imprinted gene DMR methylation is warranted to determine its efficacy as a biomarker for cervical cancer screening.
Keywords: methylation, imprinted genes, cervical cancer, neoplasia, epigenetics, HPV
Introduction
As of 2014, an estimated 250,000 women were living with cancer of the cervix in the United States (US).1 At current incidence and mortality rates, approximately 13,000 women will be diagnosed with cervical cancer in the US in 2017, resulting in over 4,000 subsequent deaths.2 Though overall rates of cervical cancer in the US have decreased over time, the highest rates of cervical cancer incidence and mortality occur in the Southern states.2
Nearly all invasive cervical cancers are caused by the human papillomavirus (HPV), a sexually transmitted infection that affects over 79 million people in the US.3–5 While most HPV infections clear spontaneously, oncogenic or high-risk HPV (hrHPV) types often lead to persistent HPV infection and subsequent high-grade cervical intraepithelial neoplasia (CIN2+), a risk factor for progression to invasive cervical cancer.6
Current cervical cancer prevention strategies include the use of cytology-based testing (Pap testing) as a primary screening tool, with the addition of HPV testing to increase screening sensitivity for the detection of CIN2+ among women 30 years and older, as well as primary hrHPV screening.7 HrHPV testing is more sensitive, although less specific, than liquid-based cytology for the detection of high-grade (CIN2+).8 A relatively small proportion of low-grade CIN cases progress to CIN2+, while most CIN1 cases regress to normal epithelia.9 Follow-up of low-grade CIN is recommended in the US until regression to normal colposcopic impression or negative cytology, leading to a high burden of cost and decreased clinical visit adherence.7, 10 Therefore, it would be advantageous to identify novel biomarkers that can differentiate CIN1 cases which progress from CIN1 cases which regress.
Epigenetic profiles have been hypothesized as potential diagnostic biomarkers for susceptibility to cervical cancer.11, 12 Modifications of the epigenome include DNA methylation at cytosine-guanine dinucleotide sequences (CpG sites) which can affect the expression of genes involved in cancer tumorigenesis.13 Genomic imprinting involves inheritance of parent-of-origin specific epigenetic modifications controlling allele-specific gene expression.12, 13 Imprinted genes often exist in clusters and are regulated by imprinting centers, which can include differentially methylated regions (DMRs) that are rich in CpG sites.13
Loss of imprinting (LOI) due to aberrant methylation at DMRs has been linked to various growth and developmental disorders,14 including Beckwith-Wiedemann Syndrome (BWS).15 In case-control studies, differential methylation of targeted imprinted genes has been associated with cancer outcomes, such as Wilms’ Tumor of the kidney,16 breast cancer,17, 18 colorectal cancer,11, 19, 20 and prostate cancer.21 Preliminary analyses have also found dysregulated expression of imprinted genes involved in tumor suppression (e.g. HYMAI, PEG3, PLAGL1, MEST, CDKN1C) in cervical cancer specimens compared to normal cervical tissue.22 Studies have examined the influence of methylation patterns on the expression of HPV E6/E7 oncogenic proteins which deactivate host cell tumor suppressor p53 and thus may promote cervical carcinogenesis.23, 24 The influence of host aberrant methylation at imprinted gene control regions on the natural history of low-grade CIN has not been assessed.
It is important to establish molecular-based methods of differentiating CIN1 cases which progress versus regress to improve clinical management. The current study examines whether aberrant DNA methylation patterns of imprinted genes influence regression of low-grade CIN in the Cervical Intraepithelial Neoplasia Cohort Study (CINCS).
Materials and Methods
Study population
From June 2010 – April 2014, women attending ten Duke University hospitals and clinics in Durham, North Carolina were invited to participate in CINCS, as previously described.25 Briefly, all clinics used Duke-affiliated pathology laboratories for cytology and histological evaluation. The CINCS cohort is comprised of 1,303 women who were referred for a colposcopy following an abnormal liquid-based cytology result. Participants were eligible if they provided written consent, were new visitors to the clinic, 21–79 years old, English or Spanish speakers, and able to give informed consent. We excluded women who had received previous treatment for cervical lesions—cold knife conization (CKC), electrosurgical excision procedure (LEEP), cryotherapy, or hysterectomy; had moved out of the study area; or did not intend to receive follow-up care at one of the 10 Duke clinics. Women who were diagnosed with CIN1 at enrollment and had at least one follow-up visit with HPV and methylation data were included in the present statistical analyses. Approval for this study was granted by the Institutional Review Boards at Duke University (Durham, NC, USA), North Carolina State University (Raleigh, NC, USA) and University of North Carolina (Chapel Hill, NC, USA).
Data collection and laboratory analyses
At enrollment, participants had a physician-directed cervical examination with a colposcopy-directed biopsy. Women diagnosed with CIN1 by colposcopic impression without biopsy (n=29) at enrollment were also included in the study, as prevalence of hrHPV (63%) was comparable to the hrHPV prevalence among those who underwent a biopsy (59%). Study participants attended a clinical visit approximately every 6 months for the first two years, and every 12 months for the final third year. During each follow-up visit, all women underwent a liquid-based cytology (LBC) test. For participants with abnormal cytology results, clinic physicians performed colposcopy examination. Directed biopsies at follow-up colposcopy visits occurred only if clinically necessary, according to the physician’s best judgement and per clinical guidelines for management of precancerous cervical lesions.7 Study staff administered a questionnaire to ascertain information on any behavioral and clinical characteristics at enrollment and follow-up visits, including age, race/ethnicity, current smoking status, history of oral contraceptive use and parity.
Ascertainment of Cervical Cytology and Histology
To conduct a LBC test, the clinic physician utilized a spatula and cytobrush to obtain exfoliated cervical cells. Cervical exfoliated cell specimens were suspended in a vial containing ThinPrep® solution (Hologic®, Malborough, MA, USA) for cytological assessment. All study clinic pathologists evaluated LBC cytology according to Bethesda criteria.26 The residual LBC cervical exfoliated cell specimens were stored at 4°C prior to HPV DNA testing.
Biopsy results were also reviewed and graded for severity by a pathologist at Duke-affiliated pathology laboratories. All histological biopsy specimens were tested for adequacy using the 2012 American Society for Colposcopy and Cervical Pathology (ASCCP) guidelines.7 Information on cytology and histology were abstracted from patient medical records.
HPV Testing and Typology
HPV typology was assessed using cervical exfoliated cells from the enrollment pelvic exam. ThinPrep® specimens were collected during the same enrollment visit and sent to Johns Hopkins University and the University of Hawaii Cancer Center for laboratory testing, as previously described.27, 28 Following DNA extraction, HPV status was determined by targeted amplification of a 450bp region of the HPV L1 genome using PGMY09/PGMY11 primers.27, 28 Amplification of the human β-globin gene was included as an internal control for sample sufficiency. Specimens identified as HPV-positive were subsequently genotyped using the HPV Linear Array® (Roche Diagnostics, Branchburg, NJ, USA). This assay is designed up to 37 high-risk and low-risk genotypes.
Assessment of DNA Methylation in Imprinted Differentially Methylated Regions (DMRs)
Nucleic acid extraction
DNA was extracted from the LBC cell pellet using a protocol for simultaneous nucleic acid extraction provided by Teltest (Friendswood, TX) for DNA Stat60 reagents. Nucleic acids were aliquoted, barcoded, and stored at −80°C until required.
DNA methylation analysis
DNA methylation was measured using genomic DNA at differentially methylated regions (DMRs) regulating genomic imprinting of IGF2/H19, IGF2AS (IGF2-antisense), MESTIT1/MEST, KvDMR, MEG3, PLAGL1/HYMAI, and PEG3, PEG10 imprinted domains, using Sequenom (San Diego, CA) MassARRAY EpiTYPER assays. Bisulfite-treated DNA was processed using the EZ-96 DNA Methylation Kit (Zymo Research Corporation, Irvine, CA) to convert unmethylated DNA cytosine bases to uracil bases, leaving methylated cytosines unchanged per manufacturer’s protocol. We used Sequenom (San Diego, CA) EpiDesigner software to design primers complementary to bisulfite-converted DNA in regions without CpG nucleotides, adding a T7 promoter site to all forward primers. Polymerase chain reaction (PCR) assays were performed on the treated DNA samples using HotStarTaq DNA Polymerase (Qiagen, Hilden, Germany). PCR products were treated with Shrimp alkaline phosphatase (SAP, Sequenom, San Diego, CA) followed by transcription and T cleavage reactions according to the protocol. Cleanup and sequencing were performed according to the EpiTYPER user guide. Matrix-assisted laser desorption/ionization and time-of-flight mass (MALDI-TOF) spectrometry analysis was performed on resulting transcripts using the MassARRAY system (Sequenom). Fragments generated from the PCR assay differed by size and mass, allowing for quantification of methylated forms of each targeted fragment.
Statistical analyses
Methylation percentage was calculated at each CpG site of each imprinted gene DMR. Because imprinted gene DMRs are characterized by having one fully methylated allele and one unmethylated allele, the expected value of methylation for an imprinted gene in a diploid cell is approximately 50%. A total of 8 imprinted DMRs were considered a priori with 5 CpG sites for the IGF2/H19 DMR; 10 CpGs for the IGF2AS DMR; 31 CpGs for the MEST/MESTIT1 DMR; 27 CpG sites for the Kv DMR; 31 CpG sites for the MEG3 DMR; 8 CpG sites for the PLAGL1/HYMAI DMR; 11 CpG sites for the PEG10 DMR; and 12 CpG sites for the PEG3 DMR. Genomic coordinates for each DMR have been previously published.29 Four DMRs were excluded from the analyses due to extensive missing data at CpG sites. As a result, DMRs in the analysis included IGF2AS, MEG3, PEG10 and the Kv DMR. Median percentages were calculated across DMRs to estimate methylation for cis-acting CpGs at a given region12.
Regression of cervical lesions was defined as a diagnosis of negative/normal cytology (or histology if applicable) at two consecutive follow-up visits. Cytology results were utilized to determine regression status if the participants had missing histology data given no biopsy was performed, per conservative clinical practice. Women with a negative/normal screening cytology or histology at one follow-up time point only (e.g. regressed to negative/normal at the first follow-up visit, and had cervical abnormalities at the subsequent visit) were not considered to have regressed for the main study analyses. Cervical lesion persistence was defined as a diagnosis of low-grade histology (CIN1) at follow-up or low-grade lesions during cytology testing (e.g. low-grade squamous epithelial lesions (LSIL), or atypical squamous cells of undetermined significance (ASC-US). Progression was defined as a follow-up histological diagnosis of CIN2+, or as a cytological diagnosis of high-grade squamous epithelial lesions (HSIL), LSIL-H (LSIL, cannot exclude HSIL), or ASC-H (ASC, cannot exclude HSIL). For women who received treatment (LEEP, CKC, cryotherapy, or hysterectomy) at a follow-up visit, the histological diagnosis from the pre-treatment specimen was utilized.
A univariate analysis was performed to assess the distribution of methylation biomarkers and covariates. Kaplan-Meier product-limit method was used to estimate the cumulative proportion of CIN1 regression, stratified by median methylation percentage at each DMR. The Log-rank test was used to assess differences between regression probabilities over time at methylation percentages below and above the median for each DMR (Figures 1a–1d). Cox proportional hazards regression models were employed to estimate unadjusted hazard ratios (HR) and 95% confidence intervals (95% CI) to examine associations between methylation at a specific CpG site, and CIN1 regression. Time-to-regression was measured from the date of enrollment to the date of the second consecutive negative/normal histological or cytological diagnosis. Participants contributed person-time to the longitudinal analyses up to the occurrence of regression or the date of the last attended clinical study visit. Participants who received treatment during the study were right censored at the date of procedure. Administrative censoring occurred at 3 years. Woman-months were calculated as the sum of person-time for all women at risk among each specific methylation exposure group.
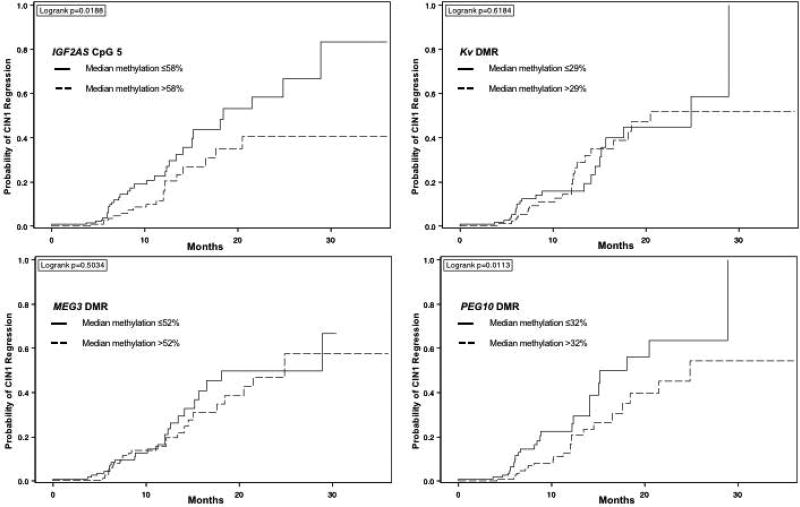
Figure 1.
a. Time to CIN1 regression for IGF2AS (at CpG 5), stratified at median methylation percentage*
*CIN = cervical intraepithelial neoplasia
b. Time to CIN1 regression for Kv DMR, stratified at median methylation percentage*
* CIN = cervical intraepithelial neoplasia; DMR = differentially methylated region (median methylation of all CpG sites)
c. Time to CIN1 regression, stratified at median MEG3 DMR methylation percentage*
* CIN = cervical intraepithelial neoplasia; DMR = differentially methylated region (median methylation of all CpG sites)
d. Time to CIN1 regression, stratified at median PEG10 DMR methylation percentage*
* CIN = cervical intraepithelial neoplasia; DMR = differentially methylated region (median methylation of all CpG sites)
We calculated a median methylation percentage to represent a summary measure of methylation across each candidate region (when applicable) to estimate HRs and 95% CIs in the univariate and multivariate Cox regression models. In the Cox models, methylation levels (treated as a continuous variable) were rescaled using the interquartile range (IQR) for each CpG site or the IQR for median methylation across the gene DMR. Confounders selected for the multivariable Cox regression model were determined a priori using conceptual models (directed acyclic graphs).30 Covariates considered for the analyses included continuous age at enrollment, race/ethnicity, current smoking status at enrollment (current vs. non-current), history of oral contraceptive use (ever vs. never), parity (continuous), and hrHPV infection at enrollment. We considered a 2-level hrHPV variable (16,18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 66, 68) infection vs. non-hrHPV infection, as well 3-level hrHPV variable (HPV-16/18 infection vs. non-HPV-16/18 hrHPV infection vs. lrHPV/no infection). Covariate modification was assessed using Akaike Information Criteria for model fit.
A sensitivity analysis was conducted to determine the change in estimate given a regression event at CIN1 regression at one follow-up visit. Further sensitivity analyses were also conducted to determine the impact of drop outs; exclusion of women who were hrHPV negative at enrollment; and the exclusion of women who had high-grade cytology (HSIL or higher) at their enrollment pap (preceding the enrollment colposcopy). All statistical analyses were conducted using SAS version 9.4 (SAS Institute, Cary, NC).
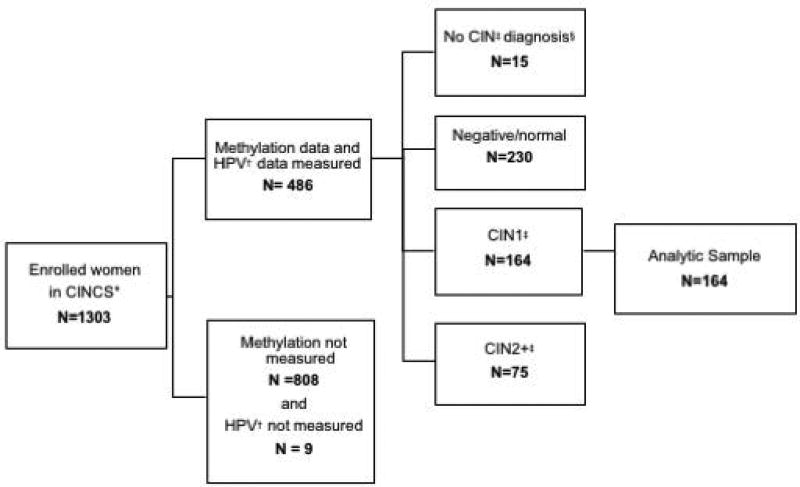
Of the 1,303 enrolled CINCS participants, 495 participant samples were tested for methylation (38%), of which 486 (98%) had HPV DNA laboratory results. Of these, 15 women with no CIN diagnosis, 230 women with a negative/normal histological diagnosis at enrollment, and 75 women with CIN2+ histological diagnosis were excluded. The remaining 164 CIN1 cases at enrollment were included in analyses.
Results
Median age of CIN1 cases at enrollment (n=164) was 26.6 years (range: 21–64.4 years; Table 1). Nearly half of participants were non-Hispanic White (47%), while over one-third were Black (39%). Only 11% of participating women were current smokers, whereas most had a history of oral contraceptive use (78%). Over 80% of participants had laboratory-confirmed infection with any hrHPV type at enrollment (81.3%). A total of 25 women had infection with either HPV16 or 18: 12% had HPV-16 infection, 5.3% with HPV-18 and 0.6% with both. The most prevalent hrHPV genotypes in the sample were HPV 66 (19.9%) and HPV 51 (13.3%; Table A1 in Appendix).
Table 1.
Characteristics of 164 women with CIN1 at enrollment in the CINCS Study*
| Enrollment characteristic | N | (Range)% | |
|---|---|---|---|
| Age (years) | |||
| Median | 26.6 | (21.0–64.4) | |
| 18–24 | 65 | 39.6 | |
| 25–29 | 56 | 34.2 | |
| 30–34 | 13 | 7.9 | |
| 35+ | 30 | 18.3 | |
| High-Risk HPV† | |||
| Negative | 32 | 19.5 | |
| Non-16/18 Positive‡ | 106 | 64.6 | |
| 16/18 Positive | 26 | 15.9 | |
| Race | |||
| Non-Hispanic White | 77 | 47.0 | |
| Black/African-American | 64 | 39.0 | |
| Other§ | 23 | 14.0 | |
| Current Smoker | |||
| No | 146 | 89.0 | |
| Yes | 18 | 11.0 | |
| Ever Use of Oral Contraceptives‖ | |||
| No | 33 | 21.7 | |
| Yes | 119 | 78.3 | |
| Parity‖ | |||
| Nulliparous | 93 | 57.4 | |
| Primiparous (1) | 29 | 17.9 | |
| Multiparous (2+) | 40 | 24.7 | |
CIN = Cervical Intraepithelial Neoplasia; CINCS = Cervical Intraepithelial Neoplasia Cohort Study
HPV=human papillomavirus
Includes high-risk HPV types 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68
Other includes Hispanic/Asian/Pacific Islander/Native American/Multiracial
Numbers do not add up to the total sample size due to missing data
Median DMR methylation levels were 58% for IGF2AS; 29% for Kv DMR, 52% for MEG3, and 32% for PEG10 (Table 2). Within the Kv and PEG10 DMRs, there was ≤30% methylation at most CpG sites. There was little variation in Kv DMR methylation (DMR IQR = 0.07) and in PEG10 methylation (DMR IQR = 0.07) among participating women with CIN1.
Table 2.
| Gene Name |
CpG site | N† | Mean | Median | SD‡ | Interquartile Range |
Range |
|---|---|---|---|---|---|---|---|
| IGF2AS | 5 | 157 | 0.47 | 0.58 | 0.32 | 0.57 | (0.00, 0.96) |
|
| |||||||
| Kv | |||||||
| DMR§ | 144 | 0.30 | 0.29 | 0.05 | 0.07 | (0.16, 0.42) | |
|
|
|||||||
| 1 | 143 | 0.25 | 0.26 | 0.06 | 0.07 | (0.12, 0.39) | |
| 6 | 144 | 0.25 | 0.26 | 0.06 | 0.08 | (0.08, 0.41) | |
| 8,9 | 144 | 0.27 | 0.27 | 0.05 | 0.07 | (0.03, 0.40) | |
| 10,11,12 | 142 | 0.23 | 0.23 | 0.06 | 0.07 | (0.03, 0.38) | |
| 15 | 144 | 0.30 | 0.31 | 0.06 | 0.08 | (0.14, 0.43) | |
| 17,18 | 144 | 0.30 | 0.31 | 0.05 | 0.07 | (0.14, 0.49) | |
| 20 | 144 | 0.31 | 0.31 | 0.06 | 0.06 | (0.10, 0.47) | |
| 21 | 144 | 0.30 | 0.30 | 0.06 | 0.05 | (0.12, 0.47) | |
| 22 | 144 | 0.24 | 0.24 | 0.04 | 0.04 | (0.12, 0.43) | |
| 24 | 144 | 0.29 | 0.30 | 0.08 | 0.10 | (0.11, 0.44) | |
| 26,27 | 144 | 0.55 | 0.55 | 0.06 | 0.07 | (0.35, 0.70) | |
|
| |||||||
| MEG3‖ | |||||||
| DMR§ | 152 | 0.49 | 0.52 | 0.11 | 0.19 | (0.26, 0.82) | |
|
|
|||||||
| 3 | 152 | 0.57 | 0.59 | 0.16 | 0.17 | (0.11, 1.00) | |
| 6 | 150 | 0.54 | 0.54 | 0.09 | 0.28 | (0.31, 0.96) | |
| 15 | 152 | 0.40 | 0.39 | 0.10 | 0.14 | (0.00, 0.80) | |
| 16 | 150 | 0.38 | 0.38 | 0.10 | 0.15 | (0.00, 0.82) | |
| 20,21 | 150 | 0.43 | 0.42 | 0.09 | 0.14 | (0.05, 0.75) | |
| 22 | 150 | 0.54 | 0.54 | 0.09 | 0.17 | (0.31, 0.96) | |
| 23 | 152 | 0.46 | 0.46 | 0.10 | 0.14 | (0.21, 0.95) | |
| 26,27,28 | 152 | 0.55 | 0.55 | 0.11 | 0.21 | (0.03, 0.89) | |
| 29,30,31 | 152 | 0.57 | 0.56 | 0.12 | 0.19 | (0.12, 1.00) | |
|
| |||||||
| PEG10 | |||||||
| DMR§ | 140 | 0.30 | 0.32 | 0.08 | 0.07 | (0.00, 0.45) | |
|
|
|||||||
| 5 | 137 | 0.31 | 0.32 | 0.10 | 0.07 | (0.02, 0.83) | |
| 6 | 138 | 0.26 | 0.27 | 0.12 | 0.14 | (0.00, 0.47) | |
| 7 | 139 | 0.32 | 0.34 | 0.10 | 0.07 | (0.00, 0.85) | |
CIN = Cervical Intraepithelial Neoplasia; CINCS = Cervical Intraepithelial Neoplasia Cohort Study
Numbers do not add up to the total sample size due to missing data
SD = Standard Deviation
DMR = Differentially methylated region; Median across all CpG loci for candidate gene
MEG3 intronic differentially methylated region
Median study follow-up time was 10.5 months (range: 0.9–30.8). Thirteen women (n=13; 8.5%) dropped out after enrollment. No differences were observed between those who dropped out (n=13) compared to those who had at least one follow-up visit (n=151; data not shown). Over the 3-year study duration, a total of 53 (35.1%) women regressed from CIN1, compared to 98 (64.9%) who did not regress (including 41 women with persistent CIN1, 20 women progressed to CIN2+, and 37 women who regressed at only one visit). Median time to CIN1 regression was 12.6 months (range: 4.5–24.0 months). Approximately 60% of follow-up diagnoses assessed for the longitudinal analysis were defined by pathology-confirmed histology (57%), as a large proportion of women (43%) with abnormal screening results had CIN1 colposcopic impression at their follow-up colposcopy visit and thus did not warrant a biopsy, per the clinician’s best judgement.
The unadjusted estimated cumulative probability of CIN1 regression plots showed that women with methylation percentages above the median at IGF2AS CpG 5, and at the PEG10 DMR had lower incidence of regression over a 3-year period as compared to women with methylation percentages below the median (Figures 1a & 1d). No notable differences were observed in CIN1 regression probability at the Kv and MEG3 DMRs (Figures 1b–1c).
There was at least a 40% decrease in the probability of CIN1 regression for women with higher methylation versus lower methylation at IGF2AS CpG 5 (unadjusted HR, 0.57; 95% CI: 0.34, 0.79; Table 3). At the PEG10 DMR, women with higher methylation had a 21% decrease in likelihood of CIN1 regression versus lower methylation (unadjusted HR, 0.79; 95% CI: 0.65–0.97). After adjusting for continuous age, hrHPV status, race, current smoking status, continuous parity and history of oral contraceptive use, estimates for CIN1 regression were similar to unadjusted estimates— the probability of CIN1 regression decreased by 59% for women with higher methylation at IGF2AS CpG 5 versus lower methylation (adjusted HR, 0.41; 95% CI, 0.23–0.76). Within the PEG10 DMR, the likelihood of CIN1 regression decreased by 20% for women who had higher methylation (adjusted HR: 0.80, 95% CI, 0.65–0.98 versus lower methylation).
Table 3.
Analysis of CIN Regression, stratified by Imprinted Gene DMR/CpG Site CIN1 cases in North Carolina*
| Gene Name | CpG Site | Woman- Months |
Regression Events (N)† |
Unadjusted HR‡ (95% CI) |
Adjusted HR (95% CI) ठ|
|---|---|---|---|---|---|
| IGF2AS | 5 | 2742 | 53 | 0.57 (0.34, 0.94) | 0.41 (0.23, 0.76) |
|
| |||||
| Kv | |||||
| DMR‖ | 2514 | 49 | 0.81 (0.55, 1.19) | 0.83 (0.55, 1.26) | |
| 1 | 2485 | 48 | 0.78 (0.54, 1.14) | ||
| 6 | 2514 | 49 | 0.75 (0.50, 1.12) | ||
| 8,9 | 2514 | 49 | 1.06 (0.74, 1.50) | ||
| 10,11,12 | 2473 | 48 | 0.92 (0.64, 1.30) | ||
| 15 | 2514 | 49 | 0.73 (0.51, 1.05) | ||
| 17,18 | 2514 | 49 | 0.87 (0.59, 1.27) | ||
| 20 | 2514 | 49 | 0.89 (0.67, 1.17) | ||
| 21 | 2514 | 49 | 0.89 (0.66, 1.19) | ||
| 22 | 2514 | 49 | 1.02 (0.76, 1.37) | ||
| 24 | 2514 | 49 | 0.70 (0.45, 0.99) | ||
| 26,27 | 2514 | 49 | 1.10 (0.80, 1.53) | ||
|
| |||||
| MEG3¶ | |||||
| DMR‖ | 2762 | 53 | 0.80 (0.52, 1.21) | 0.89 (0.57, 1.38) | |
| 3 | 2762 | 53 | 0.88 (0.58, 1.33) | ||
| 6 | 2720 | 53 | 0.90 (0.61, 1.35) | ||
| 15 | 2762 | 53 | 0.67 (0.49, 0.91) | ||
| 16 | 2741 | 52 | 0.76 (0.54, 1.06) | ||
| 20,21 | 2725 | 52 | 0.75 (0.54, 1.03) | ||
| 22 | 2720 | 53 | 0.90 (0.60, 1.35) | ||
| 23 | 2762 | 53 | 0.92 (0.68, 1.25) | ||
| 26,27,28 | 2762 | 53 | 0.78 (0.53, 1.15) | ||
| 29,30,31 | 2762 | 53 | 0.85 (0.57, 1.28) | ||
|
| |||||
| PEG10 | |||||
| DMR‖ | 2517 | 50 | 0.79 (0.65, 0.97) | 0.80 (0.65, 0.98) | |
| 5 | 2461 | 49 | 0.57 (0.42, 0.78) | ||
| 6 | 2475 | 50 | 0.66 (0.47, 0.92) | ||
| 7 | 2510 | 50 | 0.85 (0.69, 1.05) | ||
CIN = cervical intraepithelial neoplasia; 151 cases with at least one follow-up visit
Regression event defined as histologically or cytologically negative diagnosis over two consecutive follow-up visits
HR= Hazard ratio; 95% CI= 95% confidence intervals; continuous methylation levels rescaled using interquartile range for each CpG site
Adjusted for continuous age, high risk-HPV, race, smoking, continuous parity and history of oral contraceptive use
DMR = differentially methylated region (median methylation of all CpG sites)
MEG3 intronic differentially methylated region
No differences in CIN regression rates were observed stratified by hrHPV status (Table A2 in Appendix). Similarly, there was no evidence of modification by race/ethnicity or by other covariates (data not shown). Data suggest potential modification by age, where women 25 years of age and older with higher methylation at MEG3 DMR had 50% decreased likelihood of regression (HR, 95% CI: 0.54, 0.32–0.92; Table 4).
Table 4.
Analysis of CIN Regression Imprinted Gene DMRs among CIN1 cases in North Carolina: Stratified by Age at Enrollment*
| Gene | CpG Site | Overall | Age <25 years N=56 |
Age ≥25 years N=78 |
|---|---|---|---|---|
|
| ||||
| HR (95% CI)†‡ | ||||
| IGFAS | 5 | 0.41 (0.23, 0.76) | 0.41 (0.17, 0.97) | 0.65 (0.34, 1.25) |
| Kv | DMR§ | 0.83 (0.55, 1.26) | 0.73 (0.40, 1.34) | 0.86 (0.52, 1.41) |
| MEG3 | DMR§ | 0.89 (0.57, 1.38) | 1.53 (0.80, 2.94) | 0.54 (0.32, 0.92) |
| PEG10 | DMR§ | 0.80 (0.65, 0.98) | 0.60 (0.43, 0.85) | 0.88 (0.69, 1.12) |
CIN = cervical intraepithelial neoplasia; 151 cases with at least one follow-up visit
HR=Hazard ratio; 95% CI= 95% confidence intervals; continuous methylation levels rescaled using interquartile range for each CpG site
Adjusted for race, current smoking, continuous parity and history of oral contraceptive use
DMR = differentially methylated region (median of all CpG sites
MEG3 intronic differentially methylated region
A sensitivity analysis was conducted to consider time to CIN1 regression at a single follow-up visit, and increased methylation at both PEG10 and IGF2AS DMRs remained associated with a decreased probability of CIN1 regression (refer to Appendix Table A1). An increase in methylation at Kv DMR resulted in a 20% decrease in the probability of first CIN1 regression (adjusted HR: 0.81; 95% CI, 0.63–1.01). Exclusion of either hrHPV-negative participants or participants with high grade cervical cytology at their enrollment Pap test did not significantly change adjusted HR estimates (data not shown).
Discussion
This longitudinal study of 164 CIN1 patients is among the first to prospectively examine aberrant methylation patterns of regulatory regions of imprinted genes and their association with low-grade CIN regression. Women with higher levels of methylation at the IGF2AS DMR CpG 5 and the PEG10 DMR had a lower 3-year cumulative probability of CIN1 regression as compared to women with lower levels of methylation. A decrease in the probability of CIN1 regression due to increased methylation at Kv DMR was also observed over one follow-up visit. These findings may implicate these DMRs as potential epigenetic biomarkers of a lower regression potential in low-grade CIN cases, and thus higher risk of high-grade precancer or more severe disease.
IGF2AS is a paternally expressed component of a downstream imprinted center, IC1 (located on human chromosome 11p15.5) that promotes cell proliferation.31 Abnormal methylation of IGF2 may be associated with mechanisms involved in cervical tumorigenesis. Aberrant DNA methylation of the IGF2 DMRs and other sequences regulating imprinted genes have been previously associated with higher risk of cervical dysplasia and invasive cancer in cross-sectional and case studies in Tanzania and France.32, 33 In contrast with our findings, decreased methylation at the IGF2 DMR was associated with an increased risk of invasive cervical carcinoma.33 Though a notable association was found between aberrant methylation at IGF2AS CpG 5, we could not make conclusions regarding the entire regulatory IGF2AS region from these data. However, the presence of abnormal methylation patterns at IGF2 warrants additional research.
Paternally-expressed PEG10 also appears to have a role in increased cell proliferation.34 Decreased methylation was associated with overexpression of PEG10 in hepatocellular carcinoma samples.35 In contrast, our current findings demonstrated that an increase in methylation of PEG10 DMR may be associated with a higher risk of cervical precancer, as evidenced by a lower risk of CIN1 regression. It is possible that regulatory mechanisms of PEG10 may differ in CIN from its involvement in hepatocellular carcinogenesis. Further investigation on the relationship between PEG10 methylation and CIN development in a larger cohort may provide insight on methylation patterns that are indicative of the risk of CIN progression.
Interestingly, age appeared to modify regression rates for DLK1/MEG3 DMR. Relatively older women (≥ 25 years of age) were less likely to regress over time given increased methylation relative to younger women. The MEG3 DMR is maternally expressed and is reciprocally imprinted with paternally-expressed DLK1 on chromosome 14q3236. Though little research describes aberrant methylation of MEG3, hypermethylation has been implicated as a potential biomarker in cervical cancer.37 When estimating time-to-regression at a single follow-up visit, increased methylation at the Kv DMR also decreased the probability of regression relative to women who had lower levels of methylation. The Kv DMR is maternally methylated, comprised of the imprint control region IC2 located at Chr11p15.5, which regulates at least 11 imprinted genes.38 Imprinting at this region controls transcription of the long non-coding RNA KCNQ1OT1, which regulates the expression of cyclin-dependent kinase inhibitor 1C gene (CDKN1C), an inhibitor protein involved in cell proliferation and growth regulation.39 While studies on Kv DMR in the context of cervical dysplasia and ICC are limited, changes in methylation at Kv DMR/IC2 have been positively associated with colorectal cancer20 and breast cancer.17
Infection with hrHPV, the primary cause of invasive cervical cancer, may interact with regulation of imprinted genes among women with cervical dysplasia. Hypermethylation of imprinted genes PEG3 and PEG1/MEST were positively correlated with hrHPV infection, suggesting it may serve as an intermediate in CIN development.33, 40 Imprinted tumor suppressor CDKN1C, controlled by IC2/Kv DMR, was upregulated during E2 (HPV viral regulatory protein)-mediated HeLa cell senescence and concomitant repression of E6/E7 HPV viral oncogenes.41 These findings implicate downregulation of CDKN1C, leading to upregulated cervical cell proliferation and subsequent cervical tumor development.41 Inhibition of cell apoptosis due to loss of E2 expression in cervical carcinogenesis may be mediated in part by aberrant methylation and subsequent deregulation of pivotal genes, some of which are imprinted and implicated in cervical cancer development pathways.33, 41
A major advantage of this study was the ability to prospectively assess the association between methylation markers and CIN. Accounting for time in estimating the probability of CIN1 regression improved the strength of the association. The findings here, comparable to previous studies, further support the consideration of imprinted gene biomarkers as a screening tool for LSIL/CIN1 cases.33, 40
Among potential limitations, this study did not assess HPV infection at study follow-up, which would have allowed for a more stringent definition of cervical lesion regression, including HPV negative status at follow-up. However, the decision to define the cervical regression as two consecutive negative screening results, rather than one, strengthened the robustness of the outcome. We also had limited power to assess the association of methylation on CIN regression stratified by individual HPV type, although no evidence of modification was found. Future work should include capturing type-specific HPV infection status at all follow-up visits with a larger cohort to determine the extent to which persistent hrHPV infection plays a role in the association between methylation patterns and the natural history of cervical dysplasia. The current study was also limited by the possibility that women with CIN1 at enrollment may have been misclassified. To address this, a sensitivity analysis was conducted by excluding women with high-grade cervical cytology at enrollment, producing similar results. The 2012 ASCCP guideline update,7 which shifted follow-up algorithms for CIN management, likely affected the variability in the number of visits and duration between follow-up visits for each participant. Obtaining data on changes in smoking habits and other time-dependent behavioral/lifestyle factors also would broaden future research in order to investigate their influence on methylation patterns among women with CIN.42 The incorporation of RNA/gene expression data would further characterize the influence of aberrant methylation among women with and without disease.
It is critical to understand risk factors that determine the natural course of CIN in order to improve the effectiveness of current cervical cancer screening methods. These study findings indicate further investigation into IGF2 and PEG10 DMRs as diagnostic biomarkers in women with low-grade CIN is warranted. Characterization of potential cervical tumorigenesis pathways related to the dysregulation of imprinted gene networks would help to establish novel epigenetic biomarkers in CIN management to reduce cervical cancer incidence, while avoiding the unnecessary follow-up of patients at relatively lower risk of progression to high-grade precancerous lesions or more severe.
Supplementary Material
Figure 2.
CINCS* study population flowchart
* CINCS = Cervical Intraepithelial Neoplasia Cohort Study
† HPV = Human papillomavirus
‡ CIN= Cervical intraepithelial neoplasia
§ No colposcopy/unable to grade
Novelty and Impact.
Few studies have assessed DNA methylation of imprinted genes as a potential biomarker for cervical dysplasia. Successful characterization of the natural history of low-grade cervical lesions may improve current screening approaches.
Acknowledgments
National Institutes of Health 2T32AI007001-39, R01CA142983 and P30ES025128; Chris Wiesen of Odum Institute at the University of North Carolina-Chapel Hill
Financial Support: National Institutes of Health T32 5T32AI007001, R01CA142983, and R01CA142983-02S1
References
- 1.United States National Cancer Institute. SEER Cancer Stat Facts: Cervix Uteri Cancer. National Cancer Institute; Bethesda, MD: 2016. [Google Scholar]
- 2.U.S. Cancer Statistics Working Group. United States Cancer Statistics: 1999–2013 Incidence and Mortality Web-based Report. 2016 [Google Scholar]
- 3.Walboomers JMM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJF, Peto J, Meijer CJLM, Munoz N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. The Journal of pathology. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 4.Satterwhite CL, Torrone E, Meites E, Dunne EF, Mahajan R, Ocfemia MCB, Su J, Xu F, Weinstock H. Sexually transmitted infections among US women and men: prevalence and incidence estimates, 2008. Sexually transmitted diseases. 2013;40:187–93. doi: 10.1097/OLQ.0b013e318286bb53. [DOI] [PubMed] [Google Scholar]
- 5.National Center for HIV/AIDS Viral Hepatitis STD and TB Prevention: Division of STD Prevention. Genital HPV Infection–CDC Fact Sheet. 2014 [Google Scholar]
- 6.Schlecht NF, Kulaga S, Robitaille J, Ferreira S, Santos M, Miyamura RA, Duarte-Franco E, Rohan TE, Ferenczy A, Villa LL. Persistent human papillomavirus infection as a predictor of cervical intraepithelial neoplasia. Jama. 2001;286:3106–14. doi: 10.1001/jama.286.24.3106. [DOI] [PubMed] [Google Scholar]
- 7.Massad LS, Einstein MH, Huh WK, Katki HA, Kinney WK, Schiffman M, Solomon D, Wentzensen N, Lawson HW. 2012 updated consensus guidelines for the management of abnormal cervical cancer screening tests and cancer precursors. Obstetrics & Gynecology. 2013;121:829–46. doi: 10.1097/AOG.0b013e3182883a34. [DOI] [PubMed] [Google Scholar]
- 8.Molden T, Nygård JF, Kraus I, Karlsen F, Nygård M, Skare GB, Skomedal H, Thoresen SØ, Hagmar B. Predicting CIN2+ when detecting HPV mRNA and DNA by PreTect HPV-proofer and consensus PCR: A 2-year follow-up of women with ASCUS or LSIL pap smear. International Journal of Cancer. 2005;114:973–6. doi: 10.1002/ijc.20839. [DOI] [PubMed] [Google Scholar]
- 9.Bansal N, Wright JD, Cohen CJ, Herzog TJ. Natural history of established low grade cervical intraepithelial (CIN 1) lesions. Anticancer research. 2008;28:1763–6. [PubMed] [Google Scholar]
- 10.Paskett ED, McLaughlin JM, Reiter PL, Lehman AM, Rhoda DA, Katz ML, Hade EM, Post DM, Ruffin MT. Psychosocial predictors of adherence to risk-appropriate cervical cancer screening guidelines: a cross sectional study of women in Ohio Appalachia participating in the Community Awareness Resources and Education (CARE) project. Preventive medicine. 2010;50:74–80. doi: 10.1016/j.ypmed.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nature reviews genetics. 2007;8:253–62. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skaar DA, Li Y, Bernal AJ, Hoyo C, Murphy SK, Jirtle RL. The human imprintome: regulatory mechanisms, methods of ascertainment, and roles in disease susceptibility. ILAR journal. 2012;53:341–58. doi: 10.1093/ilar.53.3-4.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feinberg AP, Tycko B. The history of cancer epigenetics. Nature Reviews Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 14.Rainier S, Johnson LA, Dobry CJ, Ping AJ, Grundy PE, Feinberg AP. Relaxation of imprinted genes in human cancer. 1993 doi: 10.1038/362747a0. [DOI] [PubMed] [Google Scholar]
- 15.Choufani S, Shuman C, Weksberg R. Beckwith–Wiedemann syndrome. American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 2010;154:343–54. doi: 10.1002/ajmg.c.30267. [DOI] [PubMed] [Google Scholar]
- 16.Weksberg R, Shuman C, Smith AC. Beckwith–Wiedemann syndrome. American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 2005;137:12–23. doi: 10.1002/ajmg.c.30058. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez BA, Weng Y-I, Liu T-M, Zuo T, Hsu P-Y, Lin C-H, Cheng A-L, Cui H, Yan PS, Huang TH-M. Estrogen-mediated epigenetic repression of the imprinted gene cyclin dependent kinase inhibitor 1C in breast cancer cells. Carcinogenesis. 2011:bgr017. doi: 10.1093/carcin/bgr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larson PS, Schlechter BL, King C-L, Yang Q, Glass CN, Mack C, Pistey R, De las Morenas A, Rosenberg CL. CDKN1C/p57 kip2 is a candidate tumor suppressor gene in human breast cancer. BMC cancer. 2008;8:68. doi: 10.1186/1471-2407-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva A-L, Maia A-T, Huddleston JE, Uribe-Lewis S. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Human molecular genetics. 2008;17:2633–43. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakano S, Murakami K, Meguro M, Soejima H, Higashimoto K, Urano T, Kugoh H, Mukai T, Ikeguchi M, Oshimura M. Expression profile of LIT1/KCNQ1OT1 and epigenetic status at the KvDMR1 in colorectal cancers. Cancer science. 2006;97:1147–54. doi: 10.1111/j.1349-7006.2006.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monk D. Deciphering the cancer imprintome. Briefings in functional genomics. 2010:elq013. doi: 10.1093/bfgp/elq013. [DOI] [PubMed] [Google Scholar]
- 22.Song J, Lee J, Lee N, Jung H, Kim S, Lee K. Microarray analysis of normal cervix, carcinoma in situ, and invasive cervical cancer: identification of candidate genes in pathogenesis of invasion in cervical cancer. International Journal of Gynecological Cancer. 2008;18:1051–9. doi: 10.1111/j.1525-1438.2007.01164.x. [DOI] [PubMed] [Google Scholar]
- 23.Dueñas-González A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Molecular cancer. 2005;4:38. doi: 10.1186/1476-4598-4-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding D-C, Chiang M-H, Lai H-C, Hsiung CA, Hsieh C-Y, Chu T-Y. Methylation of the long control region of HPV16 is related to the severity of cervical neoplasia. European Journal of Obstetrics & Gynecology and Reproductive Biology. 2009;147:215–20. doi: 10.1016/j.ejogrb.2009.08.023. [DOI] [PubMed] [Google Scholar]
- 25.Vidal AC, Smith JS, Valea F, Bentley R, Gradison M, Yarnall KS, Ford A, Overcash F, Grant K, Murphy SK. HPV genotypes and cervical intraepithelial neoplasia in a multiethnic cohort in the southeastern USA. Cancer Causes & Control. 2014;25:1055–62. doi: 10.1007/s10552-014-0406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T., Jr The 2001 Bethesda System: terminology for reporting results of cervical cytology. Jama. 2002;287:2114–9. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 27.Gravitt PE, Peyton CL, Apple RJ, Wheeler CM. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. Journal of clinical microbiology. 1998;36:3020–7. doi: 10.1128/jcm.36.10.3020-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gravitt PE, Peyton CL, Alessi TQ, Wheeler CM, Coutlee F, Hildesheim A, Schiffman MH, Scott DR, Apple RJ. Improved amplification of genital human papillomaviruses. Journal of clinical microbiology. 2000;38:357–61. doi: 10.1128/jcm.38.1.357-361.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Xie C, Murphy SK, Skaar D, Nye M, Vidal AC, Cecil KM, Dietrich KN, Puga A, Jirtle RL. Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environmental health perspectives. 2016;124:666. doi: 10.1289/ehp.1408577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenland S, Pearl J, Robins JM. Causal diagrams for epidemiologic research. Epidemiology. 1999:37–48. [PubMed] [Google Scholar]
- 31.Du M, Beatty LG, Zhou W, Lew J, Schoenherr C, Weksberg R, Sadowski PD. Insulator and silencer sequences in the imprinted region of human chromosome 11p15. 5. Human molecular genetics. 2003;12:1927–39. doi: 10.1093/hmg/ddg194. [DOI] [PubMed] [Google Scholar]
- 32.Douc-Rasy S, Barrois M, Fogel S, Ahomadegbe J, Stehelin D, Coll J, Riou G. High incidence of loss of heterozygosity and abnormal imprinting of H19 and IGF2 genes in invasive cervical carcinomas. Uncoupling of H19 and IGF2 expression and biallelic hypomethylation of H19. Oncogene. 1996;12:423–30. [PubMed] [Google Scholar]
- 33.Vidal AC, Henry N, Murphy S, Oneko O, Nye M, Bartlett J, Overcash F, Huang Z, Wang F, Mlay P. PEG1/MEST and IGF2 DNA methylation in CIN and in cervical cancer. Clinical and Translational Oncology. 2014;16:266–72. doi: 10.1007/s12094-013-1067-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y, Zhang Y-l, Hu X, Deng Q, Li Q, Gao Y, Teng X-m, Han Z-g, Huang J. Molecular mechanism and function research of highly expressed PEG10 gene in hepatocellular carcinoma. Tumor. 2013;33:28–35. [Google Scholar]
- 35.Bang H, Ha SY, Hwang SH, Park C-K. Expression of PEG10 is associated with poor survival and tumor recurrence in hepatocellular carcinoma. Cancer Research and Treatment. 2015;47:844–52. doi: 10.4143/crt.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmidt JV, Matteson PG, Jones BK, Guan X-J, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes & development. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Yao T, Lin Z, Gao Y. Aberrant Methylation of MEG3 Functions as a Potential Plasma-Based Biomarker for Cervical Cancer. Scientific reports. 2017;7:6271. doi: 10.1038/s41598-017-06502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cerrato F, Sparago A, Di Matteo I, Zou X, Dean W, Sasaki H, Smith P, Genesio R, Bruggemann M, Reik W. The two-domain hypothesis in Beckwith–Wiedemann syndrome: autonomous imprinting of the telomeric domain of the distal chromosome 7 cluster. Human Molecular Genetics. 2005;14:503–11. doi: 10.1093/hmg/ddi047. [DOI] [PubMed] [Google Scholar]
- 39.Matsuoka S, Thompson JS, Edwards MC, Bartletta J, Grundy P, Kalikin LM, Harper JW, Elledge SJ, Feinberg AP. Imprinting of the gene encoding a human cyclin-dependent kinase inhibitor, p57KIP2, on chromosome 11p15. Proceedings of the National Academy of Sciences. 1996;93:3026–30. doi: 10.1073/pnas.93.7.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nye MD, Hoyo C, Huang Z, Vidal AC, Wang F, Overcash F, Smith JS, Vasquez B, Hernandez B, Swai B. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One. 2013;8:e56325. doi: 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells SI, Aronow BJ, Wise TM, Williams SS, Couget JA, Howley PM. Transcriptome signature of irreversible senescence in human papillomavirus-positive cervical cancer cells. Proceedings of the National Academy of Sciences. 2003;100:7093–8. doi: 10.1073/pnas.1232309100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeilinger S, Kühnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PloS one. 2013;8:e63812. doi: 10.1371/journal.pone.0063812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.