Abstract
Double-strand breaks (DSBs) can be repaired by homologous recombination (HR) in mammalian cells, often resulting in gene conversion. RAD51 functions with RAD52 and other proteins to effect strand exchange during HR, forming heteroduplex DNA (hDNA) that is resolved by mismatch repair to yield a gene conversion tract. In mammalian cells RAD51 and RAD52 overexpression increase the frequency of spontaneous HR, and one study indicated that overexpression of mouse RAD51 enhances DSB-induced HR in Chinese hamster ovary (CHO) cells. We tested the effects of transient and stable overexpression of human RAD51 and/or human RAD52 on DSB-induced HR in CHO cells and in human cells. DSBs were targeted to chromosomal recombination substrates with I-SceI nuclease. In all cases, excess RAD51 and/or RAD52 reduced DSB-induced HR, contrasting with prior studies. These distinct results may reflect differences in recombination substrate structures or different levels of overexpression. Excess RAD51/RAD52 did not increase conversion tract lengths, nor were product spectra otherwise altered, indicating that excess HR proteins can have dominant negative effects on HR initiation, but do not affect later steps such as hDNA formation, mismatch repair or the resolution of intermediates.
INTRODUCTION
DNA double-strand breaks (DSBs) are potentially lethal lesions that are efficiently repaired in eukaryotes by homologous recombination (HR) or non-homologous end joining (NHEJ). A specific type of HR is gene conversion, a conservative process that can occur with or without an associated crossover. DSBs stimulate gene conversion, involving the non-reciprocal transfer of information from an unbroken donor locus to a broken recipient locus. Studies with multiply marked alleles have shown that conversion typically involves the transfer of a continuous block of information, creating continuous conversion tracts. In yeast, conversion tracts reflect the formation of heteroduplex DNA (hDNA) by strand transfer (including strand invasion and branch migration) and subsequent mismatch repair processing (1,2). Gene conversion in yeast requires Rad51 and Rad52, and involves several other members of the RAD52 epistasis group (3). A non-conservative type of HR called single-strand annealing (SSA) can occur between direct repeats, leading to deletion of one repeat and sequences between repeats. Deletions can also occur by crossing over between direct repeats and by unequal sister chromatid exchange. In yeast, SSA requires Rad52 but not Rad51 (4,5).
Homologs of the yeast HR proteins are also involved in mammalian HR (6–8). RAD51 and other members of the RAD51 family (RAD51B, RAD51C, RAD51D, DMC1, XRCC2 and XRCC3) are homologs of Escherichia coli RecA (9). Similar to RecA, RAD51 is a DNA-dependent ATPase that can effect strand transfer in vitro (7). Human RAD52 binds to DSBs (10) and in vitro assays indicate that RAD52 promotes strand annealing (11) and enhances RAD51 strand exchange activity (7,12). Other proteins known or suspected to influence HR directly or indirectly include RAD54, RAD54B, XRCC2, XRCC3, BRCA1, BRCA2, replication protein A, ATM, UBL1, UBC9, XRCC9/FANCG and p53 (13). Many of these proteins interact directly or indirectly with RAD51, RAD52 or each other, suggesting that HR is mediated by one or more multi-subunit complexes.
The in vivo functions of RAD51 have been difficult to investigate because vertebrate RAD51 gene knockouts are non-viable at the cellular level (14–16). Alternative strategies to study in vivo roles of RAD51 include conditional knockouts and gene overexpression. Unlike yeast, mouse RAD52 knockouts show a minimal phenotype (8). Thus, overexpression is also a reasonable strategy for investigating in vivo roles of mammalian RAD52. In mammalian cells, the dominant DSB repair pathway is thought to be NHEJ, consistent with the idea that one or more HR proteins are rate limiting. This idea is supported by several studies showing that spontaneous HR and/or resistance to ionizing radiation is enhanced by overexpression of HR proteins, including human RAD52 in monkey cells (17), Chinese hamster RAD51 in Chinese hamster ovary (CHO) cells (18,19) and yeast RAD52 in human cells (20). These results argue for strong conservation of RAD51 and RAD52 function in evolution, and this is borne out by the highly conserved amino acid sequences of these proteins, particularly among mammalian species (21,22). Only two previous reports describe the effects of HR protein overexpression on DSB-induced HR. Yanez and Porter (23) showed that overexpression of human RAD51 enhanced HR-dependent gene targeting in human cells several-fold. Lambert and Lopez (24) targeted DSBs to a chromosomal direct repeat recombination substrate in hamster cells by using I-SceI nuclease and found that overexpression of mouse RAD51 enhanced DSB-induced gene conversion 5-fold, but saw no effect on the frequency of deletion events. In contrast, overexpression of yeast Rad51 in hamster cells reduced gene conversion 40-fold. Thus, overexpression of RAD51 from a closely related species had positive effects on DSB-induced HR in mammalian cells, whereas overexpression of the more evolutionarily distant yeast Rad51 had dominant negative effects (24).
Several studies have correlated changes in RAD51 levels with cancer and other cellular phenotypes; however, little is known about the effects of altered RAD52 levels. Increased RAD51 was reported in sporadic ductal breast cancer and levels correlated with histological grading of the tumors (25). In contrast, 30% of breast carcinomas displayed decreased levels of RAD51 (26). Two reports linked RAD51 polymorphisms with increased breast cancer risk in persons with mutations in BRCA2 (27,28), but it is not yet known whether these polymorphisms influence RAD51 expression or mRNA/protein stability. RAD51 levels were higher in immortalized human fibroblasts compared to normal diploid fibroblasts and increased RAD51 correlated with increased resistance to ionizing radiation (29). Radiation resistance of carcinoma cell lines correlated with increased RAD51 (30,31) and reducing RAD51 by antisense and ribozyme approaches decreased radiation resistance (32,33).
In the present study we examined the effects of human RAD51 and/or human RAD52 overexpression on DSB-induced HR in CHO and human cells. In contrast to the results described above, we found that DSB-induced HR was reduced by overexpression of RAD51 or RAD52, perhaps reflecting an imbalance in the stoichiometry of the HR protein complex(es). These disparate results may reflect differences in overexpression levels or recombination substrates. In an attempt to restore stoichiometric balance, we co-overexpressed human RAD51 and human RAD52, but this led to further reductions in DSB-induced HR. In light of the role of RAD51 in strand transfer and the potential for RAD52 enhancement of this activity, we were also interested in determining whether overexpression of one or both of these proteins would increase conversion tract lengths, but found no evidence for such an effect. These results indicate that overexpression of RAD51 and/or RAD52 inhibits an early step in DSB-induced HR but does not affect the later steps, involving hDNA formation, mismatch repair or resolution of recombination intermediates.
MATERIALS AND METHODS
Plasmid DNAs
Plasmid pOPUR (34) is a mammalian expression vector carrying the RSV LTR promoter and triplicated lac operators from pOPI3CAT (Stratagene, La Jolla, CA) and the puromycin resistance gene (pac) from pPUR (Clontech, Palo Alto, CA). Human RAD51 and RAD52 cDNAs (21,22) were inserted downstream of the RSV LTR promoter, creating plasmids pOPUR/RAD51 and pOPUR/RAD52. RAD52 mRNA is unstable due to the presence of mRNA degradation signals in the 3′-untranslated region (21). RAD52 mRNA stability was enhanced by deletion of these signals in pOPUR/RAD52; this change does not affect the RAD52 amino acid sequence, but does improve detection of RAD52 by western blot. When integrated into chromosomal DNA, these plasmids provide constitutive expression of RAD51 or RAD52 in the absence of the LacI repressor. pOPUR/RAD51 and pOPUR/RAD52 were also used to transiently express RAD51 and RAD52 in CHO cells with neo direct repeats; derivatives of these plasmids lacking the pac gene (called pO/RAD51 and pO/RAD52) were used to transiently express RAD51 and RAD52 in human cells carrying a pac inverted repeat recombination substrate (see below). pOPI3 is a derivative of pOPI3CAT that lacks CAT and was used as ‘empty’ control vector in some experiments. Plasmid pCMV(3×NLS)I-SceI (kindly provided by Greg Donoho, Lexicon Genetics, The Woodlands, TX) contains the I-SceI nuclease coding sequence fused at its 5′-end to a triplicated nuclear localization signal, driven by the CMV promoter, and terminated by the bovine growth hormone polyadenylation signal.
Cell lines and northern and western analyses
CHO and human cells were grown in αMEM with 10% fetal bovine serum, 100 U/ml penicillin and 100 µg/ml streptomycin in a humidified atmosphere with 5% CO2. CHO strain 33 (35) carries a single integrated copy of a neo direct repeat recombination substrate with a central SV40 promoter-driven E.coli gpt gene (Fig. 1A). Human cell line HT1080-1885 is a derivative of the HT1080 fibrosarcoma cell line with a single integrated copy of a pac inverted repeat recombination substrate (Fig. 1B). Derivatives of strain 33 carrying integrated copies of pOPUR-RAD51 or pOPUR-RAD52 were constructed by electroporating limiting amounts of either vector into strain 33 and selecting for transfectants resistant to 3.5 µg/ml puromycin. RAD51 and/or RAD52 overexpression was monitored by northern and/or western blot analysis as described (34,36). Rabbit polyclonal anti-RAD51 antibodies (Oncogene Science, Boston, MA) were used as described previously (34). Full-length RAD52 protein was expressed and purified from E.coli by using a His tag and injected into mouse for antibody production. We did not observe non-specific reactivity with pre-immune serum collected before RAD52 injection (data not shown).
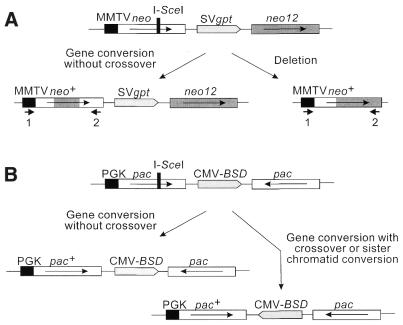
Figure 1.
Structures of recombination substrates and products. (A) 1.4 kb neo direct repeats in CHO strain 33 are shown by open and shaded boxes flanking SVgpt. The upstream neo is driven by the MMTV promoter and an I-SceI recognition site interrupts the reading frame; neo12 has 12 silent RFLP markers (see Fig. 4). Gene conversion without a crossover results in loss of the I-SceI site and transfer of none, one or more markers from neo12, but maintains the gross structure of the substrate. Deletions via SSA, crossovers or unequal sister chromatid exchanges result in loss of one neo and SVgpt. PCR primers 1 and 2 amplify the recipient allele in conversion products and the remaining allele in deletion products. (B) Structure of inverted pac repeats flanking CMV-BSD in human cell line HT1080-1885. The upstream pac is driven by the mouse PGK promoter and inactivated by insertion of an I-SceI site in an 80 bp deletion of the MscI–BssHII fragment in the pac coding sequence. Gene conversion without crossover replaces the I-SceI site and 80 bp deletion with wild-type sequence from the second pac gene. Conversion with crossover or large-scale sister chromatid conversion inverts the central sequences, including CMV-BSD.
Homologous recombination assays
DSB-induced HR frequencies were measured in CHO cells as described previously (35). Briefly, 4 × 105 cells were seeded into 3.5 cm (diameter) wells, incubated for 24 h and lipofected with 2 µg pCMV(3×NLS)I-SceI to induce DSBs. To measure spontaneous HR, we transfected cells with 2 µg negative control vector pCMV(I-SceI–) (37) or pSV2gpt (38). In transient RAD51/RAD52 overexpression experiments, 1 µg each of the RAD51 and RAD52 expression vectors were included with 1 µg pCMV(3×NLS)I-SceI or negative control vector. The total amount of transfected DNA was held constant at 3 µg since cell survival can be influenced by the amount of DNA present during lipofection (unpublished results); this was accomplished by adjusting the amount of negative control vector. Twenty-four hours post-transfection, 105 cells were seeded to each of four 10 cm dishes. After an additional 24 h, G418 was added (final concentration 250 µg/ml, 100% active) and cells were incubated for 12–14 days before colonies were stained and scored. Cell viability was measured by seeding appropriate dilutions of cell suspensions into non-selective medium, incubating for 7–10 days and scoring colony formation. DSB-induced HR frequencies were measured in HT1080-1885 cells using the same procedure, except that recombinants were selected with 0.8 µg/ml puromycin. To measure spontaneous HR in HT1080-1885, 2 × 106 cells were seeded to a 10 cm (diameter) dish, incubated for 24 h and lipofected with either 8 µg control vector (pOPI3) or 4 µg each pO/RAD51 and/or pO/RAD52, with the total amount held constant at 8 µg by adjusting the amount of negative control vector. Puromycin (0.8 µg/ml) was added 48 h post-lipofection and recombinants and cell viability were scored as above. HR frequencies were calculated as the ratio of G418-resistant (G418r) or puromycin-resistant (Purr) colonies to the total number of viable cells plated in selective medium. Gross structures of recombination products were analyzed by Southern hybridization as described (35). Gene conversion tracts in the neo recombination substrate were measured by restriction mapping of a 1.5 kb PCR fragment amplified with a MMTV promoter-specific primer (5′-CCTTCACTTTCCAGAGGGTC) and a neo-specific primer (5′-GCGAAGAACTCCAGCATGAG). All statistical analysis was performed using t-tests unless specified otherwise.
RESULTS
Experimental design
We used CHO and human cell lines carrying chromosomal recombination substrates to test the effects of RAD51 and RAD52 overexpression on DSB-induced HR. CHO strain 33 cells carry direct neo repeats and human HT1080-1885 cells carry inverted pac repeats (Fig. 1). MMTVneo is inactivated by an I-SceI recognition sequence inserted into the natural BanII site and the second neo is inactive because it lacks a promoter. The second neo has 12 phenotypically silent single base mutations at ∼100 bp intervals that create RFLPs (35). The silent RFLPs allow high resolution analysis of conversion tracts, including length, directionality and continuity. In HT1080-1885, one copy of pac is driven by the mouse PGK promoter, but is inactive because an I-SceI recognition site was inserted into an 80 bp deletion in the coding sequence. The second copy of pac has a wild-type coding sequence but is inactive because it lacks a promoter. With either recombination substrate, transfection with an I-SceI endonuclease expression vector leads to cleavage of the I-SceI site, greatly enhancing HR and yielding selectable G418r or Purr recombinants. There are several possible outcomes of HR (Fig. 1). HR can result in gene conversion without an associated crossover, which conserves the gross structure of the recombination substrates, but replaces the I-SceI sites with wild-type information from the silent (donor) alleles. In the neo direct repeat, HR with an associated crossover leads to deletion of one repeat and the central sequences; deletions can also arise by unequal sister chromatid exchange or SSA. Based on evidence from yeast, all of these events are likely to require RAD52, and all but SSA are likely to require RAD51 (3,4,39). In the inverted repeat, HR with an associated crossover leads to inversion of sequences between the two pac genes; such inversions may also arise by sister chromatid conversion (40,41). Other more complex HR events are also possible; however, gene conversions without associated crossovers are the dominant DSB-induced product in both CHO strain 33 (34,35) and in HT1080-1885 cells (data not shown). Selecting for G418r or Purr products detects HR but not NHEJ.
Both transient and stable overexpression strategies were used to test the effects of excess RAD51 and RAD52 on DSB-induced HR. Transient overexpression was examined in CHO and human cells by co-transfecting RAD51 and RAD52 expression vectors with the I-SceI expression vector. Due to incompatibilities with the various selectable markers, stable overexpression of RAD51 and RAD52 was attempted only in CHO cells. For each recombination substrate and RAD51/RAD52 overexpression condition, we measured DSB-induced HR frequencies and product spectra. Spontaneous HR can be measured by omitting the I-SceI expression vector. The spontaneous HR frequency in CHO strain 33 is very low (near the limit of detection of 10–7); this low level most likely reflects inhibition by the multiple markers in neo12 (35). Thus, strain 33 and its derivatives are not suitable for analyzing spontaneous HR and we limited our analysis with these cells to DSB-induced HR. Consistent with prior studies (17–19), spontaneous HR in HT1080-1885 was enhanced by >2-fold by overexpression of RAD51 and/or RAD52 (Table 1). Spontaneous HR was not significantly different when both RAD51 and RAD52 were overexpressed compared to values when only one of these proteins was overexpressed (both P > 0.32).
Table 1. Spontaneous HR in human cells is enhanced by transient overexpression of human RAD51 or RAD52.
| Overexpressiona |
Recombination frequency (×107)b |
Fold increasec |
P valuec |
| None | 191 ± 31 | ||
| RAD51 | 500 ± 90 | 2.6 | 0.005 |
| RAD52 | 398 ± 71 | 2.1 | 0.01 |
| RAD51 + RAD52 | 475 ± 97 | 2.5 | 0.009 |
aHT1080-1885 cells were lipofected with empty, RAD51 and/or RAD52 vectors in the absence of the I-SceI expression vector.
bAverage recombination frequencies for three determinations ± SD; plating efficiencies were not affected by overexpression (range 0.17–0.23).
cFold increases and P values from t-tests are calculated relative to no overexpression.
Overexpression of human RAD51 or RAD52 reduces DSB-induced HR in CHO cells
Our original strategy was to introduce the LacI repression system into CHO strain 33 and then stably transfect this derivative with pOPUR-RAD51 or pOPUR-RAD52 so that transgene expression could be regulated with IPTG. With this strategy it would be possible to compare DSB-induced HR in a single cell line with or without overexpression, although we found that most cell lines showed significant basal expression of RAD51 or RAD52, and upon addition of IPTG we observed little or no increase in RAD51 or RAD52 mRNA levels. However, preliminary experiments indicated that DSB-induced HR was ∼2-fold lower in these cell lines than in strain 33, and that DSB-induced HR was similar in strain 33 and the intermediate cell line expressing just LacI (data not shown).
The difficulties with the inducible expression system prompted us to switch to a constitutive overexpression strategy involving comparisons between CHO strain 33 and derivatives produced by stably integrating pOPUR-RAD51 or pOPUR-RAD52 into strain 33. Although these transgenes carry the lac operator, they are constitutively expressed in the absence of the LacI repressor. Northern (not shown) and western analyses (Fig. 2A and B) confirmed the constitutive overexpression of RAD51 and RAD52. We induced HR by transient transfection of the I-SceI expression vector and observed >2-fold lower HR frequencies in cells overexpressing either RAD51 or RAD52 compared to parallel determinations with strain 33 (both P < 0.0001; Fig. 3A).
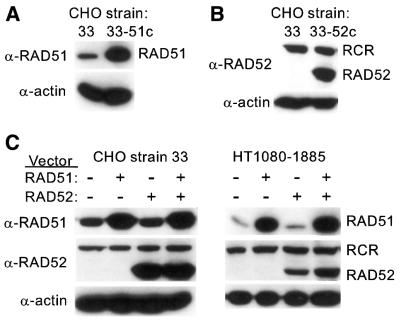
Figure 2.
Western blot analysis of overexpressed human RAD51 and RAD52. For detection of RAD51 and RAD52, we loaded 20 and 60 µg, respectively, of protein from whole cell extracts. Proteins were separated by SDS–PAGE, transferred to PVDF membranes and detected with polyclonal antibodies to RAD51 (α-RAD51) or RAD52 (α-RAD52) using ECL reagents (Amersham Pharmacia Biotech, Piscataway, NJ). Parallel lanes were probed with anti-β-actin antibodies as loading controls. (A) Endogenous RAD51 levels in CHO strain 33 (left lane) and the RAD51 constitutive overexpression strain 33-51c (right lane). (B) Endogenous RAD52 in strain 33 is too low to be detected in this exposure (left lane), but easily detected in the RAD52 constitutive overexpression strain 33-52c (right lane). An α-RAD52 cross-reacting (RCR) species of higher molecular weight was also detected. (C) RAD51 and/or RAD52 expression vectors were transfected into strain 33 or HT1080-1885 as indicated and cell extracts were prepared 48 h post-transfection for western blot analysis as above.
Figure 3.
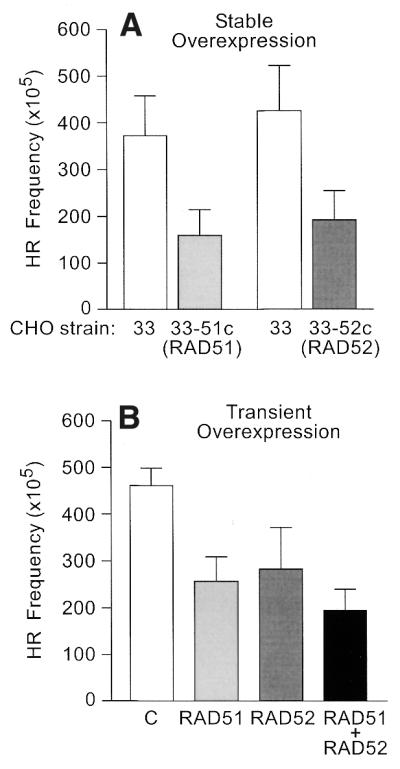
Overexpression of human RAD51 and/or RAD52 decreases DSB-induced HR in CHO cells. In all cases, cells were transfected with the I-SceI expression vector; control transfections without this vector yielded few or no recombinants (data not shown). (A) DSB-induced HR frequencies of CHO strain 33 were determined in parallel with either strain 33-51c or strain 33-52c that overexpress human RAD51 or RAD52, respectively. Values represent averages (± SD) for 10 independent determinations per strain. (B) DSB-induced HR in CHO strain 33 co-transfected with an empty vector (C) or with one or both of the RAD51 and RAD52 expression vectors. Values represent averages ± SD for six independent determinations.
Because several steps are required to construct constitutive overexpression cell lines (transfection, drug selection and population expansion), it was possible that the reduced HR described above was independent of excess RAD51 or RAD52, but instead reflected unwanted changes during cell line construction, such as altered transfection efficiency. To rule out this possibility, we performed transient overexpression assays in CHO strain 33. CHO strain 33 cells were co-transfected with the I-SceI expression vector and pOPUR-RAD51 or pOPUR-RAD52. Western analysis confirmed transient overexpression of RAD51 and RAD52 (Fig. 2C). Similar to the results above, transient overexpression of RAD51 or RAD52 significantly reduced DSB-induced HR (Fig. 3B; both P < 0.0001). Although we did not measure nuclear levels of RAD51 and RAD52, the observed effects on HR likely reflect increased nuclear protein levels since HR occurs in the nucleus.
The transient overexpression strategy provides a simple means to test the effects of simultaneous overexpression of RAD51 and RAD52 on DSB-induced HR. We hypothesized that the reduction in DSB-induced HR with excess RAD51 or RAD52 might result from an imbalance in the stoichiometry of HR proteins. Since RAD51 and RAD52 interact with each other, we reasoned that overexpression of both proteins might restore the balance and restore HR to wild-type levels. However, the opposite result was obtained: HR levels were significantly lower when both RAD51 and RAD52 were overexpressed compared to levels seen when the individual proteins were overexpressed (Fig. 3B; both P < 0.05). Together, these results indicate that overexpression of wild-type human RAD51 and/or RAD52 in CHO cells has dominant negative effects on DSB-induced HR.
We were interested in whether transcription would influence DSB-induced HR in the presence of excess RAD51 or RAD52. Transcription enhances spontaneous HR (36), but it has no effect on the frequency of DSB-induced HR or gene conversion tract lengths in CHO strain 33 (35). Since one copy of neo in CHO strain 33 is driven by the MMTV promoter, transcription can be adjusted from low to high levels by adding dexamethasone to the growth medium. RAD51 and RAD52 overexpression had similar dominant negative effects on DSB-induced HR in the presence and absence of dexamethasone (data not shown), hence, the remainder of the present study was performed without added dexamethasone.
Overexpression of RAD51 or RAD52 has little effect on HR product spectra
RAD51 is thought to play a central role in strand transfer during HR, and RAD52 is thought to augment RAD51 function. We were interested in whether overexpression of these proteins would increase hDNA formation, reflected in increased conversion tract lengths. Because of the potential connection between gene conversion tract lengths and crossing over (42), we were also interested in whether RAD51 and RAD52 overexpression would alter the relative proportion of non-deletion (gene conversion without associated crossover) and deletion events. Product spectra were generated from strain 33 with or without overexpression of RAD51 and/or RAD52 (Fig. 4 and Table 2); these spectra were also compared to a prior strain 33 spectrum obtained without overexpression of RAD51 or RAD52 (35). The new and old control spectra were similar in that most or all products resulted from gene conversion without an associated crossover and average tract lengths were ∼290 bp (Table 2). The three product spectra with RAD51 and/or RAD52 overexpression were similar to each other and to the control spectra: in each case, all products resulted from gene conversion without an associated crossover and average tract lengths were not significantly different from control values, from each other or from that obtained when both RAD51 and RAD52 were overexpressed (all P > 0.3). These spectra were also similar in that most tracts were bidirectional (Table 2), although there was a reduction in bidirectional tracts with excess RAD51 (P = 0.014, Fisher exact test), possibly reflecting a reduction in the number of conversions involving two-ended strand invasions. These results indicate that excess RAD51 and/or RAD52 affects only early steps in recombination; once an event is initiated, excess HR proteins do not appear to affect later steps such as hDNA formation, mismatch repair or the resolution of intermediates.
Figure 4.
DSB-induced product spectra from strain 33 with or without transient overexpression of RAD51 and/or RAD52. neo12, shown above, is the information donor for conversions of MMTVneo; the position of each marker in neo12 is indicated by the number in each marker name. Below, conversion tracts are indicated by black bars. The I-SceI site in MMTVneo is located at position 845 within neo and is always converted among neo+ products. The number of products of each tract type is shown for control and each overexpression condition.
Table 2. DSB-induced HR product spectra with or without excess RAD51 and RAD52.
| Spectruma |
Overexpression |
No. of products |
Conversion (%)b |
Average tract lengthc |
Percent bidirectionald |
| 1 | None | 31 | 97 | 287 ± 47 | 85 |
| 2 | None | 11 | 100 | 296 ± 50 | 91 |
| 3 | RAD51 | 15 | 100 | 264 ± 64 | 60 |
| 4 | RAD52 | 14 | 100 | 361 ± 78 | 86 |
| 5 | RAD51 + RAD52 | 14 | 100 | 343 ± 85 | 86 |
aData for spectrum 1 are from Taghian and Nickoloff (35); data for the other spectra are from the present study.
bFraction of products arising by conversion without crossover; the remainder are deletions.
cLengths of each tract type shown in Figure 4 were calculated as the mean of the longest and shortest possible tract lengths. These values were used to calculate an average tract length (± SEM) for each product spectrum.
dBidirectional tracts are those in which at least one marker converted on either side of the I-SceI site.
Overexpression of human RAD51 and RAD52 reduces DSB-induced HR in human cells
Because RAD51 and RAD52 are each highly conserved in human and hamster, it was unlikely that the dominant negative effects of human RAD51 and RAD52 overexpression in CHO cells reflected a cross-species effect. To rule out this possibility, we transiently overexpressed human RAD51 and/or RAD52 in human HT1080-1885 cells and measured the frequency of I-SceI-induced HR between inverted pac repeats. As shown in Figure 5, overexpression of human RAD51 or RAD52 significantly reduced DSB-induced HR in human cells (both P < 0.0001). Similar to the results in CHO cells, co-overexpression of RAD51 and RAD52 significantly reduced HR below that seen with overexpression of either protein alone (both P < 0.001).
Figure 5.
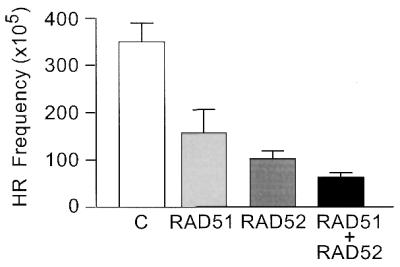
Overexpression of human RAD51 and/or RAD52 decreases DSB-induced HR in human cells. Human HT1080-1885 cells were co-transfected with the I-SceI expression vector and an empty vector (C) or with one or both of the RAD51 and RAD52 expression vectors. Values represent averages ± SD for six independent determinations.
DISCUSSION
We demonstrated that overexpression of wild-type human RAD51 and/or wild-type human RAD52 has dominant negative effects on DSB-induced HR in CHO and human cells. Similarly, overexpression of yeast Rad54 in yeast has dominant negative effects on cell survival and resistance to genotoxic agents (43). We recently showed that overexpression of yeast Rad51 in yeast reduces DSB-induced HR and increases DSB-dependent cell killing (K.Spitz, J.Clikeman, S.Palmer and J.Nickoloff, unpublished results). It is a formal possibility that excess RAD51 or RAD52 reduces DSB induction by I-SceI. However, we found no correlation between HR and DSB levels in yeast. This is a difficult issue to resolve since DSB levels reflect both DSB induction and repair, both of which may be affected by excess RAD proteins. Although we cannot rule out the possibility that excess RAD proteins reduce DSB induction, this is highly unlikely since there is no reason to suspect that RAD proteins directly interact with I-SceI nuclease or that excess RAD proteins block access of I-SceI to target sites in the genome.
In contrast to the negative effects of excess RAD proteins on DSB-induced HR, prior studies demonstrated that excess RAD51 or RAD52 enhances spontaneous HR in mammalian cells (17–19). This difference does not reflect strain or recombination substrate differences as we show that excess RAD51 and/or RAD52 reduces DSB-induced HR and enhances spontaneous HR in the same cell line (Table 1 and Fig. 5). It is not known how spontaneous recombination is initiated, but it may depend on replication, occurring when a replication fork encounters unrepaired or incompletely repaired DNA damage. For example, DSBs may arise when a replication fork encounters a single-strand break in the template strand (44) or lesions that block replication forks may be bypassed by a recombinational mechanism that is independent of DSB formation (45). The increased levels of RAD51 and RAD52 during S phase (46,47) and the fact that RAD51 is essential for cell division in higher eukaryotes (14,16) indicate that recombination plays an important role in replication. In contrast, DSB-induced HR may be partly or completely replication independent. Thus, the distinct effects of HR protein overexpression on spontaneous and DSB-induced HR may reflect differences in the roles of these proteins during replication and DSB repair. For example, excess HR proteins may stimulate spontaneous recombination by enhancing strand exchange between sister chromatids independently of blocked replication forks. Excess HR proteins may reduce DSB-induced HR by directly inhibiting one or more steps during HR or by causing a shift in the types of products produced, as outlined below.
RAD51 and RAD52 interact with each other and with several other proteins and may form transient complexes that function at different stages of recombination (48). Overexpression of one member of a complex can lead to imbalanced stoichiometry that compromises the function of the complex. This can occur through depletion of interacting partners, a mechanism proposed to account for the dominant negative effects of human MSH3 and yeast Mlh1 overexpression on mismatch repair (49,50). A similar model was advanced to account for the dominant negative effects of high concentrations of yeast Rad51 and Rad52 on in vitro strand exchange. Thus, strand exchange increases in proportion to the concentration of these proteins to a point, but is inhibited by higher levels (51,52). Since RAD51 and RAD52 both have DNA-binding activity, an alternative explanation for the dominant negative effects observed in the present study is that the individual proteins might compete with active complexes for DNA ends. In this view, an individual HR protein present in excess binds to a DNA end but is unable on its own to facilitate the repair event, blocking access of active complexes to the DNA end and reducing HR. A variation on this idea derives from findings that in vitro strand exchange is strongly affected by the order of addition of HR proteins (7,48,51,53), suggesting that efficient HR requires an optimal temporal association among DSBs, RAD51 and RAD52. The dominant negative effects we observed with HR protein overexpression may reflect disruption of temporal requirements. We showed that co-overexpression of human RAD51 and RAD52 failed to restore HR to wild-type levels, but instead reduced HR even further. This result might be interpreted in two ways: (i) the two proteins in excess are unable to sequester each other and restore stoichiometric balance of HR complexes or (ii) co-overexpression further disrupts the temporal requirements for HR, an idea supported by the finding that mixing human RAD51 and RAD52 prior to addition to DNA prevents in vitro strand exchange (7).
Each of the models above is based on the idea that excess RAD51 and RAD52 inhibits one or more steps in the HR reaction and, at least in the case of excess RAD51, this idea is supported by the observed reduction in bidirectional tracts (Table 2). We also observed a reduction in bidirectional tracts of DSB-induced products in yeast overexpressing yeast RAD51 (K. Spitz, J. Clikeman, S. Palmer and J. Nickoloff, unpublished results). In this view, excess RAD51 may reduce the efficiency of strand invasion and thereby reduce the probability of two-ended invasions. Alternatively, excess HR proteins might instead stimulate specific types of HR events leading to a shift in the product spectrum. For example, if excess RAD51 and RAD52 shifted events from unequal to equal sister chromatid exchange, the number of detectable (unequal) events would decrease. Current data do not distinguish between these different classes of models.
Gene conversion tracts reflect both hDNA formation and mismatch repair of hDNA (1,2,54,55). hDNA may form by Rad51-mediated strand exchange as well as by branch migration of Holliday junctions. We found that increasing RAD51 and RAD52 had little or no effect on gene conversion tract lengths. It is possible that increasing RAD51 and RAD52 does not increase tract lengths because these proteins have limited strand transfer activity outside of the normal HR complex(es).
Published reports suggest that HR contributes significantly to radiation resistance. RAD51 levels were higher in immortalized human fibroblasts compared to normal diploid fibroblasts and increased RAD51 correlated with increased resistance to ionizing radiation (29). Radiation resistance of carcinoma cell lines correlated with increased RAD51 (30,31) and reducing RAD51 decreased radiation resistance (32,33). In the present study we did not observe significant cell killing upon DSB induction with I-SceI, nor was cell killing enhanced by overexpression of RAD51 and RAD52 (data not shown). However, our assays are probably not sensitive enough to detect RAD51/RAD52 effects on cell killing since only a fraction of cells are transfected by the I-SceI expression vector, and these cells each suffer at most a single DSB. As mentioned above, DSB-dependent cell killing is enhanced in yeast overexpressing yeast Rad51.
The dominant negative effects of RAD51 overexpression on DSB-induced HR that we observed contrast with the results of Lambert and Lopez (24), who found that overexpression of mouse RAD51 enhanced DSB-induced HR in CHO cells. There are a number of similarities and differences in the experimental designs of these studies. In both cases, DSB-induced HR was examined at neo direct repeats in CHO cells, with a transcriptionally active neo inactivated by an I-SceI site and a second neo lacking a promoter. Under normal conditions, transient expression of I-SceI greatly enhances HR in both recombination substrates. One subtle difference is that mouse RAD51 was overexpressed in the prior study and we used human RAD51, but this is unlikely to account for the distinct results given that RAD51 amino acid sequences are highly conserved among mammals. In addition, the reduction in HR in CHO cells is unlikely to be due to cross-species effects since we found that HR was similarly reduced when human RAD51 was overexpressed in human cells. Unlike the prior study, our neo repeats had multiple markers, but this too is unlikely to account for the distinct results since we also observed dominant negative effects with the unmarked pac substrate.
There are two other differences in the two study designs that are more likely to influence the outcome: repeat lengths and the level of RAD51 expression. In the prior study, the neo repeats were 0.7 kb in length, compared to 1.4 kb in our study, and repeat length appears to strongly affect product spectra. With the 0.7 kb repeats, ∼33% of products resulted from gene conversion without an associated crossover and the remainder resulted in deletions (via conversion with crossover, SSA or unequal sister chromatid exchange). In contrast, nearly all DSB-induced HR in the 1.4 kb repeats is by conversion without an associated crossover (34,35; this study). Thus, the proportion of deletion events is inversely correlated with repeat length; a similar correlation is apparent in yeast (56,57). Overexpression of mouse RAD51 stimulated only the minor (conversion) pathway; there was no effect on deletions (24). In contrast, we observed dominant negative effects on conversions (and no shift in product spectra) upon overexpression of human RAD51. It is possible that excess RAD51 stimulated conversion in the 0.7 kb repeats because the conversion pathway is less efficient with short repeats and is therefore more easily facilitated by extra RAD51. With longer repeats, conversion is the dominant outcome and additional RAD51 does not facilitate this pathway but instead is inhibitory. Note that this model is inconsistent with the idea that excess HR protein disrupts the temporal requirements of HR since temporal disruptions would be expected to affect HR independently of repeat length. The other significant difference between the two studies is the level of RAD51 overexpression. In the prior study only modest overexpression was achieved (visual inspection of western blots suggests a 2- to 4-fold increase over endogenous levels), whereas we estimate increases at 4- to >10-fold (Fig. 2A and C). The different results of the two studies could be explained if DSB-induced HR is enhanced by a small increase in RAD51 but inhibited by a large increase, as seen with in vitro strand exchange (51,52). A definitive answer to this question will require cell lines with recombination substrates having variable length repeats (targeted to a single chromosomal locus to avoid position effects) and cell lines that allow controlled RAD51 expression; these experiments are in progress.
Most gene targeting involves interactions between a linear exogenous vector sharing sequence homology to the target locus. Because HR in mammalian cells is relatively inefficient and NHEJ is relatively efficient, this mode of gene targeting is inefficient, and there has been some success in enhancing gene targeting by overexpressing E.coli RecA and human RAD51 (23,58). An alternative way to enhance gene targeting is by creating DSBs in the target locus (59), but our results suggest that overexpression of RAD51 or RAD52 is unlikely to enhance this mode of gene targeting.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Mark Brenneman and David Chen for the human HT1080-1885 cell line, Jingmei Liu for assistance with western blots, and Mark Brenneman, Jennifer Clikeman and Cheryl Miller for helpful comments on the manuscript. This research was supported by grant CA77693 to J.A.N. from the National Cancer Institute and by grant ES08353 to Z.S. from the National Institute of Environmental Health Science.
References
- 1.Weng Y.-s. and Nickoloff,J.A. (1998) Evidence for independent mismatch repair processing on opposite sides of a double-strand break in Saccharomyces cerevisiae. Genetics, 148, 59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petes T.D., Malone,R.E. and Symington,L.S. (1991) Recombination in yeast. In Broach,J.R., Pringle,J.R. and Jones,E.W. (eds), The Molecular and Cellular Biology of the Yeast Saccharomyces: Genome Dynamics, Protein Synthesis and Energetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. I, pp. 407–521.
- 3.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ivanov E.L., Sugawara,N., Fishman-Lobell,J. and Haber,J.E. (1996) Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics, 142, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugawara N. and Haber,J.E. (1992) Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol. Cell. Biol., 12, 563–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumann P. and West,S.C. (1998) Role of the human Rad51 protein in homologous recombination and double-stranded break repair. Trends Biochem. Sci., 23, 247–251. [DOI] [PubMed] [Google Scholar]
- 7.Benson F.E., Baumann,P. and West,S.C. (1998) Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature, 391, 401–404. [DOI] [PubMed] [Google Scholar]
- 8.Rijkers T., Vandenouweland,J., Morolli,B., Rolink,A.G., Baarends,W.M., Vansloun,P.P.H., Lohman,P.H.M. and Pastink,A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol., 18, 6423–6429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thacker J. (1999) A surfeit of RAD51-like genes? Trends Genet., 15, 166–168. [DOI] [PubMed] [Google Scholar]
- 10.Van Dyck E., Stasiak,A.Z., Stasiak,A. and West,S.C. (1999) Binding of double-strand breaks in DNA by human Rad52 protein. Nature, 398, 728–731. [DOI] [PubMed] [Google Scholar]
- 11.Reddy G., Golub,E.I. and Radding,C.M. (1997) Human Rad52 protein promotes single-strand DNA annealing followed by branch migration. Mutat. Res., 377, 53–59. [DOI] [PubMed] [Google Scholar]
- 12.Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein-A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- 13.Nickoloff J.A. and Brenneman,M.A. (2001) Recombination. In Creighton,T.E. (ed.), The Encyclopedia of Molecular Medicine. Wiley, New York, NY, in press.
- 14.Tsuzuki T., Fujii,Y., Sakumi,K., Tominaga,Y., Nakao,K., Sekiguchi,M., Matsushiro,A., Yoshimura,Y. and Morita,T. (1996) Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl Acad. Sci. USA, 93, 6236–6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lim D.-S. and Hasty,P. (1996) A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol., 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonoda E., Sasaki,M.S., Buerstedde,J.M., Bezzubova,O., Shinohara,A., Ogawa,H., Takata,M., Yamaguchiiwai,Y. and Takeda,S. (1998) Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J., 17, 598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park M.S. (1995) Expression of human RAD52 confers resistance to ionizing radiation in mammalian cells. J. Biol. Chem., 270, 15467–15470. [DOI] [PubMed] [Google Scholar]
- 18.Vispe S., Cazaux,C., Lesca,C. and Defais,M. (1998) Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res., 26, 2859–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnaudeau C., Helleday,T. and Jenssen,D. (1999) The RAD51 protein supports homologous recombination by an exchange mechanism in mammalian cells. J. Mol. Biol., 289, 1231–1238. [DOI] [PubMed] [Google Scholar]
- 20.Johnson B.L., Thyagarajan,B., Krueger,L., Hirsch,B. and Campbell,C. (1996) Elevated levels of recombinational DNA repair in human somatic cells expressing the Saccharomyces cerevisiae RAD52 gene. Mutat. Res., 363, 179–189. [DOI] [PubMed] [Google Scholar]
- 21.Shen Z., Denison,K., Lobb,R., Gatewood,J.M. and Chen,D.J. (1995) The human and mouse homologs of the yeast RAD52 gene: cDNA cloning, sequence analysis, assignment to human chromosome 12p12.2-p13 and mRNA expression in mouse tissues. Genomics, 25, 199–206. [DOI] [PubMed] [Google Scholar]
- 22.Shinohara A., Ogawa,H., Matsuda,Y., Ushio,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fission yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 23.Yanez R.J. and Porter,A.C.G. (1999) Gene targeting is enhanced in human cells overexpressing hRAD51. Gene Ther., 6, 1282–1290. [DOI] [PubMed] [Google Scholar]
- 24.Lambert S. and Lopez,B.S. (2000) Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J., 19, 3090–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maacke H., Opitz,S., Jost,K., Hamdorf,W., Henning,W., Kruger,S., Feller,A.C., Lopens,A., Diedrich,K., Schwinger,E. et al. (2000) Over-expression of wild-type Rad51 correlates with histological grading of invasive ductal breast cancer. Int. J. Cancer, 88, 907–913. [DOI] [PubMed] [Google Scholar]
- 26.Yoshikawa K., Ogawa,T., Baer,R., Hemmi,H., Honda,K., Yamauchi,A., Inamoto,T., Ko,K., Yazumi,S., Motoda,H. et al. (2000) Abnormal expression of BRCA1 and BRCA1-interactive DNA-repair proteins in breast carcinomas. Int. J. Cancer, 88, 28–36. [PubMed] [Google Scholar]
- 27.Kato M., Yano,K., Matsuo,F., Saito,H., Katagiri,T., Kurumizaka,H., Yoshimoto,M., Kasumi,F., Akiyama,F., Sakamoto,G. et al. (2000) Identification of Rad51 alteration in patients with bilateral breast cancer. J. Hum. Genet., 45, 133–137. [DOI] [PubMed] [Google Scholar]
- 28.Levy-Lahad E., Lahad,A., Eisenberg,S., Dagan,E., Paperna,T., Kasinetz,L., Catane,R., Kaufman,B., Beller,U., Renbaum,P. et al. (2001) A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc. Natl Acad. Sci. USA, 98, 3232–3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xia S.J.J., Shammas,M.A. and Reis,R.J.S. (1997) Elevated recombination in immortal human cells is mediated by hsRad51 recombinase. Mol. Cell. Biol., 17, 7151–7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanagisawa T., Urade,M., Yamamoto,Y. and Furuyama,J. (1998) Increased expression of human DNA repair genes, XRCC1, XRCC3 and RAD51, in radioresistant human KB carcinoma cell line N10. Oral Oncol., 34, 524–528. [DOI] [PubMed] [Google Scholar]
- 31.Maacke H., Jost,K., Opitz,S., Miska,S., Yuan,Y., Hasselbach,L., Luttges,J., Kalthoff,H. and Sturzbecher,H.W. (2000) DNA repair and recombination factor RAD51 is over-expressed in human pancreatic adenocarcinoma. Oncogene, 19, 2791–2795. [DOI] [PubMed] [Google Scholar]
- 32.Taki T., Ohnishi,T., Yamamoto,A., Hiraga,S., Arita,N., Izumoto,S., Hayakawa,T. and Morita,T. (1996) Antisense inhibition of the Rad51 enhances radiosensitivity. Biochem. Biophys. Res. Commun., 223, 434–438. [DOI] [PubMed] [Google Scholar]
- 33.Collis S.G., Tighe,A., Scott,S.D., Roberts,S.A., Hendry,J.H. and Margison,G.P. (2001) Ribozyme-mediated RAD51 down-regulation increases radiosensitivity of human prostate cancer cells. Nucleic Acids Res., 29, 1534–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W.H., Hesabi,B., Babbo,A., Pacione,C., Liu,J.M., Chen,D.J., Nickoloff,J.A. and Shen,Z.Y. (2000) Regulation of double-strand break-induced mammalian homologous recombination by UBL1, a RAD51-interacting protein. Nucleic Acids Res., 28, 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghian D.G. and Nickoloff,J.A. (1997) Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol. Cell. Biol., 17, 6386–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nickoloff J.A. (1992) Transcription enhances intrachromosomal homologous recombination in mammalian cells. Mol. Cell. Biol., 12, 5311–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choulika A., Perrin,A., Dujon,B. and Nicolas,J.-F. (1995) Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol. Cell. Biol., 15, 1968–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mulligan R.C. and Berg,P. (1981) Factors governing the expression of a bacterial gene in mammalian cells. Mol. Cell. Biol., 1, 449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartsch S., Kang,L.E. and Symington,L.S. (2000) Rad51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol., 20, 1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W. and Jinks-Robertson,S. (1998) Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol. Cell. Biol., 18, 6525–6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rattray A.J. and Symington,L.S. (1994) Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics, 138, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilera A. and Klein,H.L. (1989) Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics, 123, 683–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clever B., Schmuckli-Maurer,J., Sigirst,M., Glassner,B.J. and Heyer,W.D. (1999) Specific negative effects resulting from elevated levels of the recombinational repair protein Rad54p in Saccharomyces cerevisiae. Yeast, 15, 721–740. [DOI] [PubMed] [Google Scholar]
- 44.Rothstein R., Michel,B. and Gangloff,S. (2000) Replication fork pausing and recombination or “gimme a break”. Genes Dev., 14, 1–10. [PubMed] [Google Scholar]
- 45.Kogoma T. (1997) Stable DNA replication: interplay between DNA replication, homologous recombination and transcription. Microbiol. Mol. Biol. Rev., 61, 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen F.Q., Nastasi,A., Shen,Z.Y., Brenneman,M., Crissman,H. and Chen,D.J. (1997) Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes RAD51 and RAD52. Mutat. Res., 384, 205–211. [DOI] [PubMed] [Google Scholar]
- 47.Yamamoto A., Taki,T., Yagi,H., Habu,T., Yoshida,K., Yoshimura,Y., Yamamoto,K., Matsushiro,A., Nishimune,Y. and Morita,T. (1996) Cell cycle-dependent expression of the mouse Rad51 gene in proliferating cells. Mol. Gen. Genet., 251, 1–12. [DOI] [PubMed] [Google Scholar]
- 48.Mazin A.V., Bornarth,C.J., Solinger,J.A., Heyer,W.D. and Kowalczykowski,S.C. (2000) Rad54 protein is targeted to pairing loci by the Rad51 nucleoprotein filament. Mol. Cell, 6, 583–592. [DOI] [PubMed] [Google Scholar]
- 49.Marra G., Iaccarino,I., Lettieri,T., Roscilli,G., Delmastro,P. and Jiricny,J. (1998) Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc. Natl Acad. Sci. USA, 95, 8568–8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shcherbakova P.V. and Kunkel,T.A. (1999) Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol. Cell. Biol., 19, 3177–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song B.W. and Sung,P. (2000) Functional interactions among yeast Rad51 recombinase, Rad52 mediator and replication protein A in DNA strand exchange. J. Biol. Chem., 275, 15895–15904. [DOI] [PubMed] [Google Scholar]
- 52.Sung P. and Robberson,D.L. (1995) DNA strand exchange catalyzed by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell, 82, 453–461. [DOI] [PubMed] [Google Scholar]
- 53.Sigurdsson S., Trujillo,K., Song,B.W., Stratton,S. and Sung,P. (2001) Basis for avid homologous DNA strand exchange by human Rad51 and RPA. J. Biol. Chem., 276, 8798–8806. [DOI] [PubMed] [Google Scholar]
- 54.Nickoloff J.A. and Hoekstra,M.F. (1998) Double-strand break and recombinational repair in Saccharomyces cerevisiae. In Nickoloff,J.A. and Hoekstra,M.F. (eds), DNA Damage and Repair, Vol. 1, DNA Repair in Prokaryotes and Lower Eukaryotes. Humana Press, Totowa, NJ, pp. 335–362.
- 55.Nickoloff J.A., Sweetser,D.B., Clikeman,J.A., Khalsa,G.J. and Wheeler,S.L. (1999) Multiple heterologies increase mitotic double-strand break-induced allelic gene conversion tract lengths in yeast. Genetics, 153, 665–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nickoloff J.A., Chen,E.Y.C. and Heffron,F. (1986) A 24-base-pair sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl Acad. Sci. USA, 83, 7831–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ray A., Siddiqi,I., Kolodkin,A.L. and Stahl,F.W. (1988) Intrachromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J. Mol. Biol., 201, 247–260. [DOI] [PubMed] [Google Scholar]
- 58.Shcherbakova O.G., Lanzov,V.A., Ogawa,H. and Filatov,M.V. (2000) Overexpression of bacterial RecA protein stimulates homologous recombination in somatic mammalian cells. Mutat. Res., 459, 65–71. [DOI] [PubMed] [Google Scholar]
- 59.Jasin M. (1996) Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet., 12, 224–228. [DOI] [PubMed] [Google Scholar]