Abstract
Background
Lipid intermediates produced during triacylglycerols (TAGs) synthesis and lipolysis in adipocytes interfere with the intracellular insulin signaling pathway and development of insulin resistance. This study aims to compare TAG species and their fatty acid composition in adipose tissues from insulin sensitive (IS), insulin resistant (IR) and type 2 diabetes mellitus (T2DM) obese individuals.
Methods
Human subcutaneous and omental adipose tissue biopsies were obtained from 64 clinically characterized obese individuals during weight reduction surgery. TAGs were extracted from the adipose tissues using the Bligh and Dyer method, then were subjected to non-aqueous reverse phase ultra-high performance liquid chromatography and full scan mass spectrometry acquisition and data dependent MS/MS on LTQ dual cell linear ion trap. TAGs and their fatty acid contents were identified and compared between IS, IR and T2DM individuals and their levels were correlated with metabolic traits of participants and the adipogenic potential of preadipocyte cultures established from their adipose tissues.
Results
Data revealed 76 unique TAG species in adipose tissues identified based on their exact mass. Analysis of TAG levels revealed a number of TAGs that were significantly altered with disease progression including C46:4, C48:5, C48:4, C38:1, C50:3, C40:2, C56:3, C56:4, C56:7 and C58:7. Enrichment analysis revealed C12:0 fatty acid to be associated with TAGs least abundant in T2DM whereas C18:3 was found in both depleted and enriched TAGs in T2DM. Significant correlations of various adipose tissue-derived TAG species and metabolic traits were observed, including age and body mass index, systemic total cholesterol, TAGs, and interleukin-6 in addition to adipogenic potential of preadipocytes derived from the same adipose tissues.
Conclusion
Pilot data suggest that adipose tissues from obese IR and T2DM individuals exhibit TAG-specific signatures that may contribute to their increased risk compared to their IS counterparts. Future experiments are warranted to investigate the functional relevance of these specific lipidomic profiles.
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1548-x) contains supplementary material, which is available to authorized users.
Keywords: Lipidomics, Adipose tissue, Triaclyglycerols, Insulin sensitivity, Insulin resistance, Type 2 diabetes mellitus
Background
Adipose tissue is the main site for storing and mobilizing energy in response to metabolic demand. Obesity is associated with changes in the structure and function of the adipose tissue, leading to progression of insulin resistance and type 2 diabetes mellitus (T2DM) [1]. However, a subset of obese individuals, known as the insulin sensitive (IS) obese, maintain insulin sensitivity and exhibit better adipose tissue functions compared to equally obese insulin resistant (IR) counterparts [2]. Obesity triggers hypertrophy of adipocytes within the subcutaneous (SC) adipose tissues to enable accumulation of excess triacylglycerols (TAGs). Additional energy intake causes further fat accumulation within the omental (OM) depot, which is associated with ectopic fat deposition in the liver, skeletal muscle and heart tissues [3]. The subsequent hyperinsulinemia inhibits hormone sensitive lipase and triggers the lipoprotein lipase causing additional glucose intolerance, hyperinsulinemia, hypertriglyceridemia and higher risk of insulin resistance in these tissues [4].
Analysis of complex biological systems has become possible by the newly emerging metabolomics techniques where metabolites serve as direct indicators of biochemical activity of complex phenotypes such as insulin resistance and T2DM [5]. In this context, lipidomics studies were utilized to study differences between SC and OM depots. These studies have revealed depot-specific enrichment of specific TAGs, glycerophospholipids, and sphingolipids and differences in the association of lipid species with body mass index, inflammation and insulin sensitivity [6, 7]. Although TAGs themselves are unlikely to be signaling molecules, an increasing body of evidence suggests that lipid intermediates produced during TAG synthesis or breakdown interfere with the intracellular insulin signaling pathway and contribute to the development of insulin resistance, including free fatty acids, diacylglycerols and ceramides [8]. Indeed, elevated fatty acid efflux from the adipose tissue stimulates TAG synthesis in the liver and triggers stress of endoplasmic reticulum and stimulation of June kinase pathway in the adipose tissues [9, 10]. This leads to an overload of TAG’s synthetic capacity, causing an increase in both diacylglycerols (DAGs) and ceramide levels and further development of insulin resistance in adipocytes [11].
Despite various studies investigating lipidomic differences in human serum and adipose tissues in relation to insulin sensitivity, no studies have compared differences in TAG signatures and their fatty acid composition in adipose tissues from IS, IR and T2DM obese individuals and their correlations with mediators of metabolic disease. Identification of the fatty acids that are enriched or depleted in tissues from insulin resistance and T2DM individuals could shed light on their functional role in disease progression, thus providing potential novel targets for therapeutic intervention. The aims of this study were to profile TAG species and measure their levels in two fat depots and to compare their fatty acid composition between IS, IR and T2DM individuals.
Methods
Materials
Interleukin 6 (IL-6) and leptin ELISAs were from R&D systems (Abingdon, UK). Insulin ELISA was from Mercodia Diagnostics (Uppsala, Sweden). 4′,6-Diamidino-2-phenylindole (DAPI), and LipidTOX Green Neutral Lipid were from Life Technologies (Warrington, UK). Other chemicals and reagents were from Sigma (Munich, Germany).
Cohort
Participants’ recruitment criteria were described previously [12]. Briefly, 64 consented obese individuals undergoing bariatric surgery at AlEmadi hosptial (Doha, Qatar) were recruited. Protocols were approved by Institutional Review Board of ADLQ (X2017000224). Blood was taken prior to operation and 1–5 g of abdominal SC and OM adipose tissues biopsies were collected during the surgery and stored at − 80 °C until use. Plasma cholesterol, fasting glucose and liver function enzymes were measured by COBAS INTEGRA (Roche Diagnostics, Basil). IL-6, leptin and insulin were determined using commercially available ELISA. Insulin resistance was computed by homeostatic model assessment (HOMA-IR) [13] using 30th percentile (HOMAIR = 2.4) as a threshold point. Accordingly, subjects were dichotomized into IS (HOMA-IR < 2.4, n = 18, 3 males and 15 females) and IR (HOMA-IR > 2.4, n = 35, 9 males and 26 females). Eleven participants were clinically diagnosed with T2DM (8 males and 3 females).
Preadipocyte culture and differentiation
Stromal vascular fraction (SVF) cells were obtained by collagenase digestion of adipose tissues as described previously [12]. Cell pellets were re-suspended in stromal medium containing Dulbecco’s modified Eagle’s Medium-F12 (DMEM-F12) supplemented with 10% fetal bovine serum (FBS) and Penicillin/Streptomycin, then maintained at 37 °C with 5% CO2 until confluence. To induce differentiation, early passaged stromal vascular fraction (SVF)-derived preadipocytes (passages 1–3) were grown at 2 × 104/cm2 in stromal medium overnight, then incubated in differentiation medium (DMEM-F12, 3% FBS, 33 μM biotin, 17 μM d-pantothenate, 1 μM dexamethasone, 250 μM of methylisobutylxanthine, 0.1 μM human insulin, 5 μM of Peroxisome proliferator-activated receptor gamma PPARγ agonist, rosiglitazone) for 7 days, followed by 12 days in maintenance medium containing the same components as the differentiation medium but omitting methylisobutylxanthine and rosiglitazone. Differentiation potential (adipogenic capacity) was determined as a percentage of lipidtox positive stained cells to total number of stained nuclei (DAPI).
Sample preparation
Human SC and OM adipose tissue specimens from IS, IR and T2DM individuals were extracted using the Bligh and Dyer Method [14]. Homogenization of tissue was carried out in the gentle MACS Dissociator (Miltenyi Biotech, Germany) with one volume of PBS for every gram of tissue. Following tissue homogenization, 1 mL of each sample solution was transferred into a separate 15 mL Falcon tube, and 3 mL of 3:1 ratio of Chloroform:MeOH were added into each tube. One microliter of PBS was added and samples were centrifuged at 3000 RPM for 20 min at room temperature. The organic layer (bottom layer) was carefully transferred into new 15 mL Falcon tubes and evaporated to dryness under a stream of high purity nitrogen. Samples were then reconstituted with 1:1:1 mixture of hexane, isopropanol, acetonitrile. Subsequently the extracts were analysed using data dependent full scan MS and MS/MS acquisition using the Thermo LTQ VelosPro dual cell linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA).
Sample analysis
Separation of TAGs was carried out using non-aqueous reverse phase UHPLC separation (NARP), on a Dionex Ultimate 3000 UHPLC system, using acetonitrile w/0.1% formic acid (eluent A), and isopropanol w/10 mM ammonium formate (eluent B) as the mobile phase. The column was a Phenomenex UHPLC C30 core shell, 150 mm × 2.1 mm and 2.7 µm particle size (Phenomenex Torrance CA, USA). Gradient conditions started with 5% B held for 2 min, then raised to 50% B at 30 min, held for 10 min and then reduced to 5% B at 45 min and held for further 5 min.
Mass spectrometry
MS analyses were conducted using the Thermo LTQ VelosPro dual cell linear ion trap mass spectrometer (Thermo Fisher Scientific, San Jose, CA, USA), acquiring both full scan MS and subsequent data dependent full scan MS/MS product ion spectra with wide band activation. Target parent ions were automatically selected from an inclusion list. The low resolution full scan analysis provides molecular parent masses (M+NH4+). These parent ion full scan MS/MS analysis provided further elucidation of possible structures represented in each lipid (fatty acid composition). Relative abundances of each identified TAG were estimated from the height values for each extracted ion current profile for parent masses of each compound (M+NH4+).
Separation by equivalent carbon number
The above UHPLC conditions (NARP) provide separations of TAGs by their equivalent carbon number (ECN). The ECN is calculated, from the total number of non-glycerol carbons in the TAG minus twice the number of the double bonds in the molecule (ECN = CN − 2DB). NARP eluted the TAGs from lower to higher ECN with increasing percent B in the eluent. NARP-HPLC is commonly used for TAG separation because it works on both the chain and absolute height or area counts for each identified TAG. As some of the TAG may not show baseline resolution, the height counts were chosen to better represent the TAG.
Statistical analysis
All statistical analyses were carried out using R version 3.2.1 and SIMCA 13.0.1 software (Umetrics, Sweden). Variables with skewed distributions were log transformed or taken the square root of as appropriate to ensure normality. An initial PCA was conducted to identify components that explain large proportion of the TAG variance. A repeated measures linear model incorporating confounders: gender, age, BMI, PC1 and PC2 (derived from earlier principle component analysis, PCA) and covariates: tissue and diabetic status (IS, IR, T2DM) was used to assess the differences in each TAG between the two tissues and amongst the insulin/diabetes groups. The model was based on repeated measures statistics since a TAG measurement from an individual was taken from two separate tissues: SC and OM. The model allows the individual inherent variation to be taken out of the total variance. Such enhanced modelling of the error structure increases the model’s ability to detect significance of covariate effects. Nonetheless, we have repeated the analysis using the standard linear model and confirmed the superiority of the repeated measures linear model counterpart. The linear model was sometimes used when fitting the repeated measures model was not possible due to missing data. False discovery rate (FDR) multiple testing correction was also performed on the differentially expressed TAG species identified between adipose tissues from IS, IR and T2DM individuals. Fatty acid enrichment amongst diabetes/tissue significant TAGs was assessed using the one tailed Wilcoxon sum of the ranks test on the list of metabolites that differed significantly between IS, IR and T2DM after correcting for covariates including gender, age, BMI, PC1 and PC2. The analysis was based on assessing the likelihood of randomly observing a given fatty acid that often amongst highly ranked TAGs along the list of all TAGs ordered by p value as follows: For each of the following contrasts: subcutaneous versus omental, IR versus IS, IR versus T2DM and IS versus T2DM, TAGs were ranked by their p values and a given fatty acid mapped to the ranks of TAGs within which it is found. The analysis proceeds by assessing the likelihood of obtaining the observed sum of fatty acid identified ranks by chance. If the fatty acid is observed amongst the significant TAG at the top of the list, the sum of the ranks would be too small to be explained by chance alone; hence the null hypothesis is rejected in favor of enrichment. Enrichment hits failed to remain significant after FDR multiple testing correction but data was reported because of agreement with literature as elaborated in “Discussion” section. A similar test was used to assess enrichment in constituent fatty acid saturation levels.
Results
General characteristics of participants
Sixty-four (44 females and 20 males) obese and morbidly obese (BMI = 43.1 ± 7.5 kg/m2) participants were recruited from amongst patients undergoing weight reduction surgery. Participants exhibited hyperleptinemia and hyperinsulinemia and were dichotomized into IS and IR groups based on their HOMA-IR index and into T2DM based on their medical records. Compared to BMI-matched IS and IR subjects, T2DM individuals were older and had higher circulating levels of TAG and lower leptin (Table 1). Compared to females, males had higher mean arterial blood pressure (MAP) (93.7 vs 84.7, p > 0.01) and lower HDL (1.1 vs 1.5, p = 0.05) and leptin (42.0 vs 67.3, p > 0.01) (Additional file 1: Table S1). IS males had lower HOMA-IR than their age and BMI-match IS females, whereas IR males had higher HOMA-IR than their age, but not BMI, matched females (Additional file 1: Table S1). Compared to obese subjects (n = 26), the morbidly obese participants (n = 46) had significantly higher BMI, SBP, IL-6, FPG and HOMA-IR (Additional file 1: Table S2).
Table 1.
General characteristics of participants
| Variables | IS | IR | T2DM | P value | IS+IR | P value | |||
|---|---|---|---|---|---|---|---|---|---|
| (N = 18) | (N = 35) | (N = 11) | ANOVA | IS vs IR | IS vs T2DM | IR vs T2DM | (N = 46) | IS+IR vs T2DM | |
| Age (years) | 32.09 (9.7) | 30.26 (9.3) | 43.57 (9.4) | 0.000 | 0.739 | 0.017 | 0.003 | 30.9 (9.4) | 0.001 |
| BMI (kg m−2) | 41.44 (7.0) | 43.31 (6.9) | 45.53 (9.6) | 0.38 | 0.583 | 0.394 | 0.731 | 42.7 (6.9) | 0.322 |
| SBP (mmHg) | 119.0 (13.8) | 122.67 (15.7) | 132.14 (11.6) | 0.13 | 0.616 | 0.111 | 0.268 | 121.5 (15.1) | 0.075 |
| DBP (mmHg) | 66.1 (9.8) | 69.98 (12.7) | 73.33 (7.0) | 0.3 | 0.422 | 0.369 | 0.783 | 68.7 (11.9) | 0.35 |
| MAP | 83.4 (8.1) | 88.08 (12.4) | 93.53 (8.9) | 0.11 | 0.288 | 0.130 | 0.499 | 86.6 (11.3) | 0.15 |
| Cholesterol (mmol/L) | 4.4 (0.9) | 4.6 (1.2) | 5.22 (0.9) | 0.24 | 0.785 | 0.215 | 0.364 | 4.5 (1.1) | 0.122 |
| LDL (mmol/L) | 2.73 (0.8) | 2.91 (0.9) | 3.1 (1.0) | 0.55 | 0.693 | 0.593 | 0.855 | 2.8 (0.8) | 0.466 |
| HDL (mmol/L) | 1.33 (0.4) | 1.46 (0.9) | 1.29 (0.3) | 0.74 | 0.793 | 0.990 | 0.844 | 1.4 (0.8) | 0.678 |
| Triglyceride (mmol/L) | 1.13 (0.6) | 1.27 (0.7) | 1.91 (1.1) | 0.04 | 0.729 | 0.031 | 0.072 | 1.2 (0.6) | 0.015 |
| Leptin (ng/mL) | 64.36 (25.5) | 60.7 (21.9) | 39.16 (31.5) | 0.06 | 0.854 | 0.055 | 0.091 | 62.1 (23.1) | 0.021 |
| Adiponetin (ng/mL) | 3.24 (2.2) | 3.62 (1.9) | 3.47 (2.4) | 0.88 | 0.866 | 0.982 | 0.992 | 3.5 (1.9) | 0.979 |
| IL-6 (pg/mL) | 3.28 (1.8) | 3.72 (1.8) | 4.03 (2.2) | 0.58 | 0.683 | 0.627 | 0.913 | 3.6 (1.8) | 0.521 |
| FBG (mmol/L) | 5.73 (2.5) | 12.76 (8.4) | 12.84 (6.5) | 0.000 | 0.001 | 0.072 | 1.000 | 10.3 (7.7) | 0.439 |
| Insulin (mIU/L) | 6.33 (1.9) | 12.6 (10.0) | 11.92 (6.4) | 0.01 | 0.010 | 0.241 | 0.976 | 10.3 (8.6) | 0.633 |
| HOMA-IR | 1.56 (0.6) | 4.86 (2.0) | 6.6 (3.2) | 0.000 | 0.000 | 0.000 | 0.080 | 3.7 (2.3) | 0.005 |
BMI body mass index, SBP systolic blood pressure, DBP diastolic blood pressure, MAP mean arterial blood pressure, LDL low density lipoprotein, HDL high density lipoprotein, IL-6 interleukin 6, FPG fasting blood glucose, HOMA-IR homeostatic model assessment of insulin resistance. Data are presented as mean (SD). Differences between IS, IR and T2DM were tested by ANOVA. Differences between (IS+IR vs T2DM) were tested by the independent-sample t test or Mann–Whitney U test. A p value significance level of 0.05 was used
Differences in TAG content between omental and subcutaneous adipose tissues
Using a non-targeted approach, a comprehensive parent mass list of 120 identified TAGs was created, of which 76 TAG species were identified (Appendix, Table 6) based on their molecular weights and peak heights. A linear model was used to assess depot-specific TAG associations after correcting for participant diabetes group, gender, PC1 and PC2 (refer to “methods” section). Analysis revealed 7 TAGs that were significantly different between SC and OM tissues. C53:5, C51:3, C50:4, C59:1, C54:6 and C50:2 were higher in OM than SC. C38:1 was higher in the SC compared to OM tissues. The full scan MS/MS analysis revealed the fatty acid composition for each identified TAG (Table 2).
Table 6.
List of identified TAG, their molecular weights (MW), fatty acid compositions and fatty acids identities
| Component name | TAG | MW | Fatty acid composition | Fatty acids identities |
|---|---|---|---|---|
| TAG2 | C38:1 | 664.7 | C10:0, C12:0, C16:1 | Capric acid, lauric acid, palmitoleic acid |
| TAG2A | C38:1 | 664.7 | C18:1, C16:0, C4:0 | Oleic acid, palmitic acid, butyric acid |
| TAG3 | C38:0 | 666.7 | C12:0, C12:0, C14:0 | Lauric acid, lauric acid, myristic acid |
| TAG7 | C40:2 | 690.7 | C6:0, C16:0, C18:2 | Caproic acid, palmitic acid, linoleic acid |
| TAG84 | C40:1 | 692.7 | C18:1, C16:0, C6:0 | Oleic acid, palmitic acid, caproic acid |
| TAG9 | C42:2 | 718.7 | C18:2, C12:0, C12:0 | Linoleic acid, lauric acid, lauric acid |
| TAG10 | C42:1 | 720.7 | C12:0, C14:0, C16:1 | Lauric acid, myristic acid, palmitoleic acid |
| TAG12 | C44:3 | 744.7 | C18:2, C16:1, C10:0 | Linoleic acid, palmitoleic acid, capric acid |
| TAG13 | C44:2 | 746.7 | C18:2, C14:0, C12:0 | Linoleic acid, myristic acid, lauric acid |
| TAG14 | C44:1 | 748.7 | C18:1, C16:0, C10:0 | Oleic acid, palmitic acid, capric acid |
| TAG16A | C46:4 | 770.7 | C18:2, C18:2, C10:0 | Linoleic acid, linoleic acid, capric acid |
| TAG17 | C46:3 | 772.7 | C18:2, C16:1, C12:0 | Linoleic acid, palmitoleic acid, lauric acid |
| TAG18 | C46:2 | 774.8 | C18:2, C16:0, C12:0 | Linoleic acid, palmitic acid, lauric acid |
| TAG19 | C46:1 | 776.8 | C18:1, C16:0, C12:0 | Oleic acid, palmitic acid, lauric acid |
| TAG20 | C46:0 | 778.8 | C16:0, C16:0, C14:0 | Palmitic acid, palmitic acid, myristic acid |
| TAG21 | C48:5 | 796.7 | C18:2, C18:3, C12:0 | Linoleic acid, linolenic acid, lauric acid |
| TAG22 | C48:4 | 798.7 | C18:2, C18:2, C12:0 | Linoleic acid, linoleic acid, lauric acid |
| TAG23 | C48:3 | 800.8 | C18:2, C16:1, C14:0 | Linoleic acid, palmitoleic acid, myristic acid |
| TAG24 | C48:2 | 802.8 | C18:2, C16:0, C14:0 | Linoleic acid, palmitic acid, myristic acid |
| TAG24A | C48:2 | 802.8 | C18:1, C16:1, C14:0 | Oleic acid, palmitoleic acid, myristic acid |
| TAG25 | C48:1 | 804.8 | C18:1, C16:0, C14:0 | Oleic acid, palmitic acid, myristic acid |
| TAG26 | C48:0 | 806.7 | C16:0, C16:0, C16:0 | Palmitic acid, palmitic acid, palmitic acid |
| TAG27 | C48: | 818.7 | C18:1, C16:0, C14: | Oleic acid, palmitic acid, pentadecylic acid |
| TAG29 | C50:6 | 822.7 | C18:3, C18:3, C14:0 | Linolenic acid, linolenic acid, myristic acid |
| TAG30 | C50:5 | 824.7 | C18:3, C16:1, C16:1 | Linolenic acid, palmitoleic acid, palmitoleic acid |
| TAG31 | C50:4 | 826.7 | C18:2, C18:2, C14:0 | Linoleic acid, linoleic acid, myristic acid |
| TAG32 | C50:3 | 828.8 | C16:1, C16:1, C18:1 | Palmitoleic acid, palmitoleic acid, oleic acid |
| TAG33 | C50:2 | 830.8 | C18:2, C16:0, C16:0 | Linoleic acid, palmitic acid, palmitic acid |
| TAG34 | C50:1 | 832.8 | C18:1, C16:0, C16:0 | Oleic acid, palmitic acid, palmitic acid |
| TAG35 | C50:0 | 834.7 | C18:0, C16:0, C16:0 | Stearic acid, palmitic acid, palmitic acid |
| TAG36 | C51:3 | 842.6 | C18:1, C16:1, C17:1 | Oleic acid, palmitoleic acid, cis-10-heptadecenoic acid |
| TAG37 | C50: | 844.7 | C18:1, C18:1, C14: | Oleic acid, oleic acid, pentadecylic acid |
| TAG38 | C51:1 | 846.7 | C18:1, C17:0, C16:0 | Oleic acid, heptadecanoic acid, palmitic acid |
| TAG39 | C52:7 | 848.7 | C18:3, C18:3, C16:1 | Linolenic acid, linolenic acid, palmitoleic acid |
| TAG39A | C51:0 | 848.7 | C18:0, C17:0, C16:0 | Stearic acid, heptadecanoic acid, palmitic acid |
| TAG40 | C52:6 | 850.7 | C18:2, C14:0, C20:4 | Linoleic acid, myristic acid, C20:4 |
| TAG40A | C52:6 | 850.7 | C18:3, C16:0, C18:3 | Linolenic acid, palmitic acid, linolenic acid |
| TAG41 | C52:5 | 852.7 | C18:2, C18:2, C16:1 | Linoleic acid, linoleic acid, palmitoleic acid |
| TAG42 | C52:4 | 854.8 | C18:2, C18:2, C16:0 | Linoleic acid, linoleic acid, palmitic acid |
| TAG43 | C52:3 | 856.8 | C18:1, C18:1, C16:1 | Oleic acid, oleic acid, palmitoleic acid |
| TAG44 | C52:2 | 858.8 | C18:1, C18:1, C16:0 | Oleic acid, oleic acid, palmitic acid |
| TAG45 | C52:1 | 860.8 | C16:0, C18:1, C18:0 | Palmitic acid, oleic acid, stearic acid |
| TAG46 | C52:0 | 862.7 | C18:0, C18:0, C16:0 | Stearic acid, stearic acid, palmitic acid |
| TAG47 | C53:5 | 866.7 | C17:0, C17:1, C19:4 | Heptadecanoic acid, cis-10-heptadecanoic acid, C19:4 |
| TAG49A | C53:1 | 870.7 | C18:1, C18:2, C17:0 | Oleic acid, linoleic acid, heptadecanoic acid |
| TAG52 | C54:7 | 876.7 | C18:2, C18:2, C18:3 | Linoleic acid, linoleic acid, linolenic acid |
| TAG52A | C54:7 | 876.7 | C18:2, C18:2, C18:3 | Linoleic acid, linoleic acid, linolenic acid |
| TAG53 | C54:6 | 878.7 | C18:2, C18:1, C18:3 | Linoleic acid, oleic acid, linolenic acid |
| TAG53A | C54:6 | 878.7 | C18:2, C18:2, C18:2 | Linoleic acid, linoleic acid, linoleic acid |
| TAG54 | C54:5 | 880.8 | C18:2, C18:2, C18:1 | Linoleic acid, linoleic acid, oleic acid |
| TAG54A | C54:5 | 880.8 | C18:2, C18:2, C18:1 | Linoleic acid, linoleic acid, oleic acid |
| TAG55 | C54:4 | 882.8 | C18:1, C18:1, C18:2 | Oleic acid, oleic acid, linoleic acid |
| TAG56A | C54:3 | 884.8 | C18:1, C18:1, C18:1 | Oleic acid, oleic acid, oleic acid |
| TAG57 | C54:2 | 886.8 | C18:1, C18:1, C18:0 | Oleic acid, oleic acid, stearic acid |
| TAG58A | C54:1 | 888.8 | C18:0, C18:0, C18:1 | Stearic acid, stearic acid, oleic acid |
| TAG59 | C54:0 | 890.8 | C18:0, C18:0, C18:0 | Stearic acid, stearic acid, stearic acid |
| TAG60 | C56:8 | 902.8 | C20:5, C18:1, C18:2 | Eicosapentaenoic acid, oleic acid, linoleic acid |
| TAG60A | C56:8 | 902.8 | C20:5, C18:1, C18:2 | Eicosapentaenoic acid, oleic acid, linoleic acid |
| TAG60B | C55:1 | 902.8 | C20:0, C18:1, C17:0 | Eicosapentaenoic acid, oleic acid, linoleic acid |
| TAG61 | C56:7 | 904.8 | C20:4, C18:1, C18:2 | Arachidonic acid, oleic acid, linoleic acid |
| TAG64A | C56:4 | 910.8 | C18:1, C18:2, C20:1 | Oleic acid, linoleic acid, gadoleic |
| TAG65 | C56:3 | 912.8 | C20:1, C18:1, C18:1 | Gadoleic, oleic acid, oleic acid |
| TAG66 | C56:2 | 914.8 | C18:1, C18:1, C20:0 | Oleic acid, oleic acid, arachidic acid |
| TAG67 | C56:1 | 916.8 | C20:0, C18:0, C18:1 | Arachidic acid, stearic acid, oleic acid |
| TAG67A | C56:1 | 916.8 | C18:0, C18:0, C20:1 | Stearic acid, stearic acid, gadoleic |
| TAG71 | C58:9 | 928.8 | C22:6, C18:1, C18:2 | Docosahexaenoic acid, oleic acid, linoleic acid |
| TAG71A | C58:9 | 928.8 | C22:6, C18:1, C18:2 | Docosahexaenoic acid, Oleic acid, linoleic acid |
| TAG71B | C58:9 | 928.8 | C22:6, C18:1, C18:2 | Docosahexaenoic acid, oleic acid, linoleic acid |
| TAG74 | C58:7 | 934.8 | C18:6, C24:0, C16:1 | C18:6, lignoceric acid, palmitoleic acid |
| TAG75 | C58:4 | 938.7 | C18:3, C24:0, C16:1 | Linolenic acid, lignoceric acid, palmitic acid |
| TAG78 | C58:2 | 942.8 | C20:1, C20:1, C18:0 | Gadoleic, gadoleic, stearic acid |
| TAG79A | C58:1 | 944.8 | C22:0, C18:0, C18:1 | Behenic acid, stearic acid, oleic acid |
| TAG80 | C64:1 | 956.8 | C23:0, C23:0, C18:1 | Tricosanoic acid, tricosanoic acid, oleic acid, |
| TAG81 | C59:1 | 958.8 | C23:0, C18:0, C18:1 | Tricosanoic acid, stearic acid, oleic acid |
| TAG82 | C60:3 | 968.8 | C24:0, C18:1, C18:2 | Lignocerric acid, oleic acid, linoleic acid |
| TAG83 | C60:1 | 970.8 | C24:0, C24:0, C18:1 | Lignocerric acid, lignocerric acid, oleic acid |
Table 2.
Differential TAG species identified between subcutaneous and omental adipose tissues
| ID | TAG | MW | Fatty acid composition | Fatty acids identities | Fold change (SC-OM) | P value |
|---|---|---|---|---|---|---|
| TAG47 | C53:5 | 866.7 | C17:0, C17:1, C19:4 | Heptadecanoic acid, cis-10-heptadecanoic acid, C19:4 | 0.17 | 0.01 |
| TAG2A | C38:1 | 664.7 | C18:1, C16:0, C4:0 | Oleic acid, palmitic acid, butyric acid | − 0.44 | 0.02 |
| TAG36 | C51:3 | 842.6 | C18:1, C16:1, C17:1 | Oleic acid, palmitoleic acid, cis-10-heptadecenoic acid | 0.32 | 0.03 |
| TAG31 | C50:4 | 826.7 | C18:2, C18:2, C14:0 | Linoleic acid, linoleic acid, myristic acid | 0.37 | 0.03 |
| TAG81 | C59:1 | 958.8 | C23:0, C18:0, C18:1 | Tricosanoic acid, stearic acid, oleic acid | 0.16 | 0.04 |
| TAG53A | C54:6 | 878.7 | C18:2, C18:2, C18:2 | Linoleic acid, linoleic acid, linoleic acid | 0.17 | 0.04 |
| TAG33 | C50:2 | 830.8 | C18:2, C16:0, C16:0 | Linoleic acid, palmitic acid, palmitic acid | 0.19 | 0.05 |
Molecular weight (MW), fatty acid composition, fatty acid identity, fold change in SC tissue compared to OM are also indicated
TAGs with varying levels between IS, IR and T2DM
A linear model was used to assess TAG associations with participant groups after correcting for possible confounders (refer to “Methods” section). A number of TAGs were significantly decreased in T2DM compared to IS and/or IR including C46:4, C48:5, C48:4, C38:1, C50:3 and C40:2 whereas a number of TAGs were increased in T2DM compared to the other two groups including C56:3, C56:4, C56:7 and C58:7. No significant differences in TAGs between IS and IR groups was detected. Table 3 summarizes the list of differentially expressed TAGs with their fatty acids compositions. When looking at gender versus group (IS, IR and T2DM) interaction, there were no FDR significant interaction effects. However, when considering BMI versus group interaction, two TAG species showed FDR significant interaction effects including C40:2 and C53:4. Whereas the former (C40:1) shows more pronounced decrease in T2DM compared to IS in low BMI than in high BMI, the latter (C53:4) shows a more pronounced increase in low BMI than in high BMI (Additional file 1: Table S3).
Table 3.
Differentially expressed TAG species identified between adipose tissues from IS, IR and T2DM individuals
| ID | TAG | MW | Fatty acid composition | Fatty acids identities | Comparison | Fold change | FDR p value |
|---|---|---|---|---|---|---|---|
| TAG16A | C46:4 | 770.7 | C18:2, C18:2, C10:0 | Linoleic acid, linoleic acid, capric acid | IS vs T2DM | − 0.62 | 0.005 |
| IR vs T2DM | − 0.53 | 0.01 | |||||
| TAG21 | C48:5 | 796.7 | C18:2, C18:3, C12:0 | Linoleic acid, linolenic acid, lauric acid | IR vs T2DM | − 0.39 | 0.0005 |
| IS vs T2DM | − 0.38 | 0.0013 | |||||
| TAG22 | C48:4 | 798.7 | C18:2, C18:2, C12:0 | Linoleic acid, linoleic acid, lauric acid | IR vs T2DM | − 0.96 | 0.002 |
| TAG2A | C38:1 | 664.7 | C18:1, C16:0, C4:0 | Oleic acid, palmitic acid, butyric acid | IS vs T2DM | − 1.00 | 0.0007 |
| TAG32 | C50:3 | 828.8 | C16:1, C16:1, C18:1 | Palmitoleic acid, palmitoleic acid, oleic acid | IR vs T2DM | − 0.78 | 1.37E–05 |
| IS vs T2DM | − 0.76 | 5.62E–05 | |||||
| TAG61 | C56:7 | 904.8 | C20:4, C18:1, C18:2 | Arachidonic acid, oleic acid, linoleic acid | IS vs T2DM | 0.81 | 0.0006 |
| IR vs T2DM | 0.74 | 0.001 | |||||
| TAG64 | C56:4 | 910.8 | C18:1, C18:2, C20:1 | Oleic acid, linoleic acid, gadoleic acid | IS and IR vs T2DM | 0.98 | 0.004 |
| TAG65 | C56:3 | 912.8 | C20:1, C18:1, C18:1 | Gadoleic acid, oleic acid, oleic acid | IR vs T2DM | 0.51 | 0.002 |
| IS vs T2DM | 0.54 | 0.002 | |||||
| TAG7 | C40:2 | 690.7 | C6:0, C16:0, C18:2 | Caproic acid, palmitic acid, linoleic acid | IS vs T2DM | − 1.07 | 1.26E–05 |
| IR vs T2DM | − 0.83 | 0.0002 | |||||
| TAG74 | C58:7 | 934.8 | C18:6, C24:0, C16:1 | C18:6, lignoceric acid, palmitoleic acid | IS and IR vs T2DM | 0.48 | 0.007 |
| TAG75 | C58:4 | 938.7 | C18:3, C24:0, C16:1 | Linolenic acid, lignoceric acid, palmitic acid | IR vs T2DM | 0.69 | 0.0005 |
| IS vs T2DM | 0.73 | 0.0005 | |||||
| TAG9 | C42:2 | 718.7 | C18:2, C12:0, C12:0 | Linoleic acid, lauric acid, lauric acid | IS vs T2DM | − 0.77 | 0.0008 |
| IR vs T2DM | − 0.71 | 0.001 |
MW molecular weight, fatty acid composition, fatty acid identity, fold change between specified groups are also indicated
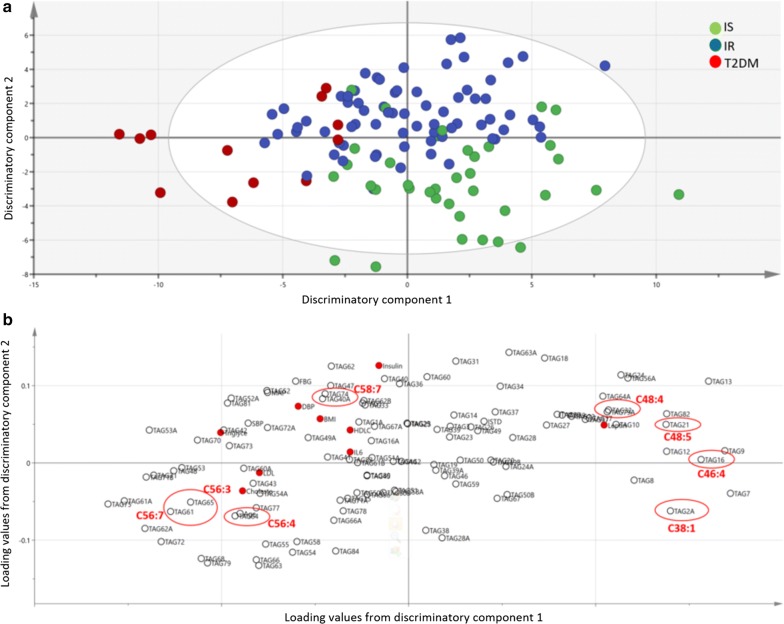
An orthogonal partial least square discriminate analysis (OPLS-DA) comparing subjects from IS, IR and T2DM revealed two significant class-discriminatory components (R2X = 0.18, R2Y = 1, R2Q2 = 0.27, CV-ANOVA p = 0.0001) (Fig. 1). The score plot in Fig. 1a indicates an x-axis differentiating the T2DM group from IS and IR; the latter two groups being rather separated along the y-axis. The corresponding loading score, shown in Fig. 1b, features similar TAG/group associations to those obtained with the linear model (shown in Table 3). Specifically, lower amounts of C38:1, C46:4, C48:5 and C48:4 as opposed to higher levels of C58:7, C56:4, C56:4 and C56:7 in the T2DM group (also circled in red, Fig. 1b).
Fig. 1.
OPLS-DA model comparing adipose tissue-derived TAGs from IS, IR and T2DM individuals. a A score plot showing the class-discriminatory component 1 (x-axis) versus class-discriminatory component 2 (y-axis). b The corresponding loading plot showing similar TAG/diabetes group associations to the linear model (circled in red)
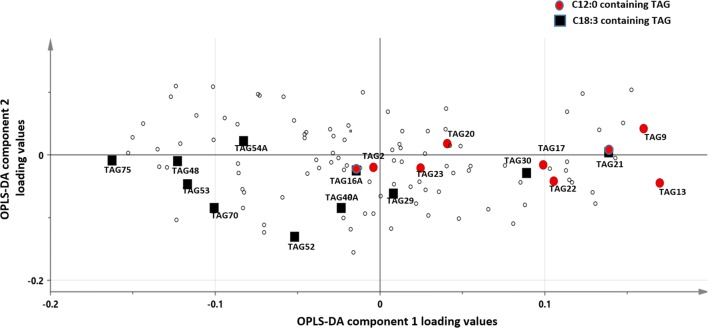
In order to study the possible enrichment/depletion of certain fatty acid constituents of TAGs in association with insulin sensitivity or diabetes, an enrichment analysis based on the Wilcoxon sum of the ranks test was conducted (refer to “Methods” section). The results of the analysis are presented in Table 4 and further illustrated on Fig. 2. Overall, C12:0 appears to be associated with TAGs least abundant in T2DM in both tissues whereas C18:3 is found in both depleted and enriched TAGs in T2DM (both sides of the x-axis in Fig. 2). This could be justified by a potentially induced flow of C18:3 in certain recipient TAGs at the expense of other TAGs with diabetes. Further supporting this are the observed negative correlations between depleted and enriched C18:3 carrying TAGs (Fig. 3). Interestingly, many of the C12:0 and C18:3 containing TAGs, including TAG21, TAG22, TAG75 and TAG9, were previously identified as significantly changing in level with diabetes by the linear model (Table 3).
Table 4.
TAG fatty acid association with tissue and diabetes/insulin sensitivity groups
| Compared groups | Fixed variable | Fatty acid | p value |
|---|---|---|---|
| IR × T2DM | SC | C12:0 | 0.045 |
| SC | C18:3 | 0.048 | |
| OM | C12:0 | 0.016 | |
| (Full model) SC+OM | C12:0 | 0.025 | |
| IS × T2DM | OM | C12:0 | 0.03 |
| OM | C18:3 | 0.048 | |
| SC × OM | (Full model) IS+IR+T2DM | C18.3 | 0.027 |
Analysis conducted using the Wilcoxon sum of the ranks test indicates fatty acids that were overrepresented amongst hit TAGs when comparing the groups specified in column 1. Comparing IS, IR and T2DM was done in individual tissues as well as when pooling data from the two tissues. Similarly, tissues were compared per group and when groups were combined (column 2). Only significant results are shown at a nominal p value of 0.05
Fig. 2.
An OPLS-DA loading plot showing the spread of C12:0 and C18:3 containing TAG along the x-axis found previously (Fig. 1a) to differentiate T2DM from IS+IR subjects. Unlike the C12:0 containing TAGs, the TAGs comprising C18:3 feature on both sides of the x-axis implying depletion of certain recipient TAGs (right side) as oppose to enrichment of others (left side) with diabetes
Fig. 3.
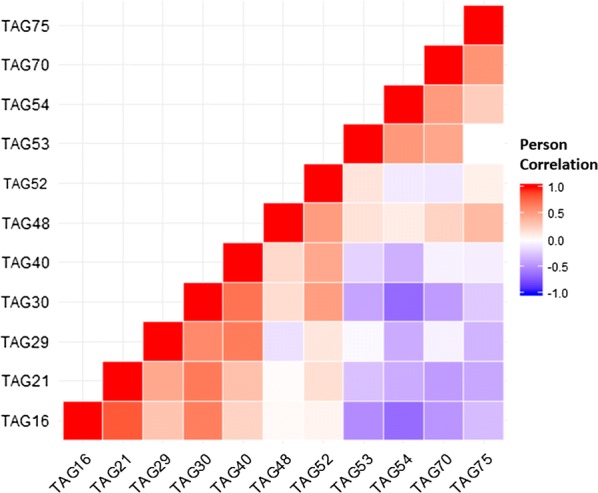
A triangular heatmap showing correlations between C18:3 carrying TAGs
Furthermore, C18:3 was also found to have a strong tissue signature featuring frequently amongst highly ranked TAGs from statistical analysis of TAG association with tissue type (data not shown). However, C18:3 does not feature amongst the TAGs found to significantly differ between tissues by the linear model (Table 2). This indicates that the collective tissue changes in C18:3 containing TAGs were rather subtle.
The Wilcoxon sum of the ranks analysis was also used to look for enrichment of fatty acid saturation level amongst the highly ranked significant TAGs from comparison of tissue/diabetes-insulin sensitivity groups but no significance was detected.
Correlation of TAG species with mediators of metabolic disease
A step-wise regression was performed to identify the best TAG predict of various traits including age, BMI, systemic TAG, total cholesterol, IL-6 and HOMA-IR, SC and OM adipogenic capacity reported previously [12, 15] and shown in Additional file 2: Figure S1. Table 5 lists TAG species identified with significant (p = 0.0001) association with various metabolic traits and shows their importance and fatty acid compositions.
Table 5.
List of TAGs associated with metabolic traits such as age, BMI, TC, TG, IL-6, HOMA-IR, SC and OM adipogenic capacity
| Metabolic trait | R2 | Importance | TAG | MW | Fatty acid composition | Fatty acids identities |
|---|---|---|---|---|---|---|
| Age | 0.4 | 0.12 | C56:1 | 916.8 | C20:0, C18:0, C18:1 | Arachidic acid, stearic acid, oleic acid |
| 0.12 | C54:8 | 874.8 | C18:3, C18:3, C18:2 | Linolenic acid, linolenic acid, linoleic acid | ||
| BMI | 0.5 | 0.12 | C57:1 | 930.8 | C17:0, C24:0, C16:1 | Heptadecanoic acid, lignoceric acid, palmitoleic acid |
| 0.1 | C48:1 | 804.8 | C18:0, C16:1, C14:0 | Stearic acid, palmitoleic acid, myristic acid | ||
| 0.09 | C54:5 | 880.8 | C18:1, C18:1, C18:3 | Oleic acid, oleic acid, linolenic acid | ||
| TAG | 0.5 | 0.12 | C52:1 | 860.8 | C16:0, C18:1, C18:0 | Palmitic acid, oleic acid, stearic acid |
| 0.11 | C54:1 | 888.8 | C18:0, C18:0, C18:1 | Stearic acid, stearic acid, oleic acid | ||
| TC | 0.4 | 0.35 | C40:2 | 690.7 | C6:0, C16:0, C18:2 | Caproic acid, palmitic acid, linoleic acid |
| IL-6 | 0.6 | 0.13 | C38:1 | 664.7 | C10:0, C12:0, C16:1 | Capric acid, lauric acid, palmitoleic acid |
| 0.1 | C42:1 | 720.7 | C16:0, C16:1, C10:0 | Palmitic acid, palmitoleic acid, capric acid | ||
| 0.07 | C56:1 | 916.8 | C18:0, C18:0, C20:1 | Stearic acid, stearic acid, gadoleic | ||
| HOMA-IR | 0.5 | 0.09 | C44:2 | 746.7 | C18:2, C14:0, C12:0 | Linoleic acid, myristic acid, lauric acid |
| 0.09 | C56:7 | 904.8 | C20:4, C18:1, C18:2 | Arachidonic acid, oleic acid, linoleic acid | ||
| SC adipogenic | 0.9 | 0.16 | C58:10 | 926.8 | C18:2, C18:2, C22:6 | Linoleic acid, linoleic acid, docosahexaenoic acid |
| 0.16 | C56:4 | 910.8 | C18:1, C18:2, C20:1 | Oleic acid, linoleic acid, gadoleic acid | ||
| 0.14 | C57:4 | 924.7 | C22:0, C19:4, C16:0 | Behenic acid, C19:4, palmitic acid | ||
| 0.09 | C40:1 | 692.7 | C18:1, C16:0, C6:0 | Oleic acid, palmitic acid, caproic acid | ||
| 0.08 | C60:1 | 970.8 | C24:0, C24:0, C18:1 | Lignoceric acid, lignocerric acid, oleic acid | ||
| 0.22 | C38:1 | 664.7 | C18:1, C16:0, C4:0 | Oleic acid, palmitic acid, butyric acid | ||
| OM adipogenic | 1 | 0.18 | C48:1 | 804.8 | C18:0, C16:1, C14:0 | Stearic acid, palmitoleic acid, myristic acid |
| 0.14 | C49:1 | 818.7 | C18:1, C17:0, C14:0 | Oleic acid, heptadecanoic acid, myristic acid | ||
| 0.11 | C56:1 | 916.8 | C18:0, C18:0, C20:1 | Stearic acid, stearic acid, gadoleic | ||
| 0.09 | C54:0 | 890.8 | C18:0, C18:0, C18:0 | Stearic acid, stearic acid, stearic acid, | ||
| 0.06 | C38:0 | 666.7 | C10:0, C14:0, C14:0 | Capric acid, myristic acid, myristic acid | ||
| 0.05 | C56:2 | 914.8 | C18:1, C18:1, C20:0 | Oleic acid, oleic acid, arachidic acid | ||
| 0.04 | C51:1 | 846.7 | C18:1, C15:0, C18:0 | Oleic acid, pentadecanoic acid, stearic acid |
A step-wise regression was performed to identify the best TAG predictors of various traits. A p value significance level of 0.001 was used
Discussion
TAGs constitute over 99% of lipid species in the adipose tissue of healthy individuals, with cholesterol and phospholipids making minor contributions [16]. TAGs are located within dynamic functional organelles known as lipid droplets that play important roles in intracellular vesicle trafficking, cell signaling and lipid homeostasis [17]. Although TAGs are not signaling molecules, fatty acids produced during their synthesis or breakdown were shown to interfere with the intracellular insulin signaling pathway and contribute to the development of insulin resistance [10]. Previous studies investigating TAG and fatty acid composition between subcutaneous and omental depots were published [6, 7]. However, this is the first study comparing TAGs and their fatty acid species in adipose tissues derived from IS, IR and T2DM obese individuals. Current technologies enable high-throughput profiling of the lipidome [18, 19]. In this study, LC/MS-based lipid profiling was performed to identify adipose signature of obesity-associated insulin sensitivity, insulin resistant and T2DM. The emerging data reveal differences in TAG species between SC and OM adipose tissues such as C38:1, C53:5, C51:3, C50:4, C59:1, C54:6 and C50:2 and among IS, IR and T2DM obese individuals including C46:4, C48:5, C48:4, C38:1, C50:3, C40:2, C56:3, C56:4, C56:7 and C58:7. The data also show differences in fatty acid compositions of TAGs associated with T2DM such as C12:0 and C18:3, suggesting a potential functional role of the identified species. Significant associations between the identified TAG species and traits of metabolic syndrome such as age, BMI, lipids (total cholesterol and circulating TAG), the inflammatory marker IL-6 and adipogenic capacity of preadipocytes derived from the same adipose tissues were identified. These associations could shed light on the molecular mechanisms contributing to the increased risk of metabolic disease.
Depot specific differences
Our data identified few TAGs that were differently expressed between SC and OM tissues. One TAG that was higher in SC compared to OM was C38:1, which contains C4:0 (butyric acid). The latter was shown before to inhibit lipolysis and increase insulin sensitivity in primary rat adipocytes [20], perhaps contributing to the greater association of insulin resistance with OM mass compared to SC mass [21]. A previous study in obese men has shown increased C50:0, C59:2, C58:2, C60:3, C64:4, C51:0 and C65:1 fatty acids in OM compared to SC adipose tissues [6]. Changes in lipid composition between the two depots were attributed to differences in adipocyte differentiation, metabolism of the lipid droplet, and extent of beta-oxidation [6]. Differences between the two studies may reflect ethnic and/or diet differences between our Asian and the other study’s Caucasian population. Variations in fatty acid composition between SC and OM fat depots confirm the specific metabolism of each depot, as selective lipolytic and lipogenic mechanisms may function in each tissue depot. Indeed, studies have shown that desaturase enzymes, regulating the number of saturated fatty acids, exhibit a depot-specific profile [22] in close association with insulin resistance [23].
IS, IR and T2DM specific differences
Systemic levels of fatty acid increase with obesity and T2DM, perhaps as a result of insulin resistance of adipose tissue and subsequent increased lipolysis; although in some obese individuals, fatty acid release from adipose tissues is reduced per kg fat in order to normalize plasma non-esterified fatty acid concentrations [24]. In our study, significant differences in levels of TAG composition were detected between IS, IR and T2DM. A number of candidates were either increased or decreased with risk of insulin resistance and T2DM, despite the predominant view of TAGs as an adverse risk factor for diabetes. Using the Wilcoxon sum of ranks statistics, fatty acids frequently occurring in highly-ranked TAGs along the list of TAGs ordered by p value from diabetes association analysis were revealed. Two fatty acids were identified: C12:0 and C18:3. The strength of this enrichment analysis approach is that, unlike the Fishers’ exact test, no arbitrary significance cut-off is applied on the list of TAGs. However, a possible weakness relates to the fact that since the TAGs are ordered by p value, no account is given to the direction of change and therefore one may not speak of depletion or increase in fatty acid TAG level but rather a dynamic in metabolic activity involving the fatty acid in association with the phenotype of interest. This was observed with C18:3, and a negative correlation was noted between C18:3 host TAGs found increased and others decreased with diabetes, effectively suggesting a metabolic link between the two sets of TAGs. Our findings confirm previous studies that showed significant correlations of specific fatty acids with insulin sensitivity. These include a cross-sectional analysis of adipose tissue biopsies from elderly obese men, which identified positive correlations between levels of C12:0, C18:2 and C18:3 and insulin sensitivity [25]. Our data also confirmed the association of C18:3 with metabolic status as shown previously in two groups of obese individuals who underwent weight loss surgery [26]. Furthermore, subjects in the most insulin-sensitive quintile showed a significantly higher percentage of circulating C18:2 (pre-cursor of C18:3) than the remaining subjects [27], further confirming our data. Functionally, previous work implicated C18:2 in the modulation of insulin signaling in rat skeletal muscle [28]. Therefore, our findings confirm previous results with regard to the association of C12:0 with insulin sensitivity [25], perhaps through triggering Glut4 translocation [27]. Our data also revealed reduction in C18:3 with T2DM incidents. This also confirms previous findings showing a negative correlation of C18:3 and its precursor with insulin resistance and positive association with insulin sensitivity [27].
Association of TAGs with mediators of metabolic syndrome
Further, our data highlight a panel of TAGs that were associated with mediators of metabolic disease in obese individuals. Increased age was associated with accumulation of C56:1 that is composed of saturated fatty acids C20:0 and C18:0 and mono-unsaturated C18:1, whereas age was negatively correlated with C54:8 that is composed of unsaturated fatty acids C18:2 and C18:3. Although participants had comparable BMI, the small increase in BMI was positively correlated with three unsaturated TAGs (C57:1, C48:1 and C54:5). Whereas circulating TAGs were associated with accumulation of C52:1 and C54:1 in the adipose tissue, total cholesterol was positively correlated with C40:2. The negative correlation between IL-6 and C38:1, C42:1 and C56:1 may suggest an anti-inflammatory effect of fatty acids that constitute these TAGs, in particular C10:0 that was shown previously to exert an anti-inflammatory properties [29]. HOMA-IR was also negatively correlated with C44:2 and C56:7, both containing C18:2 shown previously to negatively correlate with insulin resistance [25].
Association of TAGs with adipogenic capacity
Several TAGs were highly correlated with SC or OM adipogenic capacity. Previous studies have shown that the greater adipogenic capacity of SC and OM preadipocytes taken from IS obese individuals compared to IR and T2DM counterparts is partially mediated by lower IL-6 secretion and oxidative stress [12, 15, 30]. Secretion of interleukin IL-6 is significantly decreased after treatment with C18:2, C22:6 and C16:0 via inhibition of nuclear factor kappa B (NF-κB) and subsequent activation of the master regulator of adipogenesis, PPARγ [31]. Our data revealed positive correlations of C56:4 and C57:4, containing C18:2, C16:0, with SC adipogenic capacity. OM adipogenic capacity was associated with C49:1, C38:0 and C56:2, containing C16:0, C18:1 and C14:0. These fatty acids were shown previously to induce adipocyte differentiation in rodents [32–36] and potentially play a similar role in human preadipocytes.
Study limitations
One main limitation of this study is the relatively low number of participants, especially in the T2DM group. Additionally, the difference in gender distribution between IS and IR groups (predominantly females) and T2DM (predominantly males) group may have introduced bias in the study design that may have influenced the results. Despite these factors, clear TAG and fatty acid signatures were identified after correcting for potential confounders such as gender and BMI. Another limitation of the current work is its focus on association of TAGs with insulin resistance and risk of T2DM without an absolute quantitation of any specific analyte. Incorporating isotope-labeled standards would allow absolute quantitation and improve the precision of measurements. Finally, differences in TAG composition in adipose tissues among the studied groups may have been influenced by their diet. Indeed, previous studies have shown that the process of fatty acid and TAG deposition in rat adipose tissue depends on the composition of the diet [37]. Dietary linoleic acid content was shown to influence the distribution of TAG species in rat adipose tissue, particularly di- and trilinoleoyl containing TAG as a result of linoleic acid intake [38]. Other studies have shown that the composition of TAG in rat epididymal, subcutaneous and perirenal adipose tissues was broadly reflecting dietary oils such as isomeric octadecenoic acids from coriander oil and high oleic sunflower oil [39]. Taken all these limitations into account, confirmation in different populations is warranted to validate these findings.
Conclusion
In summary, our data supports the dynamic nature of adipose tissue and the complex interaction between adipose tissue physiology and its lipid composition. The TAGs and their fatty acid composition within human adipose tissues from obese subjects are markedly different, depending on the insulin sensitivity status of the donors. Our data suggest that adipose tissues from obese IR and T2DM individuals exhibit TAG-specific signatures that may contribute to their increased risk compared to their insulin-sensitive counterparts or could reflect different dietary consumption among the studied groups. Future experiments are warranted to investigate the functional relevance of these specific lipidomic profiles with reference to participants’ consumed diet.
Additional files
Additional file 1: Table S1. Comparison of participants’ characteristics by gender. Table S2. Differences between obese and morbidly obese subjects. Table S3. TAGs exhibiting BMI interaction.
Additional file 2: Figure S1. Adipogenic capacity of preadipocytes derived from subcutaneous (SC) and omental (OM) adipose tissues from insulin sensitive (IS), insulin resistant (IR) and type 2 diabetes mellitus (T2DM) patients. Representative images of SC and OM adipocytes form IS and IR individuals stained with DAPI in blue (nuclear staining) and lipidtox in green (lipid droplet staining) (A). A bar chart showing differences in the adipogenic capacity (percentage of differentiated adipocytes to total number of nuclei) in SC and OM preadipocytes derived from IS, IR and T2DM individuals (B). Significant differences in adipogenic capacity with disease progression were detected as reported previously [12, 15].
Authors’ contributions
HS carried out most of the data acquisition and analysis, helped with drafting the article and approved the final version. ID carried out the statistical analysis and helped with data interpretation. SB helped with data acquisition. ME and PA contributed to the study design and provided samples and information on participants. TMH helped with processing and interpretation of LC–MS–MS data. AD contributed to HS’s supervision and advised on experimental design. AL conceived the idea and contributed to experimental design and data interpretation. MAE was lead principle investigator, designed the experiments, supervised progress, analysed data and wrote the final version of the article. MAE is responsible for the integrity of the work as a whole. All authors read and approved the final manuscript.
Acknowledgements
We thank Qatar National Research Fund (QNRF) for funding this project (Grant no. NPRP6-235-1-048).
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
All participants were consented. Protocols were approved by Institutional Review Board of ADLQ (X2017000224).
Funding
This research was sponsored by QNRF, Grant no. NPRP6-235-1-048 (MAE, AL, ME).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- DAPI
4′,6-diamidino-2-phenylindole
- BMI
body mass index
- DBP
diastolic blood pressure
- DAGs
diacylglycerols
- ECN
effective carbon number
- FPG
fasting blood glucose
- HDL
high density lipoprotein
- HOMA-IR
homeostatic model assessment
- IS
insulin sensitive
- IR
insulin resistant
- IL-6
interleukin 6
- LDL
low density lipoprotein
- MAP
mean arterial blood pressure
- NARP
non-aqueous reverse phase UHPLC separation
- NF-κB
nuclear factor kappa B
- OM
omental
- OPLS-DA
orthogonal partial least square discriminate analysis
- PCA
principle component analysis
- RT
retention time
- SBP
systolic blood pressure
- SVF
stromal vascular fraction
- SC
subcutaneous
- TAGs
triacylglycerols
- T2DM
type 2 diabetes mellitus
- UHPLC
ultra-high performance liquid chromatography
Appendix
See Table 6.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12967-018-1548-x) contains supplementary material, which is available to authorized users.
Contributor Information
Haya Al-Sulaiti, Email: halsulaiti@adlqatar.qa.
Ilhame Diboun, Email: idibou01@mail.bbk.ac.uk.
Sameem Banu, Email: sbanu@adlqatar.qa.
Mohamed Al-Emadi, Email: dremadim@yahoo.com.
Parvaneh Amani, Email: parvaneh@alemadihospital.com.qa.
Thomas M. Harvey, Email: tmichael@adlqatar.qa
Alex S. Dömling, Email: a.s.s.domling@rug.nl
Aishah Latiff, Email: alatiff@adlqatar.qa.
Mohamed A. Elrayess, Email: melrayess@adlqatar.qa
References
- 1.Guilherme A, Virbasius JV, Puri V, Czech MP. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat Rev Mol Cell Biol. 2008;9:367–377. doi: 10.1038/nrm2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badoud F, Perreault M, Zulyniak MA, Mutch DM. Molecular insights into the role of white adipose tissue in metabolically unhealthy normal weight and metabolically healthy obese individuals. FASEB J. 2015;29:748–758. doi: 10.1096/fj.14-263913. [DOI] [PubMed] [Google Scholar]
- 3.Hocking S, Samocha-Bonet D, Milner KL, Greenfield JR, Chisholm DJ. Adiposity and insulin resistance in humans: the role of the different tissue and cellular lipid depots. Endocr Rev. 2013;34:463–500. doi: 10.1210/er.2012-1041. [DOI] [PubMed] [Google Scholar]
- 4.Snel M, Jonker JT, Schoones J, Lamb H, de Roos A, Pijl H, Smit JW, Meinders AE, Jazet IM. Ectopic fat and insulin resistance: pathophysiology and effect of diet and lifestyle interventions. Int J Endocrinol. 2012;2012:983814. doi: 10.1155/2012/983814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts LD, Koulman A, Griffin JL. Towards metabolic biomarkers of insulin resistance and type 2 diabetes: progress from the metabolome. Lancet Diabetes Endocrinol. 2014;2:65–75. doi: 10.1016/S2213-8587(13)70143-8. [DOI] [PubMed] [Google Scholar]
- 6.Jove M, Moreno-Navarrete JM, Pamplona R, Ricart W, Portero-Otin M, Fernandez-Real JM. Human omental and subcutaneous adipose tissue exhibit specific lipidomic signatures. FASEB J. 2014;28:1071–1081. doi: 10.1096/fj.13-234419. [DOI] [PubMed] [Google Scholar]
- 7.Hodson L, Skeaff CM, Fielding BA. Fatty acid composition of adipose tissue and blood in humans and its use as a biomarker of dietary intake. Prog Lipid Res. 2008;47:348–380. doi: 10.1016/j.plipres.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Zhang C, Klett EL, Coleman RA. Lipid signals and insulin resistance. Clin Lipidol. 2013;8:659–667. doi: 10.2217/clp.13.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiao P, Ma J, Feng B, Zhang H, Diehl JA, Chin YE, Yan W, Xu H. FFA-induced adipocyte inflammation and insulin resistance: involvement of ER stress and IKKbeta pathways. Obesity (Silver Spring) 2011;19:483–491. doi: 10.1038/oby.2010.200. [DOI] [PubMed] [Google Scholar]
- 10.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summers SA. Ceramides in insulin resistance and lipotoxicity. Prog Lipid Res. 2006;45:42–72. doi: 10.1016/j.plipres.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 12.Almuraikhy S, Kafienah W, Bashah M, Diboun I, Jaganjac M, Al-Khelaifi F, Abdesselem H, Mazloum NA, Alsayrafi M, Mohamed-Ali V, Elrayess MA. Interleukin-6 induces impairment in human subcutaneous adipogenesis in obesity-associated insulin resistance. Diabetologia. 2016;59:2406–2416. doi: 10.1007/s00125-016-4031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy SM, Brewer HB, Jr, Cleeman JI, Smith SC, Jr, Lenfant C, National Heart, Lung, and Blood Institute/American Heart Association Definition of metabolic syndrome: report of the National Heart, Lung, and Blood Institute/American Heart Association conference on scientific issues related to definition. Arterioscler Thromb Vasc Biol. 2004;24:e13–e18. doi: 10.1161/01.ATV.0000111245.75752.C6. [DOI] [PubMed] [Google Scholar]
- 14.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 15.Jaganjac M, Almuraikhy S, Al-Khelaifi F, Al-Jaber M, Bashah M, Mazloum NA, Zarkovic K, Zarkovic N, Waeg G, Kafienah W, Elrayess MA. Combined metformin and insulin treatment reverses metabolically impaired omental adipogenesis and accumulation of 4-hydroxynonenal in obese diabetic patients. Redox Biol. 2017;12:483–490. doi: 10.1016/j.redox.2017.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wahle KW, McIntosh G, Duncan WR, James WP. Concentrations of linoleic acid in adipose tissue differ with age in women but not men. Eur J Clin Nutr. 1991;45:195–202. [PubMed] [Google Scholar]
- 17.Arrese EL, Saudale FZ, Soulages JL. Lipid droplets as signaling platforms linking metabolic and cellular functions. Lipid Insights. 2014;7:7–16. doi: 10.4137/LPI.S11128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 19.German JB, Gillies LA, Smilowitz JT, Zivkovic AM, Watkins SM. Lipidomics and lipid profiling in metabolomics. Curr Opin Lipidol. 2007;18:66–71. doi: 10.1097/MOL.0b013e328012d911. [DOI] [PubMed] [Google Scholar]
- 20.Heimann E, Nyman M, Degerman E. Propionic acid and butyric acid inhibit lipolysis and de novo lipogenesis and increase insulin-stimulated glucose uptake in primary rat adipocytes. Adipocyte. 2015;4:81–88. doi: 10.4161/21623945.2014.960694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bjorndal B, Burri L, Staalesen V, Skorve J, Berge RK. Different adipose depots: their role in the development of metabolic syndrome and mitochondrial response to hypolipidemic agents. J Obes. 2011;2011:490650. doi: 10.1155/2011/490650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petrus P, Edholm D, Rosqvist F, Dahlman I, Sundbom M, Arner P, Ryden M, Riserus U. Depot-specific differences in fatty acid composition and distinct associations with lipogenic gene expression in abdominal adipose tissue of obese women. Int J Obes (Lond) 2017;41:1295–1298. doi: 10.1038/ijo.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Serrano S, Moreno-Santos I, Garrido-Sanchez L, Gutierrez-Repiso C, Garcia-Almeida JM, Garcia-Arnes J, Rivas-Marin J, Gallego-Perales JL, Garcia-Escobar E, Rojo-Martinez G, et al. Stearoyl-CoA desaturase-1 is associated with insulin resistance in morbidly obese subjects. Mol Med. 2011;17:273–280. doi: 10.2119/molmed.2010.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karpe F, Dickmann JR, Frayn KN. Fatty acids, obesity, and insulin resistance: time for a reevaluation. Diabetes. 2011;60:2441–2449. doi: 10.2337/db11-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iggman D, Arnlov J, Vessby B, Cederholm T, Sjogren P, Riserus U. Adipose tissue fatty acids and insulin sensitivity in elderly men. Diabetologia. 2010;53:850–857. doi: 10.1007/s00125-010-1669-0. [DOI] [PubMed] [Google Scholar]
- 26.Ni Y, Zhao L, Yu H, Ma X, Bao Y, Rajani C, Loo LW, Shvetsov YB, Yu H, Chen T, et al. Circulating unsaturated fatty acids delineate the metabolic status of obese individuals. EBioMedicine. 2015;2:1513–1522. doi: 10.1016/j.ebiom.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernandez-Real JM, Broch M, Vendrell J, Ricart W. Insulin resistance, inflammation, and serum fatty acid composition. Diabetes Care. 2003;26:1362–1368. doi: 10.2337/diacare.26.5.1362. [DOI] [PubMed] [Google Scholar]
- 28.Yu C, Chen Y, Cline GW, Zhang D, Zong H, Wang Y, Bergeron R, Kim JK, Cushman SW, Cooney GJ, et al. Mechanism by which fatty acids inhibit insulin activation of insulin receptor substrate-1 (IRS-1)-associated phosphatidylinositol 3-kinase activity in muscle. J Biol Chem. 2002;277:50230–50236. doi: 10.1074/jbc.M200958200. [DOI] [PubMed] [Google Scholar]
- 29.Huang WC, Tsai TH, Chuang LT, Li YY, Zouboulis CC, Tsai PJ. Anti-bacterial and anti-inflammatory properties of capric acid against Propionibacterium acnes: a comparative study with lauric acid. J Dermatol Sci. 2014;73:232–240. doi: 10.1016/j.jdermsci.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 30.Elrayess MA, Almuraikhy S, Kafienah W, Al-Menhali A, Al-Khelaifi F, Bashah M, Zarkovic K, Zarkovic N, Waeg G, Alsayrafi M, Jaganjac M. 4-Hydroxynonenal causes impairment of human subcutaneous adipogenesis and induction of adipocyte insulin resistance. Free Radic Biol Med. 2017;104:129–137. doi: 10.1016/j.freeradbiomed.2017.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Zhao G, Etherton TD, Martin KR, Vanden Heuvel JP, Gillies PJ, West SG, Kris-Etherton PM. Anti-inflammatory effects of polyunsaturated fatty acids in THP-1 cells. Biochem Biophys Res Commun. 2005;336:909–917. doi: 10.1016/j.bbrc.2005.08.204. [DOI] [PubMed] [Google Scholar]
- 32.Wolins NE, Quaynor BK, Skinner JR, Schoenfish MJ, Tzekov A, Bickel PE. S3-12, Adipophilin, and TIP47 package lipid in adipocytes. J Biol Chem. 2005;280:19146–19155. doi: 10.1074/jbc.M500978200. [DOI] [PubMed] [Google Scholar]
- 33.Davies JD, Carpenter KL, Challis IR, Figg NL, McNair R, Proudfoot D, Weissberg PL, Shanahan CM. Adipocytic differentiation and liver × receptor pathways regulate the accumulation of triacylglycerols in human vascular smooth muscle cells. J Biol Chem. 2005;280:3911–3919. doi: 10.1074/jbc.M410075200. [DOI] [PubMed] [Google Scholar]
- 34.McNeel RL, Mersmann HJ. Effects of isomers of conjugated linoleic acid on porcine adipocyte growth and differentiation. J Nutr Biochem. 2003;14:266–274. doi: 10.1016/S0955-2863(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 35.Ding S, Mersmann HJ. Fatty acids modulate porcine adipocyte differentiation and transcripts for transcription factors and adipocyte-characteristic proteins*. J Nutr Biochem. 2001;12:101–108. doi: 10.1016/S0955-2863(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 36.Amri EZ, Ailhaud G, Grimaldi PA. Fatty acids as signal transducing molecules: involvement in the differentiation of preadipose to adipose cells. J Lipid Res. 1994;35:930–937. [PubMed] [Google Scholar]
- 37.Perona JS, Portillo MP, Teresa Macarulla M, Tueros AI, Ruiz-Gutierrez V. Influence of different dietary fats on triacylglycerol deposition in rat adipose tissue. Br J Nutr. 2000;84:765–774. [PubMed] [Google Scholar]
- 38.Huang YS, Lin X, Smith RS, Redden PR, Jenkins DK, Horrobin DF. Effect of dietary linoleic acid content on the distribution of triacylglycerol molecular species in rat adipose tissue. Lipids. 1992;27:711–715. doi: 10.1007/BF02536030. [DOI] [PubMed] [Google Scholar]
- 39.Weber N, Schonwiese S, Klein E, Mukherjee KD. Adipose tissue triacylglycerols of rats are modulated differently by dietary isomeric octadecenoic acids from coriander oil and high oleic sunflower oil. J Nutr. 1999;129:2206–2211. doi: 10.1093/jn/129.12.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Comparison of participants’ characteristics by gender. Table S2. Differences between obese and morbidly obese subjects. Table S3. TAGs exhibiting BMI interaction.
Additional file 2: Figure S1. Adipogenic capacity of preadipocytes derived from subcutaneous (SC) and omental (OM) adipose tissues from insulin sensitive (IS), insulin resistant (IR) and type 2 diabetes mellitus (T2DM) patients. Representative images of SC and OM adipocytes form IS and IR individuals stained with DAPI in blue (nuclear staining) and lipidtox in green (lipid droplet staining) (A). A bar chart showing differences in the adipogenic capacity (percentage of differentiated adipocytes to total number of nuclei) in SC and OM preadipocytes derived from IS, IR and T2DM individuals (B). Significant differences in adipogenic capacity with disease progression were detected as reported previously [12, 15].
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.