Abstract
Soluble urokinase plasminogen activator receptor (suPAR) independently predicts chronic kidney disease (CKD) incidence and progression. Apolipoprotein L1 (APOL1) gene variants G1 and G2, but not the reference allele (G0), are associated with an increased risk of CKD in individuals of recent African ancestry. Here we show in two large, unrelated cohorts that decline in kidney function associated with APOL1 risk variants was dependent on plasma suPAR levels: APOL1-related risk was attenuated in patients with lower suPAR, and strengthened in those with higher suPAR levels. Mechanistically, surface plasmon resonance studies identified high-affinity interactions between suPAR, APOL1 and αvβ3 integrin, whereby APOL1 protein variants G1 and G2 exhibited higher affinity for suPAR-activated avb3 integrin than APOL1 G0. APOL1 G1 or G2 augments αvβ3 integrin activation and causes proteinuria in mice in a suPAR-dependent manner. The synergy of circulating factor suPAR and APOL1 G1 or G2 on αvβ3 integrin activation is a mechanism for CKD.
In individuals of recent African ancestry, variants in APOL1 have been associated with certain forms of CKD. Carriers of two risk alleles (either two of G1 or G2; G1/G1 or G2/G2, or one each of G1 and G2; G1/G2) are at heightened risk for focal segmental glomerulosclerosis (FSGS), HIV-associated nephropathy and hypertension-attributed kidney disease1–5. The molecular mechanisms underlying the association between APOL1 risk variants and CKD remain unclear. Inflammation is thought to be a major contributor to APOL1-related CKD, because APOL1 expression in podocytes is induced by components of innate immune pathways, such as those of interferons and Toll-like receptors6. Moreover, the APOL1 high-risk genotype is strongly associated with nephropathy in people with HIV, a state of immune dysregulation7,8. However, a puzzling observation is that not all individuals carrying the high-risk APOL1 genotype develop renal disease, and equally, not all transgenic animal models, including models of mice9 and zebrafish10, expressing high-risk APOL1 variants display the expected kidney phenotypes. Taken together, these findings suggest the possibility of a ‘second hit’ in the pathogenesis of APOL1-associated nephropathy10.
SuPAR is a member of the Ly-6/neurotoxin family of signaling proteins and a marker of immune activation that associates with states of systemic inflammation, such as HIV11–13. SuPAR has recently been shown to be strongly predictive of incident CKD14, and it is thought to be involved in the pathogenesis of kidney disease, specifically FSGS which is characterized by progressive scarring of the kidney that often leads to renal failure, through the activation of the αvβ3-integrin signaling pathway on podocytes15–17. Given the strong association of both suPAR and APOL1 genetic risk variants with inflammation and CKD, we sought to determine (i) whether the relationship in African Americans between the APOL1 genotype and decline in kidney function is influenced by suPAR levels, and (ii) to identify potential molecular mechanisms that would underlie the interaction between suPAR and APOL1 in the context of CKD.
RESULTS
Clinical characteristics, suPAR levels and APOL1 risk variants
We characterized APOL1 genetic variants and plasma suPAR levels in two separate cohorts of African American individuals: the Emory Cardiovascular Biobank (EmCAB) and the African American Study of Kidney Disease and Hypertension (AASK). The EmCAB cohort consisted of 487 patients with a mean age of 58 ± 12 years, of which 50% were male, 49% had diabetes and 27% had an estimated glomerular filtration rate (eGFR) of <60 ml/min/1.73 m2. A total of 73 (15%) cohort members had a high-risk APOL1 genotype (renal risk alleles on both parental chromosomes), whereas 220 (45%) and 194 (40%) had 1 and 0 copies, respectively, of a high-risk haplotype. Individuals in the APOL1 high-risk-genotype group had lower eGFRs and higher suPAR levels at baseline (Table 1). However, when adjusted for suPAR levels, APOL1 genotype was not independently associated with baseline kidney function (Supplementary Table 1). Plasma suPAR levels were approximately 20% higher in the APOL1 high-risk group than in the low-risk group (P = 0.001). In a multivariable analysis adjusted for age, gender, body-mass index, smoking history, hypertension, diabetes mellitus and baseline eGFR, those with APOL1 high-risk status remained independently associated with higher suPAR levels (Supplementary Table 1).
Table 1.
Demographics and clinical characteristics stratified by cohort and number of APOL1 risk variants
| EmCAB (n = 487) | AASK (n = 607) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Number of APOL1 risk variants | Number of APOL1 risk variants | |||||
|
|
|
|||||
| Variables | 0/1 (n = 414) | 2 (n = 73) | P valuea | 0/1 (n = 464) | 2 (n = 143) | P valueb |
| Age, years (s.d.) | 59 (12) | 56 (11) | 0.05 | 55 (10) | 52 (12) | 0.02 |
| Male, n (% of column) | 205 (50%) | 37 (51%) | 0.85 | 279 (60%) | 83 (58%) | 0.20 |
| Body-mass index, kg/m2 (s.d.) | 30 (8) | 32 (6) | 0.80 | 31 (7) | 32 (7) | 0.15 |
| History of smoking, n (%) | 251 (61%) | 44 (60%) | 0.95 | 273 (59%) | 77 (54%) | 0.29 |
| Hypertension, n (%) | 355 (90%) | 65 (94%) | 0.24 | 464 (100%) | 143 (100%) | – |
| Diabetes mellitus, n (%) | 186 (47%) | 39 (58%) | 0.10 | 0 (0%) | 0 (0%) | – |
| Obstructive coronary artery disease, n (%) | 276 (74%) | 55 (81%) | 0.20 | 242 (52%) | 60 (42%) | 0.03 |
| eGFR, ml/min/1.73 m2 (s.d.) | 76 (30) | 66 (33) | 0.01 | 50 (14) | 44 (13) | <0.001 |
| eGFR, <60 ml/min/1.73 m2, n (%) | 66 (25%) | 17 (33%) | 0.28 | 344 (74%) | 128 (90%) | <0.001 |
| SuPAR, pg/ml (25th, 75th percentiles) | 2,823 (2,127, 3,871) | 3423 (2,577, 5,055) | 0.001 | 4,338 (3,306, 5,690) | 4700 (3,685, 5,872) | 0.07 |
Values are mean (s.d.), n (%) or median (25th and 75th percentiles), as noted. eGFR, estimated glomerular filtration rate. Coronary artery disease is defined as the presence of a ≥50% obstructive lesion in any of the major vessels on coronary angiogram in EmCAB. P value represents statistical significance of a χ-squared test for categorical variables and one-way ANOVA for continuous variables comparing the groups with different APOL1 alleles.
Superscript a (a) indicates Emory Cardiovascular Biobank cohort (EmCAB);
superscript b (b) indicates African American Study of Kidney Disease and Hypertension cohort (AASK).
The AASK cohort consisted of 607 people with a mean age of 54 ± 11 years; 60% were male, and the mean baseline eGFR was 48 ml/min/1.73 m2. Individuals with two risk alleles of APOL1 genotypes (n = 143, 23.6%) were younger, had a lower prevalence of heart disease and a lower eGFR at baseline (Table 1). The difference in plasma suPAR levels between patients with one or zero and those with two risk alleles of APOL1 (4,700 pg/ml and 4,338 pg/ml, respectively) was not statistically significant (P = 0.07). There was no difference between suPAR levels among the 199 and 265 participants with no or one risk allele(s) of APOL1, respectively (4,354 pg/ml and 4,322 pg/ml; P = 0.78).
The decline in kidney function attributed to APOL1 risk variants is dependent on suPAR
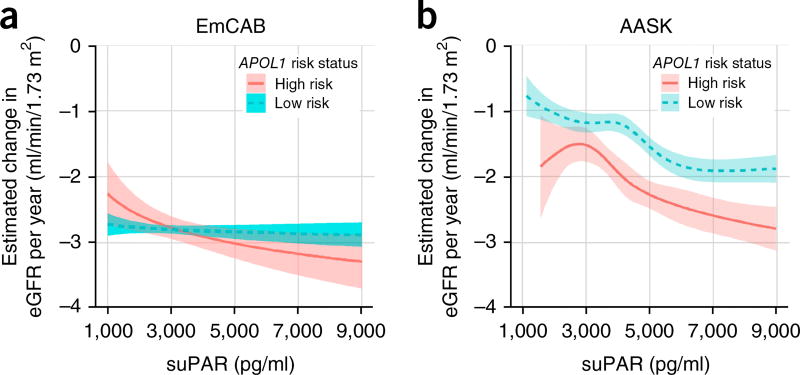
We next examined the association between suPAR, APOL1 risk variants and eGFR decline in both cohorts. In the EmCAB cohort, the mean decline in eGFR per year was −4.00 (95% CI 3.3, 4.6) and −3.4 (95% CI 2.9, 3.8) ml/min/1.73 m2, respectively, in the APOL1 high-risk group and low-risk group. In an analysis adjusted for age, gender, body-mass index, smoking history, hypertension and diabetes, the APOL1 risk genotype was associated with faster eGFR decline in the presence of suPAR >3,000 pg/ml, but not when levels were <3,000 pg/ml (P for interaction = 0.03) (Table 2). In the AASK cohort, the mean eGFR decline per year was −2.2 ml/min/1.73 m2 per year for the APOL1 two-risk-allele group, as compared to −1.5 ml/min/1.73 m2 per year for those with one or zero risk alleles. In our adjusted analysis, the APOL1 high-risk genotype was associated with faster eGFR decline in the presence of suPAR levels >3,000 pg/ml but not <3,000 pg/ml (P for interaction = 0.006), similar to the EmCAB cohort. In AASK, continuous analysis suggested that the threshold for decline in kidney function associated with suPAR was lower among people with two risk alleles of APOL1 (Fig. 1).
Table 2.
APOL1 risk variants, suPAR levels and decline in kidney function
| eGFR | ||||
|---|---|---|---|---|
|
|
||||
| EmCAB (n = 487) | AASK (n = 607) | |||
|
|
|
|||
| Variables | β, P value | 95% CI | β, P value | 95% CI |
| Age, per year | −0.35, <0.001 | −0.52, −0.19 | 0.13, 0.04 | 0.01, 0.25 |
| Female gender | −5.72, <0.001 | −9.57, −1.86 | −1.66, 0.19 | −4.18, 0.85 |
| Body-mass index, per 1 kg/m2 | 0.15, 0.25 | −0.11, 0.42 | 0.17, 0.08 | −0.02, 0.37 |
| Smoking history | 3.45, 0.08 | −0.41, 7.31 | 1.09, 0.41 | −1.48, 3.65 |
| Hypertension | −7.75, 0.01 | −13.83, −1.66 | −0.77, 0.54 | −3.22, 1.67 |
| Diabetes mellitus | −8.57, <0.001 | −12.47, −4.66 | − | − |
| SuPAR > 3,000 pg/ml | −20.83, <0.001 | −24.99, −16.68 | −10.95, <0.001 | −14.69, −7.21 |
| APOL1 high risk | −4.80, 0.24 | −12.83, 3.23 | −3.48, 0.45 | −12.54, 5.59 |
| SuPAR > 3,000 pg/ml*APOL1 high risk | −3.12, 0.57 | −13.90, 7.65 | −1.00, 0.84 | −10.56, 8.56 |
| Follow-up time, per year | −2.30, 0.004 | −3.85, −0.75 | −1.08, <0.001 | −1.25, −0.92 |
| Age, *follow-up time | 0.03, 0.17 | −0.01, 0.06 | 0.01, 0.01 | 0.00, 0.01 |
| Female gender, *follow-up time | 0.05, 0.91 | −0.82, 0.91 | −0.09, 0.10 | −0.19, 0.02 |
| Body-mass index, *follow-up time | 0.03, 0.33 | −0.03, 0.09 | −0.03, <0.001 | −0.04, −0.02 |
| Smoking history, *follow-up time | −0.18, 0.67 | −1.00, 0.64 | 0.01, 0.89 | −0.10, 0.11 |
| Hypertension, *follow-up time | −0.08, 0.92 | −1.60, 1.43 | −0.13, 0.01 | −0.23, −0.03 |
| Diabetes mellitus, *follow-up time | −1.75, <0.001 | −2.57, −0.94 | − | − |
| SuPAR >3,000 pg/mL, *follow-up time | 0.61, 0.19 | −0.29, 1.52 | −0.25, <0.001 | −0.39, −0.11 |
| APOL1 high-risk variant, *follow-up time | 0.26, 0.76 | −1.42, 1.93 | −0.18, 0.36 | −0.56, 0.20 |
| SuPAR > 3,000 pg/ml, *APOL1 high risk, *follow-up time | −2.52, 0.03 | −4.79, −0.25 | −0.57, 0.01 | −0.97, −0.16 |
The model includes all listed covariates, including the interaction terms. Linear mixed-effects model was used with both data sets to account for within-subject correlations among repeated measures of eGFR over time (random subject-specific intercept and random time effect). Age and BMI were centered at the study-specific mean, respectively. Diabetes status was not included in the AASK model, given that no cohort members had diabetes. Hypertension in the AASK cohort refers to blood pressure randomization group. The relevant interaction term, including suPAR, APOL1 and follow-up time, is highlighted in bold. P values less than 0.05 were considered to be statistically significant. Linear mixed-effects model was used.
Asterisks denote an interaction term.
Figure 1.
Synergy of an APOL1 high-risk genotype and suPAR levels determines yearly loss of kidney function. (a,b) Estimated yearly change in estimated glomerular filtration rate (eGFR) according to APOL1 status (green, low risk; red, high risk) and continuous levels of suPAR at cohort enrollment for the Emory Cardiovascular Biobank (EmCAB; n = 487) (a) and the African American Study of Kidney Disease and Hypertension cohort (AASK; n = 607) (b). Yearly change in eGFR was estimated from linear mixed-effects models of eGFR adjusted for age, sex, body-mass index, hypertension, diabetes mellitus (in EmCAB), smoking status, randomization group (in AASK) and suPAR level in both baseline levels and slope over time. In EmCAB, suPAR was log-transformed. In AASK, suPAR was log-transformed and modeled using restricted cubic splines to allow for deviations from log linearity.
In summary, suPAR levels were a substantial modifier of the association between APOL1 genotype and subsequent eGFR decline. Higher baseline suPAR levels were associated with a significantly greater yearly decline in kidney function in patients with two APOL1 risk alleles than those in patients with one or zero alleles (Fig. 1), which suggests that suPAR-related decline might be dependent on the presence of the APOL1 risk allele.
APOL1 proteins bind suPAR and active αvβ3 integrin with high affinity
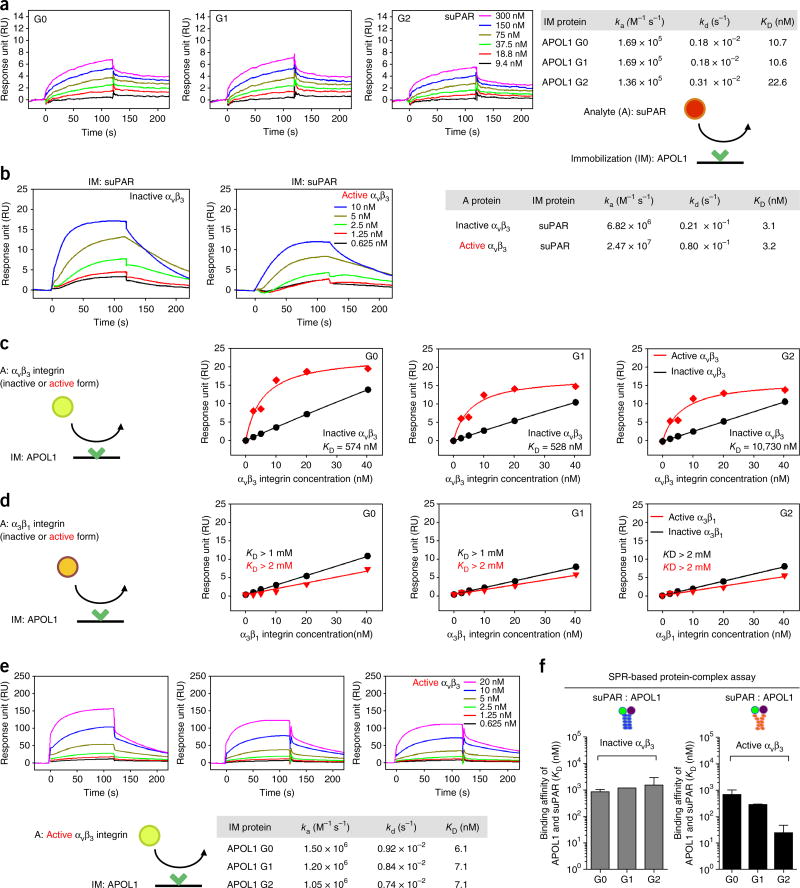
Given the strong association noted in African American individuals between suPAR levels, APOL1 risk alleles and decline in kidney function, as well as the previous evidence for a role of suPAR in kidney disease through the activation of αvβ3 integrin12,13, we next explored the possibility of a protein–protein interaction between suPAR, APOL1 and αvβ3 integrin. To this end, we performed a series of surface plasmon resonance (SPR) assays using recombinant APOL1 proteins (Supplementary Fig. 1). Given that circulating APOL1 is a component of a lipid-rich trypanolytic complex that has an electrophoretic mobility of high-density lipoprotein (HDL) particles18, we assessed the functionality of recombinant APOL1 proteins by measuring the binding affinity of APOL1 to HDL. All three variant APOL1 proteins (G0, G1 and G2) exhibited similarly high affinities for HDL (KD, ~150 nM; Supplementary Fig. 2). Next, we investigated whether APOL1 can directly bind suPAR. Of note, all three variant APOL1 proteins exhibited very high affinities for suPAR (KD, ~10–20 nM; Fig. 2a). These data also suggest that suPAR–APOL1 interactions are likely not mediated by the C-terminal domain of APOL1, given that amino acids that are altered in both APOL1 risk variants are situated in the C-terminal domain.
Figure 2.
APOL1 forms high-affinity interactions with suPAR and αvβ3 integrin. All sensorgrams were generated using SPR assays. Rate constants (ka and kd) were determined by kinetic fitting of the sensorgrams using a one-to-one binding equation, and equilibrium dissociation constant (KD) values were determined by calculating kd/ka. (a) Sensorgrams of suPAR (0–300 nM) binding to immobilized APOL1 protein variants (G0, G1 and G2), and calculated KD values. (b) Sensorgrams of αvβ3 integrin (0–10 nM) binding to immobilized suPAR in the absence (inactive form) and presence (active form) of Mn2+ (0.5 mM), and calculated KD values. (c,d) Steady-state affinity fitting curves of αvβ3 (c) or α3β1 (d) integrin (0–40 nM) binding to immobilized APOL1 (G0, G1 and G2) in the absence (inactive) and presence (active) of Mn2+ (0.5 mM). (e) Sensorgrams of αvβ3 integrin (0–20 nM) binding to immobilized APOL1 (G0, G1 and G2) in the presence of Mn2+ (0.5 mM), and calculated KD values. (f) Bar graphs showing the determined KD values for suPAR in the presence of αvβ3 (5 nM), increasing concentrations of suPAR (0–300 nM) and immobilized APOL1 proteins (G0, G1 and G2) in the absence (inactive) or presence (active) of Mn2+ (0.5 mM). The average KD values were determined from two independent experiments, and error bars for KD measurements indicate s.e.m. (n = 2) (a–f). A, analyte; IM, immobilization.
We next tested whether the variant APOL1 proteins can also bind αvβ3 and/or α3β1, two major integrins in podocytes. Consistently with a previous report17, suPAR exhibited a high affinity for αvβ3 integrin (KD = ~3 nM; Fig. 2b). The affinity of suPAR for αvβ3 was not considerably altered in the presence of Mn2+, a potent, nonspecific activator of integrins19 (Fig. 2b; in the presence of Mn2+, integrin is defined to be in the active form). In contrast to suPAR, all three variant APOL1 proteins exhibited moderate to low affinities for inactive αvβ3 integrin (KD = 574 nM (G0); KD = 528 nM (G1); KD = 10,730 nM (G2); Fig. 2c), and very low affinity for α3β1 integrin (KD >1 mM; Fig. 2d). Unexpectedly, the affinity of all three variant APOL1 proteins for αvβ3 but not α3β1 integrin was at least 100-fold higher in the presence of Mn2+ (KD = ~7 nM; Fig. 2c,e). The effect was most pronounced for the G2 APOL1 variant, which bound the active form of αvβ3 integrin with a more than 1,000-fold greater affinity than that of the nonactive form. Taken together, these data suggest that APOL1 might exert a synergistic interaction with suPAR in potentiating the activation of αvβ3 integrin. To examine the formation of such a potential triple complex, we performed a SPR assay in which the concentration of suPAR was increased in the presence of constant concentrations of αvβ3 and APOL1. Although all three variant APOL1 proteins exhibited similarly low affinities for suPAR in the presence of αvβ3 (Fig. 2f and Supplementary Fig. 2b), the APOL1 risk variants G1 and G2 exhibited a dramatically higher affinity (KD = 277 nM, KD = 24 nM, respectively) for suPAR in the presence of αvβ3 with Mn2+, comparing with APOL1 G0 (KD = 665 nM) (Fig. 2f and Supplementary Fig. 2b). We detected increased binding affinities for both the APOL1 G1 and G2 risk variants, and, to a greater extent, for the APOL1 G2 risk-variant protein (Supplementary Fig. 2b). Together, these data using SPR identify high-affinity interactions between recombinant APOL1 and suPAR, and a synergistic effect between APOL1 and suPAR in the activation of αvβ3 integrin, as well as a difference between the nonrisk and risk-variant APOL1 proteins in such activation.
SuPAR and APOL1 risk variants synergize to activate αvβ3 integrin on podocytes
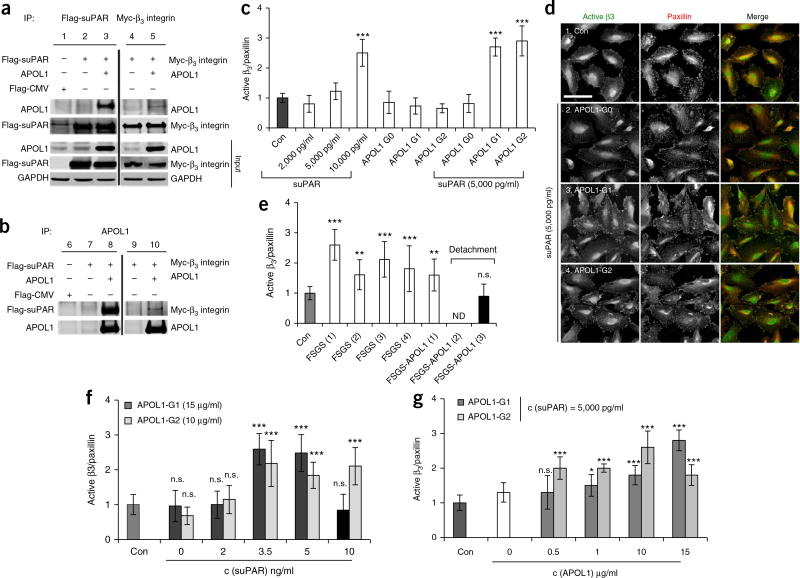
Experiments using recombinant proteins suggested the formation of a triple complex between suPAR, αvβ3 integrin and APOL1. To examine whether such a complex could be formed on the cell membrane, we expressed the reference allele APOL1 (i.e., G0), suPAR and β3 integrin in human embryonic kidney (HEK) 293T cells, and used cell lysates for immunoprecipitation experiments. Both suPAR and β3 integrin co-immunoprecipitated with APOL1 G0 (Fig. 3a), and APOL1 G0 co-immunoprecipitated with both suPAR and, to a lesser degree, β3 integrin (Fig. 3b). Taken together with previous studies15, showing that suPAR interacts with β3 integrin, these data suggest that the tripartite-protein complex might form on the cell membrane.
Figure 3.
High levels of suPAR are needed to synergize with APOL1 G1 and G2 to induce αvβ3 integrin activation on human podocytes. (a,b) Representative western blot images for immunoprecipitation assay using HEK293T cells transfected with plasmids expressing APOL1 G0, Flag-tagged suPAR and Myc-tagged β3 integrin in different combinations, as indicated. Data are representative of at least three independent experiments. The empty pFlag-CMV3 vector was used as a negative control. (a) Cell lysates were immunoprecipitated using a mouse monoclonal anti-Flag (M2) or monoclonal anti-Myc antibody. (b) Cell lysates were immunoprecipitated using a rabbit polyclonal anti-APOL1. The cell lysates and immunoprecipitates were analyzed by immunoblotting with antibodies, as indicated. Analysis of the input (1%) indicates that the levels of APOL1, β3 integrin and suPAR are overexpressed in the transfected cells. IP, immunoprecipitation. (c) Bar graph representing level of activated β3 integrin detected in human podocytes cultured in healthy serum without (Con) or with added proteins: suPAR (2,000, 5,000 and 10,000 pg/ml), APOL1 G0, G1 and G2 (15 µg/ml) or a combination of both proteins. Data represent measurements of >50 cells and are plotted as means ± s.d. (n = 3). ***P < 0.001. Statistical significance was determined by unpaired two-tailed Student’s t-test. (d) Immunofluorescence analysis of human podocytes grown in human serum in the absence (Con) or presence of suPAR (5,000 pg/ml) and three APOL1 variant proteins (15 µg/ml). Cells were stained with AP5 antibody that specifically recognizes the active form of β3 integrin, and with anti-paxillin antibody to mark focal adhesions. Scale bar, 25 µm. (e) Bar graph representing the level of β3-integrin activation detected in human podocytes cultured in healthy serum without (Con) or with three serum samples from individuals with FSGS carrying the nonrisk APOL1 genotype, and three different serum samples from individuals with FSGS carrying two APOL1 risk-allele genotypes. FSGS-APOL1 serum #2, and to a substantial degree, serum #3, detached the cells such that the quantification of β3-integrin activation was either not determined (n.d.) or not accurate (striped bar graph). Data represent measurements of >50 cells and are plotted as means ± s.d. (n = 3). Statistical significance was determined by unpaired two-tailed Student’s t-test. (f,g) Bar graph representing the level of β3-integrin activation detected in human podocytes cultured in healthy serum without (Con) or with the addition of the indicated concentrations of the proteins. In g, the concentration of suPAR was 5,000 pg/ml. Data represent measurements of >50 cells and are plotted as means ± s.d. (n = 3). *P < 0.01; ***P < 0.001. Statistical significance was determined by unpaired two-tailed Student’s t-test.
We next tested whether APOL1 G0 synergizes with suPAR in the activation of β3 integrin on human cultured podocytes by using sera from individuals with FSGS as an exogenous source. We modified the original assay17 to measure the ratio of activated β3 integrin level (as determined by staining for AP5, a monoclonal antibody that specifically recognizes the active conformation of β3 integrin) over the total amount of focal adhesion, as determined by staining with an anti-paxillin antibody (Supplementary Fig. 3a–c). We added Mn2+ to determine the maximal level of β3-integrin activation in the assay (Supplementary Fig. 3b,c). In all subsequent experiments, we compared the level of activation to that of serum from a healthy individual, normalized to 1.
Although suPAR levels similar to those of healthy individuals (2,000 pg/ml) did not induce significant β3 integrin activation, high suPAR levels (10,000 pg/ml) did (Fig. 3c), as previously reported17. Importantly, the presence of APOL1 proteins at concentrations similar to those in plasma (15 µg/ml)20 did not result in significant β3-integrin activation (Fig. 3c). By contrast, the addition of both suPAR and APOL1 G1 and G2 proteins, but not of the reference protein APOL1 G0, resulted in potent activation of β3 integrin (Fig. 3c,d). In addition to β3-integrin activation, a combination of both suPAR and APOL1 G1 and G2 proteins also resulted in substantial cell detachment (Supplementary Fig. 4a), which suggests that a combination of these two proteins results in toxic effects with regard to podocytes in culture. The ability of suPAR, as well as a combination of suPAR and APOL1 risk-variant proteins, to induce β3-integrin activation is consistent with the integrin activation observed in the presence of sera from individuals with FSGS carrying or not carrying APOL1 risk alleles (Fig. 3e). Indeed, two of the serum samples from individuals who have FSGS and carry two risk alleles resulted in such dramatic cell detachment (striped column, Fig. 3e) that it impaired our ability to accurately quantify β3-integrin activation.
More detailed concentration analysis showed that APOL1 G1 and G2 required higher levels of suPAR (≥3,500 pg/ml; Fig. 3f) to induce integrin activation. For a given suPAR concentration (5,000 pg/ml), APOL1 G2 induced potent integrin activation at lower concentrations (0.5 µg/ml) than were required for induction by APOL1 G1 protein (15 µg/ml) (Fig. 3g), consistent with its higher binding affinity, as determined by SPR. Of note, physiological concentrations of APOL1 proteins in circulation range from 0.4 to 16 µg/ml (median, 4 µg/ml)20. Taken together, these data provide further support for the hypothesis that high suPAR levels synergize with APOL1 G1 and G2 proteins to induce not only potent β3-integrin activation, but also podocyte detachment.
Given that integrin activation is linked to the regulation of autophagy in many cells, and because APOL1 risk alleles have been linked to the induction of nonfunctional autophagosomes21,22, we next examined the effects of suPAR and APOL1 proteins on the formation of autophagosomes. To this end, we determined the percentage of cells that stained positive for the presence of autophagosomes, using anti-LC3B antibody as an endogenous autophagic marker23. Potent integrin activation by Mn2+ or high suPAR concentrations that also induced integrin activation (Fig. 3c) resulted in a significant increase in the number of cells with autophagosomes (Supplementary Fig. 4b; P ≤ 0.003). Indeed, suPAR-induced autophagosome formation was blocked by the anti-suPAR antibody (Supplementary Fig. 4c). We used rapamycin, an mTOR inhibitor, as a positive control. In addition, APOL1 G1 and G2 proteins, but not the G0 protein, also increased the number of cells with autophagosomes in the absence of exogenous suPAR (Supplementary Fig. 4d). The presence of both APOL1 G1 or G2 and suPAR at a concentration that results in integrin activation (Fig. 3c) led to significant cell detachment (Supplementary Fig. 4a; P < 0.001), thus impairing the quantification of cells with autophagosomes (striped black graphs in Supplementary Fig. 4b). Taken together, these data suggest that autophagosome formation in podocytes can be induced by suPAR-dependent αvβ3-integrin activation, as well as by APOL1 G1 and G2 mutant proteins.
Expression of APOL1 G1 and G2 but not the G0 allele induces kidney injury in mice
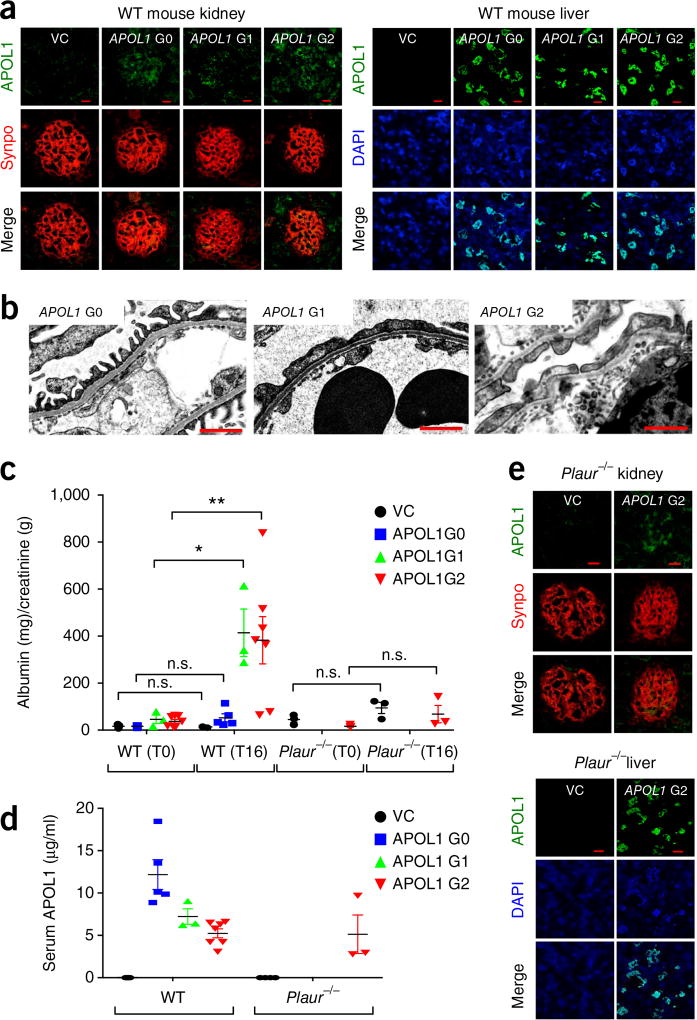
The foregoing association data in human cohorts, taken together with experiments using recombinant proteins and podocytes in culture, suggest that expression of the APOL1 G1 and G2 risk alleles might result in suPAR-dependent podocyte injury. To this end, we injected DNA encoding each of three variant APOL1 proteins into the tail vein of wild-type (WT) mice, as previously described24,25. 16 h after APOL1 gene delivery, we were able to detect APOL1 proteins, using immunofluorescence, in both the liver and glomerulus (Fig. 4a), as well as in the circulation (Fig. 4b). Of note, mice do not have an APOL1-equivalent gene. Animals expressing human APOL1 G1 and G2, but not G0, protein exhibited significant levels of proteinuria (Fig. 4c). There was also foot-process effacement—as determined by electron microscopy of the glomerulus (Fig. 4d)—and induction of β3-integrin activation as evidenced by increased staining with AP5 in the glomerulus (Supplementary Fig. 5a). In addition, the ability of APOL1 proteins to induce proteinuria required the presence of Plaur, the gene encoding both uPAR and suPAR. The expression of APOL1 G2 protein using a gene-delivery technique24,25 in uPAR-deficient animals (Plaur−/−) did not cause significant levels of proteinuria (Fig. 4c), despite the presence of APOL1 protein in the circulation, glomerulus and liver (Fig. 4d,e). This finding further supports a role for suPAR (or uPAR) in APOL1-driven kidney injury.
Figure 4.
In vivo gene delivery of APOL1 G1 and G2, but not of G0, gene constructs caused proteinuria and podocyte foot-process effacement in wild-type mice, but not in Plaur−/− mice. To express APOL1 in C57BL/6 mice or Plaur−/− mice, in vivo gene delivery was performed and the kidney phenotype evaluated 16 h after APOL1 gene (G0, G1 and G2) injection. (a) APOL1 protein could be detected in both the glomerulus and liver of injected animals. Double immunostaining of APOL1 (green) and synaptopodin (synpo, red), a podocyte marker. VC, pcDNA 3.1 vector control. Scale bars, 10 µm. (b) Transmission electron microscopy (TEM) analysis of the kidney glomeruli in APOL1 gene (G0, G1 and G2) engineered mice. TEM images display podocyte foot processes. Scale bars, 1 µm. (c,d) Examination of proteinuria (c) and serum APOL1 levels (d) in animals into which APOL1 genes were injected (n = 7 for VC; n = 5 for APOL1 G0; n = 3 for APOL1 G1; n = 7 for APOL1 G2; n = 3 for all Plaur−/− mice). Analysis of urinary protein is shown as albumin (mg)/creatinine (g) ratio before (T0) and 16 h after (T16) gene delivery. Data are shown as means ± s.e.m. One-way ANOVA, followed by Tukey′s multiple-comparison test in which values at T16 were compared to those at T0; *P < 0.05, **P < 0.01. Welch’s ANOVA was used when unequal variances were noted between groups (P = 0.014). (e) APOL1 G2 protein was detected in the glomerulus and liver of Plaur−/− mice 16 h after the gene was injected into a tail vein. Double immunostaining of Plaur−/− (green) and synaptopodin (synpo, red). VC, pcDNA 3.1 vector control. Scale bars, 10 µm.
DISCUSSION
This study provides a mechanism for APOL1-associated kidney disease in African Americans. We uncover an interaction between suPAR and APOL1 as they relate to kidney-function decline, and we describe a potential mechanism that underlies this association. We have measured suPAR levels and determined the APOL1 genotypes in two differing cohorts of African American individuals, and found that patients with the high-risk APOL1 variant genotypes exhibited a steeper decline in renal function, than individuals carrying low-risk variants, particularly in the setting of high suPAR levels; this remained true even after adjusting for the relevant covariates. Biochemically, we identify high-affinity interactions between APOL1 and suPAR, as well as between APOL1 and αvβ3 integrin. In addition, we show that the G1 and G2 risk variants of APOL1, but not the reference protein G0, synergize with suPAR in the activation of αvβ3 integrin on podocytes. Finally, the expression of APOL1 G1 and G2 but not G0 proteins in mice resulted in uPAR-dependent proteinuria and foot-process effacement. Taken together, our findings identify a biological link between suPAR and APOL1 interaction in the mediation of kidney disease (Supplementary Fig. 6).
The observation that African American individuals with a high-risk APOL1 genotype experience a more rapid decline in eGFR is consistent with what has previously been reported in various cohorts1–5. We found the association to be highly dependent on baseline suPAR levels, and to involve a rapid increase in the magnitude of estimated yearly eGFR decline as suPAR levels increase beyond 3,000 pg/ml. At low suPAR levels (<3,000 pg/ml), there were no notable differences in eGFR decline between the APOL1 high-risk genotypes and the remainder of the cohort. Together, these findings suggest that the presence of elevated suPAR levels is likely to be a major contributor underlying the association between APOL1 genotype and kidney disease. The replication of the APOL1–suPAR interaction in the AASK cohort strengthens our confidence in our findings, given that EmCAB and AASK are cohorts with distinct selection criteria and study design. The EmCAB is an ongoing prospective observational study of patients undergoing cardiac catheterization for various indications, and represents a heterogeneous, high-risk population with less than one-third of the cohort having an eGFR of <60 ml/min/1.73 m2. The AASK cohort is an extension of a randomized, controlled trial of individuals with CKD attributed to hypertension, for which enrollment occurred in the late 1990s. Follow-up eGFR was measured for clinical indications in EmCAB-cohort participants, and at semi-annual protocol visits for the AASK cohort. In addition, suPAR levels were measured in the two cohorts using two different kits (Quantikine in EmCAB, and Virogates in AASK) by off-site technicians blinded to the clinical data.
The protein–protein interaction studies identified high-affinity interactions between all three APOL1 protein variants and suPAR. APOL1 exhibited almost ten-fold higher affinity for suPAR than for high-density lipoprotein (HDL). In addition, we show that APOL1 G1, and especially G2, also bind αvβ3 integrin with high affinity, but only if αvβ3 integrin is in the active conformation (i.e., Mn2+ is present). In keeping with studies using recombinant proteins, higher levels of suPAR were needed to synergize with APOL1 G1 and G2 proteins to induce substantial αvβ3-integrin activation within cultured human podocytes. Given that concentrations of suPAR in the circulation are fairly low (2,000–10,000 pg/ml) and that APOL1 proteins circulate with HDL, it seems reasonable to assume that the protein complex between suPAR and APOL1, as well as APOL1 and αvβ3 integrin, is confined to the podocyte membrane (Supplementary Fig. 6). Although APOL1 risk-variant proteins were linked to the induction of nonfunctional autophagosomes21,22, transgenic animal models expressing high-risk APOL1 variants did not develop the expected kidney phenotypes9,10. Indeed, whereas the addition of both suPAR and APOL1 G1 and G2 proteins resulted in an increase in the number of cells that contained measurable levels of autophagosomes, the most dramatic effect observed when both proteins were present was substantial cell detachment. In addition, whereas APOL1 G1 and G2 proteins could induce the formation of autophagosomes even in the absence of a high concentration of suPAR and resultant β3-integrin activation, the presence of suPAR either in circulation or on the podocyte membrane was required for cell detachment. Accordingly, mice lacking the Plaur gene have normal kidneys and do not develop proteinuria after expression of APOL1 G2, which suggests that uPAR is needed to allow for the development of clinically significant APOL1-induced autophagosme dysfunction.
The current experiments do not identify the origin, whether hepatic26 or from local secretion by podocytes, of the APOL1 proteins involved in suPAR–αvβ3 integrin interactions (Supplementary Fig. 7). Regardless of the origin of APOL1, our study identifies a molecular mechanism whereby APOL1 risk-variant proteins lead to podocyte injury by synergizing with high levels of suPAR to induce potent αvβ3-integrin activation on the podocyte membrane. The formation of such a triple complex results in the activation of multiple αvβ3-integrin signaling pathways, which leads to the formation of autophagosomes, dysregulation of the actin cytoskeleton and, ultimately, cell detachment. Therefore, our results do not challenge a concurrent mechanism for cell injury emanating from podocyte-generated APOL1 proteins, as was previously suggested27–30. Rather, they suggest that, for the podocyte injury to occur, the secreted APOL1 risk-variant protein needs to be internalized by the cell in a urokinase receptor and β3-integrin-dependent manner (Supplementary Fig. 6)31–34. This rationale is supported by the fact that the APOL1 gene encodes a signal peptide sequence that targets newly synthesized protein for secretion, by the fact that exogenously added APOL1 risk-variant proteins, but not the non-risk-variant, affected endocytic machinery within podocytes (formation of autophagosomes)—the mechanism through which APOL1 risk proteins lyse trypanosomes29—and lastly, by studies describing suPAR–β3 integrin-dependent endocytosis of hantaviruses35,36. Interestingly, a previous study showed that podocyte-specific expression of APOL1 risk variants in mice resulted in moderate podocyte depletion later in life and occasional preeclampsia9. It is worth noting that endothelial cells, similarly to podocytes, also express αvβ3 integrin, and therefore, the formation of the triple complex between APOL1, suPAR and αvβ3 integrin might also occur on endothelial cells. In a recent study, podocyte-specific expression of APOL1 risk variants in mice induced albuminuria and effacement of foot processes37, in contrast to the findings of another study9. Podocyte injury was attributed to altered endosomal trafficking and blocked autophagic flux, ultimately resulting in podocyte loss and glomerular scarring. Given that APOL1 risk-variant proteins did not alter exocytosis of the newly synthesized proteins, it is reasonable on the basis of our study to suggest that observed impairment in the autophagic flux required endocytosis of podocyte-secreted APOL1 proteins through urokinase-receptor-dependent αvβ3-integrin activation (Supplementary Fig. 6)—a hypothesis that deserves direct testing.
In summary, our study describes clinical and molecular interactions between suPAR and APOL1, both novel and strong predictors of kidney disease. Because the nephropathologic effect of APOL1 is based on suPAR-mediated αvβ3-integrin activation, modification of suPAR could be a safe and therapeutic approach to treat CKD in individuals of recent African ancestry.
METHODS
Methods, including statements of data availability and any associated accession codes and references, are available in the online version of the paper.
ONLINE METHODS
Study design and population
We measured plasma suPAR levels and determined the APOL1 genotype in two separate cohorts of African American adult individuals with available plasma, DNA samples and measures of kidney function: the Emory Cardiovascular Biobank (EmCAB, n = 487) and the African American Study of Kidney Disease and Hypertension (AASK, n = 607). We first determined whether suPAR levels differed according to APOL1 genotype, then examined the interaction between suPAR and APOL1 genotype in predicting eGFR decline in individuals with at least 3 months follow-up. Studies were approved by the Emory University and Johns Hopkins University institutional review boards, and all enrolled patients provided written informed consent.
The Emory Cardiovascular Biobank (EmCAB) is a prospective cohort of patients undergoing cardiac catheterization for suspected or confirmed coronary artery disease (CAD) at Emory Healthcare sites in Atlanta, GA, between 2003 and 2015 (refs. 38,39). Patients with congenital heart disease, severe anemia, recent blood transfusion, myocarditis, history of active inflammatory disease or cancer were excluded. Serum creatinine measurements at enrollment and all subsequent values acquired during routine follow-up clinic visits or hospitalizations within the Emory Healthcare system were collected. eGFR was calculated using the CKD–EPI equation40. A subset of 487 patients without acute kidney injury at enrollment had follow-up eGFR measures, with a median number of eGFR measures per person of 6 and a median follow-up time of 1.5 years (range 0–9 years).
The African American Study of Kidney Disease and Hypertension (AASK) is a randomized clinical trial which tested the effect of three antihypertensives (ramipril, metoprolol and amlodipine) and level of blood pressure control on CKD progression in self-identified adult (18–70 years of age) African American patients with CKD (with an iothalamate-derived eGFR between 20 and 65 ml/min/1.73 m2) attributed to hypertension; this was followed by a 5-year prospective, observational extension. Patients with secondary or malignant hypertension, diabetes mellitus or a fasting glucose of >140 mg/dl, or causes of CKD other than hypertension, were excluded. A total of 607 patients had suPAR measures and APOL1 genotype available. The median number of eGFR measures was 19, with a median follow-up of 9 years (range 1–12).
SuPAR measurement and APOL1 genotype determination
Fasting arterial blood samples from EmCAB cohort members were collected at the time of catheterization and stored at −80 °C for a mean duration of 4.9 years. Plasma suPAR was measured by technicians blinded to APOL1 genotype and clinical data using human uPAR Quantikine ELISA kits (R&D systems, DUP00) according to the manufacturer’s instructions. The minimum detectable amount of suPAR is less than 33 pg/ml. Both the intra- and inter-assay variations were less than 5%.
Blood samples of AASK cohort members were obtained at enrollment and stored at −80 °C for a mean duration of 19 years. Plasma levels of suPAR were measured by ELISA (suPARnostic kit; ViroGates, Copenhagen, Denmark), with a lower detection limit of 100 pg/ml and intra- and inter-assay variation of 2.75% and 9.17%, respectively.
Genomic DNA from patients in both cohorts was isolated from whole blood using a Puregene Kit (Qiagen, 158389), quantified and quality-checked with Picogreen assay (Quant-iT PicoGreen dsDNA Assay Kit, Thermo Scientific, P11496). APOL1 G1 SNPs (rs73885319 and rs60910145) and the G2 (rs717185313)8 were genotyped by TaqMan SNP Genotyping Assays (Life Technologies) using CFX96TM Real-Time System (Bio-Rad). EmCAB samples were additionally validated by capillary sequencing with sequencing primers (forward: 5′-GCCAATCTCAGCTGAAAGCG-3′; reverse: 5′-TGCCAGGCATATCTCTCCTGG -3′) from 50 ng of genomic DNA, which was used as a template for PCR amplification (Primer set forward: 5′-AGACGAGCCAGAGCCAATC-3′; reverse: 5′-CTGCCAGGCATATCTCTCCT-3′).
Generation of APOL1 constructs
APOL1 G0 full-length (transcript variant 1 (NM_001136540) or 3 (NM_003661)), G1 and G2 cDNAs were cloned into pcDNA3.1 (ref. 21). Full-length of human uPAR cDNA was cloned in pFLAG-CMV-3 vector (FLAG-CMV). Human integrin β3 (ITGB3; Myc-DDK-tagged) clones were purchased from OriGene (RC221606).
APOL1 recombinant protein expression and purification
The APOL1 coding sequence (G0, G1 or G2) was amplified through PCR from the pcDNA3.1-APOL1 constructs with primers (forward, 5′-ATGCAGCTCGAGGAGGGAGCTGCTTTGCTGAGAGT-3′; reverse, 5′-ATGGATGGATCCCAGTTCTTGGTCCGCCTGCAGAA-3′), creating a 5′ XhoI site and a 3′ BamHI site. The PCR fragments were cloned in XhoI and BamHI sites of a pET-15b expression vector carrying an N-terminal His-Tag sequence (Novagen)41, verified by sequencing. The plasmid constructs were transformed into Escherichia coli Rosetta2 (DE3) (Novagen, 71397-4). 1 liter of Rosetta2 (DE3) cells containing the recombinant APOL1 G0, G1 or G was grown to an optical density (OD)600 of 0.65–0.70 at 37 °C in Luria Bertani (LB) broth medium (Sigma-Aldrich, L3022). The cells were then induced from an exponentially growing culture by 0.5-mM isopropyl β-d-1-thiogalactopyranoside (IPTG, Sigma-Aldrich, 16758) for 15–16 h at 22 °C, harvested and lysed by sonication in lysis buffer (1 mg/ml lysozyme and protease inhibitor cocktail in Ni-Affinity buffer: 50-mM NaH2PO4, 500-mM NaCl, 6-M urea, 20-mM imidazole and 5-mM β-mercaptoethanol, pH 7.8). The His-tag fused APOL1 protein was purified by HisTrap HP column (GE Healthcare, 17-5248-02) chromatography with a series of stepwise gradient of Ni-Elution buffer: 50-mM NaH2PO4, 500-mM NaCl, 6-M urea, 500-mM imidazole and 5-mM β-mercaptoethanol, pH 7.8). The purified APOL1 protein was then further dialyzed to remove urea in dialysis buffer (50-mM NaH2PO4, 50-mM NaCl, 1-mM DTT, 15% glycerol, 0.1% Triton X-100, pH 7.8) for 2 h. The dialysis procedure was repeated three times by transferring the dialysis bag (Slide-A-Lyzer Dialysis Cassettes, Thermo Scientific, 66810) into fresh dialysis buffer each time, and the process was continued overnight at 4 °C. The structure of protein was restored by gradual dialysis and then concentrated using an Amicon-Ultra-15 (10,000 NMWL centrifugal filter device, Millipore, UFC901008). Purity was estimated by SDS–PAGE (sodium dodecyl sulfate–polyacrylamide gel) electrophoresis and subsequent protein staining (GelCode blue stain, Thermo Scientific, 24590), and the concentration of the purified protein sample was determined using a Nano-drop spectrophotometer (Thermo Scientific). Finally, the purified APOL1 protein G0, G1 or G2 (Supplementary Fig. 1) was flash frozen in liquid nitrogen and stored at −80 °C.
Immunoprecipitation assay and western blotting
HEK293T human embryonic kidney cells were transiently transfected with a total of 1 µg of plasmid DNA using either Lipofectamine 2000 (Thermo Scientific, 11668019) or Fugene 6 reagent (Promega, E2692) in 100 mm × 20–mm tissue culture dishes (BD Falcon, 353003). Cells were seeded at a density of 1 × 105 cells/dish the night before. Cells were incubated in 1 ml of OptiMEM (Gibco, 31985062) and 250 µl of lipofectamine mix containing plasmid DNA to initiate transfection. Following 24–48 h of incubation, cells were washed with sterile PBS once, and the medium was replaced with regular culture medium. 48 h after transfection, cells were harvested and cell pellets were resuspended in modified RIPA lysis buffer with protease-inhibitor cocktail (Roche, 04693116001) and Phosphatase Inhibitor Cocktail 2 & 3 (Sigma-Aldrich, P5726 and P0044). Co-immunoprecipitation was performed using Pierce Protein A/G Agarose (Thermo Scientific, 20421) according to the manufacturer’s instructions. Briefly, the Myc mouse monoclonal (OriGene, TA150121), Flag-tagged mouse (M2, Sigma-Aldrich, F1804), APOL1 rabbit antibody (Abcam, ab108315) or normal rabbit IgG (Santa Cruz, sc-2027), as an isotype control, was added to the precleared cell lysate, rotated overnight at 4 °C and then incubated with A/G Agarose slurry. The samples were analyzed by western blotting with rabbit or goat anti-APOL1 (Sigma-Aldrich, SAB2500087), anti-FLAG (for detecting suPAR), rabbit anti-uPAR (Santa Cruz, sc-10815), goat anti-integrin β3 (Santa Cruz, sc-6627) or anti-Myc (for detecting integrin β3) antibodies as primary antibodies. Western blots were probed with appropriate IRDye 680RD donkey anti-mouse IgG (H+L) (926-68072), IRDye 680RD donkey anti-rabbit IgG (H+L) (926-68073), and 680RD donkey anti-goat IgG (H+L) (926-68074) as secondary antibodies from LI-COR Biosciences (Lincolon, NE). Blotted proteins were detected using an Odyssey CLx imaging system (LI-COR Biosciences).
Determination of dissociation equilibrium constant (KD) by SPR
All experiments were performed by using SPR technology with a Biacore T200 instrument (GE Healthcare, Piscataway, NJ) according to the manufacturer’s instructions. Human recombinant protein integrin αvβ3 (Millipore, cc1018, R&D, 3050-AV), human integrin α3β1 (R&D, 2840-A3), human HDL full-length protein (Abcam, ab77897) and human suPAR (R&D, 807-UK-100/CF) were purchased. The proteins that required buffer exchange were applied by Zeba Spin desalting columns (Thermo Scientific, 89882). In brief, to measure the binding affinities of the potential binding proteins (integrin αvβ3, integrin α3β1, HDL and suPAR) to APOL1 protein (G0, G1 or G2), the purified APOL1 proteins (G0, G1, G2) were immobilized to flow channels 2, 3 and 4, respectively, on a CM5 sensor chip using the standard amine-coupling method. The dextran surface was activated by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC)/N-hydroxy succinimide (NHS) mixture, and the activated surface was blocked by ethanolamine (pH 8.5) as a control on flow channel 1. The APOL1 protein was diluted in 10-mM sodium acetate (pH 4.0) and immobilized after sensor-surface activation with EDC/NHS by a 7-min injection, followed by ethanolamine blocking on unoccupied surface area. Protein (αvβ3/α3β1 integrin, HDL or suPAR) with a series of increasing concentrations (0–300 nM or as indicated in the figures) as an analyte, was applied to all four channels at a 15 µl/min flow rate at 25 °C. The binding experiments were run with 10-mM HEPES buffered saline (10 mM HEPES, 150-mM NaCl, 1-mM MgCl2, 0.5-mM MnCl2, 0.05% N-octyl-β-d-glucose, pH 7.1 for the study of αvβ3 integrin, α3β1 integrin and HDL) or (10-mM HEPES, 150-mM NaCl, 3-mM EDTA, 0.05% Tween 20, pH 7.5 for a few sets with suPAR). HDL was used as a positive control18. Data were referenced with blank (ethanolamine) response units. Response unit values at each concentration were measured during the equilibration phase for steady-state affinity fittings using either Biacore T200 evaluation software 2.0.3 or Sigmaplot 11.0 with an embedded equation, y = (ymax × x)/(KD + x), where y is the response, ymax is the maximum response and x is the concentration of the analyte protein. Kinetic rate constants were determined by fitting globally to the 1:1 Langmuir (one-to-one binding) model (or two-state reaction) embedded in the Biacore T200 evaluation software v3.0. Three-protein-complex experiments for suPAR binding to the APOL1-immobilized surfaces were performed using the same experimental settings as for the suPAR binding analyses except that increasing concentrations of suPAR (0–300 nM) were mixed with αvβ3 integrin (5 nM, an average binding affinity between APOL1 protein and αvβ3 integrin). The sensor surface was regenerated between each experiment with a 30-s injection of 10-mM glycine-HCl (pH 2.5) at a flow rate of 30 µl/min.
Blood collection from human participants for in vitro experiments
Permission to obtain whole blood from healthy volunteers and African American patients with CKD was obtained from the Institutional Review Boards of Rush University Medical Center in Chicago. The volunteers and patients were enrolled in the study according to IRB-approved protocols (#14030304-IRB01) under informed consent. Serum or plasm sample was isolated from the whole blood. The whole blood was used for APOL1 genotyping, as described above.
Human podocyte cell cultures
Immortalized human podocyte cell lines were previously described42 and cultured in RPMI-1640 medium (Gibco, 11875) enriched with 10% FBS (FBS) (US certified FBS, Gibco, 16000), 1% Penicillin/Streptomycin (Gibco, 15140) and 1% ITS supplement (Sigma-Aldrich, I-3146) in a humidified atmosphere of 5% CO2 at 33 °C (permissive conditions) or 37 °C (nonpermissive conditions). Cells routinely tested negative for the presence of mycoplasma and were allowed to differentiate for 10 d before experiments.
AP5 staining
Human podocytes were proliferated at 33 °C and differentiated at 37 °C for 10 d on coverslips in a six-well plate. On day 10, the podocytes were serum-starved overnight (RPMI 1640 + anti-anti). Cells were subsequently incubated with 10% healthy serum or FSGS serum for 24 h at 37 °C. Healthy serum was used as the negative control, and healthy serum supplemented with MnCl2 (750 µM) served as the positive control. After treatment, samples were fixed with 4% paraformaldehyde for 20 min, washed and permeabilized with 0.3% Triton-X for 3 min and blocked (5% donkey serum, 5% goat serum, in 1× PBS) for 45 min. The cells were incubated with the primary antibody (anti-Paxillin; Abcam, cat. #ab32084, 1:300 Rabbit) for 1 h, followed by the secondary antibody goat-anti-rabbit 568 (1:2,000, Invitrogen). The cells were incubated with the second primary antibody (AP5; Blood Center of Wisconsin, 1:50 Mouse) for 1 h, followed by the secondary antibody goat-anti-mouse 488 (1:1,000, Invitrogen). The coverslips were mounted with Flouroshield using DAPI (Abcam, ab104139). Images were acquired using Zeiss LSM 5 Pascal. The detector gain, amplifier offset and laser power settings were kept consistent for the collection of all images. The images were analyzed using ImageJ. Additional information for image analysis: the files were opened in ImageJ, and the channels were separated and images inverted. The threshold was set for each channel and kept consistent for the analysis of all images. For β3-integrin images, the nuclear staining was excised from each cell. Using the same freehand selection function, the cell was circled and the integrity density was measured for each channel. The data were exported into Microsoft Excel, and the integrity-density value of β3 staining of each cell was divided by the integrity-density value of focal adhesion staining of each cell, and normalized to the negative control.
Autophagosome staining with LC3B antibody
Human podocytes were treated with 10% FBS and, when indicated, recombinant suPAR or APOL1 proteins (G0, G1 and G2) for 24 h. Rapamycin treatment for 24 h (100 ng/ml; Tocris Bioscience, 1292)43 was used as a control. When indicated, 20 µg/ml of anti-suPAR antibody (ATN615)44 was added. Cells were subsequently fixed with 4% PFA for 15 min at room temperature (RT) and permeabilized with 0.3% TritonX-100 for 5 min. Unspecific binding was blocked with a blocking buffer (5% goat serum, 5% donkey serum in PBS) for 45 min at RT, followed by overnight staining with a primary antibody specific for LC3B (1:200, Cell Signaling Technology). The next day, after sufficient wash with PBS (3 min, three times), the secondary antibody, AlexaFluor 568 Goat Anti-Rabbit IgG (Invitrogen, 1:1,000) was added and incubated for 45 min. Slides were washed and mounted with Fluoroshield with DAPI (Sigma-Aldrich). Images were acquired by a Zeiss LSM 5 PASCAL laser-scanning microscope at 40× objective. A consistent threshold was set using ImageJ software (1.64r; NIH, Bethesda, Maryland, USA) to remove background, and cells with morphologically defined autophagosomes were scored as positive. At least 100 cells were scored in each experiment, with at least two experiments. The percentage of podocytes deemed positive for LC3B staining was shown. Data were further analyzed using GraphPad Prism (v 4.03) for Windows (GraphPad Software, San Diego, CA) to perform statistical analysis using two-tailed Student’s t-tests. On the basis of this analysis, *P ≤ 0.05, **P ≤ 0.01 and ***P ≤ 0.001 were considered to be statistically significant, and P > 0.05 was considered not significant.
Immunofluorescence of kidney and liver tissue
Frozen mouse kidney and liver tissues were cut at 4-µm thickness and fixed with ice-cold acetone for 3 min. After blocking with blocking solution (2.5% donkey normal serum and 2.5% FBS (in PBS)) for 1 h, samples were stained with rabbit anti-human APOL1 (1:100; Sigma-Aldrich, HPA018885) and/or goat anti-mouse synaptopodin (1:300; Santa Cruz Biotechnology, sc-21537) and/or, followed by AlexaFluor-488 conjugated donkey anti-rabbit IgG (1:1,000; Molecular Probes, A-21206) and/or AlexaFluor-568 conjugated donkey anti-goat IgG (1:1,000; Molecular Probes, A-11057), respectively. After washing, stained samples were mounted with ProLong Gold antifade reagent with DAPI (Molecular Probes, P36935). Images were obtained and analyzed using a Carl Zeiss LSM 700 confocal microscope.
Transient in vivo gene delivery
In vivo gene-delivery assays were performed using the TransIT-EE hydrodynamic in vivo gene-delivery system according to the manufacturer’s instructions (Mirus Bio, MRI5340), as described previously24,25. APOL1 G0, G1 and G2 constructs cloned with pcDNA 3.1 vector were introduced into C57BL/6 and Plaur−/− (uPAR KO) mice obtained from the Jackson Laboratory (Bar Harbor, Maine) n = 7 for vector control, n = 5 for APOL1 G0, n = 3 for APOL1 G1, n = 7 for APOL1 G2, n = 3 for all Plaur−/− mice, (female, 10 weeks old). Briefly, C57BL/6 or uPAR KO mice were injected with 45 µg DNA per each mouse. On the basis of body weight (17–21 g) of each mouse, injection volume was determined. 16 h after gene delivery, mice were killed. Mouse urine and blood were collected, and kidneys and liver were removed for further study. Animal experiments were carried out according to the National Institutes of Health Guide for Care and Use of Experimental Animals and were approved by the Rush University Institutional Animal Care and Use Committee (IACUC).
Albumin–creatinine ratio (ACR) and human APOL1 measurement
Urinary albumin and creatinine were determined using mouse albumin ELISA (Bethyl Labs, E99-134) and creatinine assay (Cayman Chemical, 500701) kits according to the manufacturers’ protocols. Serum APOL1 (1:2,000 dilution) obtained from mouse blood samples was measured using a human APOL1 ELISA kit (Proteintech, KE00047) according to the manufacturer’s protocol.
Transmission electron microscopy
Transmission electron microscopy was performed as previously described15. In brief, renal tissues were collected and dissected into 2 × 2–mm pieces. The tissues were fixed in 4% PFA O/N, washed three times in PBS and postfixed in 1% OsO4. Tissues were washed in 0.1-M cacodylate buffer, dehydrated and embedded in Epon812 (EMS). 70-nm sections were mounted onto Formvar-coated Ni slot grids (EMS). Grids were stained for 15 min in 5% uranyl acetate followed by 0.1% lead citrate for 5 min. Electron micrographs were obtained and analyzed using the Sigma HDVP Electron Microscope (Zeiss).
Statistical analysis
APOL1 genotype risk status was characterized as high-risk (two copies of high-risk haplotype) and low-risk (0 or 1 copy of high-risk haplotype), because the association with kidney-disease risk is thought to follow a recessive pattern5,45. Patient characteristics were summarized with means, s.d., quartiles, frequencies and percentages. Differences between the two APOL1 risk groups were assessed using Student’s t-test, Wilcoxon test and the χ-squared test when appropriate. The association of APOL1 risk status with eGFR decline, as well as whether suPAR-related eGFR decline varied by APOL1 risk status, were evaluated in linear mixed-effects models with random subject-specific intercepts and random time effects in each cohort separately. Covariates for EmCAB included age, sex, body-mass index, hypertension, diabetes and smoking status; covariates for the AASK model included a randomization arm rather than hypertension status, and did not include diabetes status, given that all AASK participants had hypertension and none had diabetes mellitus. Each covariate, APOL1 risk status, suPAR level and interaction term for APOL1 and suPAR, baseline eGFR and eGFR slope were included in the model. SuPAR was categorized as above or below the median value (3,000 pg/ml) in EmCAB, and, for replication, at the same cutpoint in AASK. So that data could be displayed graphically, suPAR was log-transformed and, in AASK, given model fit, characterized as a restricted cubic spline with 5 knots46. The statistical analysis was performed using SAS 9.4, STATA and R 3.2.2.
For animal studies, sample sizes were estimated on the basis of sample availability and previous experimental studies17. Animals were allocated at random to treatment groups. Investigators were not blinded during animal experiments. For multiple group comparisons, we performed one-way ANOVA with Tukey’s multiple-comparison test. Welch’s ANOVA was used when unequal variances were noted between groups. For in vivo and in vitro studies, statistical analyses were done by using Prism 5.0 software (GraphPad Software). Two-tailed P-values of ≤0.05 were considered to be statistically significant.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We would like to acknowledge the members of the Emory Biobank Team, Emory Clinical Cardiovascular Research Institute (ECCRI) and Atlanta Clinical and Translational Science Institute for the recruitment of participants, compilation of data and preparation of samples. Specifically, we would like to acknowledge M. Awad, T. Varghese, W. Schultz, J. Patrick, M. Namara and A. Samman Tahhan (Emory University Department of Medicine) for assisting in data collection. We also appreciate A. Arnaout and B. Adair for the discussion of APOL1 and suPAR interaction. We are grateful to A.P. Mazar for providing anti-suPAR antibody ATN615. We thank R. Shemer, (Technion, Israel Institute of Technology) and A. Ofir (Rambam Medical Center, Haifa, Israel) for discussions and technical help.
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
S.S.H., K.H.K. and C.W. designed and performed experiments, and wrote the paper; M.E.G. and Y.-A.K. contributed to data analysis; J.L., B.S., H.L., R.R.D., H.W.L., E.H., V.P., M.T., N.J.T., G.G. and N.S. contributed to experiments; K.S. participated in formulating aspects of study design relating to suPAR–APOL1 interaction and developing novel enabling reagents, and collaborated in the writing of the manuscript; V.G., M.M.A., C.A.W., M.S.L., A.T., L.A.I., A.S.L., M.Z., B.I.F., J.B.K. and J.C. contributed to the interpretation of data and manuscript revision; A.A.Q. established the Emory Cardiovascular Biobank that contributed all clinical samples and assisted in manuscript preparation; J.R. and S.S. designed and supervised the study, and wrote the paper. J.R. is supported by grants 5R01DK101350-03 (NIDDK) and 1R01DK106051 (NIDDK). A.A.Q. is supported by grants 5P01HL101398-02, 1P20HL113451-01, 1R56HL126558-01, 1RF1AG051633-01, R01 NS064162-01, R01 HL89650-01, HL095479-01, 1U10HL110302-01, 1DP3DK094346-01, 2P01HL086773-06A1 (NIH). M.E.G., J.C., A.S.L., L.A.I. and A.T. are supported by grants R01DK108803 (NIH) and U01DK085689 (NIDDK). J.C., A.S.L. and L.A.I. are supported by grant R01DK087961 (NIH). E.H. was supported by grant 1RO1 DK106051. Funding for the collection and management of samples was received from the Robert W. Woodruff Health Sciences Center Fund (Atlanta, GA), Emory Heart and Vascular Center (Atlanta, GA), Katz Family Foundation Preventive Cardiology Grant (Atlanta, GA), and National Institutes of Health (NIH) Grants UL1 RR025008 from the Clinical and Translational Science Award program and the Intramural Research Programs of NCI and NIDDK, NIH. This project has also been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Chen TK, Estrella MM, Parekh RS. The evolving science of apolipoprotein-L1 and kidney disease. Curr. Opin. Nephrol. Hypertens. 2016;25:217–225. doi: 10.1097/MNH.0000000000000222. [DOI] [PubMed] [Google Scholar]
- 2.Tzur S, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum. Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Genovese G, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kao WH, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat. Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsa A, et al. APOL1 risk variants, race, and progression of chronic kidney disease. N. Engl. J. Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nichols B, et al. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int. 2015;87:332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papeta N, et al. APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J. Am. Soc. Nephrol. 2011;22:1991–1996. doi: 10.1681/ASN.2011040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopp JB, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J. Am. Soc. Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruggeman LA, et al. APOL1-G0 or APOL1-G2 transgenic models develop preeclampsia but not kidney disease. J. Am. Soc. Nephrol. 2016;27:3600–3610. doi: 10.1681/ASN.2015111220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson BR, et al. In vivo modeling implicates APOL1 in nephropathy: evidence for dominant negative effects and epistasis under anemic stress. PLoS Genet. 2015;11:e1005349. doi: 10.1371/journal.pgen.1005349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thunø M, Macho B, Eugen-Olsen J. suPAR: the molecular crystal ball. Dis. Markers. 2009;27:157–172. doi: 10.3233/DMA-2009-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sidenius N, et al. Serum level of soluble urokinase-type plasminogen activator receptor is a strong and independent predictor of survival in human immunodeficiency virus infection. Blood. 2000;96:4091–4095. [PubMed] [Google Scholar]
- 13.Ostrowski SR, et al. High plasma levels of intact and cleaved soluble urokinase receptor reflect immune activation and are independent predictors of mortality in HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 2005;39:23–31. doi: 10.1097/01.qai.0000157950.02076.a6. [DOI] [PubMed] [Google Scholar]
- 14.Hayek SS, et al. Soluble urokinase receptor and chronic kidney disease. N. Engl. J. Med. 2015;373:1916–1925. doi: 10.1056/NEJMoa1506362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahm E, et al. Bone marrow–derived immature myeloid cells are a main source of circulating suPAR contributing to proteinuric kidney disease. Nat. Med. 2017;23:100–106. doi: 10.1038/nm.4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei C, et al. Modification of kidney barrier function by the urokinase receptor. Nat. Med. 2008;14:55–63. doi: 10.1038/nm1696. [DOI] [PubMed] [Google Scholar]
- 17.Wei C, et al. Circulating urokinase receptor as a cause of focal segmental glomerulosclerosis. Nat. Med. 2011;17:952–960. doi: 10.1038/nm.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weckerle A, et al. Characterization of circulating APOL1 protein complexes in African Americans. J. Lipid Res. 2016;57:120–130. doi: 10.1194/jlr.M063453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ni H, Li A, Simonsen N, Wilkins JA. Integrin activation by dithiothreitol or Mn2+ induces a ligand-occupied conformation and exposure of a novel NH2-terminal regulatory site on the beta1 integrin chain. J. Biol. Chem. 1998;273:7981–7987. doi: 10.1074/jbc.273.14.7981. [DOI] [PubMed] [Google Scholar]
- 20.Bruggeman LA, et al. Plasma apolipoprotein L1 levels do not correlate with CKD. J. Am. Soc. Nephrol. 2014;25:634–644. doi: 10.1681/ASN.2013070700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan X, et al. APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am. J. Physiol. Renal Physiol. 2014;307:F326–F336. doi: 10.1152/ajprenal.00647.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wan G, et al. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J. Biol. Chem. 2008;283:21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol. Biol. 2008;445:77–88. doi: 10.1007/978-1-59745-157-4_4. [DOI] [PubMed] [Google Scholar]
- 24.Sever S, et al. Proteolytic processing of dynamin by cytoplasmic cathepsin L is a mechanism for proteinuric kidney disease. J. Clin. Invest. 2007;117:2095–2104. doi: 10.1172/JCI32022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yaddanapudi S, et al. CD2AP in mouse and human podocytes controls a proteolytic program that regulates cytoskeletal structure and cellular survival. J. Clin. Invest. 2011;121:3965–3980. doi: 10.1172/JCI58552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shukha K, et al. Most ApoL1 is secreted by the liver. J. Am. Soc. Nephrol. 2016;28:1079–1083. doi: 10.1681/ASN.2016040441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu Y, et al. APOL1–G1 in nephrocytes induces hypertrophy and accelerates cell death. J. Am. Soc. Nephrol. 2017;28:1106–1116. doi: 10.1681/ASN.2016050550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma L, et al. APOL1 renal-risk variants induce mitochondrial dysfunction. J. Am. Soc. Nephrol. 2017;28:1093–1105. doi: 10.1681/ASN.2016050567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman DJ, Pollak MR. Apolipoprotein L1 and kidney disease in African Americans. Trends Endocrinol. Metab. 2016;27:204–215. doi: 10.1016/j.tem.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kruzel-Davila E, et al. APOL1-mediated cell injury involves disruption of conserved trafficking processes. J. Am. Soc. Nephrol. 2017;28:1117–1130. doi: 10.1681/ASN.2016050546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortese K, Sahores M, Madsen CD, Tacchetti C, Blasi F. Clathrin and LRP-1-independent constitutive endocytosis and recycling of uPAR. PLoS One. 2008;3:e3730. doi: 10.1371/journal.pone.0003730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Czekay RP, Kuemmel TA, Orlando RA, Farquhar MG. Direct binding of occupied urokinase receptor (uPAR) to LDL receptor-related protein is required for endocytosis of uPAR and regulation of cell surface urokinase activity. Mol. Biol. Cell. 2001;12:1467–1479. doi: 10.1091/mbc.12.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu CH, et al. Integrin-β3 clusters recruit clathrin-mediated endocytic machinery in the absence of traction force. Nat. Commun. 2015;6:8672. doi: 10.1038/ncomms9672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi N, et al. Podocyte injury-driven intracapillary plasminogen activator inhibitor type 1 accelerates podocyte loss via uPAR-mediated β1-integrin endocytosis. Am. J. Physiol. Renal Physiol. 2015;308:F614–F626. doi: 10.1152/ajprenal.00616.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Outinen TK, et al. Urine soluble urokinase-type plasminogen activator receptor levels correlate with proteinuria in Puumala hantavirus infection. J. Intern. Med. 2014;276:387–395. doi: 10.1111/joim.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gavrilovskaya IN, Shepley M, Shaw R, Ginsberg MH, Mackow ER. β3 Integrins mediate the cellular entry of hantaviruses that cause respiratory failure. Proc. Natl. Acad. Sci. USA. 1998;95:7074–7079. doi: 10.1073/pnas.95.12.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beckerman P, et al. Transgenic expression of human APOL1 risk variants in podocytes induces kidney disease in mice. Nat. Med. 2017;23:429–438. doi: 10.1038/nm.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel RS, et al. Circulating CD34+ progenitor cells and risk of mortality in a population with coronary artery disease. Circ. Res. 2015;116:289–297. doi: 10.1161/CIRCRESAHA.116.304187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eapen DJ, et al. Soluble urokinase plasminogen activator receptor level is an independent predictor of the presence and severity of coronary artery disease and of future adverse events. J. Am. Heart Assoc. 2014;3:e001118. doi: 10.1161/JAHA.114.001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levey AS, et al. A new equation to estimate glomerular filtration rate. Ann. Intern. Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee H, et al. Reducing agents affect inhibitory activities of compounds: results from multiple drug targets. Anal. Biochem. 2012;423:46–53. doi: 10.1016/j.ab.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saleem MA, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J. Am. Soc. Nephrol. 2002;13:630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 43.Wu MJ, et al. Rapamycin promotes podocyte migration through the up-regulation of urokinase receptor. Transplant. Proc. 2014;46:1226–1228. doi: 10.1016/j.transproceed.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 44.Li Y, et al. An anti-urokinase plasminogen activator receptor (uPAR) antibody: crystal structure and binding epitope. J. Mol. Biol. 2007;365:1117–1129. doi: 10.1016/j.jmb.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 45.Foster MC, et al. APOL1 variants associate with increased risk of CKD among African Americans. J. Am. Soc. Nephrol. 2013;24:1484–1491. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrell F. Regression Modeling Strategies: with Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis. Springer; 2015. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.