Abstract
Emerging and re-emerging viruses pose a significant public health challenge around the world, among which RNA viruses are the cause of many major outbreaks of infectious diseases. As one of the early lines of defense in the human immune system, RIG-I-like receptors (RLRs) play an important role as sentinels to thwart the progression of virus infection. The activation of RLRs leads to an antiviral state in the host cells, which triggers the adaptive arm of immunity and ultimately the clearance of viral infections. Hence, RLRs are promising targets for the development of pan-antivirals and vaccine adjuvants. Here, we discuss the opportunities and challenges of developing RLR agonists into antiviral therapeutic agents and vaccine adjuvants against a broad range of viruses.
Keywords: RIG-I-like receptor, pan-antivirals, vaccine adjuvants, interferon, interferon-stimulated genes, RNA therapeutics
Introduction
RNA viruses account for a third of all emerging and re-emerging infections (1). Due to the changes of abiotic and biotic landscape encountered by RNA viruses and the error-prone nature of viral replication, RNA viruses evolve quickly and contribute to the outbreak of infectious diseases (2). Many recent outbreaks of emerging and re-emerging viruses involve RNA viruses, and thus, there is an urgent need to develop antivirals against these viruses.
The innate immune system confronts viral infection via a specialized group of receptors known as pattern recognition receptors (PRRs) (3). Some PRRs recognize RNA viral infections including toll-like receptors 3 and 8 (TLRs), NOD-like receptors NLRP6 and 9 (NLRs), certain DDX/DHX helicases, and RLRs (retinoic acid inducible gene 1 (RIG-I) and melanoma differentiation associated protein 5 (MDA5)) (4–9). These PRRs usually activate interferon production and the secretion of pro-inflammatory cytokines (10). Interferon activates the Janus kinase/signal transducers and activators of transcription (JAK-STAT) signaling pathway in surrounding cells and the expression of interferon-stimulated genes (ISGs). ISGs inhibit virus replication and spread to surrounding cells by degrading viral nucleic acids and inhibiting viral gene expression (11, 12). Here, we focus on RLRs, the major sensors for pathogenic RNA species which trigger antiviral responses and discuss how modulation of RLRs may lead to broad-spectrum antivirals and new vaccine adjuvants.
RIG-I-Like Receptors
RIG-I-like receptors are a class of DExD/H box RNA helicases which recognizes double-stranded RNA (dsRNA) (13–17). RLRs consist of RIG-I, MDA5, and laboratory of genetics and physiology 2 (LGP2) (18). RIG-I and MDA5 have similar structural domains with N-terminal caspase activation and recruitment domains (CARDs), central helicase domain, and C-terminal domain, which recognizes viral RNA ligands (19–21). RIG-I recognizes short dsRNA and binds to blunt-ended RNA with 5′ triphosphate moiety (22–27). In contrast, MDA5 binds to the stem region of longer dsRNA in a cooperative manner (28–30). LGP2, on the other hand, only have the helicase and C-terminal domain and are involved in the regulatory function of RIG-I and MDA5 (31, 32).
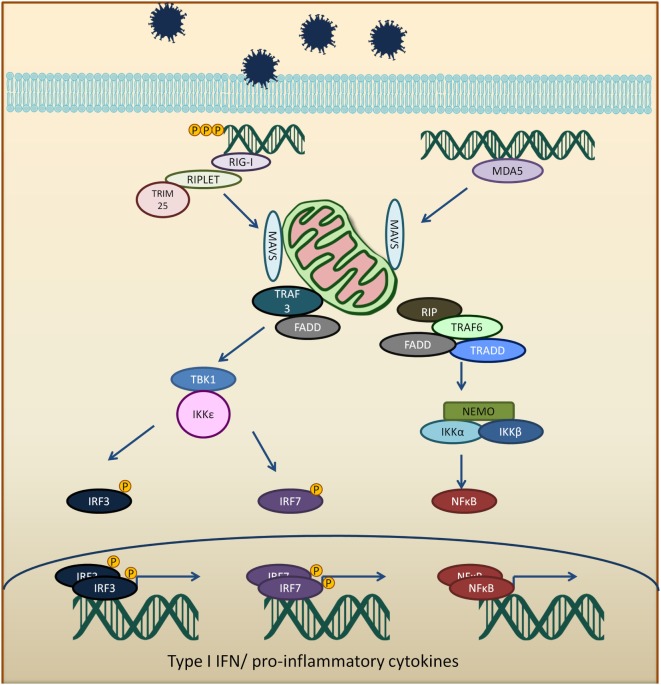
The CARD domains of RIG-I and MDA5 are involved in the activation of downstream signaling event via a protein known as mitochondria antiviral signaling protein (MAVS) (33–36). RIG-I binds to unanchored lysine-63 polyubiquitin chains and promotes efficient interaction with the CARD domain on MAVS (37, 38). MAVS protein polymerizes and forms fibrils when activated and will be polyubiquitinated and phosphorylated (38–42). The MAVS oligomer act as a platform to promote downstream antiviral signaling by recruiting several different proteins, such as tumor necrosis factor receptor type-1-associated death domain (TRADD), receptor interacting serine/threonine-protein kinase 1 (RIP1), Fas-associated protein with death domain (FADD), tumor necrosis factor receptor-associated factors (TRAF6, TRAF2, and TRAF3), as well as caspase 8 and caspase 10 (43, 44). TRAF3 activates TANK binding kinase 1/IκB kinase ε/IκB kinase γ/TANK (TBK1/IKKε/IKKγ/TANK) complex which phosphorylates and dimerizes interferon regulatory factors 3 and 7 (IRF3 and IRF7). The activated IRF3 and IRF7 translocate into the nucleus and activate IFN production (45, 46). TRAF 2 and 6 activate the IKKα/β/γ (also known as NEMO) by ubiquitination and resulting in activation of NFκB and the expression of pro-inflammatory cytokines (Figure 1) (41, 47).
Figure 1.
Viral RNA is recognized by RIG-I-like receptors (RLRs), RIG-I, or melanoma differentiation-associated protein 5 (MDA5). Activated RLRs interacts with mitochondria antiviral signaling protein (MAVS) adapter protein via CARD–CARD interactions. Activated MAVS then interacts with tumor necrosis factor receptor-associated factors 3 (TRAF3), tumor necrosis factor receptor-associated factors 6 (TRAF6), tumor necrosis factor receptor type-1-associated death domain (TRADD), receptor interacting serine/threonine-protein kinase 1 (RIP1), Fas-associated protein with death domain (FADD), and other signaling molecules. TRAF3 activates TANK binding kinase 1 (TBK1) and IκB kinase ε (IKKε), which phosphorylates interferon regulatory factors 3 and 7 (IRF3 and IRF7). The phosphorylated IRF3 and IRF7 dimerize and translocate into the nucleus to induce type 1 interferon response. On the other hand, MAVS interaction with receptor interacting serine/threonine-protein kinase 1, FADD, TRAF6, and TRADD. TRAF 6 ubiquitinate NF-kappa-B essential modulator (NEMO) which then activates IκB kinase and activates NF-κB. NF-κB transcription factor drives the expression of type 1 interferon and proinflammatory cytokines.
Pan-Antivirals Targeting RIG-I
Since RLRs are the key component for the antiviral immune response, these sensors are targets for antiviral therapeutics development. Current antiviral interventions focus on the use of direct-acting antivirals (DAAs), which target the essential components in the life cycle of a virus and thus are virus-specific (48). Although DAAs are highly effective, the low fidelity replication of the RNA virus genome could ultimately lead to the emergence of DAA therapies escape mutant (49). To circumvent this problem, broadly targeting antiviral therapeutics need to be used synergistically with DAAs. To this end, RIG-I agonists or RIG-I pathway activators represent a novel group of promising antiviral candidates. Lists of the antiviral candidates are discussed below as three categories based on their chemical nature (Table 1).
Table 1.
Pan-antivirals targeting RLRs.
| Pan-antivirals | Formula | Target pathway | Reference |
|---|---|---|---|
| Nucleotide-based | |||
| SB9200 | 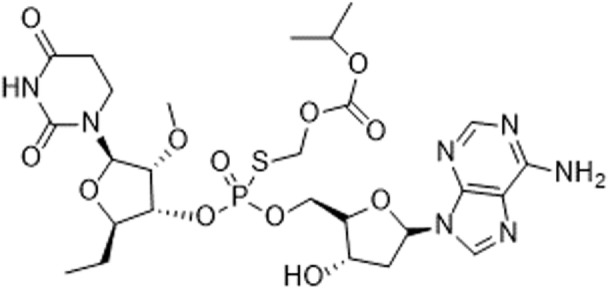 |
RLR and NLR | (50–52) |
| RNA-based | |||
| 5′ppp RNA derived from the 5′ and 3′ UTRs of the negative-strand RNA virus Vesicular Stomatitis Virus | GACGAAGACC ACAAAACCAG AUAAAAAAUA AAAUUUAAUG AUAAUAAUGG UUUGUUUGUC UUCGUC |
RLR | (53, 54) |
| 5′ppp RNA (M8) | GACGAAGACCACAAAACCAGAUAAAAAAAAAAAAAAAAAAAAAAAAAAUAAU UUUUUUUUUUUUUUUUUUUUUUUUUUAUCUGGUUUUGUGGUCUUCGUC |
RLR | (55) |
| 5′OH RNA with kink (CBS-13-BPS) | GGUAGACGAAACCAGAUAUAAUAUCUGGUUUCGUUUGCC | RIG-I, ISG56 | (56) |
| 5′PPP SLR | Stem loop RNA with the length of 10 and 14 base pair (GGACGUACGUUUC GACGUACGUCC) and (GGAUCGAUCGAUCGUUCGCGAUCGAUCGAUCC) |
RIG-I | (57) |
| Small molecular compounds | |||
| KIN 100 | 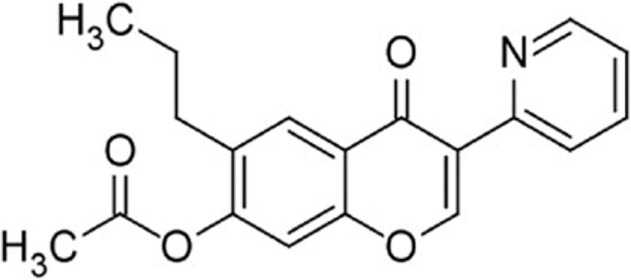 |
IRF 3 | (58) |
| KIN101 | 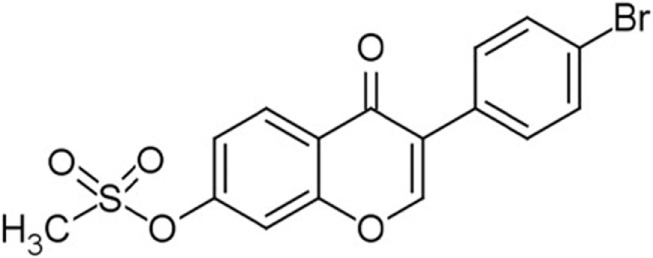 |
IRF3 | (58) |
| KIN 1000 | 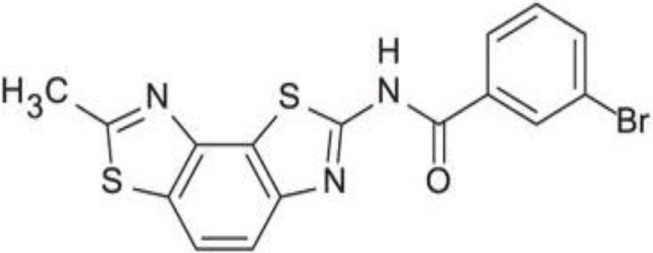 |
IRF 3 | (59) |
| KIN1400 | 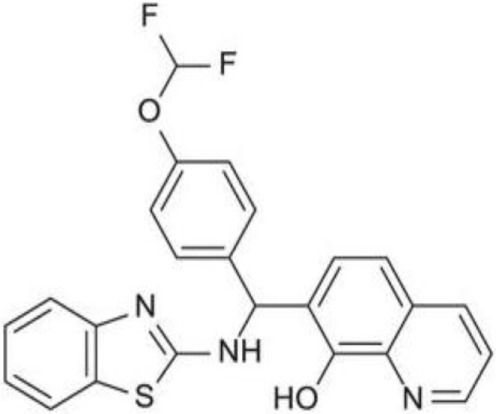 |
IRF 3 | (59) |
| KIN1408 | 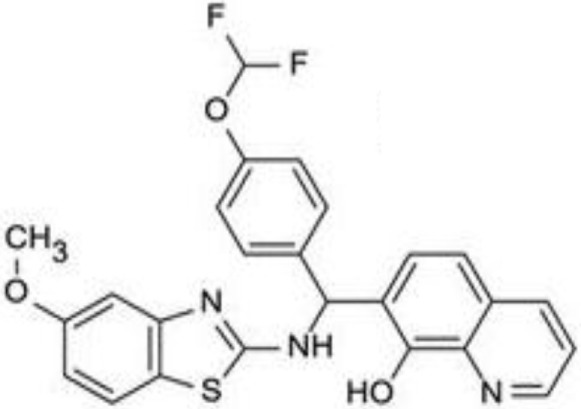 |
IRF3 | (59) |
| KIN1409 | 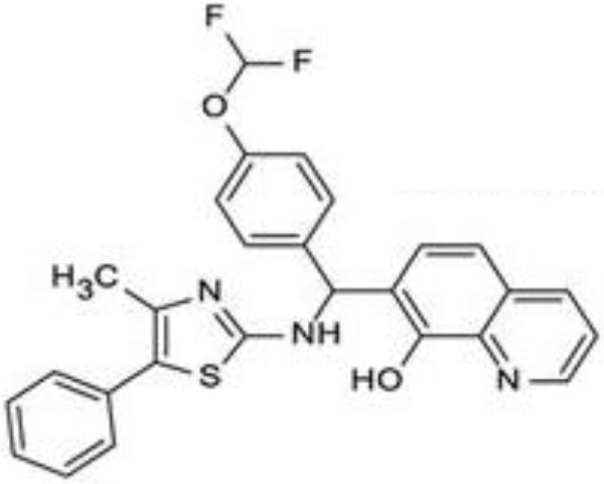 |
IRF3 | (59) |
RLRs, RIG-I-like receptors.
Nucleotide-Based Antivirals
A dinucleotide-derived small molecule compound, SB9200, has been shown to induce IFN via RIG-I and nucleotide-binding oligomerization domain containing protein 2 (NOD2). SB9200 is believed to interact with RIG-I and NOD2 that are associated with pre-genomic RNA thus blocking the HBV viral polymerase from replicating the genomic RNA (60). SB9200 was shown to confer dose-dependent and long-lasting induction of IFNα, IFNβ, and ISGs in liver tissue (50). Treatment of this compound in woodchucks infected with Woodchuck Hepatitis Virus (WHV) showed no sign of toxicity with reduced hepatic WHV antigen and nucleic acid. The sequential treatment of WHV-infected woodchuck with SB9200 followed by entecavir (ETV), a currently used antiviral to treat Hepatitis B (HBV), showed a reduction in viremia and delayed recrudescence of viral replication. The viral reduction from the treatment of SB9200 was comparable with current antivirals, such as Emtricitabine, Tenofovir, and Adefovir, when administered for up to 12 weeks (51). Drugs such as Emtricitabine, Tenofovir, and Adefovir are commonly used for the treatment of HBV. These drugs, however, may cause side effects such as lactic acidosis and possible liver, and kidney failure. SB9200 is effective for HCV patients with relapse after DAA and interferon treatment and could serve as a promising treatment option for patients who are not responding to the current regimen of DAA therapy (52). The phase 1 clinical trial on naïve adult with chronic hepatitis C showed an association between the decline in viral RNA and the peak of SB9200 detection in plasma (Clinical trial no NCT01803308). Currently, SB9200 is being tested in phase 2 clinical trials for treating subjects chronically infected with the HBV.
RNA-Based Antiviral Candidates
5′ triphosphorylated and diphosphorylated short dsRNAs are RIG-I specific ligands (22, 26, 61, 62). Goulet et al. showed that 5′pppRNA could activate a broad spectrum of antiviral and inflammatory genes such as IRF3, IRF7, NFkB, and the downstream ISGs. Treatment of lung epithelial cells A549 with 5′pppRNA confers protection against vesicular stomatitis virus (VSV), vaccinia virus, and dengue virus (DENV). The antiviral effect of 5′pppRNA was also detected against HIV in CD4+ T cells and HCV in Huh 7.5 cells (53). Besides that, 5′pppRNA was also an effective antiviral against influenza virus infection in vitro and in vivo. Treating mice with 5′pppRNA prior to influenza virus challenge also reduces pneumonia related to influenza virus infection (53). In another study, 5′pppRNA was shown to stimulate host antiviral response and reduce the infectivity of DENV and chikungunya virus (CHIKV) in human myeloid, fibroblast, and epithelial cells via RIG-I specific activation (54).
Several studies were also carried out to determine factors such as sequence, length, and structure of 5′pppRNA to enhance the antiviral activities of RIG-I (55, 56). Chiang et al. showed that 5′pppRNA with uridine-rich sequence with 99 nucleotides hairpin (M8) triggered higher interferon response when compared to other RIG-I aptamer and poly(I:C). M8 specifically activates RIG-I without triggering MDA5 or TLR3 activation. Prophylactic and therapeutic treatment using M8 protect cells from dengue and influenza viral infections. Furthermore, administration of M8 followed by influenza virus challenge improves the survival rate of mice with low lung virus titer detected at day 3 post-infection (55). In another study carried out by Lee et al., different RNA fold was shown to elicit different antiviral properties via RIG-I. Short hairpin RNA with a bent in the stem structure with phosphorothioate backbone was used as antiviral and was more potent than oseltamivir against influenza A H1N1 virus in vitro (56). Linehan et al. recently showed that short RNA with stable tetraloop at one end of duplex RNA triggers a robust IFN 1 response in vivo. These short stem-loop RNA (SLR) induces a subset of genes involved in antiviral and effector responses as well as represses gene involved in T cell maturation and could potentially be developed into a highly effective antiviral or vaccine adjuvant (57).
Small Molecular Compounds
High throughput screening (HTS) of small molecule compounds identified a group of novel agonists of the innate immune pathway. The isoflavone-like compound confers protection against HCV and Influenza A virus in vitro. These compounds were also shown to activate a narrower subset of genes and thus have potential to be useful antiviral without causing cytokine toxicity (58).
Another class of small molecule compounds, hydroxyquinolines, identified via HTS of compound library in cell culture induces the expression of innate immune antiviral genes such as RIG-I, IFIT1, IFIT2, IFITM1, OAS3, and MX1. Remarkably, although these compounds were able to induce high expression of antiviral genes, the expression of type I and III interferon remains low, suggesting the activation of distinct antiviral pathway than that of RIG-I agonists. The specific target(s) of these hydroxyquinoline compounds are not known. These hydroxyquinoline compounds were effective antivirals against a broad range of RNA viruses from the families Flaviviridae, Filoviridae, Paramyxoviridae, Arenaviridae, and Orthomyxoviridae. Interestingly, these compounds exhibit both prophylactic and therapeutic activity against infection and could be used in combination with other antivirals (59).
Innate Immune Potentiator as Vaccine Adjuvants
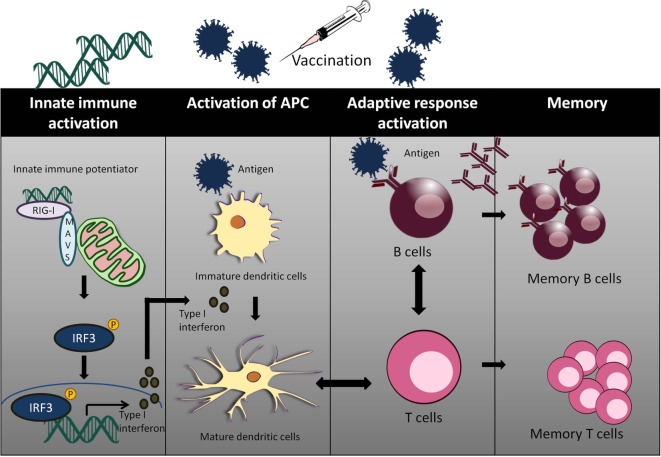
Adjuvants act as an immune enhancer to a vaccine. Several different classes of adjuvant had been approved for use in human vaccines such as alum and oil in emulsion MF59, AS03 (oil in water emulsion), virosomes, and AS04 (aluminum with monophosphoryl lipid A) (63). Alum and MF59 act by increasing antigen uptake at the injection site and activates pro-inflammatory responses (64–66). Alum mainly acts via Th2 cellular immune response, which does not confer the best protection for viral infections such as HCV and HIV. Moreover, there are safety concerns with the use of alum as an adjuvant with reported cases of hypersensitivity and erythema (67, 68). Well characterized agonists of innate immunity may serve as a better candidate of targeted vaccine adjuvants (Figure 2).
Figure 2.
The use of innate immune potentiator as adjuvant triggers the stimulation of adaptive immune responses. Innate immune potentiator stimulates RIG-I-like receptors (RIG-I) and interacts with MAVS adapter. This results in the activation of downstream signaling pathways and release of type I interferon. Type I interferon couple with the presence of antigen trigger DC maturation by enhancing surface marker expression and antigen presentation. The activated DCs interact with CD4+ T cells and thus stimulate Type 1T helper (TH1) cells. TH1 cells in turn interact with B cells to produce antibodies and trigger clonal expansion of B cells and T helper cells.
A small molecule compound named KIN 1148, discovered via HTS, was shown to activate IRF3 nuclear translocation. When this compound was tested with influenza split virus vaccine H1N1 A/California/07/2009, it confers protection from lethal challenge of influenza virus strain A/California/04/2009. KIN1148 together with the vaccine confers protection via IL-10 and Th-2 response to T cells in lung and lung-draining lymph nodes. Immunization with vaccine and KIN 1148 showed a significant increase in IgG antibodies with serum from mice receiving prime-boost immunization conferring protection to naïve mice from influenza challenge. KIN1148 was shown to be able to work alongside the vaccine to boost protective immunity and protect against influenza strain A/California/04/2009 (Table 2) (69).
Table 2.
Innate immune potentiator as virus vaccine adjuvants.
| Adjuvant | Target | Status | Virus vaccine | Reference |
|---|---|---|---|---|
KIN 1148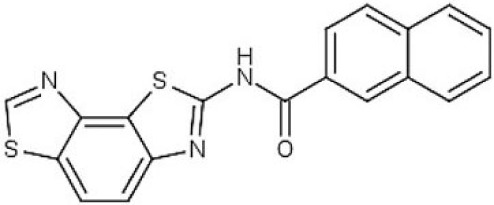 |
IRF3 | Laboratory testing | Influenza H1N1 A/California/07/2009 | (69) |
| M8 5′pppRNA GACGAAGACCACAAAACCAGAUAAA AAAAAAAAAAAAAAAAAAAAAAAUA AUUUUUUUUUUUUUUUUUUUUUU UUUUUAUCUGGUUUUGUGGUCUUCGUC |
RLR | Laboratory testing | H5N1 influenza | (70) |
| 5′pppRNA Derived from SeV DI RNA |
RLR | Laboratory testing | H1N1 2009 | (71) |
RLRs, RIG-I-like receptors.
5′triphosphorylated duplex RNA was tested as an adjuvant by Beljanski et al. M8 a potent triphosphorylated RNA was used in conjunction with virus-like particle (VLP) expressing H5N1 influenza hemagglutinin and neuraminidase. The combination of VLP and RNA increases the survival rate of mice infected with H5N1 influenza virus and induces higher antibody titer against influenza virus as compared to other adjuvants such as alum, addavax and poly(I:IC). Furthermore, vaccination with VLP and RNA stimulates TH1-biased CD4 T cells response in mouse sera (70). Another 5′triphosphorylated RNA derived from Sendai virus defective interfering RNA (SeV DI RNA) was also tested as adjuvant together with the H1N1 2009 pandemic vaccine and was shown to enhance production of influenza-specific IgG antibodies and influenza-specific IgA antibodies indicating that this 5′triphosphorylated RNA could potentially be used as influenza vaccine adjuvant (71).
Delivery of RNA-Based Agents Remains Challenging
To be effective as therapeutics, functional RNA species must be internalized into targeted cells. The delivery methods commonly used for RNA-based or nucleotide specific antivirals includes the lipid-oligo complexes, nanoparticle-based delivery, and viral-based delivery. The hydrophilic, negatively charge nature of RNA hinders the direct uptake of naked oligos into cells. The administration of RNA via inhalation was poorly efficacious in lung macrophages and only intratracheal administration leads to efficient delivery of RNA into the targeted site (72). Besides suffering from poor uptake, naked RNA is often prone to degradation in plasma (73).
One favored method of delivery for RNA is lipid-oligo complexes. This delivery method is more efficient due to the tendency of lipid to interact with the cell membrane and improve the uptake of RNA (74). A biocompatible lipid-based carrier can further reduce undesired immunogenic activation. However, the cationic nature of lipid is reported to interact with proteins in serum, and these aggregates are cleared by organs such as the spleen, lung and liver (75).
Nanoparticle-based delivery is a versatile method for RNA delivery with many organic or inorganic materials available as nanocarriers. Nanoparticle requires less RNA material with a large surface area for interaction with cells (76). The material used as a carrier could also be tailored for application such as dose, circulation time, as well as passive or active release of RNA (77). To overcome the issue of toxicity or uncontrolled immune activation, multistage delivery of RNA from nanoparticles could be carried put (78). The downside of this strategy is the need to test various materials for RNA delivery and this would drive up the cost of therapeutics or vaccine.
The Danger of Uncontrolled Immune Activation Always Exists
The therapeutic and prophylactic use of pan-antivirals was previously demonstrated in viral infection of influenza and dengue (55, 69, 71). Hotz et al. demonstrated that the pre-exposure of murine APC to synthetic poly(I:C) inhibits RLR activation while augmenting the sensitivity of TLRs. This would also imply a narrow therapeutic window for the use of pan-antiviral RNA targeting RIG-I (79). For clinical usage, the dosage of therapeutics is important to minimize side effects such as exacerbated cytokine storms and toxicity. Prater et al. showed that the injection of pregnant C57BL/6 mice with a high dose of CpG-ODN resulted in high fetal resorptions and craniofacial/limb defects (80). RIG-I agonists face similar concerns.
RIG-I SNPs may Lead to Poor or Hyper-Responsiveness
There are 324 RIG-I SNPs identified from NCBI SNPs database with 8 resulting in amino acid changes or truncation. The S183I mutation on RIG-I weakened the antiviral signaling and produces a low level of IFNβ and NFκB upon IAV and SeV challenges in a cell-based assay. Another SNP of RIG-I at P229 resulted in frameshift mutation at the CARDs domain and triggers constitutive expression of IFNβ suggesting that individual with this mutation could be linked to hyper-responsiveness in the immune system or autoimmune diseases (81). Another commonly found SNP (rs10813831) of RIG-I resulted in the substitution of R7C which could alter RIG-I interaction with MAVS (82). Individuals with these SNPs have a lower rubella-specific IgG titer when immunized with live measles-mumps-rubella (MMR-II) vaccine (83). This SNPs mutation also increased the IFNβ level and RIG-I transcription in human dendritic cells when infected with Newcastle disease virus (NDV) (82). Individuals with these SNPs were also shown to have complications of brainstem encephalitis when infected with enterovirus 71 (84). Several intronic SNPs also alter the cellular and humoral response to the measles vaccine. Genotype of individual carrying the SNP in minor allele of RIG-I (for rs12555727, rs12006123, and rs17289116) also showed less virus-specific IFN-γ secretion against measles. These findings imply that genetic variants are also involved in initial antiviral responses to vaccination (85) The haplotype of RIG-I rs3739674 which is located in the 5′UTR is associated with higher EV71 HFMD risk possibly by altering the expression level of the gene (86). In order to target RIG-I as pan-antiviral or vaccine adjuvant, the different haplotypes affecting the disease outcome should be considered. Dosage concern should be taken into account to enhance the effectiveness of RIG-I as a broadly targeting antiviral or vaccine adjuvant.
Conclusion
Emerging and re-emerging viruses present a significant public health concern, and there is an urgent need for novel vaccination and treatment strategies. RIG-I agonists as new adjuvant candidates may work alone or couple to vaccine agents such as VLPs or recombinant proteins to improve the safety and efficacy of conventional vaccines. Antivirals targeting the innate arm of immunity (host-directed therapy) would be useful to confer protection against emerging and re-emerging viruses (87). However, the development of such vaccines and antivirals is still in its infancy and many challenges related to the production and safety evaluation of vaccines and antivirals. Several key issues still need to be addressed including production platform, formulation, delivery, safety, and the ability of such class of the vaccine adjuvant or antivirals to be used in immunocompromised and elderly.
Author Contributions
DL and HY discussed, wrote, and revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by (1) a start-up grant from Lee Kong Chian School of Medicine, Nanyang Technological University, (2) MOE2016-T2-2-097 grant, and (3) National Research Foundation grant NRF2016NRF-CRP001-063.
References
- 1.Howard CR, Fletcher NF. Emerging virus diseases: can we ever expect the unexpected? Emerg Microbes Infect (2012) 1:e46. 10.1038/emi.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nichol ST, Arikawa J, Kawaoka Y. Emerging viral diseases. Proc Natl Acad Sci U S A (2000) 97:12411–2. 10.1073/pnas.210382297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol (2012) 13:214–22. 10.1038/ni.2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol (2006) 27:352–7. 10.1016/j.it.2006.06.003 [DOI] [PubMed] [Google Scholar]
- 5.Higgins SC, Mills KH. TLR, NLR agonists, and other immune modulators as infectious disease vaccine adjuvants. Curr Infect Dis Rep (2010) 12:4–12. 10.1007/s11908-009-0080-9 [DOI] [PubMed] [Google Scholar]
- 6.Hornung V. SnapShot: nucleic acid immune sensors, part 1. Immunity (2014) 41:868, 868.e1. 10.1016/j.immuni.2014.10.005 [DOI] [PubMed] [Google Scholar]
- 7.Hornung V. SnapShot: nucleic acid immune sensors, part 2. Immunity (2014) 41:1066–1066.e1. 10.1016/j.immuni.2014.10.006 [DOI] [PubMed] [Google Scholar]
- 8.Kumar H, Kawai T, Akira S. Pathogen recognition in the innate immune response. Biochem J (2009) 420:1–16. 10.1042/BJ20090272 [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Chen ZJ. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol (2014) 32:461–88. 10.1146/annurev-immunol-032713-120156 [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science (2010) 327:291–5. 10.1126/science.1183021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borden EC, Williams BR. Interferon-stimulated genes and their protein products: what and how? J Interferon Cytokine Res (2011) 31:1–4. 10.1089/jir.2010.0129 [DOI] [PubMed] [Google Scholar]
- 12.Schoggins JW, Rice CM. Interferon-stimulated genes and their antiviral effector functions. Curr Opin Virol (2011) 1:519–25. 10.1016/j.coviro.2011.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruns AM, Horvath CM. Activation of RIG-I-like receptor signal transduction. Crit Rev Biochem Mol Biol (2012) 47:194–206. 10.3109/10409238.2011.630974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loo YM, Gale M, Jr. Immune signaling by RIG-I-like receptors. Immunity (2011) 34:680–92. 10.1016/j.immuni.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo D. Toward a crystal-clear view of the viral RNA sensing and response by RIG-I-like receptors. RNA Biol (2014) 11:25–32. 10.4161/rna.27717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo D, Kohlway A, Pyle AM. Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol (2013) 10:111–20. 10.4161/rna.22706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlee M. Master sensors of pathogenic RNA – RIG-I like receptors. Immunobiology (2013) 218(11):1322–35. 10.1016/j.imbio.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol (2005) 175:2851–8. 10.4049/jimmunol.175.5.2851 [DOI] [PubMed] [Google Scholar]
- 19.Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M, Jr, Patel SS, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature (2011) 479:423–7. 10.1038/nature10537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell (2011) 147:423–35. 10.1016/j.cell.2011.09.039 [DOI] [PubMed] [Google Scholar]
- 21.Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell (2011) 147:409–22. 10.1016/j.cell.2011.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, et al. 5’-Triphosphate RNA is the ligand for RIG-I. Science (2006) 314:994–7. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- 23.Kohlway A, Luo D, Rawling DC, Ding SC, Pyle AM. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep (2013) 14:772–9. 10.1038/embor.2013.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science (2006) 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- 25.Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, et al. Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity (2009) 31:25–34. 10.1016/j.immuni.2009.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, et al. 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A (2009) 106:12067–72. 10.1073/pnas.0900971106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, Yong HY, Panutdaporn N, Liu C, Tang K, Luo D. High-resolution HDX-MS reveals distinct mechanisms of RNA recognition and activation by RIG-I and MDA5. Nucleic Acids Res (2015) 43:1216–30. 10.1093/nar/gku1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, et al. Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci U S A (2012) 109:E3340–9. 10.1073/pnas.1208618109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, et al. Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci U S A (2011) 108:21010–5. 10.1073/pnas.1113651108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, et al. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell (2013) 152:276–89. 10.1016/j.cell.2012.11.048 [DOI] [PubMed] [Google Scholar]
- 31.Bruns AM, Horvath CM. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine (2015) 74:198–206. 10.1016/j.cyto.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, et al. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A (2010) 107:1512–7. 10.1073/pnas.0912986107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol (2005) 6:981–8. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- 34.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature (2005) 437:1167–72. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- 35.Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell (2005) 122:669–82. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- 36.Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell (2005) 19:727–40. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- 37.Oshiumi H, Matsumoto M, Seya T. Ubiquitin-mediated modulation of the cytoplasmic viral RNA sensor RIG-I. J Biochem (2012) 151:5–11. 10.1093/jb/mvr111 [DOI] [PubMed] [Google Scholar]
- 38.Wu B, Peisley A, Tetrault D, Li Z, Egelman EH, Magor KE, et al. Molecular imprinting as a signal-activation mechanism of the viral RNA sensor RIG-I. Mol Cell (2014) 55:511–23. 10.1016/j.molcel.2014.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell (2011) 146:448–61. 10.1016/j.cell.2011.06.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science (2015) 347:aaa2630. 10.1126/science.aaa2630 [DOI] [PubMed] [Google Scholar]
- 41.Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife (2013) 2:e00785. 10.7554/eLife.00785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peisley A, Wu B, Xu H, Chen ZJ, Hur S. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature (2014) 509:110–4. 10.1038/nature13140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shu HB, Takeuchi M, Goeddel DV. The tumor necrosis factor receptor 2 signal transducers TRAF2 and c-IAP1 are components of the tumor necrosis factor receptor 1 signaling complex. Proc Natl Acad Sci U S A (1996) 93:13973–8. 10.1073/pnas.93.24.13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang D, Lin J, Han J. Receptor-interacting protein (RIP) kinase family. Cell Mol Immunol (2010) 7:243–9. 10.1038/cmi.2010.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, et al. A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res (2011) 21:895–910. 10.1038/cr.2011.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J (1998) 17:1087–95. 10.1093/emboj/17.4.1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol (2009) 27:693–733. 10.1146/annurev.immunol.021908.132641 [DOI] [PubMed] [Google Scholar]
- 48.Halfon P, Sarrazin C. Future treatment of chronic hepatitis C with direct acting antivirals: is resistance important? Liver Int (2012) 32(Suppl 1):79–87. 10.1111/j.1478-3231.2011.02716.x [DOI] [PubMed] [Google Scholar]
- 49.Asselah T, Marcellin P. Interferon free therapy with direct acting antivirals for HCV. Liver Int (2013) 33(Suppl 1):93–104. 10.1111/liv.12076 [DOI] [PubMed] [Google Scholar]
- 50.Korolowicz KE, Iyer RP, Czerwinski S, Suresh M, Yang J, Padmanabhan S, et al. Antiviral efficacy and host innate immunity associated with SB 9200 treatment in the woodchuck model of chronic hepatitis B. PLoS One (2016) 11:e0161313. 10.1371/journal.pone.0161313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suresh M, Korolowicz KE, Balarezo M, Iyer RP, Padmanabhan S, Cleary D, et al. Antiviral efficacy and host immune response induction during sequential treatment with SB 9200 followed by entecavir in woodchucks. PLoS One (2017) 12:e0169631. 10.1371/journal.pone.0169631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones M, Cunningham ME, Wing P, DeSilva S, Challa R, Sheri A, et al. SB 9200, a novel agonist of innate immunity, shows potent antiviral activity against resistant HCV variants. J Med Virol (2017) 89:1620–8. 10.1002/jmv.24809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, et al. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog (2013) 9:e1003298. 10.1371/journal.ppat.1003298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olagnier D, Scholte FE, Chiang C, Albulescu IC, Nichols C, He Z, et al. Inhibition of dengue and chikungunya virus infections by RIG-I-mediated type I interferon-independent stimulation of the innate antiviral response. J Virol (2014) 88:4180–94. 10.1128/JVI.03114-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiang C, Beljanski V, Yin K, Olagnier D, Ben Yebdri F, Steel C, et al. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J Virol (2015) 89:8011–25. 10.1128/JVI.00845-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Park EB, Min J, Sung SE, Jang Y, Shin JS, et al. Systematic editing of synthetic RIG-I ligands to produce effective antiviral and anti-tumor RNA immunotherapies. Nucleic Acids Res (2018) 46(4):1635–47. 10.1093/nar/gky039 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57.Linehan MM, Dickey TH, Molinari ES, Fitzgerald ME, Potapova O, Iwasaki A, et al. A minimal RNA ligand for potent RIG-I activation in living mice. Sci Adv (2018) 4:e1701854. 10.1126/sciadv.1701854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bedard KM, Wang ML, Proll SC, Loo YM, Katze MG, Gale M, Jr, et al. Isoflavone agonists of IRF-3 dependent signaling have antiviral activity against RNA viruses. J Virol (2012) 86:7334–44. 10.1128/JVI.06867-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pattabhi S, Wilkins CR, Dong R, Knoll ML, Posakony J, Kaiser S, et al. Targeting innate immunity for antiviral therapy through small molecule agonists of the RLR pathway. J Virol (2015) 90:2372–87. 10.1128/JVI.02202-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sato S, Li K, Kameyama T, Hayashi T, Ishida Y, Murakami S, et al. The RNA sensor RIG-I dually functions as an innate sensor and direct antiviral factor for hepatitis B virus. Immunity (2015) 42:123–32. 10.1016/j.immuni.2014.12.016 [DOI] [PubMed] [Google Scholar]
- 61.Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, et al. Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature (2014) 514:372–5. 10.1038/nature13590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kato H, Takahasi K, Fujita T. RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev (2011) 243:91–8. 10.1111/j.1600-065X.2011.01052.x [DOI] [PubMed] [Google Scholar]
- 63.Lee S, Nguyen MT. Recent advances of vaccine adjuvants for infectious diseases. Immune Netw (2015) 15:51–7. 10.4110/in.2015.15.2.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dupuis M, Murphy TJ, Higgins D, Ugozzoli M, van Nest G, Ott G, et al. Dendritic cells internalize vaccine adjuvant after intramuscular injection. Cell Immunol (1998) 186:18–27. 10.1006/cimm.1998.1283 [DOI] [PubMed] [Google Scholar]
- 65.Morefield GL, Sokolovska A, Jiang D, HogenEsch H, Robinson JP, Hem SL. Role of aluminum-containing adjuvants in antigen internalization by dendritic cells in vitro. Vaccine (2005) 23:1588–95. 10.1016/j.vaccine.2004.07.050 [DOI] [PubMed] [Google Scholar]
- 66.Mosca F, Tritto E, Muzzi A, Monaci E, Bagnoli F, Iavarone C, et al. Molecular and cellular signatures of human vaccine adjuvants. Proc Natl Acad Sci U S A (2008) 105:10501–6. 10.1073/pnas.0804699105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Clements CJ, Griffiths E. The global impact of vaccines containing aluminium adjuvants. Vaccine (2002) 20(Suppl 3):S24–33. 10.1016/S0264-410X(02)00168-8 [DOI] [PubMed] [Google Scholar]
- 68.Trollfors B, Bergfors E, Inerot A. Vaccine related itching nodules and hypersensitivity to aluminium. Vaccine (2005) 23:975–6. 10.1016/j.vaccine.2004.08.024 [DOI] [PubMed] [Google Scholar]
- 69.Probst P, Grigg JB, Wang M, Munoz E, Loo YM, Ireton RC, et al. A small-molecule IRF3 agonist functions as an influenza vaccine adjuvant by modulating the antiviral immune response. Vaccine (2017) 35:1964–71. 10.1016/j.vaccine.2017.01.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Beljanski V, Chiang C, Kirchenbaum GA, Olagnier D, Bloom CE, Wong T, et al. Enhanced influenza virus-like particle vaccination with a structurally optimized RIG-I agonist as adjuvant. J Virol (2015) 89:10612–24. 10.1128/JVI.01526-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martinez-Gil L, Goff PH, Hai R, Garcia-Sastre A, Shaw ML, Palese P. A Sendai virus-derived RNA agonist of RIG-I as a virus vaccine adjuvant. J Virol (2013) 87:1290–300. 10.1128/JVI.02338-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moschos SA, Frick M, Taylor B, Turnpenny P, Graves H, Spink KG, et al. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol Ther (2011) 19:2163–8. 10.1038/mt.2011.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Deering RP, Kommareddy S, Ulmer JB, Brito LA, Geall AJ. Nucleic acid vaccines: prospects for non-viral delivery of mRNA vaccines. Expert Opin Drug Deliv (2014) 11:885–99. 10.1517/17425247.2014.901308 [DOI] [PubMed] [Google Scholar]
- 74.Xue HY, Guo P, Wen WC, Wong HL. Lipid-based nanocarriers for RNA delivery. Curr Pharm Des (2015) 21:3140–7. 10.2174/1381612821666150531164540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simoes S, Filipe A, Faneca H, Mano M, Penacho N, Duzgunes N, et al. Cationic liposomes for gene delivery. Expert Opin Drug Deliv (2005) 2:237–54. 10.1517/17425247.2.2.237 [DOI] [PubMed] [Google Scholar]
- 76.Miele E, Spinelli GP, Miele E, Di Fabrizio E, Ferretti E, Tomao S, et al. Nanoparticle-based delivery of small interfering RNA: challenges for cancer therapy. Int J Nanomedicine (2012) 7:3637–57. 10.2147/IJN.S23696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan JM, Zhang L, Tong R, Ghosh D, Gao W, Liao G, et al. Spatiotemporal controlled delivery of nanoparticles to injured vasculature. Proc Natl Acad Sci U S A (2010) 107:2213–8. 10.1073/pnas.0914585107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tasciotti E, Liu XW, Bhavane R, Plant K, Leonard AD, Price BK, et al. Mesoporous silicon particles as a multistage delivery system for imaging and therapeutic applications. Nat Nanotechnol (2008) 3:151–7. 10.1038/nnano.2008.34 [DOI] [PubMed] [Google Scholar]
- 79.Hotz C, Roetzer LC, Huber T, Sailer A, Oberson A, Treinies M, et al. TLR and RLR signaling are reprogrammed in opposite directions after detection of viral infection. J Immunol (2015) 195:4387–95. 10.4049/jimmunol.1500079 [DOI] [PubMed] [Google Scholar]
- 80.Prater MR, Johnson VJ, Germolec DR, Luster MI, Holladay SD. Maternal treatment with a high dose of CpG ODN during gestation alters fetal craniofacial and distal limb development in C57BL/6 mice. Vaccine (2006) 24:263–71. 10.1016/j.vaccine.2005.07.105 [DOI] [PubMed] [Google Scholar]
- 81.Pothlichet J, Burtey A, Kubarenko AV, Caignard G, Solhonne B, Tangy F, et al. Study of human RIG-I polymorphisms identifies two variants with an opposite impact on the antiviral immune response. PLoS One (2009) 4:e7582. 10.1371/journal.pone.0007582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu J, Nistal-Villan E, Voho A, Ganee A, Kumar M, Ding Y, et al. A common polymorphism in the caspase recruitment domain of RIG-I modifies the innate immune response of human dendritic cells. J Immunol (2010) 185:424–32. 10.4049/jimmunol.0903291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ovsyannikova IG, Haralambieva IH, Dhiman N, O’Byrne MM, Pankratz VS, Jacobson RM, et al. Polymorphisms in the vitamin A receptor and innate immunity genes influence the antibody response to rubella vaccination. J Infect Dis (2010) 201:207–13. 10.1086/649588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pang L, Gong X, Liu N, Xie G, Gao W, Kong G, et al. A polymorphism in melanoma differentiation-associated gene 5 may be a risk factor for enterovirus 71 infection. Clin Microbiol Infect (2014) 20:O711–7. 10.1111/1469-0691.12618 [DOI] [PubMed] [Google Scholar]
- 85.Haralambieva IH, Ovsyannikova IG, Umlauf BJ, Vierkant RA, Shane Pankratz V, Jacobson RM, et al. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine (2011) 29:8988–97. 10.1016/j.vaccine.2011.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li YP, Li M, Jia XL, Deng HL, Wang WJ, Wu FP, et al. Association of gene polymorphisms of pattern-recognition receptor signaling pathway with the risk and severity of hand, foot, and mouth disease caused by enterovirus 71 in Chinese Han population. J Med Virol (2018) 90:692–8. 10.1002/jmv.25000 [DOI] [PubMed] [Google Scholar]
- 87.Kaufmann SHE, Dorhoi A, Hotchkiss RS, Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov (2018) 17:35–56. 10.1038/nrd.2017.162 [DOI] [PMC free article] [PubMed] [Google Scholar]