Abstract
We isolated 65 rhizobial strains from faba bean (Vicia faba L.) from Panxi, China, studied their plant growth promoting ability with nitrogen free hydroponics, genetic diversity with clustered analysis of combined ARDRA and IGS-RFLP, and phylogeny by sequence analyses of 16S rRNA gene, three housekeeping genes and symbiosis related genes. Eleven strains improved the plant shoot dry mass significantly comparing to that of not inoculated plants. According to the clustered analysis of combined ARDRA and IGS-RFLP the isolates were genetically diverse. Forty-one of 65 isolates represented Rhizobium anhuiense, and the others belonged to R. fabae, Rhizobium vallis, Rhizobium sophorae, Agrobacterium radiobacter, and four species related to Rhizobium and Agrobacterium. The isolates carried four and five genotypes of nifH and nodC, respectively, in six different nifH-nodC combinations. When looking at the species-nifH-nodC combinations it is noteworthy that all but two of the six R. anhuiense isolates were different. Our results suggested that faba bean rhizobia in Panxi are diverse at species, plant growth promoting ability and symbiosis related gene levels.
Keywords: faba bean, rhizobia, genetic diversity, multilocus sequence analysis, symbiosis gene, lateral gene transfer
Introduction
Legumes like faba bean (Vicia faba L.) and rhizobial bacteria can form a symbiotic relationship in which the legume host provides the rhizobia with nutrients and niche while rhizobia provide the host with fixed atmospheric dinitrogen in the form of ammonia. Owing to symbiosis, legumes can act as pioneer plants in nitrogen deficient areas and improve soil fertility (Graham and Vance, 2003; Gentzbittel et al., 2015). Nitrogen fertilization affects the environment; however, applying biological N fixation (BNF) has some advantages over synthetic N fertilizers. If incorporated into the soil, legumes do not acidify the soil like ammonium-based fertilizers (Crews and Peoples, 2004). Unlike the production of synthetic N fertilizers, BNF does not rely on non-renewable energy sources (Crews and Peoples, 2004).
In legumes, nitrogen fixation takes place in a specific root or stem organ called nodule. The formation of plant growth promoting symbiosis requires that the legume and the rhizobia are compatible, and that the rhizobia fix nitrogen efficiently. Inoculating the legume with suitable rhizobia increases growth when compatible rhizobia are not present or when the compatible rhizobia are not efficient (Thilakarathna and Raizada, 2017).
Faba bean, a grain legume grown worldwide, is a good resource of protein, starch, cellulose and minerals. Its high yield and great adaption to different environments makes faba bean very popular among farmers, feed and food manufacturers (Haciseferoǧullar et al., 2003). Moreover, the capacity for biological nitrogen fixation with rhizobial bacteria makes faba bean a renewable resource for sustainable agriculture (Köpke and Nemecek, 2010). Thus, it is common that faba bean is grown as an intercrop or in rotation with non-legume plants (Song et al., 2007; Mei et al., 2012). However, in China faba bean frequently receive synthetic N fertilizer, resulting in over fertilization (Li et al., 2016).
Panxi region in Sichuan, southwestern China, is on the western margin of Yangtze Block, between Tibet Plateau, Yunnan-Guizhou Plateau and Sichuan basin. Panxi is within the South-West China mountains biodiversity hotspot (Wu et al., 2006; www.cepf.net/resources/hotspots/Asia-Pacific/Pages/Mountains-of-Southwest-China.aspx). Mountains occupy 80% of the total area of Panxi and the altitude differences in this area reach 5,600 m. Panxi receives plenty of rainfall and strong solar radiation, and the climate ranges from southern Asian semitropical climate to northern temperate climate with xerothermic climate as the main characteristic of the arid-hot river valley area. Faba bean is one of the main crops in Panxi. Cultivation relies on seeds produced by farmers themselves. N fertilizers would be unnecessary if the soils hosted compatible, plant growth promoting rhizobia.
The species range of rhizobia nodulating legumes in Panxi differs from that in other parts of China. For example, in Panxi Leucaena leucocaphala and Pueraria lobate were mostly nodulated by Ensifer and Rhizobium strains, respectively, while in subtropical China L. leucocaphala was nodulated by Mesorhizobium strains and in other parts of Sichuan P. lobate by Bradyrhizobium strains (Chen et al., 2004; Wang et al., 2006; Xie et al., 2009; Xu et al., 2013). Since faba bean rhizobia in Panxi have not been studied systematically prior to this study, our aim was to assess if faba bean rhizobia in the area were diverse and unique. Thus, we isolated rhizobia from faba bean growing in the special arid-hot environment of Panxi in diverse soil types, and studied their plant growth promoting ability, genetic diversity and phylogeny based on molecular methods.
Materials and methods
Isolation of strains
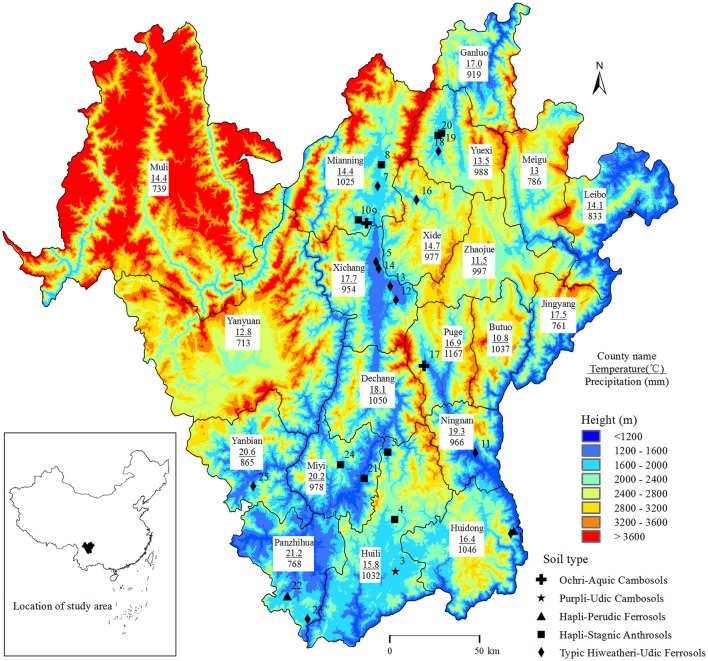
Local variety faba bean samples were taken in 25 sites in Panxi, Sichuan, China (Figure 1) to collect root nodules. Nodules were surface sterilized in 95% ethanol for 3 min and 0.1% HgCl2 for 5 min, followed by rinsing six times in sterile distilled water. The sterilized nodules were crushed individually and streaked on yeast extract mannitol (YEM) medium (Vincent, 1970) containing 25 mg L−1 congo red at 28°C. The purified strains were stored on YEM slants at 4°C for short term and in 25% glycerol at −80°C for long term storage.
Figure 1.
The 25 sampling sites in Panxi. The location of Panxi in China is shown in the inset. The maps were drawn using ArcGIS 10.0 software. Temperature and precipitation refer to the average annual values.
Nodulation assays
The nodulation ability and symbiotic efficiency of the isolates was tested on the local faba bean (V. faba L.) cultivar Hanyuan dabaidou. Seeds of faba bean were immersed in 95% ethanol for 5 min, rinsed for 5 min with 0.2% mercury bichloride (HgCl2) and 8 times (10 min per time) with sterilized water. After surface sterilization, the seeds were soaked in sterilized water overnight to soften the thick and hard seed coat. The seeds were transferred on 0.5% water-agar for germination. The seedlings were transplanted in sterile 250 ml infusion bottles containing Jensen's solution (Vincent, 1970) in all inoculation assays. The seedlings were inoculated with 1.5 ml of the culture containing ca 109 bacterial cells per milliliter and grown under a 16 h light and 8 h dark regime at 25°C in greenhouse. The assays were done in triplicate with one seedling per bottle, including the uninoculated controls. After 50 days, the plants were harvested and the numbers of nodules and the plant shoot dry mass were measured. One-way analysis of variance with a least significant difference (LSD) analysis (P = 0.05) was done using Excel 2010 (Microsoft, Redmond, USA) and SPSS 17.0 (SPSS Inc., Chicago, USA).
PCR-RFLP and CACAI
Total DNA was extracted by GUTC (Guanidinium-Tris-CDTA buffer with celite) method (Terefework et al., 2001) from purified bacteria. 16S rDNA and intergenic spacer region (IGS) of the strains were amplified for restriction fragment length polymorphism analysis. Primer pairs P1, P6 and pHr(F), p23SR01(R) (Table 1) were used for polymerase chain reaction (PCR) amplification. Amplification products (5 μl) were digested separately by four restriction enzymes HinfI, TaqI, MspI, and HaeIII following the manufacturer's instructions (Fermentas, EU). The fragments were separated by gel electrophoreses in 2% agarose with 0.5 μg ml−1 ethidium bromide at 80 V for 3 h and photographed. Amplified ribosomal DNA restriction analysis (ARDRA) and IGS-RFLP were done by combining the results from the four restrictions. Clustered analysis of combined ARDRA and IGS-RFLP (CACAI) was conducted by UPGM clustering algorithm in the NTSYS program (Rohlf, 1990).
Table 1.
PCR primers and reaction procedures applied in this study.
| Gene | Primers | Reaction procedure | References |
|---|---|---|---|
| 16S rDNA | P1:5′-AGAGTTTGATCCTGGCTCAGAACGAACGCT-3′; P6: 5′- TACGGCTACCTTGTTACGACTTCACCCC-3′ | 92°C for 3 min, 30 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 2 min, final extension for 72°C 8 min | Tan et al., 1997 |
| IGS | pHr (F):5′-TGCGGCTGGATCACCTCCTT-3′; p23SR01(R):5′-GGCTGC TTCTAAGCCAAC-3′ | 92°C for 3 min, 30 cycles of 94°C for 1 min, 60°C for 1 min, 72°C for 2 min, final extension for 72°C 8 min | Navarro et al., 1992; Massol-Deya et al., 1995 |
| atpD | atpD255F: 5′-GCTSGGCCGCATCMTSAACGTC-3′; atpD782R: 5′-GCCGACACTTCMGAACCNGCCTG-3′ | 95°C for 2 min; 30 cycles of 94°C for 45 s, 59°C for 1 min, 72°C for 1.5 min; final extension 72°C for 10 min | Vinuesa et al., 2005 |
| glnII | glnII12F:YAAGCTCGAGTACATYTGGCT; glnII689R: TGCATGCCSGAGCCGTTCCA | 95°C for 5 min; 30 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 1 min; final extension 72°C for 10 min | Vinuesa et al., 2005 |
| recA | recA 41F: 5′-TTCGGCAAGGGMTCGRTSATG-3′; recA 640R: 5′-ACATSACRCCGATCTTCATGC-3′ | 95°C for 5 min; 30 cycles of 94°C for 1 min, 58°C for 1 min, 72°C for 1 min; final extension 72°C for 10 min | Vinuesa et al., 2005 |
| nifH | nifHctg:5′-CTCATCGTCGGCTGTGACCC-3′; nifHI: 5′-AGCATGTCYTCSAGYTCNTCCA-3′ | 95°C for 3 min; 40 cycles of 94°C for 1 min, 59°C for 1 min, 72°C for 1 min; final extension 72°C for 5 min | Laguerre et al., 2001; Gurkanli et al., 2014 |
| nifH1F:5′-GTCTCCTATGACGTGCTCGG-3′; nifH1R:5′-GCTTCCATGGTGATCGGGGT-3′ | 94°C for 3 min; 30 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 1 min; final extension 72°C for 5 min | Rivas et al., 2002 | |
| nodC | nodC540: 5′-TGATYGAYATGGARTAYTGGYT-3′; nodC1160: 5′-CGYGACAGCCANTCKCTATTG-3′ | 95°C for 5 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 1 min; final extension 72°C for 10 min | Sarita et al., 2005 |
| nofCf: 5′-GCTGCCTATGCAGACGATG-3′; nodCr: 5′-GGTTACTGGCTTTCATTTGGC-3′ | 94°C for 5 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, 72°C for 3 min; final extension 72°C for 7 min | Moschetti et al., 2005 |
Sequencing of housekeeping and symbiotic genes
According to the results of CACAI, representative strains were selected for sequencing of housekeeping and symbiotic genes. To facilitate the comparison of faba bean nodulating diversity in Panxi and other parts of Sichuan, we applied the same methods as in our earlier study on rhizobia from Sichuan hilly areas (Xu et al., 2015). 16S rDNA was amplified as described above. Three housekeeping genes atpD, glnII, and recA and two symbiotic genes nifH and nodC were amplified as described in Table 1. The PCR products were sequenced directly at BGI Tech (Shenzhen, China). Sequences have been deposited to NCBI (National Center for Biotechnology Information research database) nucleotide database under the accession numbers of KU947312-KU947400.
The sequences of the housekeeping and symbiotic genes were compared with sequences in NCBI, and the 16S rDNA sequences were compared with sequences in EzTaxon (http://www.ezbiocloud.net/) using BLASTN. Phylogenetic analyses of sequences from our isolates and reference sequences from databases were done using a Neighbor-Joining method in MEGA 6.0 (Tamura et al., 2013) with 1,000 bootstrapped replicates. Genospecies were defined by multilocus sequence analysis (MLSA) using concatenated sequence of three housekeeping genes applying 97% average nucleotide identity as the threshold (Cao et al., 2014).
Results
Nodulation, plant growth promoting ability, and genetic diversity of faba bean isolates
We isolated 65 strains from root nodules of faba bean growing in Panxi, China (Table 2). All but two of the strains formed nodules on the roots of faba bean with the average nodule numbers ranging from 3.0 to 98.5 per plant. No nodules were detected on the roots of the uninoculated plants (Table 2). The plant growth promoting ability of the isolates was assessed by measuring the dry masses of the inoculated plants. The eleven strains that significantly increased the plant shoot dry mass (p < 0.05) were considered as potential inoculant strains (Table 3).
Table 2.
Rhizobial isolates from faba bean in Panxi, their genetic and symbiotic characteristics and phylogenetic affiliation.
| Isolatea | Sampling siteb | 16S rDNA RFLP genotypec | IGS RFLP genotypec | CACAI genotype | CACAI groupd | MLSAe | Plant shoot dry mass (g plant−1)f | No. of nodules per plant |
|---|---|---|---|---|---|---|---|---|
| SCAUf129 | 21 | a | 1 | 1 | A | 1.593 ± 0.340↑* | 34.3 | |
| SCAUf141 | 17 | a | 17 | 5 | A | 1.523 ± 0.020↑* | 41.7 | |
| SCAUf142 | 4 | a | 1 | 1 | A | 1.457 ± 0.247↑* | 93.0 | |
| SCAUf110 | 11 | a | 21 | 9 | B | 1.397 ± 0.301↑* | 18.7 | |
| SCAUf126 | 12 | a | 1 | 1 | A | 1.377 ± 0.074↑* | 25.0 | |
| SCAUf118 | 7 | a | 1 | 1 | A | 1.375 ± 0.025↑* | 62.0 | |
| SCAUf114 | 12 | a | 1 | 1 | A | 1.327 ± 0.186↑* | 40.3 | |
| SCAUf122 | 14 | a | 1 | 1 | A | 1.287 ± 0.156↑* | 39.7 | |
| SCAUf123 | 9 | g | 22 | 24 | C | 1.283 ± 0.063↑* | 43.3 | |
| SCAUf113 | 22 | a | 1 | 1 | A | 1.280 ± 0.265↑* | 94.3 | |
| SCAUf148 | 16 | h | 20 | 25 | D | 1.267 ± 0.201↑* | 52.0 | |
| SCAUf131 | 25 | a | 1 | 1 | A | R. anhuiense CCBAU 23252T (98.5%) | 1.215 ± 0.365 | 98.5 |
| SCAUf125 | 18 | a | 23 | 10 | E | 1.135 ± 0.095 | 61.5 | |
| SCAUf136 | 9 | a | 1 | 1 | A | 1.130 ± 0.160 | 58.7 | |
| SCAUf111 | 21 | a | 1 | 1 | A | 1.127 ± 0.109 | 72.3 | |
| SCAUf133 | 11 | a | 19 | 7 | B | R. sophorae CCBAU 03386T (96.6 %) | 1.120 ± 0.210 | 17.0 |
| SCAUf102 | 2 | a | 1 | 1 | A | 1.097 ± 0.138 | 28.3 | |
| SCAUf106 | 1 | a | 5 | 13 | F | R. vallis CCBAU 65647T (93.0%) | 1.095 ± 0.088 | 48.0 |
| SCAUf109 | 1 | a | 4 | 12 | G | R. vallis CCBAU 65647T (93.0%) | 1.094 ± 0.087 | 46.7 |
| SCAUf139 | 4 | a | 1 | 1 | A | 1.087 ± 0.121 | 40.3 | |
| SCAUf150 | 16 | h | 20 | 25 | D | A. radiobacter NCPPB 2437T (97.3%) | 1.080 ± 0.131 | 4.0 |
| SCAUf121 | 23 | a | 18 | 6 | A | 1.050 ± 0.044 | 44.7 | |
| SCAUf134 | 15 | a | 19 | 7 | B | 1.017 ± 0.153 | 27.0 | |
| SCAUf115 | 7 | a | 1 | 1 | A | 0.950 ± 0.410 | 22.5 | |
| SCAUf87 | 19 | c | 8 | 19 | H | A. radiobacter NCPPB 2437T (97.3%) | 0.934 ± 0.146 | 23.0 |
| SCAUf137 | 23 | a | 18 | 6 | A | 0.933 ± 0.206 | 35.0 | |
| SCAUf101 | 2 | a | 1 | 1 | A | 0.930 ± 0.021 | 27.7 | |
| SCAUf143 | 15 | a | 19 | 7 | B | 0.923 ± 0.228 | 22.5 | |
| SCAUf147 | 15 | a | 19 | 7 | B | 0.893 ± 0.173 | 22.0 | |
| SCAUf146 | 12 | a | 1 | 1 | A | 0.870 ± 0.150 | 4.5 | |
| SCAUf99 | 20 | a | 6 | 14 | E | R. sophorae CCBAU 03386T (96.6 %) | 0.864 ± 0.041 | 59.7 |
| SCAUf92 | 1 | a | 2 | 8 | A | 0.856 ± 0.099 | 23.7 | |
| SCAUf132 | 21 | a | 1 | 1 | A | 0.833 ± 0.231 | 16.7 | |
| SCAUf103 | 19 | f | 15 | 22 | I | 0.820 ± 0.066 | 68.0 | |
| SCAUf107 | 13 | a | 12 | 4 | J | 0.815 ± 0.066 | 3.0 | |
| SCAUf149 | 11 | i | 25 | 26 | K | A. radiobacter NCPPB 2437T (97.3%) | 0.805 ± 0.155 | 35.0 |
| CK | – | – | – | 0.801 ± 0.139 | 0.0 | |||
| SCAUf95 | 5 | a | 1 | 1 | A | 0.790 ± 0.169 | 20.0 | |
| SCAUf120 | 17 | a | 17 | 5 | A | 0.790 ± 0.030 | 25.0 | |
| SCAUf90 | 10 | a | 11 | 3 | B | R. sophorae CCBAU 03386T (96.6 %) | 0.785 ± 0.051 | 60.3 |
| SCAUf94 | 8 | a | 9 | 16 | L | R. vallis CCBAU 65647T (93.0%) | 0.780 ± 0.060 | 18.7 |
| SCAUf86 | 13 | b | 12 | 17 | J | R. sophorae CCBAU 03386T (99.3 %) | 0.770 ± 0.121 | 12.7 |
| SCAUf124 | 14 | a | 1 | 1 | A | 0.753 ± 0.067 | 36.0 | |
| SCAUf138 | 14 | a | 1 | 1 | A | 0.750 ± 0.057 | 38.0 | |
| SCAUf145 | 18 | a | 17 | 5 | A | 0.750 ± 0.110 | 35.5 | |
| SCAUf140 | 17 | a | 18 | 6 | A | R. anhuiense CCBAU 23252T (98.9%) | 0.743 ± 0.170 | 23.0 |
| SCAUf117 | 22 | a | 1 | 1 | A | 0.737 ± 0.177 | 71.7 | |
| SCAUf127 | 22 | a | 1 | 1 | A | R. anhuiense CCBAU 23252T (98.9%) | 0.733 ± 0.065 | 41.3 |
| SCAUf128 | 9 | a | 1 | 1 | A | 0.707 ± 0.188 | 13.3 | |
| SCAUf100 | 10 | e | 13 | 21 | M | R. vallis CCBAU 65647T (99.0%) | 0.689 ± 0.034 | 11.0 |
| SCAUf104 | 24 | f | 16 | 23 | I | R. anhuiense CCBAU 23252T (99.2%) | 0.680 ± 0.082 | 15.0 |
| SCAUf89 | 20 | a | 10 | 2 | B | 0.680 ± 0.060 | 21.3 | |
| SCAUf119 | 4 | a | 1 | 1 | A | 0.667 ± 0.188 | 20.0 | |
| SCAUf135 | 4 | a | 1 | 1 | A | 0.663 ± 0.114 | 16.7 | |
| SCAUf105 | 3 | a | 3 | 11 | A | R. anhuiense CCBAU 23252T (99.3%) | 0.660 ± 0.020 | 5.0 |
| SCAUf108 | 3 | a | 7 | 15 | A | 0.650 ± 0.031 | 6.3 | |
| SCAUf116 | 25 | a | 1 | 1 | A | 0.647 ± 0.189 | 47.0 | |
| SCAUf112 | 21 | a | 1 | 1 | A | 0.647 ± 0.099 | 11.7 | |
| SCAUf96 | 5 | a | 1 | 1 | A | 0.643 ± 0.070 | 21.3 | |
| SCAUf88 | 19 | a | 2 | 8 | A | 0.632 ± 0.073 | 15.0 | |
| SCAUf144 | 4 | b | 24 | 18 | C | R. fabae CCBAU 33202T (99.9 %) | 0.605 ± 0.090 | 24.0 |
| SCAUf97 | 19 | a | 1 | 1 | A | 0.573 ± 0.032 | 31.0 | |
| SCAUf91 | 6 | a | 2 | 8 | A | R. anhuiense CCBAU 23252T (99.8%) | 0.533 ± 0.087 | 13.0 |
| SCAUf130 | 23 | a | 18 | 6 | A | 0.532± 0.086 | 23.0 | |
| SCAUf93 | 19 | d | 14 | 20 | N | A. radiobacter NCPPB 2437T (97.3%) | 0.532± 0.069 | 0 |
| SCAUf98 | 19 | a | 1 | 1 | A | 0.517± 0.059 | 0 |
CK: uninoculated treatment in the symbiotic efficiency test. Representative isolates for sequencing in bold.
Sampling sites are the same as Figure 1.
Genotype: the combination of the restriction patterns obtained by enzymes MspI, HaeIII, TaqI, and HinfI.
CACAI: clustered analysis of combined ARDRA and IGS-RFLP, Groups were defined at 94.5% similarity level.
MLSA, multilocus sequence analysis of combined recA, atpD, and glnII. The percentages are sequence similarities to the closely related species or the closest type strain.
Type strain.
Significantly higher shoot dry mass than that in CK treatment according to the LSD test (P = 0.05). Data presented as mean value ± standard deviation (n = 3, except n = 2 for SCAUf93 and SCAUf98).
Table 3.
The nodC and nifH types in the representative strains isolated from faba bean.
| Representative strain | nodCa | nifHa |
|---|---|---|
| R. anhuiense SCAUf104 | R. fabae | HRsp1 |
| R. anhuiense SCAUf127 | R. fabae | HRsp1 |
| R. fabae SCAUf144 | R. fabae | HRsp1 |
| R. sophorae SCAUf86 | R. fabae | HRsp2 |
| R. anhuiense SCAUf131 | R. fabae | HRsp2 |
| R. anhuiense SCAUf140 | R. laguerreae | HRsp1 |
| R. anhuiense SCAUf105 | CRla1 | HRsp2 |
| R. vallis SCAUf100 | CRsp1 | HRsp3 |
| R. anhuiense SCAUf91 | CRsp2 | HRsp4 |
| Rhizobium sp.I f99 | CRsp2 | HRsp4 |
| Rhizobium sp.I SCAUf90 | CRsp2 | HRsp4 |
| Rhizobium sp.I SCAUf133 | CRsp2 | HRsp4 |
| Rhizobium sp.II SCAUf94 | CRsp2 | HRsp4 |
| Rhizobium sp.II SCAUf106 | CRsp2 | HRsp4 |
| Rhizobium sp.II SCAUf109 | CRsp2 | HRsp4 |
Closest type strain or clade.
Amplification of the 16S rDNA gene resulted in an approximately 1,500 bp band from all the isolates. In the 16S rDNA PCR-RFLP, nine fragment pattern types (a-i) were observed: type a included 54 strains, types b, f, and h included two strains each, and types c, d, e, g, and i included one strain each (Table 2).
For the majority of strains, IGS PCR resulted in a single band ranging from 1,900 to 2,200 bp, whereas for strains SCAUf90 and SCAUf99 IGS PCR resulted in two and three bands, respectively (Table 2). The strains were divided to 25 IGS-RFLP types. In the combined analysis of 16S rDNA RFLP and IGS-RFLP (CACAI) the strains were divided into 14 CACAI groups at 94.5% similarity level and 26 CACAI genotypes (Table 2). CACAI group A was the largest group including 40 isolates with CACAI genotypes 1, 5, 6, 8, and 15. Seven of the plant growth promoting strains represented genotype 1, and the other four were assigned to genotypes 5, 9, 24, and 25.
16S rDNA phylogeny
Based on CACAI groups as well as considering the sites of isolation of the strains, 19 representative strains were selected for sequencing. In the 16S rDNA phylogenetic tree (Figure 2), the strains clustered into six distinct clades with the reference strains. Four clades were related to Rhizobium (R group) and two to Agrobacterium (A group). Four strains clustered with Agrobacterium radiobacter type strain with 98.3–99.8% similarities. SCAUf144 clustered with R. fabae with 100% similarity, SCAUf100 clustered with R. vallis with 98.6% similarity, and SCAUf86, SCAUf90, SCAUf94, SCAUf99, and SCAUf133 clustered with Rhizobium sophorae into clade R2 with 97.9–99.9% similarities. The other eight strains clustered into a distinct clade with R. gallicum, Rhizobium anhuiense, R. laguerreae, and Rhizobium leguminosarum with similarities ranging from 99.8 to 100%.
Figure 2.
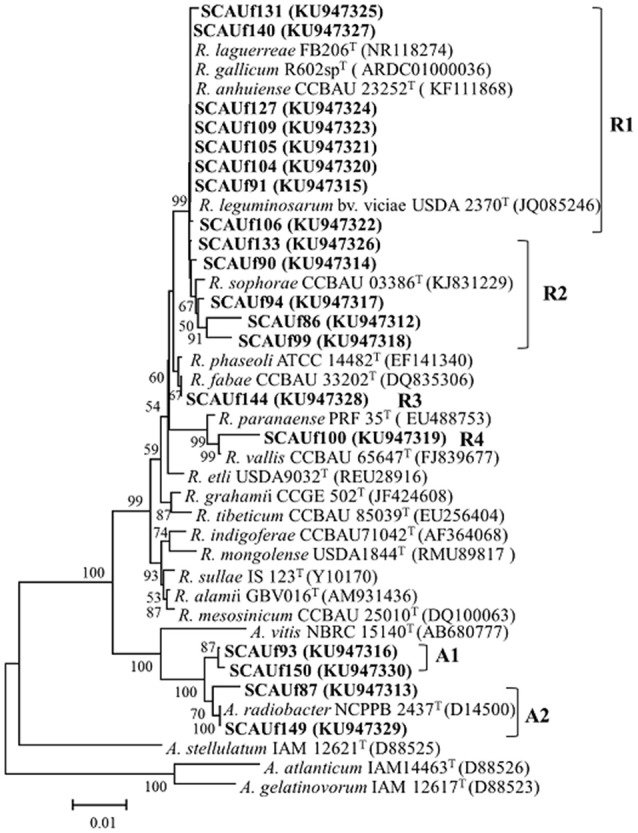
Neighbor-joining tree based on 16S rDNA (1,336 nt) presenting the phylogenetic relationship among the representative strains isolated from faba bean (in bold) and reference strains. Bootstrap values ≥50% are shown on the branches. Genbank accession numbers are in parentheses. Scale bar = 1% substitutions per site. R, Rhizobium; A, Agrobacterium.
Multilocus sequence analysis
In the multilocus sequence analysis (MLSA) based on housekeeping genes atpD, glnII and recA, the 19 representative strains clustered into nine distinct clades related to Rhizobium and Agrobacterium species (Figure 3). SCAUf86 was 99.3% similar to R. sophorae CCBAU 03386T, thus assigned as R. sophorae. SCAUf90, SCAUf99 and SCAUf133 clustered separately and were assigned as Rhizobium sp. I, as did SCAUf94, SCAUf106 and SCAUf109 that were assigned as Rhizobium sp. II. SCAUf91, SCAUf104, SCAUf105, SCAUf127, SCAUf131, and SCAUf140 were 98.5–99.8% similar to R. anhuiense type strain, thus assigned as R. anhuiense strains. SCAUf144 and SCAUf100 clustered with R. fabae CCBAU 33202T and R. vallis CCBAU 65647T, respectively, thus assigned as R. fabae and R. vallis, respectively. As in 16S rDNA analysis, four strains clustered with Agrobacterium in the MLSA. Because no glnII sequences of the relevant Agrobacterium type strains except A. radiobacter type strain were available in the GenBank sequence database, the relationships between Agrobacterium strains were studied based on non-type strains (Supplementary Figure S1). SCAUf87 clustered separately, and was assigned as Agrobacterium sp. I. SCAUf93 and SCAUf150 clustered separately and were assigned as Agrobacterium sp. II. SCAUf149 was 97.3% similar to A. radiobacter NCPPB 2437T with, thus assigned as A. radiobacter.
Figure 3.
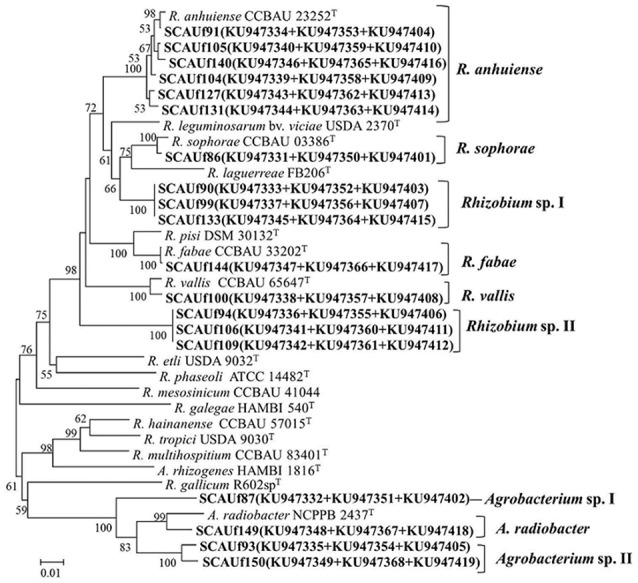
Neighbor-joining tree based on multilocus sequence analysis using concatenated sequence of atpD (395 nt), glnII (483 nt), and recA (344 nt) genes presenting the phylogenetic relationship among the representative strains isolated from faba bean (in bold) and reference strains. Bootstrap values ≥50% are shown on the branches. Genbank accession numbers are in parentheses. Scale bar = 1% substitutions per site. R, Rhizobium, A, Agrobacterium.
Diversity of symbiosis genes
For both nifH and nodC amplification was not successful with one primer pair only, possibly due to differences in primer binding sites. Approximately 700 bp fragments were obtained using primer pair nifHctg/nifHI (13 representative strains), and 400 bp products using primer pair nifH1F/ nifH1R (SCAUf87, SCAUf105, SCAUf133, SCAUf140). Amplification of nifH from SCAUf149 and SCAUf150 was not successful. Seventeen strains clustered into four clades in the nifH phylogenetic tree (Figure 4, Table 3). The nifH of Agrobacterium sp. II SCAUf93 and R. anhuiense SCAUf104 were 99.7 and 99.6%, respectively, similar to that of R. anhuiense CCBAU 23252T. The nifH of R. anhuiense SCAUf127 and R. fabae SCAUf144 were 100% similar to that of R. fabae type strain. The nifH of R. sophorae SCAUf86, R. anhuiense SCAUf105 and SCAUf131 clustered with that of R. leguminosarum USDA 2370T with 98.4% similarity. R. vallis SCAUf100 clustered with R. leguminosarum CCBAU 43200 with 100% similarity. The strains R. anhuiense SCAUf91, Rhizobium sp. I SCAUf90, Rhizobium sp. II SCAUf94, SCAUf106 and SCAUf109 carried nifH 100% similar to that of R. leguminosarum CCBAU 71124.
Figure 4.
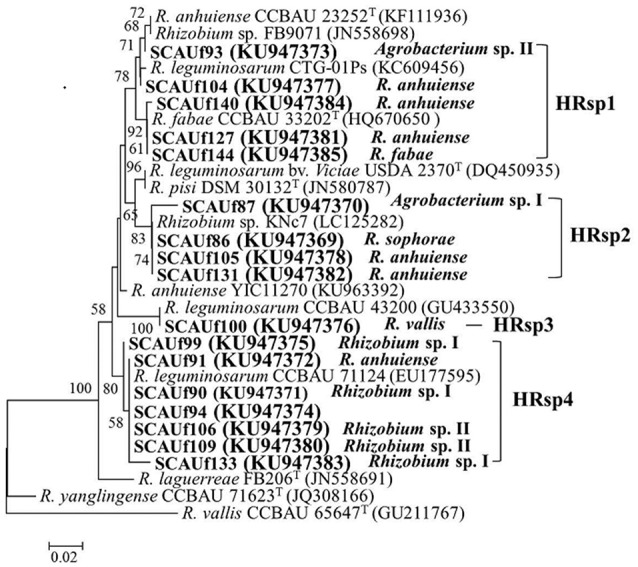
Neighbor-joining tree based on nifH (329 nt) gene of 17 representative strains isolated from faba bean (in bold) and reference strains. Genbank accession numbers are in parentheses. Bootstrap values ≥50% are shown on the branches. Scale bar = 2% substitutions per site. R, Rhizobium.
Nearly 600 bp nodC fragments were amplified from thirteen representative strains using primer pair nodC540/nodC1160. Amplification from strains SCAUf105 and SCAUf140 was successful only while using R. leguminosarum sv. viciae nodC specific primer pair nodCf/nodCr. Amplification of nodC from strains assigned as Agrobacterium was not successful. Fifteen strains clustered into five clades in the nodC phylogenetic tree (Figure 5, Table 3). The nodC of R. anhuiense SCAUf131, SCAUf127, and SCAUf104, R. fabae SCAUf144 and R. sophorae SCAUf86 were 100% similar to that of R. fabae type strain (Figure 5A). Similarly to the nifH analysis, the nodC of R. vallis SCAUf100 clustered with nodC from non-type strains. Seven strains carried nodC 100% similar to that of R. leguminosarum non-type strain. The nodC of R. anhuiense SCAUf140 and SCAUf105 (Figure 5B) were 100 and 97.2% similar to that of R. laguerreae.
Figure 5.
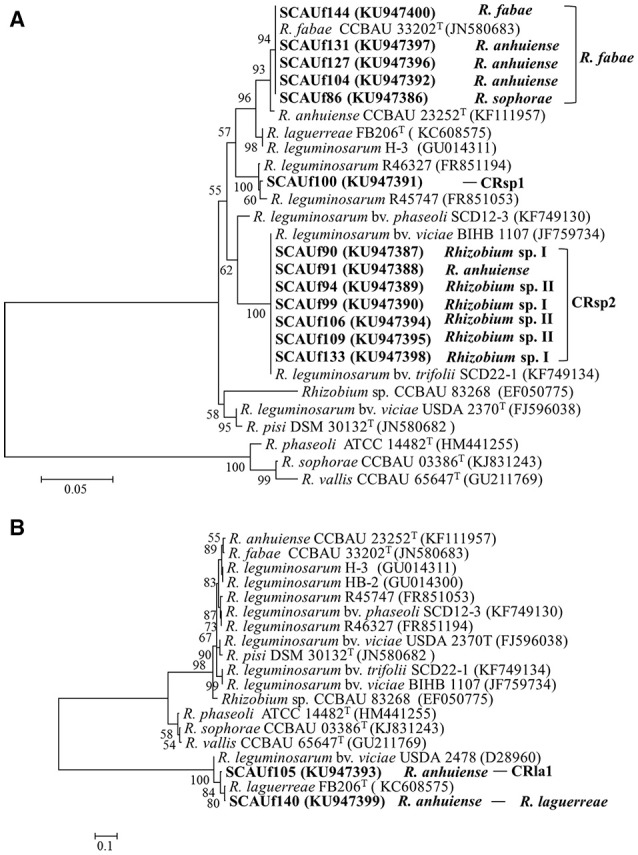
Neighbor-joining tree based on nodC gene of 13 representative strains (499 nt) (A) and 2 representative strains (230 nt) (B) isolated from faba bean (in bold) and reference strains. Genbank accession numbers are in parentheses. Bootstrap values ≥50% are shown on the branches. Scale bar = 5% substitutions per site. R, Rhizobium.
Discussion
Due to overcutting and mining Panxi in Southwestern China has suffered serious soil degradation and heavy metal contamination (Xu et al., 2013; Yu et al., 2014). Reclaiming the soils requires sustainable and efficient yet low economic input methods, for example utilization of biological nitrogen fixation (BNF) by legume-rhizobium symbiosis instead of relatively cheap nitrogen fertilizer. To facilitate the utilization of BNF we tested the plant growth promoting ability of rhizobial isolates from faba bean in search of locally adapted, potential inoculant strains. In Ethiopia, chickpea nodulating rhizobia showed big differences in efficiency and nodule numbers, and strains with similar efficiencies did not necessarily induce similar numbers of nodules and vice versa (Tena et al., 2017). Similarly, in our study variations in plant growth promoting ability and nodule numbers were large, and only 11 strains increased faba bean shoot dry mass significantly. Similar to Leucaena leucocephala isolates from Panxi (Xu et al., 2013), for many of the strains inoculation resulted in dry mass lower than that in uninoculated plants, highlighting the need to apply selected inocula to promote BNF.
The diversity and identity of the strains were assessed using molecular methods. Faba bean is nodulated by R. fabae, R. leguminosarum, R. anhuiense, R. laguerraeae and A. radiobacter strains, and the dominant species is different in different regions (Tian et al., 2007, 2008; Youseif et al., 2014; Xu et al., 2015; Zhang et al., 2015; Xiong et al., 2017). In our study, the isolated strains were related to genera Rhizobium and Agrobacterium. The Rhizobium strains were assigned as representing R. anhuiense, R. fabae, R. sophorae, and R. vallis, and two putative new species in the genus Rhizobium. Similar to subtropical provinces in East China (Xiong et al., 2017), R. anhuiense was the dominant species among the faba bean nodulating rhizobia in Panxi. To our knowledge, R. sophorae, a symbiont of medicinal legume Sophora flavescens (Jiao et al., 2015) and R. vallis, a symbiont of Phaseolus vulgaris (Wang et al., 2011), have not earlier been shown to nodulate faba bean.
The rhizobia-legume symbiosis benefits sustainable agriculture due to the symbiotic nitrogen fixation capacity that needs two key points: nodule infection and nitrogen fixation, both of which need the regulation of symbiosis related genes (Masson-Boivin et al., 2009). In the present study, nifH gene that is the structural gene encoding the nitrogenase Fe protein (Masson-Boivin et al., 2009), and nodC that is the gene encoding enzymes involved in the synthesis of the core structure of the Nod-factor (Geremia et al., 1994) were selected for sequencing to analyze the symbiotic phylogeny of the faba bean rhizobia in Panxi region. The symbiotic genes are commonly located on a symbiotic plasmid or island which may be transferred (Laranjo et al., 2012; Bakhoum et al., 2014). Faba bean nodulating R. leguminosarum strains that carried four different types of nodulation gene nodD had all similar nodC (Table 4) (Tian et al., 2007). The five types of nodC detected in this study suggest higher diversity at symbiosis related gene level. However, considering the six different nifH-nodC combinations in our study, the faba bean isolates from Yunnan (Tian et al., 2007) and Panxi were approximately equally diverse.
Table 4.
Rhizobial species, nodD and nodC diversity in subtropical China.
| Province | Climate | Species | nodD types | nodC types | Dominant species | References |
|---|---|---|---|---|---|---|
| Yunnan | Subtropical highland, humid subtropical | 5a | 3c | nd | R. laguerraeae | Xiong et al., 2017 |
| Yunnan | Subtropical highland, humid subtropical | 1b | 4d | 1d | R. leguminosarum | Tian et al., 2007 |
| Anhui, Jiangxi, Henan, Zhejiang | Humid subtropical | 3a | 5c | nd | R. anhuiense | Xiong et al., 2017 |
| Anhui, Jiangxi | Humid subtropical | 2b | 3d | 1d | R. leguminosarum | Tian et al., 2007 |
| Sichuan | Humid subtropical | 5b | nd | 3e | R. leguminosarum | Xu et al., 2015 |
| Panxi | See Introduction | 6b | nd | 5e | R. anhuiense | This study |
Nd, not determined.
Determined by amplicon sequencing targeting rpoB.
Determined by multilocus sequence analysis.
Determined by amplicon sequencing targeting nodD.
Determined by RFLP.
Determined by Sanger sequencing nodC.
The Desmodium nodulating rhizobium strains in Panxi region were quite different from those in other places such as temperate and subtropical region of China and Central and North America, possibly due to the special environmental conditions (Xu et al., 2016). The faba bean rhizobia in this area were approximately as diverse as in Sichuan hilly areas and in Yunnan (Table 4) (Xu et al., 2015; Xiong et al., 2017), yet more diverse than in other parts of subtropical China (Tian et al., 2007; Xiong et al., 2017). When looking at the species-nifH-nodC combinations it is noteworthy that all but two of the six R. anhuiense isolates were different. The symbiosis and nitrogen fixation related genes of rhizobia can be transferred laterally (Sullivan et al., 1995). However, whether the increase in diversity in Panxi was caused by lateral transfer cannot be concluded based on our data.
In conclusion, eleven out of 65 faba bean strains in Panxi area could significantly promote plant growth, and were thus considered as potential inoculants. The nodule isolates in this area were diverse belonging to nine species. R. anhuiense, the dominant faba bean nodulating species in this area, was diverse both at plant growth promoting ability and symbiosis related gene levels.
Author contributions
KX and YC conceived and designed the experiments. KX supervised the experiments. YC, LZ, PP, and KX contributed to discussion of the results, and writing and revising the manuscript. LZ performed most of the experiments and analyzed data. QC and CW participated in collecting faba bean nodules and relevant soil information, and relevant meteorological information from the Sichuan meteorological bureau. QL created the map in Figure 1 and revised the manuscript. All authors contributed to writing the article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Mr. Zhi Heng Liu for collecting faba bean nodules in Panxi.
Footnotes
Funding.This work was supported by the National Key Research and Development Program of China (2016YFD0300300).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01338/full#supplementary-material
References
- Bakhoum N., Galiana A., Le Roux C., Kane A., Duponnois R., Ndoye F., et al. (2014). Phylogeny of nodulation genes and symbiotic diversity of Acacia senegal (L.) Willd. and A. seyal (Del.) Mesorhizobium strains from different regions of Senegal. Microb. Ecol. 69, 641–651. 10.100/7/s00248-014-0507-1 [DOI] [PubMed] [Google Scholar]
- Cao Y., Wang E. T., Zhao L., Chen W. M., Wei G. H. (2014). Diversity and distribution of rhizobia nodulated with Phaseolus vulgaris in two ecoregions of China. Soil Biol. Biochem. 78, 128–137. 10.1016/j.soilbio.2014.07.026 [DOI] [Google Scholar]
- Chen Q., Chen W. X., Zhang X. P., Li D. Y., Lindstrom K. (2004). Genetic diversity of rhizobia isolated from Pueraria spp. in Sichuan, China. Sci. Agric. Sin. 37, 1641–1646. 10.3321/j.issn:0578-1752.2004.11.010 [DOI] [Google Scholar]
- Crews T. E., Peoples M. B. (2004). Legume versus fertilizer sources of nitrogen: ecological tradeoffs and human needs. Agr. Ecosyst. Environ. 102, 279–297. 10.1016/j.agee.2003.09.018 [DOI] [Google Scholar]
- Gentzbittel L., Andersen S. U., Ben C., Rickauer M., Stougaard J., Young N. D. (2015). Naturally occurring diversity helps to reveal genes of adaptive importance in legumes. Front. Plant Sci. 6:269. 10.3389/fpls.2015.00269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia R. A., Mergaert P., Geelen D., Van Montagu M., Holsters M. (1994). The NodC protein of Azorhizobium caulinodans is an N-acetylglucosaminyltransferase. Proc. Natl. acad. Sci. U.S.A. 91, 2669–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham P. H., Vance C. P. (2003). Legumes: importance and constraints to greater use. Plant physiol. 131, 872–877. 10.1104/pp.017004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurkanli C. T., Ozkoc I., Gunduz I. (2014). Genetic diversity of Vicia faba L. and Pisum sativum L. nodulating rhizobia in the central Black Sea region of Turkey. Ann. Microbiol. 64, 99–112. 10.1007/s13213-013-0638-5 [DOI] [Google Scholar]
- Haciseferoǧullar,i H., Gezer I., Bahtiyarca Y., Menge,ş H. O. (2003). Determination of some chemical and physical properties of Sakiz faba bean (Vicia faba L. Var. major). J. Food Eng. 60, 475–479. 10.1016/S026 0-8774(03)00075-X [DOI] [Google Scholar]
- Jiao Y. S., Yan H., Ji Z. J., Liu Y. H., Sui X. H., Wang E. T., et al. (2015). Rhizobium sophorae sp. nov. and Rhizobium sophoriradicis sp. nov., nitrogen-fixing rhizobial symbionts of the medicinal legume Sophora flavescens. Int. J. Syst. Evol. Microbiol. 65(Pt 2), 497–503. 10.1099/ijs.0.068916-0068916-0 [DOI] [PubMed] [Google Scholar]
- Köpke U., Nemecek T. (2010). Ecological services of faba bean. Field Crops Res. 115, 217–233. 10.1016/j.fcr.2009.10.012 [DOI] [Google Scholar]
- Laguerre G., Nour S. M., Macheret V., Sanjuan J., Drouin P., Amarger N. (2001). Classification of rhizobia based on nodC and nifH gene analysis reveals a close phylogenetic relationship among Phaseolus vulgaris symbionts. Microbiology 147, 981–993. 10.1099/00221287-147-4-981 [DOI] [PubMed] [Google Scholar]
- Laranjo M., Young J. P. W., Oliveira S. (2012). Multilocus sequence analysis reveals multiple symbiovars within Mesorhizobium species. Syst. Appl. Microbiol. 35, 359–367. 10.1016/j.syapm.2012.06.002 [DOI] [PubMed] [Google Scholar]
- Li L., Yang T., Redden R., He W., Zong X. (2016). Soil fertility map for food legumes production areas in China. Sci. Rep. 6:26102. 10.1038/srep26102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massol-Deya A. A., Odelson D. A., Hickey R. F., Tiedje J. M. (1995). Bacterial community fingerprinting of amplified 16S and 16–23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA), in the Molecular Microbial Ecology Manual, eds Akkermans A. D. L., Van Elsas J. D., De Bruijn F. J. (Dordrecht: Springer; ), 289–296. [Google Scholar]
- Masson-Boivin C., Girau D. E., Perret X., Batut J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. 10.1016/j.tim.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Mei P. P., Gui L. G., Wang P., Huang J. C., Long H. Y., Christie P., et al. (2012). Maize/faba bean intercropping with rhizobia inoculation enhances productivity and recovery of fertilizer P in a reclaimed desert soil. Field Crops Res. 130, 19–27. 10.1016/j.fcr.2012.02.007 [DOI] [Google Scholar]
- Moschetti G., Peluso A., Protopapa A., Anastasio M., Pepe O., Defez R. (2005). Use of nodulation pattern, stress tolerance, nodC gene amplification, RAPD-PCR and RFLP−16S rDNA analysis to discriminate genotypes of Rhizobium leguminosarum biovar viciae. Syst. Appl. Microbiol. 28, 619–631. 10.1016/j.syapm.2005.03.009 [DOI] [PubMed] [Google Scholar]
- Navarro E., Simonet P., Normand P., Bardin R. (1992). Characterization of natural populations of Nitrobacter spp. using PCR/RFLP analysis of the ribosomal intergenic spacer. Arch. Microbiol. 157, 107–115 [DOI] [PubMed] [Google Scholar]
- Rivas R., Velázquez E., Willems A., Vizcaíno N., Subba-Rao N. S., Mateos P. F., et al. (2002). A new species of Devosia that forms a unique nitrogen-fixing root-nodule symbiosis with the aquatic legume Neptunia natans (Lf) Druce. Appl. Environ. Microbiol. 68, 5217–5222. 10.1128/AEM.68.11.5217-5222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlf F. J. (1990). NTSYS-pc, Numerical taxonomy and Multivariate Analysis System, Version 2.02. New York, NY: Exeter Software. [Google Scholar]
- Sarita S., Sharma P. K., Priefer U. B., Prell J. (2005). Direct amplification of rhizobial nodC sequences from soil total DNA and comparison to nodC diversity of root nodule isolates. FEMS Microbiol. Ecol. 54, 1–11. 10.1016/j.femsec.2005.02.015 [DOI] [PubMed] [Google Scholar]
- Song Y. N., Zhang F. S., Marschner P., Fan F. L., Gao H. M., Bao X. G., et al. (2007). Effect of intercropping on crop yield and chemical and microbiological properties in rhizosphere of wheat (Triticum aestivum L.), maize (Zea mays L.), and faba bean (Vicia faba L.). Biol. Fertil. Soils 43, 565–574. 10.1007/s00374-006-0139-9 [DOI] [Google Scholar]
- Sullivan J. T., Patrick H. N., Lowther W. L., Scott D. B., Ronson C. W. (1995). Nodulating strains of Rhizobium loti arise through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. U.S.A. 92, 8985–8989. 10.1073/pnas.92.19.8985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z. Y., Xu X. D., Wang E. T., Gao J. L., Martinez-Romero E., Chen W. X. (1997). Phylogenetic and genetic relationships of Mesorhizobium tianshanense and related rhizobia. Int J. Syst. Bacteriol. 47, 874–879. 10.1099/00207713-47-3-874 [DOI] [PubMed] [Google Scholar]
- Tena W., Wolde-Meskel E., Degefu T., Walley F. (2017). Genetic and phenotypic diversity of rhizobia nodulating chickpea (Cicer arietinum L.) in soils from southern and central Ethiopia. Can. J. Microbiol. 63, 690–707. 10.1139/cjm-2016-0776 [DOI] [PubMed] [Google Scholar]
- Terefework Z., Kaijalainen S., Lindström K. (2001). AFLP fingerprinting as a tool to study the genetic diversity of Rhizobium galegae isolated from Galega orientalis and Galega officinalis. J. Biotechnol. 91, 169–180. 10.1016/S0168-1656(01)00338-8 [DOI] [PubMed] [Google Scholar]
- Thilakarathna M. S., Raizada M. N. (2017). A meta-analysis of the effectiveness of diverse rhizobia inoculants on soybean traits under field conditions. Soil. Biol. Biochem. 105, 177–196. 10.1016/j.soilbio.2016.11.022 [DOI] [Google Scholar]
- Tian C. F., Wang E. T., Han T. X., Sui X. H., Chen W. X. (2007). Genetic diversity of rhizobia associated with Vicia faba in three ecological regions of China. Arch. Microbiol. 188, 273–282. 10.1007/s00203-007-0245-6 [DOI] [PubMed] [Google Scholar]
- Tian C. F., Wang E. T., Wu L. J., Han T. X., Chen W. F., Gu C. T., et al. (2008). Rhizobium fabae sp. nov., a bacterium that nodulates Vicia faba. Int. J. Syst. Evol. Microbiol. 58, 2871–2875. 10.1099/ijs.0.2008/000703-0 [DOI] [PubMed] [Google Scholar]
- Vincent J. M. (1970). A Manual for the Practical Study of Root Nodule Bacteria. International Biological Programme Handbook no. 15. Oxford: Blackwell Scientific Publications. [Google Scholar]
- Vinuesa P., Silva C., Werner D., Martínez-Romero E. (2005). Population genetics and phylogenetic inference in bacterial molecular systematics: the roles of migration and recombination in Bradyrhizobium species cohesion and delineation, Mol. Phylogenet. Evol. 34, 29–54. 10.1016/j.ympev.2004.08.020 [DOI] [PubMed] [Google Scholar]
- Wang F. Q., Wang E. T., Zhang Y. F., Chen W. X. (2006). Characterization of rhizobia isolated from Albizia spp. in comparison with microsymbionts of Acacia spp. and Leucaena leucocephala grown in China. Syst. Appl. Microbiol. 29, 502–517. 10.1016/j.syapm.2005.12.010 [DOI] [PubMed] [Google Scholar]
- Wang F., Wang E. T., Wu L. J., Sui X. H., Li Y., Chen W. X. (2011). Rhizobium vallis sp. nov., isolated from nodules of three leguminous species. Int. J. Syst. Evol. Microbiol. 61, 2582–2588. 10.1099/ijs.0.026484-0 [DOI] [PubMed] [Google Scholar]
- Wu B., Zhu C., Li D., Dong K., Wang X., Shi P. (2006). Setting biodiversity conservation priorities in the Forests of the Upper Yangtze Ecoregion based on ecoregion conservation methodology. Biodiver. Sci. 14, 87–97. 10.1360/biodiv.050232 [DOI] [Google Scholar]
- Xie H., Zhang X. P., Lindstrom K. (2009). RFLP genetic diversity of rhizobia isolated from Pueraria lobata in Panxi. J. Sichuan Agric. Univ. 27, 314–320. 10.3969/j.issn.1000-2650.2009.03.010 [DOI] [Google Scholar]
- Xiong H. Y., Zhang X. X., Guo H. J., Ji Y. Y., Li Y., Wang X. L., et al. (2017). The epidemicity of facultative microsymbionts in faba bean rhizosphere soils. Soil. Biol. Biochem. 115, 243–252. 10.1016/j.soilbio.2017.08.032 [DOI] [Google Scholar]
- Xu K. W., Penttinen P., Chen Y. X., Chen Q., Zhang X. (2013). Symbiotic efficiency and phylogeny of the rhizobia isolated from Leucaena leucocephala in arid-hot river valley area in Panxi, Sichuan, China. Appl. Microbiol. Biotechnol. 97, 783–793. 10.1007/s00253-012-4246-2 [DOI] [PubMed] [Google Scholar]
- Xu K. W., Zou L., Penttinen P., Wang K., Heng N. N., Zhang X. P., et al. (2015). Symbiotic effectiveness and phylogeny of rhizobia isolated from faba bean (Vicia faba L.) in Sichuan hilly areas, China. Syst.Appl. Microbiol. 38, 515–523. 10.1016/j.syapm.2015.06.009 [DOI] [PubMed] [Google Scholar]
- Xu K. W., Zou L., Penttinen P., Zeng X., Liu M., Zhao K., et al. (2016). Diversity and phylogeny of rhizobia associated with Desmodium spp. in Panxi, Sichuan, China. Syst. Appl. Microbiol. 39, 33–40. 10.1016/j.syapm.2015.10.005 [DOI] [PubMed] [Google Scholar]
- Youseif S. H., El-Megeed F. H. A., Ageez A., Cocking E. C., Saleh S. A. (2014). Phylogenetic multilocus sequence analysis of native rhizobia nodulating faba bean (Vicia faba L.) in Egypt. Syst. Appl. Microbiol. 37, 560–569. 10.1016/j.syapm.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Yu X., Li Y., Zhang C., Liu H., Liu J., Zheng W., et al. (2014). Culturable heavy metal-resistant and plant growth promoting bacteria in V-Ti magnetite mine tailing soil from Panzhihua, China. PloS ONE 9:e106618. 10.1371/journal.pone.0106618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. J., Zheng W. T., Everall I., Young J. P. W., Zhang X. X., Tian C. F., et al. (2015). Rhizobium anhuiense sp. nov., isolated from effective nodules of Vicia faba and Pisum sativum. Int. J. Syst. Evol. Microbiol. 65, 2960–2967. 10.1099/ijs.0.000365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.