Abstract
Overall growth and development of a plant is regulated by complex interactions among various hormones, which is critical at different developmental stages. Some of the key aspects of plant growth include seed development, germination and plant survival under unfavorable conditions. Two of the key phytohormones regulating the associated physiological processes are gibberellins (GA) and abscisic acid (ABA). GAs participate in numerous developmental processes, including, seed development and seed germination, seedling growth, root proliferation, determination of leaf size and shape, flower induction and development, pollination and fruit expansion. Despite the association with abiotic stresses, ABA is essential for normal plant growth and development. It plays a critical role in different abiotic stresses by regulating various downstream ABA-dependent stress responses. Plants maintain a balance between GA and ABA levels constantly throughout the developmental processes at different tissues and organs, including under unfavorable environmental or physiological conditions. Here, we will review the literature on how GA and ABA control different stages of plant development, with focus on seed germination and selected abiotic stresses. The possible crosstalk of ABA and GA in specific events of the above processes will also be discussed, with emphasis on downstream stress signaling components, kinases and transcription factors (TFs). The importance of several key ABA and GA signaling intermediates will be illustrated. The knowledge gained from such studies will also help to establish a solid foundation to develop future crop improvement strategies.
Keywords: gibberellins, abscisic acid, hormone signaling, seed germination, abiotic stresses, crosstalk of hormone signaling
Introduction
Overall growth and different developmental stages of plants are under strict regulation by several classes of plant hormones. Hormone molecules are present at low concentrations in plants, and they function either at the sites of synthesis or after they are transported to different tissues (Santner et al., 2009; Li et al., 2016). In the last two decades, there has been rapid progress in the understanding of the biosynthetic pathways, transport, signaling and mode of action of various plant hormones. Studies related to hormone signaling have established the fact that besides acting on their own, various plant hormones interact in a highly intricate manner (Stamm et al., 2012; Kohli et al., 2013; Kumar, 2013a,b; Stamm and Kumar, 2013; Verma et al., 2015, 2016; Ravindran et al., 2017). These findings clearly indicate that plants maintain the availability and level of hormones in different parts of the plant body at different developmental stages in an intricate and balanced manner.
A convenient step for us to study plant growth may begin with seed germination. Successful germination depends on the ability of the plant embryo to gain its metabolic activity (Rajjou et al., 2012). Several molecular cues have been revealed by different genetic and proteomic investigations of various Arabidopsis mutants, showing distinct germination-related phenotypes (Achard et al., 2006, 2009; Magome et al., 2008). Germination has been found to be under strict regulation of plant hormones, including gibberellic acid (GA), abscisic acid (ABA), auxin and ethylene (Han and Yang, 2015). Germination is also significantly affected by several environmental factors, such as various abiotic stresses (Rajjou et al., 2012; Han and Yang, 2015). These factors mainly affect the metabolism and different signaling pathways of GA and ABA (Holdsworth et al., 2008).
The constantly changing external factors that most affect plant growth and development are abiotic stresses. Highly variable abiotic stresses affecting plant growth are salinity, drought, and cold. The above-mentioned stresses significantly affect yield (average yield reduction >50%) of crop plants (Mahajan and Tuteja, 2005). Plants exhibit a range of tolerance levels toward these stresses that are ultimately regulated by complex signaling pathways. Abiotic stresses trigger ABA biosynthesis, which mediates stress adaptive responses by activating several specific signaling cascades and regulating different physiological and growth-related processes.
In the past decade, several genetic, molecular and proteomic studies related to germination and abiotic stresses have been carried out. In this review, we discuss the roles of GA and ABA independently and with the possible crosstalk of these two phytohormones with respect to seed germination and abiotic stresses in various plant species, including crop plants.
ABA and abiotic stress
Plants are capable of maintaining their internal environment fairly stable within the desired range. Two important factors that are crucial for the maintenance of such homeostasis are internal water level and osmotic state, which are mainly regulated by ABA (Zhang et al., 1987; Zhu, 2002). ABA acts as a molecular signal in response to various abiotic stresses, which alter the two important physiological functions mentioned above. These abiotic stress responsive signals are the basis of the various physiological as well as growth-related processes of plants, culminating in their unique ranges of tolerance toward these stresses (Finkelstein et al., 2002; Lee and Luan, 2012; Vu et al., 2015).
ABA metabolism and abiotic stress
ABA which is reported in both primitive and higher organisms seems to have different biosynthesis pathways. In primitive organisms ABA biosynthesis is not well characterized, however, in plant-associated fungi, ABA is reported to be synthesized by the direct cytosolic pathway. In contrast, great progress has been made in identifying and characterizing the genes involved in ABA metabolism in land plants (Hauser et al., 2011). ABA biosynthesis in plants follows the organelle-specific indirect pathway. The pathway involves the key precursor compound zeaxanthin, which is synthesized by the β-carotene pathway involving pyruvate. Further, zeaxanthin is converted to xanthoxin by the enzymatic reaction catalyzed by ZEP enzyme (zeaxanthin epoxide) and 9-cis-epoxy carotenoid dioxygenase (NCED) enzyme (Hauser et al., 2011; Chan, 2012; Ruiz-Sola and Rodriguez-Concepcion, 2012). Subsequently, xanthoxin is transferred from the plastid to cytosol and converted to its aldehyde intermediate and then to ABA by short-chain-dehydrogenase reductase (SDR/ABA2 in Arabidopsis) and abscisic aldehyde oxidase (AAO), respectively (Cheng et al., 2002; González-Guzmán et al., 2014). Abiotic stresses and ABA treatment are reported to alter the transcript levels of key ABA biosynthesis genes, which in turn modulate the level of ABA in plants. Upon ABA treatment, expression levels of the genes encoding ZEP (ZEP/ABA1/LOS6) and AAO3 (AAO3/ABA3/LOS5) were upregulated in Arabidopsis. Furthermore, transcript levels of NCED3, ABA3/LOS5, and AAO3 were induced by abiotic stresses (Xiong et al., 2002; Chan, 2012). Additionally, in crop plants improved tolerance toward various abiotic stresses has been reported by introducing or inducing expression of genes encoding key enzymes of ABA biosynthesis (Table 1). Among the NCED genes, NCED3 expression level increased upon water stress, which is also reflected in the water-stress response of nced3 mutants (Table 1).
Table 1.
Summary of regulation of ABA metabolism and signaling genes with respect to abiotic stress and seed germination in different plant species.
| Gene/protein encoded | Mutation/Overexpression | Plant studied | Effect on abiotic stress and/ or germination | Altered compounds/pathways/processes involved | References | |
|---|---|---|---|---|---|---|
| I. ABA RELATED TRANSCRIPTION FACTORS | ||||||
| 1. | OsbZIP46CA1 | Overexpression | Rice | Drought tolerance | Positive regulator of ABA signaling | Tang et al., 2012 |
| 2. | OsbZIP71 | Overexpression | Rice | Drought tolerance | Positive regulator of ABA signaling | Liu et al., 2014 |
| 3. | OsbZIP52 | Overexpression | Rice | Cold and drought sensitivity | Negative regulator of ABA signaling | Liu et al., 2012 |
| 4. | OsbZIP23 | Overexpression | Rice | Salinity and drought tolerance | Positive regulator of ABA signaling | Xiang et al., 2008 |
| 5. | OsABF2 | Mutant | Rice | Sensitive to salinity, drought, and oxidative stress | Modulates transcript levels of abiotic stress-responsive genes | Hossain et al., 2010 |
| 6. | DIG1 (Dynamic Influencer of Gene expression 1), DIG2 | DEX Inducible expression | Arabidopsis | Salinity sensitivity | Differential ABA signaling | Song et al., 2016 |
| 7. | OsAP2-39 (AP2-domain containing Transcription) | Overexpression | Rice | Low germination rate, ABA sensitivity, increased endogenous ABA level | Direct activation of ABA biosynthesis gene OsNCED1, whereas directly activating GA-inactivating gene OsEUI (Elongated Uppermost Internode) | Yaish et al., 2010; Shu et al., 2018 |
| 8. | ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) | Overexpression | Arabidopsis | Insensitive to wounding and water stress treatments, ABA insensitive | Direct regulation of ABA biosynthesis genes (NCED3 and NCED9) and the ABA-responsive gene RESPONSIVE TO DESICCATION 26 (RD26) under normal and wounding conditions but not under drought stress | Chen et al., 2016 |
| 9. | ABA-INSENSITIVE 4 (ABI4) | Mutant | Arabidopsis | Reduced primary seed dormancy, Resistant to paclobutrazol PAC (GA biosynthesis inhibitor) | ABI4 negatively regulates GA biosynthesis and by inhibits ABA catabolic genes expression (CYP707A1 and CYP707A2) | Shu et al., 2013, 2016 |
| Overexpression | Arabidopsis | Sensitive to PAC | ||||
| II. ABA METABOLISM GENES | ||||||
| 1. | AtABA2 (Encodes ABA biosynthesis gene, short-chain-dehydrogenase reductase) | Overexpression | Arabidopsis | Delayed germination, Salinity tolerance | Increased ABA level, Altered primary metabolite level upon stress | Lin et al., 2007 |
| 2. | SgNCED1 (from Stylosanthes guianensis) | Heterologous expression | Tobacco | Drought tolerance | Increase in the antioxidant enzyme activities | Bao et al., 2016 |
| 3. | LeNCED1 (from tomato) | Heterologous expression | Petunia | Drought tolerance | Decreases in stomatal conductance, transpiration, and photosynthesis and increased concentrations of proline | Estrada-Melo et al., 2015 |
| 4. | Cytochrome P450 CYP707A encodes ABA 8′- hydroxylases | Mutant (cyp707a2) | Arabidopsis | Enhanced seed dormancy | Altered ABA levels in seeds | Asano et al., 2011 |
| 5. | AtBG1 (β-glucosidase) | Mutant (atbg1) | Arabidopsis | Dehydration sensitive and early germination | Defective stomatal movement | Lee et al., 2006 |
| 6. | AtBG2 (β-glucosidase) | Mutant (atbg2) | Arabidopsis | Dehydration and salinity sensitive | Altered ABA level | Xu et al., 2012 |
| Overexpression | Arabidopsis | Dehydration and salinity tolerance | ||||
| 7. | BGLU10 (β-glucosidase) | Mutant (bglu10) | Arabidopsis | Drought sensitive | Increased rate of water loss, Reduced ABA content and expressions of ABA-and drought-responsive genes | Wang et al., 2011 |
| Overexpression | Arabidopsis | Drought tolerance | Reduced rate of water loss, Increased ABA content and expressions of ABA-and drought-responsive genes | |||
| 8. | AtDTX50 (DTX/Multidrug and Toxic Compound Extrusion (MATE) family member) | Mutant (atdtx50) | Arabidopsis | Drought tolerance | ABA efflux carrier in guard cells | Zhang et al., 2014 |
| 9. | AtPDR12/ABCG40 (a ATP-binding cassette (ABC) transporter) | Mutant (atabcg40) | Arabidopsis | Drought sensitive, Low rate of ABA-induced stomatal closer, impaired ABA regulation of seed germination and root development | Altered stomatal regulation, Altered cellular uptake of ABA | Kang et al., 2010 |
| 10. | AtABCG25 (a ATP-binding cassette (ABC) transporter) | Overexpression | Arabidopsis | less transpiration from the leaves (High leaf temperature) | AtABCG25 acts as an ABA exporter Which delivers ABA to guard cells. | Kuromori et al., 2010 |
| 11. | AtABCG22 (a ATP-binding cassette (ABC) transporter) | Mutant (atabcg22) | Arabidopsis | Drought sensitive, Increased transpiration through altered stomatal regulation | ABA signaling and ABA biosynthesis | Kuromori et al., 2011 |
| 12. | XERICO (encodes a RING-H2 zinc-finger protein) | Overexpression | Arabidopsis | Hypersensitivity to osmotic and salinity stress during germination and early seedling stage; and increased drought tolerance in the adult stage | Altered accumulation of ABA and differential expression GA, ethylene and ABA biosynthesis genes | Ko et al., 2006; Verma et al., 2016 |
| Rice | Salinity and drought tolerance | Increased ABA level and ABA-mediated stress response | Zeng et al., 2015 | |||
| III. ABA SIGNALING GENES | ||||||
| 1. | PYL8/RCAR3 | Overexpression | Arabidopsis | Hypersensitive to Salt and Osmotic Stresses | Positively regulates ABA signaling | Saavedra et al., 2010 |
| 2. | PYL5 and PYL7 (from tomato) | Heterologous expression | Arabidopsis | Drought tolerance | Not defined | González-Guzmán et al., 2014 |
| 3. | SAPK9 | Overexpression | Rice | Drought tolerance | Modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression | Dey et al., 2016 |
| 4. | SnRKD,E,I | srk2d/e/i triple mutant | Arabidopsis | Drought sensitivity | Decreased expression of ABA- and stress-inducible genes | Fujita et al., 2009 |
| 5. | ZmSAPK8 (from corn) | Heterologous expression | Arabidopsis | Salinity tolerance | Proline accumulation and low relative electrolyte leakage | Ying et al., 2011 |
| 6. | Abscisic acid-insensitive(abi) | Mutant (abi3, abi4, and abi5) | Arabidopsis | ABA-mediated inhibition of seed germination and low water potential-induced ABA and proline accumulation | Altered ABA or Proline accumulation | Finkelstein, 1994; Verslues and Bray, 2006 |
| 7. | SIZ1 (SUMO E3 ligase) | Mutant (siz1–2 and siz1–3) | Arabidopsis | Increased ABA inhibition of seed germination and seedling primary root growth. | Induced expression of genes that are ABA-responsive (ABI5-dependent signaling) (e.g., RD29A, Rd29B, AtEm6, RAB18, ADH1) | Miura et al., 2009 |
The expression of ABA biosynthesis genes is reported to show a direct impact on seed germination along with abiotic stresses. The identification and characterization of NCED genes revealed that the tissue-specific expression of these genes and the resultant modulation of endogenous ABA level at different developmental stages are responsible for the regulation of specific processes, such as seed maturation and seed germination, besides response to abiotic stresses (Lefebvre et al., 2006; Martínez-Andújar et al., 2011). Within the seeds, NCED6 was shown to express in the endosperm whereas NCED9 is expressed in both embryo and endosperm during Arabidopsis seed development. The induction of NCED6 inhibits seed germination by increasing the endogenous level of ABA. These and similar findings have clearly established a causal role for ABA in regulating the physiological and developmental processes studied.
It is known that ABA accumulates under specific conditions, such as abiotic stresses. Therefore, the endogenous concentration of biologically active ABA at the site of perception has to be regulated. Apart from biosynthesis, ABA catabolism and transport are the two key essential processes that control ABA-mediated stress regulation. Cytochrome P450 type enzymes (CYP707As) catalyze the deactivation reaction resulting in phaseic acid (PA) and dihydro phaseic acid (DPA) as the main ABA catabolites (Ng et al., 2014; Sah et al., 2016), which do not appear to have any significant biological activity (Sharkey and Raschke, 1980; Kepka et al., 2011). ABA and its catabolites (hydroxylated) can be conjugated to glucose, catalyzed by ABA glucosyl ester (ABA-GE) and become inactivated (Zeevaart and Creelman, 1988; Lim et al., 2005). However, ABA-GE could be converted to ABA upon induction of different abiotic stresses (Ye et al., 2012; Sah et al., 2016). Two β-glucosidases, AtBG1 and AtBG2 localized in the vacuole and endoplasmic reticulum, respectively, hydrolyze ABA-GE (Burla et al., 2013). ABA is a weak acid (pKa ~ 4.7), which can be protonated to become membrane permeable so that it can diffuse passively across the cell membrane (Wilkinson and Davies, 2010; Ng et al., 2014; Sah et al., 2016). Several transporters have been identified in different species of plants, which regulate the accumulation and translocation of active ABA along the plant body involving different organelles (Kang et al., 2010; Kanno et al., 2012; Ye et al., 2012). Also, several genes related to ABA metabolism and transport in different plant species are reported to alter abiotic stress tolerance summarized in Table 1.
ABA signaling genes, abiotic stress, and germination
The identification of ABA receptors in Arabidopsis and other plant species is one of the key findings in ABA signaling. The PYR/PYL/RCAR family of proteins are established as the most plausible ABA receptors. Expression profile study of these receptors revealed their role in ABA signaling as well as in the regulation of abiotic stresses (Park et al., 2009). Triple and quadruple mutants of pyl showed altered ABA sensitivity with respect to seed germination and growth, while overexpression lines conferred tolerance toward abiotic stress (Santiago et al., 2009; Saavedra et al., 2010). Overexpression of RCAR gene resulted in altered ABA-dependent germination and seedling growth (Ma et al., 2009). PYR/PYL/RCAR receptors in the presence of ABA form a complex and deactivate PP2C, which otherwise inactivates the SnRK2s, a central regulator of ABA signaling. Subclass III of SnRK2 in Arabidopsis and rice are shown to be involved in ABA signaling (Kobayashi et al., 2005). Their expressions were induced in the presence of ABA. Furthermore, they are responsible for the activation several ABRE binding factors (ABFs). ABFs belong to basic leucine zipper (bZIP) transcription factor family, which is one of the key regulators of ABA responses in plants. In general, they interact with the cis-acting conserved regulatory element, ABREs (ABA-responsive elements) and in turn regulate transcription of several downstream ABA-responsive genes (Choi et al., 2000; Kim et al., 2002; Lopez-Molina et al., 2002). Table 1 summarizes the effect of the genes related to ABA signaling with respect to various abiotic stresses in different plant species.
GA biosynthesis genes, abiotic stress, and seed germination
The discovery of bioactive gibberellic acid (GA) was the result of an investigation of fungal (Gibberella fujikuroi) infection in rice by Teijiro Yabuta and co-workers (Yabuta and Sumiki, 1938). Since then, more than a hundred GAs have been identified from different sources, (from bacteria to plants). However, only a few of them have been shown to have biological activity (Yamaguchi, 2008; Hedden and Thomas, 2012). Gibberellins control different stages of plant development, including seed germination, seedling growth, stem elongation, root extension, leaf size and shape, flower and fruit development, pollination (García-Martínez et al., 1997; Yamaguchi, 2008; Hedden and Thomas, 2012).
In plants three classes of enzymes are required for the biosynthesis of bioactive GAs (GA1, GA3, and GA4) from the precursor compound geranylgeranyl diphosphate (GGDP), which is aided by terpene synthases (TPSs), cytochrome P450 monooxygenases (P450s), and 2-oxoglutarate–dependent dioxygenases (2ODDs) (Yamaguchi, 2008; Hedden and Thomas, 2012). Two TPSs, ent-copalyl diphosphate synthase (CPS) and ent-kaurene synthase (KS), which are located in the plastids are responsible for the first few steps of GA biosynthesis (conversion of GGDP to ent-kaurene). Then two P450 enzymes, namely, ent-kaurene oxidase (KO) and ent-kaurenoic acid oxidase (KAO) convert ent-kaurene to GA12. Finally, three active GAs are formed by reactions catalyzed by GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox), that belong to 2ODDs (Yamaguchi and Kamiya, 2000; Hedden, 2001; Yamaguchi, 2008; Hedden and Thomas, 2012). In plants, deactivation of the GAs is critical for maintaining the levels of bioactive GAs, which is regulated by GA 2-oxidases (GA2oxs), belonging to 2ODDs (Yamaguchi and Kamiya, 2000; Yamaguchi, 2008). Additionally, 16α,17-epoxidation (Luo et al., 2006; Zhu et al., 2006) and methylation of the C-6 carboxyl group of GAs (Varbanova et al., 2007) are involved in the deactivation of GAs in different plant species.
Several GA biosynthesis genes are expressed in growing tissues during Arabidopsis development (Silverstone et al., 1997) and also in crop plants such as wheat (Aach et al., 1997), rice (Kaneko et al., 2003), and tobacco (Itoh et al., 1999). This suggests that biologically active GAs are synthesized at the site of their action in several cases. However, in rice, it has been shown that GA biosynthesis genes are not expressed in the aleurone layer, but GA signaling event occurs there, which suggests paracrine signaling by GAs (Kaneko et al., 2002, 2003). In addition, in Arabidopsis, GA-dependent gene expressions have been shown in the sites where bioactive GAs are not produced (Yamaguchi et al., 2001). It has also been shown that early and late steps of GA biosynthesis take place in provascular tissue and, cortex and endodermis, respectively (Yamaguchi and Kamiya, 2000; Yamaguchi et al., 2001). This suggests the existence of intercellular movement/transport of GA biosynthesis intermediates. Lack or absence of GA leads to altered GA signaling and germination related phenotype, which has been revealed by different studies done in mutants of GA metabolism (Table 2). The relationship between expressions of GA metabolism-related genes and tolerance toward abiotic stresses have been shown. Mutants in GA biosynthesis genes (GA20ox and GA3ox) showed drought tolerance phenotype and overexpression of GA20ox confers drought sensitivity in Arabidopsis (Colebrook et al., 2014).
Table 2.
Summary of regulation of GA metabolism and signaling genes with respect to abiotic stress and seed germination in different plant species.
| Gene | Protein encoded | Genotype | Plant studied | Effect on abiotic stress and/ or germination | Altered compound/pathways/processes | References | |
|---|---|---|---|---|---|---|---|
| I. GA METABOLISM AND REGULATORS OF GA SIGNALING | |||||||
| 1. | GAI, RGA, RGL1, and RGL2 | DELLA Protein | DELLA quadruple-mutant gai-t6 rga-t2 rgl1-1 rgl2-1 | Arabidopsis | Salt sensitivity, Early flowering | DELLA-dependent reduced accumulation of bioactive GAs | Achard et al., 2006 |
| 2. | GA-INSENSITIVE (GAI) | DELLA Protein | Mutant (gai) | Arabidopsis | Salinity tolerance | DELLA-dependent growth restraint | Achard et al., 2006 |
| 3. | GA-deficient GA1-3 | DELLA Protein | Mutant (ga1-3) | Arabidopsis | Salinity tolerance | DELLA-dependent growth restraint | Achard et al., 2006 |
| 4. | PROCERA | DELLA Protein | Mutant (pro) | Tomato | Enhanced stomatal conductance and rapid wilting under drought stress | Negative regulator of GA signaling | Nir et al., 2017 |
| 5. | NUCLEAR FACTOR Y C proteins (NF-YC) homologs NF-YC3, NF-YC4 and NF-YC9 Nuclear factor Y (NF-Y) family proteins | Mutant nf-yc3 nf-yc4 nf-yc9 (nf-ycT) | Arabidopsis | Higher germination rates than the wild type in the presence of a of paclobutrazol PAC (GA biosynthesis inhibitor) | Altered GA and ABA signaling | Liu et al., 2016 | |
| Overexpression (35S:NF-YC3 and 35S:NF-YC9) | Arabidopsis | Lower germination rates than the wild type in the presence of a of PAC | |||||
| 6. | GA1-3 and GA2 | GA biosynthesis genes | Mutant (ga1-3 and ga2) | Arabidopsis | Unable to germinate in the absence of exogenous GA | Absence or the changed composition of endogenous GA's | Koornneef and Van Der Veen, 1980 |
| 7. | GIB-1 | Gibberellin biosynthesis gene | Mutant (gib-1) | Tomato | Unable to germinate without exogenous GA | Lack of GA | Karssen et al., 1989; Benson and Zeevaart, 1990 |
| 8. | GID1, GID2, and GID3 | GA INSENSITIVE DWARF (GID) GA receptor | Mutant (gid1a gid1b gid1c) | Arabidopsis | Unable to germinate | The absence of DELLAs destruction by gibberellins | Willige et al., 2007 |
| 9. | DAG1 | Transcription factor DOF AFFECTING GERMINATION 1 (DAG1) | dag1 | Arabidopsis | Reduced GA requirement for the seeds to germinate | Negative regulation GA biosynthesis by acting downstream of PIL5 (PHYTOCHROME INTERACTING FACTOR 3-LIKE 5) | Gabriele et al., 2010 |
| 10. | SPY-1 and SPY-3 | O-linked N-acetylglucosamine transf Erase | Mutants (spy-1 and spy-3) | Arabidopsis | Drought and salinity tolerance | Altered environmental stress signals via Gibberellin and Cytokinin cross talk | Qin et al., 2011 |
| Overexpression | Arabidopsis | Drought sensitive | |||||
| 11. | KYP/SUVH4 | Histone methyltransferase | Mutant | Arabidopsis | Increased seed dormancy and sensitivity to ABA | Altered ABA and GA regulation, and altered expression of dormancy-related genes | Zheng et al., 2012 |
| Overexpression | Arabidopsis | Reduced seed dormancy and sensitivity to ABA | |||||
| II. GA-RELATED TFS | |||||||
| 1. | DDF1 | AP2 transcription factor of the DREB1/CBF subfamily | Overexpression | Arabidopsis | Salinity tolerance | Upregulates expression of a gibberellin-deactivating gene, GA2ox7 | Magome et al., 2008 |
| 2. | ERF6 | Ethylene response factor (ERF) | Gain-of-function | Arabidopsis | Hypersensitive to osmotic stress | Inhibits growth through a GA/DELLA dependent mechanism | Dubois et al., 2013 |
| 3. | CBF1 | C-repeat/drought-responsive element binding factor | Overexpression | Arabidopsis | Cold tolerance, Inhibit seed germination | Accumulation of DELLAs | Achard et al., 2008 |
| 4. | PIL5 | PHYTOCHROME INTERACTING FACTOR 3-LIKE 5 | Overexpression | Arabidopsis | Inhibits seed germination | Activation of a GA catabolic gene (GA2ox2) and repression of GA biosynthesis genes GA3ox1 and GA3ox2 | Oh et al., 2007 |
| 5. | CHOTTO1 | Double APETALA2 Repeat Transcription Factor | Overexpression | Arabidopsis | Sensitive to ABA during seed germination processes | Inactivates GA biosynthesis | Yano et al., 2009; Shu et al., 2018 |
| 6. | SNORKEL1 and SNORKEL2 | Ethylene response factor (ERF) domain proteins | Overexpression | Rice | Submergence-tolerance | Increases in bioactive GA levels | Hattori et al., 2009 |
| 7. | SUB1A | Ethylene response factor (ERF) domain proteins | Overexpression | Rice | Submergence-tolerance | Negatively regulates GA responses by accumulation of the GA signaling repressors Slender Rice-1 (SLR1) and SLR1 Like-1 (SLRL1) | Fukao and Bailey-Serres, 2008 |
Characterization of mutants and genetic studies revealed several GA signaling components (Hedden and Phillips, 2000; Stamm et al., 2012; Davière and Achard, 2013). DELLA proteins, belonging to the GRAS family of transcription factors, are identified as a major repressor of GA signaling. DELLA proteins restrict cell proliferation and expansion by negatively regulating gibberellin signaling and hence inhibit the plant growth (Peng et al., 1997, 1999; Fleet and Sun, 2005). DELLA proteins such as RGL2 complexed with DOF6 transcription factor has also been shown to have positive effect on target genes such as GATA12 in regulating seed germination (Ravindran et al., 2017). DELLAs are degraded by a signal cascade involving GA and its positive regulators (Hedden and Phillips, 2000; Achard and Genschik, 2009; Wang et al., 2009; Figure 1). The GA signaling components are reported to affect various aspects of germination and abiotic stresses as well (Table 2). DELLAs are also reported to confer salt tolerance in Arabidopsis by altering the duration of vegetative growth. Also, two of the DELLA proteins, RGA and GAI have a major role in salt-induced plant growth regulation (Achard et al., 2006).
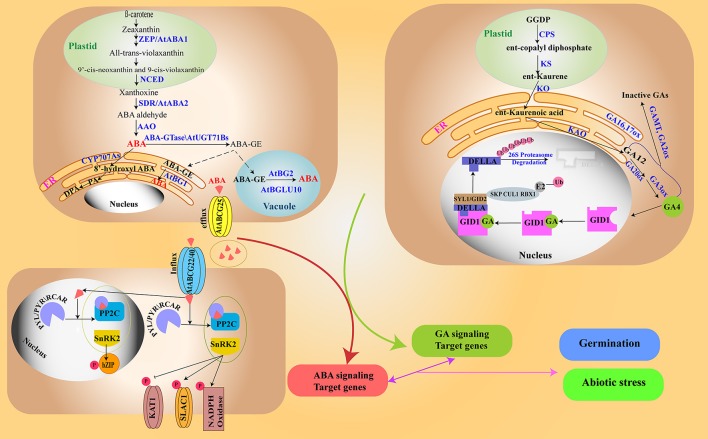
Figure 1.
ABA and GA metabolism and signaling/ABA is synthesized from carotenoids in a series of reactions in the plastids and cytoplasm (top left). ABA is catabolized to form phaseic acid. ABA transport occurs through different transporters, and ABA elicits distinct signaling cascades (in the nucleus and cytoplasm) (bottom left). GA biosynthesis starts from GGDP in the plastid and a portion of it is catabolized to inactive forms (top right). In the GA signaling pathway, GA causes destruction of DELLAs (negative regulator of GA) via the 26S proteasome machinery. The two signaling pathways crosstalk to regulate seed germination and abiotic stresses (bottom right).
GA and ABA crosstalk
Hormones regulate plant growth and development either synergistically or antagonistically, involving a series of complex pathways and networks (Liu et al., 2010; Dong et al., 2016; Rowe et al., 2016). In the preceding sections, we described the individual roles of GA and ABA in two important aspects affecting plant development; germination and abiotic stresses. The information summarized in Tables 1, 2 along with the preceding description show that ABA and GA antagonistically mediate plant developmental processes including seed dormancy and germination. Hence, it is essential to maintain an optimal balance between the endogenous levels of ABA and GA for plant development.
In response to different developmental stages and environmental conditions, various changes occur in the metabolism and signal transductions of these two plant hormones which keep a correct balance between GA and ABA and hence plant homeostasis. In the following sections we will summarize how genes, components and network involving crosstalk of GA and ABA participate in the regulatory processes.
In many instances, possible crosstalk events have been shown between ABA and GA with respect to various abiotic stresses and plant growth. Unfavorable conditions lead to high ABA and low GA levels in seeds whereas favorable conditions cause the reverse situation. Seed dormancy is maintained by ABA whose level is found to progressively increase from embryogenesis to embryo maturation (Karssen et al., 1983). ABA restricts embryo growth potential by inhibiting water uptake (imbibition) and hence cell-wall loosening, which is a key step to start germination (Schopfer and Plachy, 1984; Gimeno-Gilles et al., 2009). ABA also leads to induction of Late Embryogenesis Abundant (LEA) genes and growth arrest by activating a basic leucine zipper transcription factor, ABSCISIC ACID INSENSITIVE 5 (ABI5) (Finkelstein and Lynch, 2000). Many LEA genes are reported to confer abiotic stress tolerance in plants (Lopez-Molina and Chua, 2000; Lee et al., 2005). Synergistic repression of germination has been reported through ABRE and RY elements by ABI5 and ABI3 (activated by ABA) (Lopez-Molina et al., 2002; Park et al., 2011). Under favorable conditions (light, temperature and moisture) GA biosynthesis and associated pathways are activated, which results in the release from the inhibitory effect of ABA. Cold stratification and light lead to an increase in bioactive GAs via transcription factors PIF3-like 5 (PIL5), Blue Micropylar End3 (BME3) and SPATULA (SPT) (Liu et al., 2005; Penfield et al., 2005; Oh et al., 2006; Figure 1). Thus, it is clear that various interactions between ABA and GA in seeds help to regulate dormancy and germination.
Several recent studies showed the regulation of GA and ABA in light- and temperature-mediated seed germination and dormancy. PIL5, a light-labile transcription factor, regulates both GA and ABA signaling and thereby inhibits seed germination. It indirectly regulates GA biosynthesis genes and directly regulates GA signaling genes. Thus, PIL5 represses GA biosynthesis genes (GA3ox1 and GA3ox2) and activates a GA catabolic gene (GA2ox2) indirectly (Gabriele et al., 2010). However, it binds to the promoter region of the GA signaling repressor genes, GAI and RGA and regulates their transcription (Oh et al., 2007). On the other hand, PIL5 has the opposite effect on the ABA biosynthesis genes. It activates ABA biosynthesis genes (ABA1, NCED6, and NCED9) and represses an ABA catabolic gene (CYP707A2) (Finkelstein et al., 2008). Furthermore, increased expression of DELAY OF GERMINATION 1 (DOG1) which acts downstream to PIL5, leads to repression of GA biosynthesis and activation of ABI3 and ABI5 (Bentsink et al., 2006; Skubacz and Daszkowska-Golec, 2017). Similarly, a CCCH-Type zinc finger protein, SOMNUS (SOM) is reported to act downstream of PIL5 in order to negatively regulate light-dependent seed germination in Arabidopsis (Kim et al., 2008). Several other CCCH zinc finger proteins (AtTZF4, 5, and 6) negatively regulate GA- and light-mediated seed germination and positively regulate ABA-mediated seed germination. Expression patterns of genes regulating GA and ABA metabolism have been reported to be well coordinated with seasonal seed dormancy in Arabidopsis. Thus, upregulation of GA catabolism and ABA biosynthesis genes was observed during low temperature (winter) which leads to increased dormancy (Footitt et al., 2011). Consistent with that, upregulation of GA biosynthesis ABA catabolism genes have been reported during high temperature (spring and summer) and decreased dormancy (Footitt et al., 2011). The transcription factor SPT controls the germination response to cold and light. It can repress the GA biosynthesis genes (GA3ox1 and GA3ox2) (Penfield et al., 2005) as well as the expression of ABI4 and a DELLA gene RGA, but it promotes expression of ABI5 and RGL3, another DELLA gene (Vaistij et al., 2013).
Various abiotic stresses (external environment) lead to changes in the plant response and therefore alter the balance of endogenous levels of GA and ABA. High temperature induces ABA biosynthesis genes (ZEP, NCED2, NCED5, and NCED9) and hence increases the ABA level whereas it decreases the GA level by repressing GA biosynthesis genes in Arabidopsis seeds (Toh et al., 2008). The transcription factor FUS3 leads to delayed germination at high temperature by activating seed-specific, ABA biosynthetic and ABA signaling genes (Chiu et al., 2012). ABI3, ABI5, and DELLAs form a complex to directly activate SOM expression at high temperature, which results in altered expression of ABA and GA metabolism genes (Lim et al., 2013).
DELLA-dependent salt-induced growth inhibition in the DELLA quadruple-mutant was also shown to be associated with DELLA accumulation and ABA signaling. Also, upon ABA treatment accumulation of GFP-RGA was not observed in the abi1-1 roots, but only seen in the untreated WT control (Peng et al., 1997; Fleet and Sun, 2005; Achard et al., 2006), showing the crosstalk between ABA and GA signaling. Furthermore, quadruple-DELLA mutant was also shown to have ABA insensitive phenotype. In addition, ABA-induced delay in flowering was shown to be DELLA dependent (Achard et al., 2006). PROCERA (a DELLA protein in tomato) promotes stomatal closure in an ABA-dependent manner by increasing ABA sensitivity (Nir et al., 2017). Another study showed that NUCLEAR FACTOR-Y C (NF-YC) homologs (NF-YC3, NF-YC4, and NF-YC9) interact with the DELLA protein RGL2 and target ABI5 (Liu et al., 2016), thus regulating germination by modulating GA- and ABA-responsive genes in Arabidopsis. In addition, NF-YC9 was also reported to regulate ABA signaling via direct interaction with ABI5 (Bi et al., 2017). Therefore, NF-YC family members could integrate GA and ABA antagonistic crosstalk involving DELLA protein and ABA signaling TFs. Global analysis of DELLA targets revealed several downstream targets and responsive genes (Zentella et al., 2007). XERICO which has a key role in mediating various abiotic stresses by modulation of ABA level and expression of ABA-responsive genes is a target of DELLA (Ko et al., 2006; Zentella et al., 2007; Zeng et al., 2015). Other reports have also shown that DELLA contributes toward upregulation of ABA level by increasing the XERICO transcript levels (Zentella et al., 2007; Figure 2). This represents another example of how DELLA proteins can control plant growth and abiotic stress tolerance through specific crosstalk with ABA signaling pathway.
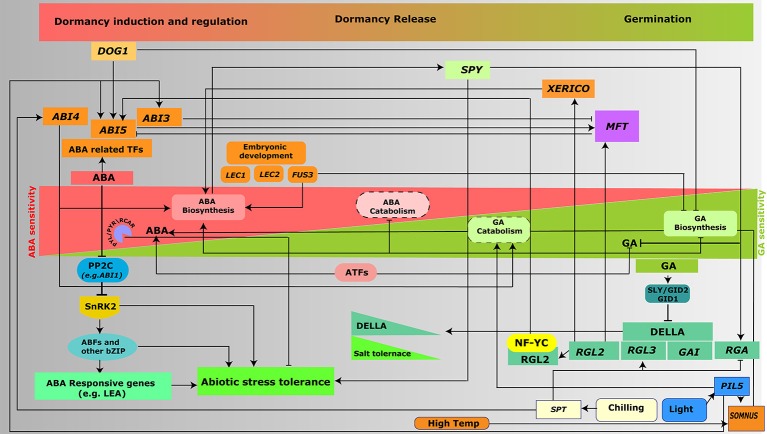
Figure 2.
Interplay of ABA and GA signaling in the regulation of seed germination and abiotic stresses. Switch from seed dormancy to germination is controlled by the intricate balance between ABA and GA levels. ABA- and GA-signaling and metabolism genes regulate the expression of various genes (as mentioned in the text) and hence control two of the major aspects of plant development, germination and response to abiotic stresses.
DELLA repressors are mainly degraded through the ubiquitin-proteasome system involving recruitment of Skip, Cullin, and F-box E3 ubiquitin ligase to the GA-GID1-DELLA complex by SLEEPY1 (SLY1) (Steber et al., 1998; Hedden, 2001; Murase et al., 2008; Achard and Genschik, 2009; Wang et al., 2009; Figure 1). In addition to ubiquitination, the DELLA signaling components are regulated by SUMOylation (small ubiquitin-related modifier). E3 SUMO ligase AtSIZ1 negatively regulates ABA signaling by SUMOylation of ABI5 in Arabidopsis during germination (Miura et al., 2009; Liu and Hou, 2018). In addition, AtSIZ1 was reported to positively regulate GA signaling by SUMOylating SLY1 (Kim et al., 2015; Liu and Hou, 2018). Therefore, SIZ1 could be another direct link between GA and ABA signaling by regulating ABI5 and SLY1.
Another E3 ligase, ANAPHASE-PROMOTING COMPLEX/CYCLOSOME (APC/C) has a link between GA and ABA signaling in rice via SnRK2-APC/CTE(Tiller Enhancer/activator) module (Lin et al., 2015; Liu and Hou, 2018). Loss-of-function of TE, leads to hyposensitivity to GA and hypersensitivity to ABA. Furthermore, ABA inhibits APC/CTE activity by phosphorylation of TE through the activation of rice SnRK2s. This event interrupts the association between TE and OsPYL/RCARs (ABA receptor), which results in stabilization of the receptor. Conversely, opposite effect has been shown by GA by inhibiting rice SnRK2 (Lin et al., 2015).
Several TFs other than DELLAs have been reported to act as potential mediators between ABA and GA metabolism and signaling. Two APETALA 2 (AP2)-domain containing transcription factors (ATFs), Arabidopsis ABA-INSENSITIVE 4 (ABI4) and rice OsAP2-39, have key roles in the antagonistic crosstalk between ABA and GA. ABI4 positively regulates primary seed dormancy by downregulating GA biosynthesis and by inhibiting ABA catabolic genes (CYP707A1 and CYP707A2) (Shu et al., 2013). Further, GA represses the expression level of ABA biosynthesis gene, NCED6 and increases expression of the GA-deactivating gene GA2ox7 in an ABI4-dependent manner (Shu et al., 2016). In rice, OsAP2-39 induces ABA level by directly activating ABA biosynthesis gene OsNCED1, whereas it reduces GA level by directly activating GA-inactivating gene OsEUI (Elongated Uppermost Internode) (Shu et al., 2018). Further, enhanced ABA level due to activation of OsNCED1 induces the OsEU1 expression, which ultimately decreases GA accumulation (Yaish et al., 2010; Shu et al., 2018). Another study showed that CHOTTO1, a double-AP2 domain-containing TF regulates seed germination in Arabidopsis through ABA-mediated repression of GA biosynthesis (Yano et al., 2009).
MYB96 TF controls primary seed dormancy by directly activating ABA biosynthesis genes (NCED2, NCED5, NCED6, and NCED9) and indirectly repressing GA biosynthesis genes (GA3ox1 and GA20ox1) (Lee et al., 2015). Another key regulator of seed dormancy Mother of FT and TFL 1 (MFT), controls ABA and GA signaling pathways (Xi et al., 2010). MFT promotes germination by downregulating ABA signal via repression of ABI5 expression. MFT expression is induced by RGL2 and ABI5, but downregulated by ABI3 and MFT (Xi et al., 2010; Skubacz and Daszkowska-Golec, 2017; Figure 2). Another three transcriptional regulators involved in regulating embryonic development are the LEC genes; LEAFY COTYLEDON1 (LEC1), B3 domain factors LEC2, and FUSCA3 (FUS3) (Keith et al., 1994; Gazzarrini et al., 2004). Loss-of-function of these genes leads to the alteration of embryonic leaves (cotyledons) to take on the appearance of vegetative leaves (Gazzarrini et al., 2004). One of them, FUS3 is known to positively regulate ABA biosynthesis, and negatively regulate GA biosynthesis (Gazzarrini et al., 2004). Another B3 TF GERMINATION DEFECTIVE 1 (GD1) regulates seed germination by suppressing a LEC2/FUS3-like gene of rice (OsLFL1) and modulating expression of GA metabolic genes (OsGA3ox, OsGA20ox, and OsGA2ox) (Guo et al., 2013).
These examples clearly show the crosstalk between ABA and GA in controlling seed development as well as germination. Such crosstalk has been predicted based on earlier studies. With the limited number of definitive studies on such signal crosstalk, we are just beginning to gain valuable insights regarding the regulation of specific growth and developmental processes.
Conclusions and future perspectives
It is evident from the foregoing review that the signaling interactions among several phytohormones are common in regulating various stages and processes of plant development. Such regulatory crosstalk can occur at multiple stages of biosynthesis or signaling for different hormones. Biosynthesis of bioactive hormones and their transport (passive and/or active) as well as signaling cascades that regulate downstream target genes (of different classes) further add to the complexity of the already elaborate cellular communication network. This has been highlighted here with the examples of ABA and GA metabolism and their regulation. Selected genes that play significant roles in the regulation of seed dormancy and germination and various abiotic stresses were also discussed. It is evident that several positive and negative regulators of ABA and GA have direct or indirect impacts on germination and abiotic stresses. Many transcription factors and signaling components of these two phytohormones help to maintain an intricate balance between endogenous levels of bioactive ABA and GA. Furthermore, studies have identified several ABA and GA crosstalk points showing positive and negative regulation of different molecular modules associated with their metabolism and signaling. There are a few open questions that can help in formulating the future research directions. Despite the fact that there are some studies on ABA transport in different cell types and tissues, there might be many unknown pathways/transporters that are yet to be explored. Moreover, very few reports on the transport mechanism of GA are available. The antagonistic roles of GA and ABA in controlling developmental processes have been established by several pieces of evidence; however, there could be synergistic crosstalk between GA and ABA in some instances whose underlying molecular mechanisms remain undiscovered. Although several target genes of a few TFs have been established (eg. MYB96, ABI4, OsAP2-39) (Yaish et al., 2010; Shu et al., 2013; Lee et al., 2015) identification of direct targets/genes of several TFs and components of GA and ABA signaling modules are worth investigating. The detailed analyses of direct targets involved in GA and ABA metabolism and signaling at different developmental stages will provide us with more insights into GA and ABA crosstalk. Recent studies revealed several new cues associated with GA and ABA signaling. A few epigenetic modifiers have been documented to be involved in GA and ABA signaling cascade (Ryu et al., 2014; Liu et al., 2016; Peirats-Llobet et al., 2016). However, the mechanisms by which these epigenetic regulators mediate crosstalk between GA and ABA need to be investigated. It is known that complexes of TFs regulate downstream target genes (Kepka et al., 2011; Lim et al., 2013; Heyman et al., 2016; Iwata et al., 2017), and therefore, future investigations into new protein complexes associated with GA and ABA signaling will reveal interesting molecular mechanisms of developmental regulation. Although several signaling components controlling various aspects of germination and abiotic stresses have been identified, the nature of the underlying mechanisms of many of the events remain to be clarified. Nevertheless, such specific interaction points that have been identified for these two phytohormones will offer potential genetic intervention strategies to control growth and abiotic stress remediation in future crop breeding programs.
Author contributions
BV and PK conceived the idea and wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Research work in our laboratory was supported by National Research Foundation, Singapore under its Competitive Research Program (CRP award no. NRF-CRP 7-2010-02). We thank Dr. Pratibha Ravindran for useful discussions.
References
- Aach H., Bode H., Robinson D. G., Graebe J. E. (1997). ent-Kaurene synthase is located in proplastids of meristematic shoot tissues. Planta 202, 211–219. 10.1007/s004250050121 [DOI] [Google Scholar]
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. 10.1126/science.1118642 [DOI] [PubMed] [Google Scholar]
- Achard P., Genschik P. (2009). Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J. Exp. Bot. 60, 1085–1092. 10.1093/jxb/ern301 [DOI] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20, 2117–2129. 10.1105/tpc.108.058941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., et al. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19, 1188–1193. 10.1016/j.cub.2009.05.059 [DOI] [PubMed] [Google Scholar]
- Asano T., Hakata M., Nakamura H., Aoki N., Komatsu S., Ichikawa H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. 10.1007/s11103-010-9717-1 [DOI] [PubMed] [Google Scholar]
- Bao G., Zhuo C., Qian C., Xiao T., Guo Z., Lu S. (2016). Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol. J. 14, 206–214. 10.1111/pbi.12374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson J., Zeevaart J. (1990). Comparison of ent-kaurene synthase A and B activities in cell-free extracts from young tomato fruits of wild-type and gib-1, gib-2, and gib-3 tomato plants. J. Plant Growth Regul. 9, 237–242. 10.1007/BF02041969 [DOI] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C. J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 17042–17047. 10.1073/pnas.0607877103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi C., Ma Y., Wang X.-F., Zhang D.-P. (2017). Overexpression of the transcription factor NF-YC9 confers abscisic acid hypersensitivity in Arabidopsis. Plant Mol. Biol. 95, 425–439. 10.1007/s11103-017-0661-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burla B., Pfrunder S., Nagy R., Francisco R. M., Lee Y., Martinoia E. (2013). Vacuolar transport of abscisic acid glucosyl ester is mediated by ATP-binding cassette and proton-antiport mechanisms in Arabidopsis. Plant Physiol. 163, 1446–1458. 10.1104/pp.113.222547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan Z. (2012). Expression profiling of ABA pathway transcripts indicates crosstalk between abiotic and biotic stress responses in Arabidopsis. Genomics 100, 110–115. 10.1016/j.ygeno.2012.06.004 [DOI] [PubMed] [Google Scholar]
- Chen H. Y., Hsieh E. J., Cheng M. C., Chen C. Y., Hwang S. Y., Lin T. P. (2016). ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 211, 599–613. 10.1111/nph.13914 [DOI] [PubMed] [Google Scholar]
- Cheng W. H., Endo A., Zhou L., Penney J., Chen H. C., Arroyo A., et al. (2002). A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14, 2723–2743. 10.1105/tpc.006494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu R. S., Nahal H., Provart N. J., Gazzarrini S. (2012). The role of the Arabidopsis FUSCA3 transcription factor during inhibition of seed germination at high temperature. BMC Plant Biol. 12:15 10.1186/1471-2229-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.-I., Hong J.-H., Ha J.-O., Kang J.-Y., Kim S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. 10.1074/jbc.275.3.1723 [DOI] [PubMed] [Google Scholar]
- Colebrook E. H., Thomas S. G., Phillips A. L., Hedden P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. 10.1242/jeb.089938 [DOI] [PubMed] [Google Scholar]
- Davière J.-M., Achard P. (2013). Gibberellin signaling in plants. Development 140, 1147–1151. 10.1242/dev.087650 [DOI] [PubMed] [Google Scholar]
- Dey A., Samanta M. K., Gayen S., Maiti M. K. (2016). The sucrose non-fermenting 1-related kinase 2 gene SAPK9 improves drought tolerance and grain yield in rice by modulating cellular osmotic potential, stomatal closure and stress-responsive gene expression. BMC Plant Biol. 16:158 10.1186/s12870-016-0845-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Z., Yu Y., Li S., Wang J., Tang S., Huang R. (2016). Abscisic acid antagonizes ethylene production through the ABI4-mediated transcriptional repression of ACS4 and ACS8 in Arabidopsis. Mol. Plant 9, 126–135. 10.1016/j.molp.2015.09.007 [DOI] [PubMed] [Google Scholar]
- Dubois M., Skirycz A., Claeys H., Maleux K., Dhondt S., De Bodt S., et al. (2013). Ethylene Response Factor6 acts as a central regulator of leaf growth under water-limiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. 10.1104/pp.113.216341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada-Melo A. C., Chao, Reid M. S., Jiang C.-Z. (2015). Overexpression of an ABA biosynthesis gene using a stress-inducible promoter enhances drought resistance in petunia. Hortic. Res. 2:15013. 10.1038/hortres.2015.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R. (1994). Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 5, 765–771. 10.1046/j.1365-313X.1994.5060765.x [DOI] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Gampala S. S., Rock C. D. (2002). Abscisic acid signaling in seeds and seedlings. Plant Cell 14(Suppl.), S15–S45. 10.1105/tpc.010441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Lynch T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. 10.1105/tpc.12.4.599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet C. M., Sun T. P. (2005). A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr. Opin. Plant Biol. 8, 77–85. 10.1016/j.pbi.2004.11.015 [DOI] [PubMed] [Google Scholar]
- Footitt S., Douterelo-Soler I., Clay H., Finch-Savage W. E. (2011). Dormancy cycling in Arabidopsis seeds is controlled by seasonally distinct hormone-signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 108, 20236–20241. 10.1073/pnas.1116325108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Nakashima K., Yoshida T., Katagiri T., Kidokoro S., Kanamori N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. 10.1093/pcp/pcp147 [DOI] [PubMed] [Google Scholar]
- Fukao T., Bailey-Serres J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 16814–16819. 10.1073/pnas.0807821105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele S., Rizza A., Martone J., Circelli P., Costantino P., Vittorioso P. (2010). The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 61, 312–323. 10.1111/j.1365-313X.2009.04055.x [DOI] [PubMed] [Google Scholar]
- García-Martínez J. L., Lopez-Diaz I., Sanchez-Beltran M. J., Phillips A. L., Ward D. A., Gaskin P., et al. (1997). Isolation and transcript analysis of gibberellin 20-oxidase genes in pea and bean in relation to fruit development. Plant Mol. Biol. 33, 1073–1084. 10.1023/A:1005715722193 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S., Tsuchiya Y., Lumba S., Okamoto M., McCourt P. (2004). The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev. Cell 7, 373–385. 10.1016/j.devcel.2004.06.017 [DOI] [PubMed] [Google Scholar]
- Gimeno-Gilles C., Lelièvre E., Viau L., Malik-Ghulam M., Ricoult C., Niebel A., et al. (2009). ABA-mediated inhibition of germination is related to the inhibition of genes encoding cell-wall biosynthetic and architecture: modifying enzymes and structural proteins in medicago truncatula embryo axis. Mol. Plant 2, 108–119. 10.1093/mp/ssn092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Guzmán M., Rodriguez L., Lorenzo-Orts L., Pons C., Sarrion-Perdigones A., Fernandez M. A., et al. (2014). Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 65, 4451–4464. 10.1093/jxb/eru219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Hou X., Fang J., Wei P., Xu B., Chen M., et al. (2013). The rice GERMINATION DEFECTIVE 1, encoding a B3 domain transcriptional repressor, regulates seed germination and seedling development by integrating GA and carbohydrate metabolism. Plant J. 75, 403–416. 10.1111/tpj.12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Yang P. (2015). Studies on the molecular mechanisms of seed germination. Proteomics 15, 1671–1679. 10.1002/pmic.201400375 [DOI] [PubMed] [Google Scholar]
- Hattori Y., Nagai K., Furukawa S., Song X. J., Kawano R., Sakakibara H., et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. 10.1038/nature08258 [DOI] [PubMed] [Google Scholar]
- Hauser F., Waadt R., Schroeder J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–355. 10.1016/j.cub.2011.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. (2001). Gibberellin metabolism and its regulation. J. Plant Growth Regul. 20, 317–318. 10.1007/s003440010039 [DOI] [PubMed] [Google Scholar]
- Hedden P., Phillips A. L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5, 523–530. 10.1016/S1360-1385(00)01790-8 [DOI] [PubMed] [Google Scholar]
- Hedden P., Thomas S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. 10.1042/BJ20120245 [DOI] [PubMed] [Google Scholar]
- Heyman J., Cools T., Canher B., Shavialenka S., Traas J., Vercauteren I., et al. (2016). The heterodimeric transcription factor complex ERF115–PAT1 grants regeneration competence. Nat. Plants 2:16165. 10.1038/nplants.2016.165 [DOI] [PubMed] [Google Scholar]
- Holdsworth M. J., Bentsink L., Soppe W. J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179, 33–54. 10.1111/j.1469-8137.2008.02437.x [DOI] [PubMed] [Google Scholar]
- Hossain M. A., Cho J.-I., Han M., Ahn C.-H., Jeon J.-S., An G., et al. (2010). The ABRE-binding bZIP transcription factor OsABF2 is a positive regulator of abiotic stress and ABA signaling in rice. J. Plant Physiol. 167, 1512–1520. 10.1016/j.jplph.2010.05.008 [DOI] [PubMed] [Google Scholar]
- Itoh H., Tanaka-Ueguchi M., Kawaide H., Chen X., Kamiya Y., Matsuoka M. (1999). The gene encoding tobacco gibberellin 3β-hydroxylase is expressed at the site of GA action during stem elongation and flower organ development. Plant J. 20, 15–24. 10.1046/j.1365-313X.1999.00568.x [DOI] [PubMed] [Google Scholar]
- Iwata A., Durai V., Tussiwand R., Briseño C. G., Wu X., Grajales-Reyes G. E., et al. (2017). Quality of TCR signaling determined by differential affinities of enhancers for the composite BATF–IRF4 transcription factor complex. Nat. Immunol. 18:563 10.1038/ni.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M., Itoh H., Inukai Y., Sakamoto T., Ueguchi-Tanaka M., Ashikari M., et al. (2003). Where do gibberellin biosynthesis and gibberellin signaling occur in rice plants? Plant J. 35, 104–115. 10.1046/j.1365-313X.2003.01780.x [DOI] [PubMed] [Google Scholar]
- Kaneko M., Itoh H., Ueguchi-Tanaka M., Ashikari M., Matsuoka M. (2002). The α-amylase induction in endosperm during rice seed germination is caused by gibberellin synthesized in epithelium. Plant Physiol. 128, 1264–1270. 10.1104/pp.010785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Hwang J.-U., Lee M., Kim Y.-Y., Assmann S. M., Martinoia E., et al. (2010). PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. U.S.A. 107, 2355–2360. 10.1073/pnas.0909222107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., et al. (2012). Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. U.S.A. 109, 9653–9658. 10.1073/pnas.1203567109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karssen C., Brinkhorst-Van Der Swan D., Breekland A., Koornneef M. (1983). Induction of dormancy during seed development by endogenous abscisic acid: studies on abscisic acid deficient genotypes of Arabidopsis thaliana (L.) Heynh. Planta 157, 158–165. 10.1007/BF00393650 [DOI] [PubMed] [Google Scholar]
- Karssen C. M., Zagorski S., Kepczynski J., Groot S. (1989). Key role for endogenous gibberellins in the control of seed germination. Ann. Bot. 63, 71–80. 10.1093/oxfordjournals.aob.a087730 [DOI] [Google Scholar]
- Keith K., Kraml M., Dengler N. G., McCourt P. (1994). fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell 6, 589–600. 10.1105/tpc.6.5.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepka M., Benson C. L., Gonugunta V. K., Nelson K. M., Christmann A., Grill E., et al. (2011). Action of natural abscisic acid precursors and catabolites on abscisic acid receptor complexes. Plant Physiol. 157, 2108–2119. 10.1104/pp.111.182584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., et al. (2008). SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell 20, 1260–1277. 10.1105/tpc.108.058859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Park B. S., Yeu S. Y., Song S. I., Song J. T., Seo H. S. (2015). E3 SUMO ligase AtSIZ1 positively regulates SLY1-mediated GA signalling and plant development. Biochem. J. 469, 299–314. 10.1042/BJ20141302 [DOI] [PubMed] [Google Scholar]
- Kim S. Y., Ma J., Perret P., Li Z., Thomas T. L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130, 688–697. 10.1104/pp.003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J. H., Yang S. H., Han K. H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47, 343–355. 10.1111/j.1365-313X.2006.02782.x [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., et al. (2005). Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44, 939–949. 10.1111/j.1365-313X.2005.02583.x [DOI] [PubMed] [Google Scholar]
- Kohli A., Sreenivasulu N., Lakshmanan P., Kumar P. P. (2013). The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abiotic stresses. Plant Cell Rep. 32, 945–957. 10.1007/s00299-013-1461-y [DOI] [PubMed] [Google Scholar]
- Koornneef M., Van Der Veen J. (1980). Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 58, 257–263. 10.1007/BF00265176 [DOI] [PubMed] [Google Scholar]
- Kumar P. P. (2013a). Regulation of biotic and abiotic stress responses by plant hormones. Plant Cell Rep. 32:943 10.1007/s00299-013-1460-z [DOI] [PubMed] [Google Scholar]
- Kumar P. P. (2013b). Plant hormones and their intricate signaling networks: unraveling the nexus. Plant Cell Rep. 32, 731–732. 10.1007/s00299-013-1435-0 [DOI] [PubMed] [Google Scholar]
- Kuromori T., Miyaji T., Yabuuchi H., Shimizu H., Sugimoto E., Kamiya A., et al. (2010). ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. U.S.A. 107, 2361–2366. 10.1073/pnas.0912516107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Sugimoto E., Shinozaki K. (2011). Arabidopsis mutants of AtABCG22, an ABC transporter gene, increase water transpiration and drought susceptibility. Plant J. 67, 885–894. 10.1111/j.1365-313X.2011.04641.x [DOI] [PubMed] [Google Scholar]
- Lee H. G., Lee K., Seo P. J. (2015). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 87, 371–381. 10.1007/s11103-015-0283-4 [DOI] [PubMed] [Google Scholar]
- Lee K. H., Piao H. L., Kim H.-Y., Choi S. M., Jiang F., Hartung W., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120. 10.1016/j.cell.2006.07.034 [DOI] [PubMed] [Google Scholar]
- Lee S. C., Lee M. Y., Kim S. J., Jun S. H., An G., Kim S. R. (2005). Characterization of an abiotic stress-inducible dehydrin gene, OsDhn1, in rice (Oryza sativa L.). Mol. Cells 19, 212–218. [PubMed] [Google Scholar]
- Lee S. C., Luan S. (2012). ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 35, 53–60. 10.1111/j.1365-3040.2011.02426.x [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., et al. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. 10.1111/j.1365-313X.2005.02622.x [DOI] [PubMed] [Google Scholar]
- Li C., Li J., Chong K., Harter K., Lee Y., Leung J., et al. (2016). Toward a molecular understanding of plant hormone actions. Mol. Plant 9, 1–3. 10.1016/j.molp.2015.12.006 [DOI] [PubMed] [Google Scholar]
- Lim E.-K., Doucet C. J., Hou B., Jackson R. G., Abrams S. R., Bowles D. J. (2005). Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron 16, 143–147. 10.1016/j.tetasy.2004.11.062 [DOI] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., et al. (2013). ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25, 4863–4878. 10.1105/tpc.113.118604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. C., Hwang S. G., Endo A., Okamoto M., Koshiba T., Cheng W. H. (2007). Ectopic expression of ABSCISIC ACID 2/GLUCOSE INSENSITIVE 1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol. 143, 745–758. 10.1104/pp.106.084103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Wu F., Sheng P., Zhang Z., Zhang X., Guo X., et al. (2015). The SnRK2-APC/C TE regulatory module mediates the antagonistic action of gibberellic acid and abscisic acid pathways. Nat. Commun. 6:7981. 10.1038/ncomms8981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Mao B., Ou S., Wang W., Liu L., Wu Y., et al. (2014). OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 84, 19–36. 10.1007/s11103-013-0115-3 [DOI] [PubMed] [Google Scholar]
- Liu C., Wu Y., Wang X. (2012). bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235, 1157–1169. 10.1007/s00425-011-1564-z [DOI] [PubMed] [Google Scholar]
- Liu J., Mehdi S., Topping J., Tarkowski P., Lindsey K. (2010). Modelling and experimental analysis of hormonal crosstalk in Arabidopsis. Mol. Syst. Biol. 6:373. 10.1038/msb.2010.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P. P., Koizuka N., Martin R. C., Nonogaki H. (2005). The BME3 (Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination. Plant J. 44, 960–971. 10.1111/j.1365-313X.2005.02588.x [DOI] [PubMed] [Google Scholar]
- Liu X., Hou X. (2018). Antagonistic regulation of ABA and GA in metabolism and signaling pathways. Front. Plant Sci. 9:251. 10.3389/fpls.2018.00251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Hu P., Huang M., Tang Y., Li Y., Li L., et al. (2016). The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 7:12768. 10.1038/ncomms12768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N.-H. (2000). A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 41, 541–547. 10.1093/pcp/41.5.541 [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D. T., Chait B. T., Chua N. H. (2002). ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 32, 317–328. 10.1046/j.1365-313X.2002.01430.x [DOI] [PubMed] [Google Scholar]
- Luo A., Qian Q., Yin H., Liu X., Yin C., Lan Y., et al. (2006). EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 47, 181–191. 10.1093/pcp/pci233 [DOI] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. 10.1126/science.1172408 [DOI] [PubMed] [Google Scholar]
- Magome H., Yamaguchi S., Hanada A., Kamiya Y., Oda K. (2008). The DDF1 transcriptional activator upregulates expression of a gibberellin-deactivating gene, GA2ox7, under high-salinity stress in Arabidopsis. Plant J. 56, 613–626. 10.1111/j.1365-313X.2008.03627.x [DOI] [PubMed] [Google Scholar]
- Mahajan S., Tuteja N. (2005). Cold, salinity and drought stresses: an overview. Arch. Biochem. Biophys. 444, 139–158. 10.1016/j.abb.2005.10.018 [DOI] [PubMed] [Google Scholar]
- Martínez-Andújar C., Ordiz M. I., Huang Z., Nonogaki M., Beachy R. N., Nonogaki H. (2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. U.S.A. 108, 17225–17229. 10.1073/pnas.1112151108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J. B., Yoo C. Y., Miura T., Hasegawa P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 5418–5423. 10.1073/pnas.0811088106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase K., Hirano Y., Sun T.-P., Hakoshima T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459. 10.1038/nature07519 [DOI] [PubMed] [Google Scholar]
- Ng L. M., Melcher K., Teh B. T., Xu H. E. (2014). Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin. 35:567 10.1038/aps.2014.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nir I., Shohat H., Panizel I., Olszewski N., Aharoni A., Weiss D. (2017). The tomato DELLA protein PROCERA acts in guard cells to promote stomatal closure. Plant Cell 29, 3186–3197. 10.1105/tpc.17.00542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Hu J., Yusuke J., Jung B., Paik I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. 10.1105/tpc.107.050153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W. I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139. 10.1111/j.1365-313X.2006.02773.x [DOI] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23, 1404–1415. 10.1105/tpc.110.080721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. 10.1126/science.1173041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirats-Llobet M., Han S.-K., Gonzalez-Guzman M., Jeong C. W., Rodriguez L., Belda-Palazon B., et al. (2016). A direct link between abscisic acid sensing and the chromatin-remodeling ATPase BRAHMA via core ABA signaling pathway components. Mol. Plant 9, 136–147. 10.1016/j.molp.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.-M., Kannangara R., Gilday A. D., Halliday K. J., Graham I. A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15, 1998–2006. 10.1016/j.cub.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D. E., King K. E., Cowling R. J., Murphy G. P., et al. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11, 3194–3205. 10.1101/gad.11.23.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Richards D. E., Hartley N. M., Murphy G. P., Devos K. M., Flintham J. E., et al. (1999). ‘Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. 10.1038/22307 [DOI] [PubMed] [Google Scholar]
- Qin F., Kodaira K. S., Maruyama K., Mizoi J., Tran L. S., Fujita Y., et al. (2011). SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 157, 1900–1913. 10.1104/pp.111.187302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajjou L., Duval M., Gallardo K., Catusse J., Bally J., Job C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. 10.1146/annurev-arplant-042811-105550 [DOI] [PubMed] [Google Scholar]
- Ravindran P., Verma V., Stamm P., Kumar P. P. (2017). A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant 10, 1307–1320. 10.1016/j.molp.2017.09.004 [DOI] [PubMed] [Google Scholar]
- Rowe J. H., Topping J. F., Liu J., Lindsey K. (2016). Abscisic acid regulates root growth under osmotic stress conditions via an interacting hormonal network with cytokinin, ethylene and auxin. New Phytol. 211, 225–239. 10.1111/nph.13882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Sola M. A., Rodriguez-Concepcion M. (2012). Carotenoid biosynthesis in Arabidopsis: a colorful pathway. Arabidopsis Book 10:e0158. 10.1199/tab.0158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H., Cho H., Bae W., Hwang I. (2014). Control of early seedling development by BES1/TPL/HDA19-mediated epigenetic regulation of ABI3. Nat. Commun. 5:4138. 10.1038/ncomms5138 [DOI] [PubMed] [Google Scholar]
- Saavedra X., Modrego A., Rodriguez D., Gonzalez-Garcia M. P., Sanz L., Nicolas G., et al. (2010). The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 152, 133–150. 10.1104/pp.109.146381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah S. K., Reddy K. R., Li J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. 10.3389/fpls.2016.00571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S. Y., Jamin M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462, 665–668. 10.1038/nature08591 [DOI] [PubMed] [Google Scholar]
- Santner A., Calderon-Villalobos L. I., Estelle M. (2009). Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5, 301–307. 10.1038/nchembio.165 [DOI] [PubMed] [Google Scholar]
- Schopfer P., Plachy C. (1984). Control of seed germination by abscisic acid. Plant Physiol. 76, 155–160. 10.1104/pp.76.1.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey T. D., Raschke K. (1980). Effects of phaseic Acid and dihydrophaseic Acid on stomata and the photosynthetic apparatus. Plant Physiol. 65, 291–297. 10.1104/pp.65.2.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Chen Q., Wu Y., Liu R., Zhang H., Wang P., et al. (2016). ABI4 mediates antagonistic effects of abscisic acid and gibberellins at transcript and protein levels. The Plant Journal 85, 348–361. 10.1111/tpj.13109 [DOI] [PubMed] [Google Scholar]
- Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. 10.1371/journal.pgen.1003577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K., Zhou W., Yang W. (2018). APETALA 2-domain-containing transcription factors: focusing on abscisic acid and gibberellins antagonism. New Phytol. 217, 977–983. 10.1111/nph.14880 [DOI] [PubMed] [Google Scholar]
- Silverstone A. L., Chang C. W., Krol E., Sun T. P. (1997). Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 12, 9–19. 10.1046/j.1365-313X.1997.12010009.x [DOI] [PubMed] [Google Scholar]
- Skubacz A., Daszkowska-Golec A. (2017). Seed dormancy: the complex process regulated by abscisic acid, gibberellins, and other phytohormones that makes seed germination work, in Phytohormones - Signaling Mechanisms and Crosstalk in Plant Development and Stress Responses, ed El-Esawi M. (InTech), 77–100. 10.5772/intechopen.68735 [DOI] [Google Scholar]
- Song L., Huang S. C., Wise A., Castanon R., Nery J. R., Chen H., et al. (2016). A transcription factor hierarchy defines an environmental stress response network. Science 354:aag1550. 10.1126/science.aag1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamm P., Kumar P. P. (2013). Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep. 32, 759–769. 10.1007/s00299-013-1406-5 [DOI] [PubMed] [Google Scholar]
- Stamm P., Ravindran P., Mohanty B., Tan E. L., Yu H., Kumar P. P. (2012). Insights into the molecular mechanism of RGL2-mediated inhibition of seed germination in Arabidopsis thaliana. BMC Plant Biol. 12:179 10.1186/1471-2229-12-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C. M., Cooney S. E., Mccourt P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149, 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang N., Zhang H., Li X., Xiao J., Xiong L. (2012). Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 158, 1755–1768. 10.1104/pp.111.190389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh S., Imamura A., Watanabe A., Nakabayashi K., Okamoto M., Jikumaru Y., et al. (2008). High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 146, 1368–1385. 10.1104/pp.107.113738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij F. E., Gan Y., Penfield S., Gilday A. D., Dave A., He Z., et al. (2013). Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proc. Natl. Acad. Sci. U.S.A. 110, 10866–10871. 10.1073/pnas.1301647110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varbanova M., Yamaguchi S., Yang Y., McKelvey K., Hanada A., Borochov R., et al. (2007). Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell 19, 32–45. 10.1105/tpc.106.044602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., Ramamoorthy R., Kohli A., Kumar P. P. (2015). Rice research to break yield barriers. COSMOS 11, 37–54. 10.1142/S0219607715500032 [DOI] [Google Scholar]
- Verma V., Ravindran P., Kumar P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86 10.1186/s12870-016-0771-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verslues P. E., Bray E. A. (2006). Role of abscisic acid (ABA) and Arabidopsis thaliana ABA-insensitive loci in low water potential-induced ABA and proline accumulation. J. Exp. Bot. 57, 201–212. 10.1093/jxb/erj026 [DOI] [PubMed] [Google Scholar]
- Vu N.-T., Kang H.-M., Kim Y.-S., Choi K.-Y., Kim I.-S. (2015). Growth, physiology, and abiotic stress response to abscisic acid in tomato seedlings. Hortic. Environ. Biotechnol. 56, 294–304. 10.1007/s13580-015-0106-1 [DOI] [Google Scholar]
- Wang F., Zhu D., Huang X., Li S., Gong Y., Yao Q., et al. (2009). Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21, 2378–2390. 10.1105/tpc.108.065433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu H., Hua H., Wang L., Song C.-P. (2011). A vacuole localized β-glucosidase contributes to drought tolerance in Arabidopsis. Chin. Sci. Bull. 56, 3538–3546. 10.1007/s11434-011-4802-7 [DOI] [Google Scholar]
- Wilkinson S., Davies W. J. (2010). Drought, ozone, ABA and ethylene: new insights from cell to plant to community. Plant Cell Environ. 33, 510–525. 10.1111/j.1365-3040.2009.02052.x [DOI] [PubMed] [Google Scholar]
- Willige B. C., Ghosh S., Nill C., Zourelidou M., Dohmann E. M., Maier A., et al. (2007). The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19, 1209–1220. 10.1105/tpc.107.051441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W., Liu C., Hou X., Yu H. (2010). MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis. Plant Cell 22, 1733–1748. 10.1105/tpc.109.073072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Tang N., Du H., Ye H., Xiong L. (2008). Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 148, 1938–1952. 10.1104/pp.108.128199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L., Lee H., Ishitani M., Zhu J. K. (2002). Regulation of osmotic stress-responsive gene expression by the LOS6/ABA1 locus in Arabidopsis. J. Biol. Chem. 277, 8588–8596. 10.1074/jbc.M109275200 [DOI] [PubMed] [Google Scholar]
- Xu Z.-Y., Lee K. H., Dong T., Jeong J. C., Jin J. B., Kanno Y., et al. (2012). A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24, 2184–2199. 10.1105/tpc.112.095935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta T., Sumiki Y. (1938). On the crystal of gibberellin, a substance to promote plant growth. J. Agric. Chem. Soc. Jpn 14:1526 10.1271/nogeikagaku1924.14.12_1526 [DOI] [Google Scholar]
- Yaish M. W., El-Kereamy A., Zhu T., Beatty P. H., Good A. G., Bi Y.-M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. 10.1371/journal.pgen.1001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. 10.1146/annurev.arplant.59.032607.092804 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y. (2000). Gibberellin biosynthesis: its regulation by endogenous and environmental signals. Plant Cell Physiol. 41, 251–257. 10.1093/pcp/41.3.251 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Kamiya Y., Sun T. P. (2001). Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 28, 443–453. 10.1046/j.1365-313X.2001.01168.x [DOI] [PubMed] [Google Scholar]
- Yano R., Kanno Y., Jikumaru Y., Nakabayashi K., Kamiya Y., Nambara E. (2009). CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 151, 641–654. 10.1104/pp.109.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye N., Jia L., Zhang J. (2012). ABA signal in rice under stress conditions. Rice (N. Y). 5:1. 10.1186/1939-8433-5-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying S., Zhang D. F., Li H. Y., Liu Y. H., Shi Y. S., Song Y. C., et al. (2011). Cloning and characterization of a maize SnRK2 protein kinase gene confers enhanced salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 30, 1683–1699. 10.1007/s00299-011-1077-z [DOI] [PubMed] [Google Scholar]
- Zeevaart J. A. D., Creelman R. A. (1988). Metabolism and physiology of abscisic acid. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 439–473. 10.1146/annurev.pp.39.060188.002255 [DOI] [Google Scholar]
- Zeng D.-E., Hou P., Xiao F., Liu Y. (2015). Overexpression of Arabidopsis XERICO gene confers enhanced drought and salt stress tolerance in rice (Oryza Sativa L.). J. Plant Biochem. Biotechnol. 24, 56–64. 10.1007/s13562-013-0236-4 [DOI] [Google Scholar]
- Zentella R., Zhang Z. L., Park M., Thomas S. G., Endo A., Murase K., et al. (2007). Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19, 3037–3057. 10.1105/tpc.107.054999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Schurr U., Davies W. J. (1987). Control of stomatal behaviour by abscisic acid which apparently originates in the roots. J. Exp. Bot. 38, 1174–1181. 10.1093/jxb/38.7.1174 [DOI] [Google Scholar]
- Zhang H., Zhu H., Pan Y., Yu Y., Luan S., Li L. (2014). A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates ABA sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7, 1522–1532. 10.1093/mp/ssu063 [DOI] [PubMed] [Google Scholar]
- Zheng J., Chen F., Wang Z., Cao H., Li X., Deng X., et al. (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 193, 605–616. 10.1111/j.1469-8137.2011.03969.x [DOI] [PubMed] [Google Scholar]
- Zhu J. K. (2002). Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 53, 247–273. 10.1146/annurev.arplant.53.091401.143329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Nomura T., Xu Y., Zhang Y., Peng Y., Mao B., et al. (2006). ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18, 442–456. 10.1105/tpc.105.038455 [DOI] [PMC free article] [PubMed] [Google Scholar]