Abstract
CYP2C19 genotype-guided antiplatelet therapy following percutaneous coronary intervention is increasingly implemented in clinical practice. However, challenges such as selecting a testing platform, communicating test results, building clinical decision support processes, providing patient and provider education, and integrating methods to support the translation of emerging evidence to clinical practice are barriers to broad adoption. In this report, we compare and contrast implementation strategies of 12 early adopters, describing solutions to common problems and initial performance metrics for each program. Key differences between programs included the test result turnaround time and timing of therapy changes which are both related to CYP2C19 testing model and platform used. Sites reported the need for new informatics infrastructure, expert clinicians such as pharmacists to interpret results, physician champions, and ongoing education. Consensus lessons learned are presented to provide a path forward for those seeking to implement similar clinical pharmacogenomics programs within their institutions.
Keywords: pharmacogenomics, implementation, CYP2C19, percutaneous coronary intervention
Introduction
Cytochrome p450 2C19 (CYP2C19) genotyping for antiplatelet therapy selection after percutaneous coronary intervention (PCI) is one of the leading clinical pharmacogenomics implementation scenarios in the United States.1 Consensus standard-of-care pharmacotherapy post-PCI consists of dual antiplatelet therapy with aspirin and a P2Y12 receptor inhibitor, namely clopidogrel, prasugrel, or ticagrelor.2 Clopidogrel still remains the most commonly prescribed due to lower cost, better accessibility, and lower risk of non-coronary artery bypass graft major bleeding compared to newer agents.3–5
Clopidogrel is a prodrug that requires bioactivation to an active metabolite for therapeutic effect. Although multiple enzymes are involved in clopidogrel metabolism, only genetic variation in CYP2C19 has been consistently associated with alterations in clopidogrel pharmacokinetics and pharmacodynamic responses.6–8CYP2C19 loss-of-function alleles that produce a nonfunctional enzyme, now described as nonfunctional alleles by the Clinical Pharmacogenetics Implementation Consortium (CPIC),9 are common occurring in approximately 30% of Europeans, 30% of Africans, and 60% of Asians.10 Retrospective analyses of clinical trial and patient registry data have shown that compared with clopidogrel-treated individuals with two functional alleles of CYP2C19, similarly-treated patients carrying at least one nonfunctional allele have an increased risk of cardiovascular events after PCI.11,12 Clinical outcomes during treatment with the third generation P2Y12 inhibitors prasugrel and ticagrelor are unaffected by CYP2C19 genotype.13,14
The Food and Drug Administration added a boxed warning to the clopidogrel labeling in 2010 stating that clopidogrel effectiveness is reduced in CYP2C19 poor metabolizers (PMs), who have two no-function variant alleles, and alternative treatments should be considered in these patients.15 CPIC guidelines recommend alternative therapy in PMs as well as intermediate metabolizers (IMs), who have a single nonfunctional allele; however, they do not address whether the test should be routinely performed.10 Joint guidelines for PCI by the American Heart Association (AHA) and American College of Cardiology (ACC) state that CYP2C19 genetic testing, and tailoring antiplatelet therapy based on the result might be considered in high-risk patients; however, these guidelines recommend against the routine clinical use of genetic testing for PCI patients, based on the absence of data from large randomized controlled trials.2 Since the publication of these guidelines, recent data from the IGNITE (Implementing GeNomics in PracTice) Network (www.ignite-genomics.org) demonstrate that among patients genotyped at the time of PCI, there is a lower risk of major adverse cardiovascular events in patients with a CYP2C19 nonfunctional allele treated with prasugrel or ticagrelor compared with nonfunctional allele carriers treated with clopidogrel.16
A number of institutions now clinically test for CYP2C19 genotype in patients undergoing PCI, with more institutions likely to follow. Challenges that arise when offering a pharmacogenomic test include the selection of a testing platform, communication of test results, developing clinical decision support (CDS), patient education, and methods to support the translation of emerging evidence to clinical practice. We have gathered strategies to address these challenges from early adopters of genotype-guided antiplatelet therapy. The purpose of this paper is to summarize pathways to operationalizing CYP2C19 genotype-guided antiplatelet therapy and approaches to overcome key obstacles likely to arise during this implementation.
Results
Baseline institutional landscape and CYP2C19 implementation planning
Twelve large academic institutions in the IGNITE Network Pharmacogenetics Working Group that had implemented CYP2C19 genotype-guided antiplatelet therapy after PCI are included in this analysis (Table 1).1CYP2C19–clopidogrel was the first clinical pharmacogenomic implementation launched at 9 of the 12 institutions. The University of Illinois at Chicago had previously implemented CYP2C9/VKORC1–warfarin pharmacogenomic testing17, Vanderbilt University had launched the PREDICT program that focused on six gene–drug pairs, including CYP2C19–clopidogrel18, and Sanford Health had launched the Imagenetics program that tested eight pharmacogenes, including CYP2C19.19
Table 1.
Implementation design
| Precision Medicine | Cardiology/Cath Lab | Pharmacy/Pharmacology | IM/Primary Care | Path/Genetics/Lab | IT/EHR teams | CTSI | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Institution | Year of Launch |
Model | Leadership (shaded) and collaborators* (X) |
Testing Mode | Indication for testing | Approx # PCI per yrˆ |
Test ordering process | ||||||
| Vanderbilt University | 2010 | Clinical | X | X | X | X | X | X | X | Reactive and Preemptive | PCI and/or high clinical risk of future need for clopidogrel (predictive model) | 2000† | Unchecked orderable on post-PCI order set |
| University of North Carolina at Chapel Hill | 2012 | Clinical | X | X | X | Reactive | PCI for ACS or stable CAD (with high-risk anatomic findings) | 600 | Unchecked orderable on post-PCI order set | ||||
| University of Florida Health, Shands Hospital, Gainesville | 2012 | Clinical | X | X | X | X | X | X | Reactive | PCI for ACS or stable CAD | 400 | Pre-checked on the post-PCI order set | |
| University of Illinois at Chicago | 2014 | Research | X | X | X | X | X | Reactive | PCI for ACS or stable CAD (with high-risk anatomic findings) | 100 | Research protocol | ||
| University of Pennsylvania | 2014 | Research | X | X | X | X | X | Reactive | PCI for ACS or stable CAD | 800 | Research protocol | ||
| Indiana University | 2014 | Research | X | X | X | X | X | X | X | Reactive and Preemptive‡ | Prescribed a targeted medication | 1000† | Research protocol |
| Icahn School of Medicine at Mount Sinai and the Mount Sinai Hospital | 2014 | Research | X | X | X | X | X | Preemptive | Future need for clopidogrel or other PGx meds (predictive model) | N/A | Research protocol | ||
| Sanford Health | 2014 | Clinical | X | X | X | X | X | Reactive | PCI for ACS or stable CAD | 600 | Unchecked orderable on post-PCI order set | ||
| University of Pittsburgh, UPMC Presbyterian Hospital | 2015 | Clinical | X | X | X | X | X | X | Reactive | PCI for ACS or stable CAD (with high-risk anatomic findings) | 750 | Pre-checked post-PCI order set | |
| University of Alabama at Birmingham | 2015 | Clinical | X | X | X | X | X | X | Reactive | PCI for ACS or stable CAD (with high-risk anatomic findings) | 1000 | Unchecked orderable on post-PCI order set | |
| University of Florida Health, Jacksonville | 2016 | Research | X | X | X | X | X | Reactive and Preemptive | PCI for ACS or stable CAD; Left heart catheterization with intent for PCI | 1200 | Research protocol | ||
| University of Maryland, Baltimore | 2016 | Clinical | X | X | X | X | X | Reactive | PCI for ACS or stable CAD | 550 | Unchecked orderable on post-PCI order set | ||
Collaborators that played a key role in the design of the implementation. Gray-shaded indicates who initiated and led the effort. CTSI = Clinical and Translational Sciences group;
Meeting institution criteria for CYP2C19 testing,
includes all for the panel testing, not just CYP2C19–clopidogrel.
Reactive for the initial drug and preemptive for rest of panel.
The majority of programs were designed primarily as clinical versus research implementations (7 of 12 programs) with all but one of these submitting bills to third-party payers or patients for test reimbursement. Program oversight involved multiple collaborating stakeholders led by a formal precision medicine group at 9 of 12 institutions. Individual champions, including pharmacists, the cardiac catheterization laboratory director, and cardiology or primary care providers, led initiatives at the University of Illinois at Chicago, University of North Carolina at Chapel Hill, and Sanford Health, respectively. Program development time varied considerably across institutions and ranged from 6 to 24 months.
CYP2C19 targeted populations, ordering procedures, and testing
Eight of 12 institutions employed a reactive genotype testing model, in which the test was ordered in response to a PCI procedure with test results available soon after to guide antiplatelet medication therapy. In these instances, the indication for CYP2C19 testing generally included all patients undergoing PCI, with some sites further focusing on higher-risk populations (i.e., presence of acute coronary syndrome (ACS) or high-risk anatomical features). Four institutions implemented a preemptive model with CYP2C19 testing performed in advance of any immediate need for test results. These programs used a predictive model to identify targeted populations with high probability of future PCI or clopidogrel use. Regardless of the testing model, all institutions included inpatients and outpatients [patients undergoing elective procedures with short (<24 hour) hospitalization stays] in their study populations.
The size of the target patient population varied considerably among sites based on cardiac catheterization laboratory PCI volume. Some research-based programs focused on narrow populations, with exclusion criteria applied based on research aims. CYP2C19 test ordering processes for sites with clinical implementation programs varied. The majority of programs (5 of 7 sites) depended on a prescriber to select CYP2C19 test on the post-PCI order set (opt-in), with the remainder incorporating a pre-selected test order on the post-PCI order set so that all patients undergoing PCI were genotyped unless the test was deselected (opt-out).
CYP2C19 assay and reporting methodologies are shown in Table 2. All tests were performed using validated laboratory developed tests in College of American Pathologists/Clinical Laboratory Improvement Amendment (CAP/CLIA)-certified laboratories as necessary for performing testing within clinical care. Genotyping was performed using a variety of platforms, with all institutions testing and reporting allele-defining variants for *2, *3, and *17 at minimum. All but three institutions had protocols to allow for biobanking of genetic samples as part of the implementation program (either to an institutional biobank or as part of a research-based implementation).
Table 2.
Pharmacogenomic testing
| Institution | Gene(s) tested | Platform | CYP2C19 Alleles reported |
|---|---|---|---|
| University of Alabama at Birmingham | CYP2C19 only | Spartan RX, Spartan Biosciences (Ottawa, ON) | *2, *3, *17 |
| University of Pennsylvania | CYP2C19 only | Spartan RX, Spartan Biosciences (Ottawa, ON) | *2, *3, *17 |
| University of Florida Health, Jacksonville | CYP2C19 only | Spartan RX, Spartan Biosciences (Ottawa, ON) | *2, *3, *17 |
| University of Illinois at Chicago | CYP2C19 only | eSensor XT-8, GenMark Diagnostics (Carlsbad, CA) | *2, *3, *4,*5, *6, *8,, *9, *10, *13,*17 |
| University of Florida Health, Shands Hospital, Gainesville | CYP2C19 only* | eSensor XT-8, GenMark Diagnostics (Carlsbad, CA) | *2, *3, *4,*5, *6, *8,*9, *10, *13,*17 |
| University of Pittsburgh, UPMC Presbyterian Hospital | CYP2C19 only | eSensor XT-8, GenMark Diagnostics (Carlsbad, CA) | *2, *3, *4,*5, *6, *7,*8, *9, *10,*17 |
| University of North Carolina at Chapel Hill | CYP2C19 only | Custom Taqman® assay, ThermoFisher (Waltham, MA) | *2, *3, *17 |
| University of Maryland, Baltimore | CYP2C19 only | Custom Taqman® assay, ThermoFisher (Waltham, MA) | *2, *3, *4,*6, *8, *17 |
| Sanford Health | CYP2C19 only | BeadXpress ADME Panel, Illumina (San Diego, CA) | *2, *3, *4,*5, *6, *8,*12, *17 |
| Indiana University | Panel | Custom Taqman® assay, ThermoFisher (Waltham, MA) | *2, *3, *4,*4B, *6, *8,*10, *17 |
| Icahn School of Medicine at Mount Sinai and the Mount Sinai Hospital | Panel | MassARRAY, Agena Biosciences (San Diego, CA) | *2, *3, *4,*5, *6, *7,*8, *17 |
| Vanderbilt University | Panel | Custom QuantStudio® assay, ThermoFisher (Waltham, MA) | *2, *3, *4,*5, *6, *7,*8, *17 |
Initially launched with multi-gene panel based testing, but changed to CYP2C19 testing only after the first year
Communication of results, approach to therapy modification, and education strategies
All institutions reported CYP2C19 test results in the electronic health record (EHR). Results were documented in the “laboratory results” section of the EHR, often as discrete genotype and phenotype results (e. g. CYP2C19 *1/*2 and CYP2C19 Intermediate Metabolizer, respectively) with a linked text-based full report (9 of 12 sites). At three institutions, the patient genotype or phenotype was also recorded on the perpetual patient “problem list” (Table 3). The laboratory report included CYP2C19 genotype and predicted phenotype (metabolizer status) without patient-specific genotype-guided drug recommendations at 11 of 12 institutions (Indiana University provided a drug therapy recommendation on the laboratory report).
Table 3.
Reporting of CYP2C19 genotyping results and education strategies
| Institution | How are results stored in the EHR? | Is there a designated clinician/service to respond to results? | How are therapy recommendations communicated? | Are there CDS alerts? | Are results actively and routinely provided to external providers?* | Are results actively and routinely provided to patients? | Provider Education Strategy deemed most successful |
|---|---|---|---|---|---|---|---|
| University of Alabama at Birmingham | In lab section (discrete results) | Yes | Pharmacogenomics research group communicates with cardiologist; EHR messaging; | Yes | No | No | Focused meetings with providers; in-services; Grand Rounds |
| University of Pennsylvania | In lab section (discrete results) | No | Research coordinator communicates results to Interventionalist | No | Yes; By letter | No | Focused meetings with providers |
| University of Florida Health, Jacksonville | In lab section (text-based report) | No | Study investigator provides interpretation and recommendations when asked | No | No | No | Personal communication |
| University of Illinois at Chicago | In lab section (discrete results) | Yes | Pharmacist on Clinical PGx Service; consult notes on all patients; EHR messaging | No | No | Yes; Letter | Focused meetings with providers |
| University of Florida Health, Shands Hospital, Gainesville | In lab section (discrete results) | Yes | Pharmacists receive genotype results and provide recommendations for actionable genotypes via EHR consult notes | Yes | No | No | Grand rounds; personal communications; report of data |
| University of Pittsburgh, UPMC Presbyterian Hospital | In lab section (discrete results) | Yes | Pharmacist on Clinical PGx Service; consult notes on all patients; EHR messaging | Yes | No | No | Focused meetings with providers |
| University of North Carolina at Chapel Hill | In lab section (discrete results) | No | Pharmacist communicates with Interventionalist; EHR documentation | No | Yes | No | In-services |
| University of Maryland, Baltimore | In lab section (text-based report); in problem list (discrete results) | Yes | EHR messaging by multidisciplinary team; Elective PGx service consultation | Yes | Yes; By letter | Yes; genotype card and after visit summary | Personal communication |
| Indiana University | In lab section (discrete results); in problem list (discrete results) | No | Service, consultation notes; EHR messaging | Yes | No | No | Continual training model (in-services) |
| Icahn School of Medicine at Mount Sinai and the Mount Sinai Hospital | In lab section (text-based report) | No | CDS in EHR | Yes | No | No | Training sessions; CDS documents |
| Sanford Health | In lab section (discrete results) | No | CDS in EHR | Yes | No | No | In-services |
| Vanderbilt University | In lab section (discrete results); in problem list (discrete results) | Yes | Pharmacist sends EHR messaging; CDS in EHR | Yes | No | Yes; all results in web portal | Grand Rounds; clinical pharmacist/program champion interactions |
To downstream provider outside of institution (i.e., to a different EHR system)
A variety of different methods were used to communicate test results to providers and patients (Figure 1). Institutions with clinical implementation models generally relied on pharmacists or dedicated teams/services to provide genotype-informed drug therapy recommendations. Institutions with primarily research testing models also included study staff in this process. Four institutions relied solely on CDS or messaging for therapeutic recommendations and only Vanderbilt University used interruptive CDS delivered immediately after the test result. Eight of 12 sites provided downstream alerting or CDS triggered with future clopidogrel orders within the EHR. Three programs actively reported test results to downstream providers outside of their institution (e.g., a letter with the test result was sent to the primary care practitioner or cardiologist). Three programs routinely provided patients with their test results via a letter/report, patient web portal, an “ID” card with genotype results, and/or personal communication. Nine programs (including one of the above that also provided patients with their test results) introduced patients to CYP2C19 testing through in-person education and/or disseminated brochures, pamphlets, or flyers. Although a number of provider education strategies were used, focused discussions with providers or in-services were perceived by most sites as being the most effective strategies.
Figure 1. Modalities of communication and education.
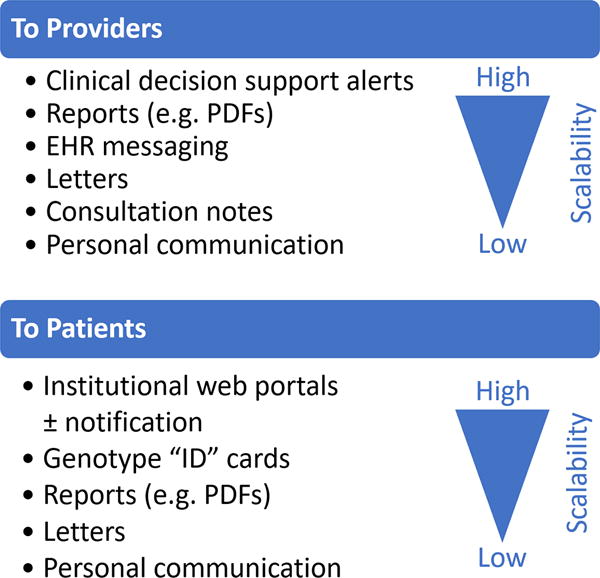
Approaches for providers and patients that were reported by sites are listed in order of decreasing scalability.
Performance metrics
Median length of hospital stay for genotyped patients was one day (range 1-3 days) across all institutions. Testing process metrics and phenotype frequencies are provided in Table S1. Median genotype turnaround time (TAT), defined as each institution’s median time from genotype order to result appearing in the EHR, varied widely among institutions, from very short (1 to 5 hours with rapid testing platforms such as Spartan RX), to 1-3 days with standard single-gene test platforms in reactive models (GenMark eSensor XT-8 or custom Taqman® assays), to longer (6 to 8 days) for those institutions who used preemptive panel-based or send-out testing strategies. Real-world CYP2C19 phenotype frequencies were similar across all sites, and consistent with those reported in clinical studies and established guidelines.16 All institutions followed CPIC guidelines that recommend alternative therapy (prasugrel or ticagrelor) for CYP2C19 IMs or PMs, and clopidogrel for CYP2C19 normal, rapid, and ultrarapid metabolizers (*1/*1, *1/*17, and *17/*17 genotypes, respectively).10 Triple dose clopidogrel (225 mg/day) was rarely used.16
Eight of 12 institutions provided data on downstream medication use for 1858 total patients who underwent genetic testing. Figure 2 depicts the proportions of patients with and without nonfunctional alleles who were on alternative therapy at time of discharge from the hospitalization associated with genetic testing and at the first follow-up appointment after discharge. Overall, 48.0% and 75.5% of IMs and PMs, respectively, were on alternative therapy at first follow-up. At the five institutions using testing strategies with >1 day TAT (“Standard Testing”: University of Pittsburgh, University of Florida Health - Shands Hospital, University of North Carolina at Chapel Hill, University of Illinois at Chicago, and Indiana University), a significantly higher proportion of nonfunctional allele carriers (PMs/IMs) were receiving alternative therapy at the first follow-appointment versus at time of discharge (58% vs 38%, p < 0.0001), whereas the proportion remained consistent (~19%) among those without a nonfunctional allele at these same institutions. This is in contrast to the institutions that used rapid testing solutions (University of Pennsylvania, University of Alabama at Birmingham, and University of Florida – Jacksonville) where similar proportions remained on alternative therapy across time-periods. The time from PCI to alternative antiplatelet therapy in patients with a nonfunctional allele occurred in less than 1 day at sites with rapid testing and in a median of one to 10 days at institutions with standard single-gene or panel-based testing, respectively. Notably, the proportion of nonfunctional allele carriers on alternative therapy at first follow-up in the standard testing group (58%) was similar to the proportion of nonfunctional allele carriers on alternative therapy at discharge (56%) and at first follow-up (54%) in the rapid testing groups, respectively. The proportion of nonfunctional allele carriers on alternative therapy at first follow-up varied widely across institutions, ranging from 13% to 100%. Use of alternative therapy in patients without a nonfunctional allele was low, ranging from 19% to 30% in both testing groups.
Figure 2. Proportion of patients carrying nonfunctional alleles on alternative (ALT) therapy at discharge and first follow-up.
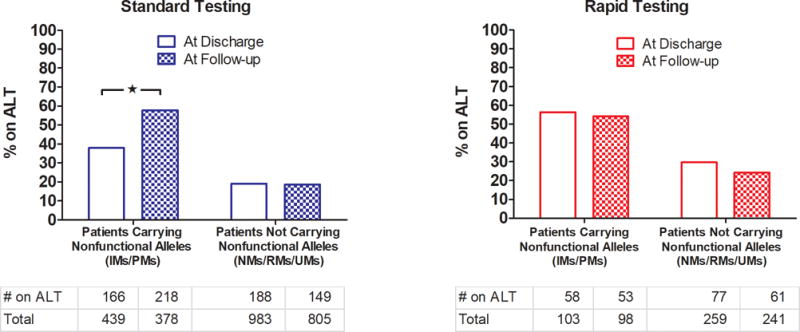
Sites using standard (single gene- and panel- based testing with >1 day TAT) and rapid (right panel) testing are grouped in the left and right panels, respectively. Tables below indicate number of patients in each group. * p < 0.0001 by Chi-square.
Implementation challenges
Sites reported multiple implementation challenges (Table 4), including obtaining provider and stakeholder support, establishing hospital contracts for testing equipment, creating laboratory reports, and building CDS systems. Logistical challenges were also commonly reported and involved sample processing, return of results to providers and patients, and timing of therapy modifications. Some specific challenges varied according to testing strategy and implementation model used. For example, the length of time before changes in antiplatelet therapy were made after PCI was an issue for institutions that did not use a rapid testing platform, while institutions using rapid testing platforms were challenged by the requirement to run testing assays within an hour of sample collection with the Spartan RX platform, storage of temperature sensitive testing reagents, and the need to recollect approximately 10% of samples for repeat genotyping due to initial sample failure. For research models, operationalizing the study design and patient recruitment difficulties were reported as barriers to implementation. Provider acceptance of clinical recommendations based on CYP2C19 genotype and reimbursement for preemptive testing were identified as specific challenges in clinical models.
Table 4.
Primary Implementation Challenges
| • Provider/stakeholder buy-in |
| • Establishing laboratory contracts with hospital |
| • EHR formatting of laboratory reports |
| • Development of clinical decision support |
| • Logistics of: ○ sample collection in cardiac catheterization laboratory or as an outpatient ○ location of rapid testing devices and freezers ○ getting results to appropriate providers ○ returning of results prior to patient discharge due to testing TAT ○ implementation of therapy changes after patient discharge ○ delays in therapy changes until after clinical decision alert fires in the EHR system ○ notifying patients they were tested and of their results |
| • Acceptance of recommendations based on testing results |
| • Number of samples that needed to be recollected (for rapid testing platforms) |
| • Study design (for research implementations) |
| • Patient recruitment (for research implementations) |
| • Billing/reimbursement for pre-emptive testing |
Lessons learned
Table 5 lists the top lessons learned from clinical implementation of CYP2C19 testing reported across sites. In the authors’ experience, clinical pharmacogenomics implementation programs require a physician champion and multidisciplinary engagement from key stakeholders. If a pre-emptive testing strategy is planned, developing a clear definition of the targeted testing population ahead of time is crucial to success. Education efforts need to be deployed early and on an ongoing basis to both implementing clinicians and to providers at all levels including nurses, nurse practitioners, pharmacists, physicians, and laboratory medicine personnel. Establishing close working relationships with informatics groups to create test orders, return and store genetic test results appropriately, and build tailored CDS alerts was also deemed critical to achieve program success and sustainability. Early adopters also emphasized the value of having a designated person or team to act on results and in particular, clinical pharmacists, to provide interpretation and increase adherence to genotype-informed drug therapy recommendations. Finally, the opportunity to share experiences and learn from leaders at other sites who were building similar programs and the availability of guidelines/scientific data (e.g., CPIC, PharmGKB) were also valued.
Table 5.
Lessons Learned
| • Design pharmacogenomics implementation program for the health system patient population |
| • Identify a physician champion and engage key stakeholders |
| • Target the right patient population for preemptive testing |
| • Preemptive testing reduced issues around TAT, but introduced challenges with patient identification and reimbursement |
| • Engage and educate clinicians early and repeatedly throughout the implementation process |
| • Partner with hospital informatics to create clinical decision support tools and solve ongoing EHR challenges |
| • A designated person or team to respond to results improves efficiency of therapy changes |
| • Integrate clinical pharmacists to ensure adherence to the implementation algorithm and appropriate follow-up |
| • Provide ongoing education programs for all health care providers |
| • Learn from published experiences of early implementers, domain expert groups (CPIC) |
Discussion
CYP2C19 genotype-guided antiplatelet prescribing after PCI is increasing in large academic medical centers across the U.S. and beyond. Although early experiences with CYP2C19 testing have been reported,18,20–25 the current data provide a unique opportunity to compare and contrast features of 12 implementation programs. Similarly, the specific challenges faced by program leaders and lessons learned in overcoming these obstacles provide a valuable perspective for those seeking to implement similar pharmacogenomics programs within their institutions.
While several different testing platforms were used among institutions in our study, all sites used in-house testing solutions within CAP/CLIA laboratories versus outsourcing to a commercial laboratory. The CYP2C19 *2, *3, and *17 alleles were consistently tested, and the CPIC guideline phenotype translation tables were used across all institutions. Nearly all sites using clinical models submitted bills to third party payers for reimbursement. These similarities in the way testing was set up, despite being independent decisions at each site, suggests these approaches may be broadly applicable to institutions designing new programs. Further, emerging data showing frequent reimbursement across multiple payers26 and cost-effectiveness of genotype testing at the time of PCI support this strategy.27
A simple metric of implementation program success is whether therapy changes were made following pharmacogenomic testing. Data on the frequency of alternative therapy prescribing at first follow-up in patients with and without a CYP2C19 nonfunctional allele (55-60% versus 20-25%) illustrate, as expected, that genotype results were routinely interpreted and used to guide antiplatelet therapy selection. Differences in alternative therapy prescribing frequencies within PMs and IMs (75.5% versus 48.0%, respectively) at first follow-up further indicate a result predicted PM phenotype was acted upon more frequently. Moreover, implementation performance metrics were specific to testing model, and platform such that the timing of expected therapy changes was consistent with each institution’s testing TAT. For instance, implementation models that employ rapid testing solutions observed similar rates in alternative therapy use in nonfunctional allele carriers at discharge and follow-up, indicating that these sites often accomplished therapy changes prior to discharge. The Spartan RX system (Spartan Biosciences, Ottawa, ON) offers advantages in terms of a rapid TAT; however, centers utilizing this method observed a re-testing rate of up to 10%. In contrast, sites that used standard single-gene and platform-based tests often reported genotype TAT that exceeded patient length of stay for PCI, and had a lower frequency of alternative therapy use in PMs/IMs at discharge. Although frequency of alternative therapy use at first follow-up was similar to sites with rapid testing, the logistics of achieving drug therapy modifications based on genotype after patient discharge is more difficult. Challenges in re-establishing contact with the patient, communication with additional providers, and changing drug therapy regimens, especially when patients have already received an initial prescription for clopidogrel, can complicate the care process. Furthermore, the impact of the delay in therapy changes on clinical outcomes remain unclear. However, it should be assumed that delays in changes of antiplatelet therapy in nonfunctional allele carriers may be associated with an increase in risk of acute and subacute stent thrombosis. Preemptive genotyping has the noted advantage of avoiding the concerns of test result TAT, but adds challenges with timely identification of which patient populations are likely to be prescribed drugs impacted by targeted variants.28 Similarly, reimbursement may be a bigger obstacle within preemptive testing models.29 Overall, while it is evident that therapy modifications were accomplished following testing, the considerable variability in the proportion of nonfunctional allele carriers on alternative therapy at first follow-up across sites (13-100%) underscores the complexity of integrating genetic results with patient-specific clinical factors and challenges of implementing genotype-guided algorithms in clinical practice.
There was near consensus among sites that return of results to the EHR should include discrete data variables such as the gene tested, genotype, and predicted phenotype versus scanned “paper” reports or only free text. The availability of consensus nomenclature determined by the CPIC term standardization project and common dictionaries will likely further encourage the use of standardized, interoperable terminology.9 Together, discrete results and standardized terms represent the current gold standard for driving CDS. The use of the laboratory reports section of the EHR was ubiquitous, but only a few sites stored results in a “problem list” or another location to make the data persistent beyond the current patient encounter. Partnering with hospital informatics at the outset of the clinical implementation program to establish these requirements, streamline clinical workflows, and maintain consistent practices beyond the initial implementation period was deemed crucial. While a few sites did post testing results to web portals or provided genotype cards, few actively returned results to patients or to downstream providers outside of the institution. This remains a future opportunity to further leverage pharmacists and for services optimization to improve patient care transitions and adherence to protocols. It will also be important to assess patients’ understanding of pharmacogenomics and their overall experience with return of test results, as these factors may influence medication adherence and can be used to develop educational materials to assist patients who undergo pharmacogenomic testing in the future.30
The summary data and reported lessons learned indicate that a dedicated clinician or team to receive the genotype test results is advantageous. This makes sense given the test reports from the laboratory rarely included specific therapy recommendations. Personnel with expertise in pharmacogenomics such as pharmacists and formal clinical pharmacogenomics services are being used to bridge the gap between pharmacogenomic test ordering and medication selection.17,31–33 In the current analysis, five sites specifically integrated clinical pharmacy experts to receive results, make recommendations, or educate patients and providers. As programs grow and are scaled to other sites, dedicated clinical services and/or an increased reliance on CDS may be expected.
The need of a physician champion and persistent, ongoing education was reported as both a challenge to overcome and a lesson learned. Program leaders thought a champion and active dissemination approaches such as in-services to be critical to the launch and ongoing success of the implementation, test adoption, and adherence to genotype guided recommendations. Ongoing education may be especially important in academic and other centers where resident physicians and other trainees rotate through the cardiac catheterization laboratory. Genetic literacy is highly variable across providers and is understood to influence pharmacogenomic testing adoption.34,35 Educational strategies have been reviewed elsewhere36 and their importance should not be undervalued.
As with any new research protocol or clinical program, there is an inherent inefficiency in being an early adopter. Whether it was through NIH networks such as IGNITE, eMERGE, or the Pharmacogenomics Research Network; professional societies; or even direct communication; program leaders emphasized the value they found in collaboration and learning from others working to overcome common challenges. To help catalyze efforts, by institutions involved in this work are available in the IGNITE Network SPARK Toolbox at www.ignite-genomics.org.
Ultimately, decisions to develop a CYP2C19 genotype-guided prescribing program and its design are driven by clinical evidence of value of these services. Recent guidelines state a preference for alternative therapy over clopidogrel in patients with ACS treated with PCI based on clinical trial data showing improved outcomes with prasugrel and ticagrelor over clopidogrel in this setting, albeit at an increased bleeding risk.2 However, approximately 30% of clopidogrel-treated patients in these ACS trials would have had a nonfunctional CYP2C19 allele and reduced response to clopidogrel, as has been demonstrated in post-hoc genetic sub-studies.12,14,37–39 Even with increased use of alternative therapies early after PCI, there remains an argument that genotyping has an important role in informing chronic antiplatelet therapy. In particular, clopidogrel may be more appropriate for a patient with a normal metabolizer phenotype and when the patient could not afford or tolerate one of the newer agents. Prasugrel and ticagrelor are also commonly prescribed in patients with stable coronary artery disease undergoing PCI, even though these agents are only indicated for ACS and there are no randomized clinical trial data showing evidence of superiority of alternative therapy in non-ACS indications. Clinical guidelines do not endorse use of alternative therapy after PCI for stable coronary artery disease, yet a finding of a nonfunctional allele in patients with stable coronary disease prescribed clopidogrel may provide support the use alternative therapy in these cases.2 Recent real-world data suggest clopidogrel still remains the most commonly prescribed antiplatelet agent after PCI and that there may be a role for genotyping in this scenario.5,16 Among patients with genotype results available early after PCI, initiation of alternative antiplatelet therapy over clopidogrel in those with a nonfunctional CYP2C19 allele resulted in a significant reduction in major adverse cardiovascular events.16 These data and similarly positive results from other clinical implementation studies and small randomized controlled trials may prompt others to move toward a genotype-guided approach to prescribing antiplatelet therapy in the PCI setting.20–23,26,40
Several limitations to the analysis should be noted. Selection bias is possible as all institutions were recruited from a common research network and approaches to implementation may therefore be similar for this reason. Similarly, while most institutions were university hospitals, the experience at Sanford Health indicates that implementation is feasible and well-accepted within a large group of private multispecialty practices. As genotype-based prescribing is further expanded to more diverse or non-academic settings, preemptive testing or rapid genotyping platforms may be considered so that results are available at the time of or soon after PCI. In the current analysis, data availability was also not consistent as not all sites tracked medication prescribing downstream of the genetic testing, and the number of eligible patients who were not genotyped was not recorded as a metric of algorithm use. Future work will focus on collecting these data, as well as the sustainability of genotype testing and genotype-guided medication selection over time, to understand which program features are necessary to drive optimal prescribing. Finally, findings may be specific to pharmacogenomics implementation in the PCI population and may not be broadly applicable to other populations.
Overall, clinical implementation of CYP2C19 genotype-guided antiplatelet prescribing after PCI is becoming increasingly common. Data and experiences from 12 early-adopter institutions presented herein provide real-world insight for institutions seeking to implement genotype-guided antiplatelet therapy, and strategies to overcome barriers likely to be encountered. Similarly, lessons learned may be applicable to clinical pharmacogenomics implementation efforts for additional gene-drug pairs and to genomic medicine more broadly.
Methods
Our goal was to summarize and compare implementation experiences of early adopters of CYP2C19 pharmacogenomic testing to guide antiplatelet medication selection. The study population included 12 large academic institutions within the IGNITE Network Pharmacogenetics Working Group (five funded institutions and seven affiliate members)1 who have tested 6340 patients for CYP2C19 alleles (see Table S1 for a breakdown by site).
Data collection was completed at each site through a structured electronic spreadsheet disseminated to each site leader. Specific data elements were selected and definitions were refined through open discussions at several in-person meetings and conference calls from September 2016 to May 2017. The tool was then pilot-tested for feasibility prior to dissemination to all sites. Areas of focus included the baseline genetics testing landscape at each site, stakeholder involvement, the design of each implementation program, testing approaches, informatics setup, return of results procedures, and any education provided. Eight of the 12 sites also provided data on antiplatelet medication use after genotyping (1858 total patients). Data cleaning was accomplished iteratively through direct follow-up communications. All data elements collected were reported.
Program performance metrics including testing turnaround times, reported predicted phenotype frequencies, and drug prescribing patterns were also sought from local EHRs or research study data sources when available. All data abstraction and reporting was approved by local institutional review board at each site. Descriptive statistics were reported by institution and proportions of patients with specific test results prescribed alternative therapy were compared using chi square testing. Finally, common challenges that must be overcome and recommendations (lessons learned) for those considering similar implementations in the future were solicited from site investigators and aggregated to a consensus lists through multiple rounds of telephone conference call discussions.
Supplementary Material
Table S1: Testing metrics
Study Highlights.
What is the current knowledge on the topic?
While recent data supports CYP2C19 genotype-guided prescribing of antiplatelet therapy following percutaneous coronary intervention and genetic testing is increasingly deployed in clinical practice, pharmacogenomic testing programs are challenging to implement.
What questions did this study address?
What are the common challenges, pathways to operationalization, initial performance metrics, and lessons learned among 12 sites that are early adopters of pharmacogenomics testing?
What does this study add to our knowledge?
Common and differentiating features (e.g. testing approaches, how results are communicated, clinical decision support, provider and patient education, and methods to support the translation of emerging evidence to clinical practice) are presented along with a discussion of implementations challenges faced and lessons learned.
How might this change clinical pharmacology or translational science?
Disseminating the implementation experiences of early-adopters of pharmacogenomics testing provides valuable perspectives for those seeking to implement similar pharmacogenomics programs within their institutions.
Acknowledgments
Work supported by the National Institutes of Health (NIH) grants U01 HG007269 (LHC, KWW, JAJ), U01 HG007253 (JFP, RAW), U01 HG007762 (TCS, VMP, RPK), and U01HG007278 (AOO) as part of the NIH IGNITE network (www.ignite-genomics.org). Additional support provided by American Society of Health System Pharmacists, NIH UL1TR0000005, and by an Anonymous Donor for PE and JS; American Heart Association 17MCPRP33400175 for JS; Penn Center for Precision Medicine at the Perelman School of Medicine at the University of Pennsylvania for ST; NIH U01 GM074492 and U01 HL105198 (both part of the NIH Pharmacogenomics Research Network) UL1 TR000064 and UL1 TR001427, and by substantial institutional support from the University of Florida and its Clinical Translational Science Institute for LHC, KW, and JAJ; NIH U01 HL105198 and support from the University of Maryland Medical Center and University of Maryland School of Medicine Program for Personalized and Genomic Medicine for ALB and LJ; University of Illinois at Chicago Offices of the Vice President for Health Affairs and Vice Chancellor for Research for JDD; NIH U01 HG008701-02S1 (eMERGE Network) for AOO; NIH R01HL092173, 1K24HL133373, University of Alabama at Birmingham, Birmingham Health Service Foundations’ General Endowment Fund, and NIH UL1TR000165 for NAL; NIH (U01HL122904), substantial institutional support from Vanderbilt University Medical Center, and the National Center for Advancing Translational Sciences (UL1TR000445) for JFP.
Conflict of Interest Disclosures: The Indiana University Pharmacogenomics Laboratory and Sema4 are fee-for-service clinical laboratories. RK served as consultant for Haemonetics. JC served as consultant for Medicure and has research support from United Therapeutics. NL serves on the Admera advisory board. DA reports receiving payments as an individual for: a) Consulting fee or honorarium from Amgen, Aralez, AstraZeneca, Bayer, Biosensors, Bristol-Myers Squibb, Chiesi, Daiichi-Sankyo, Eli Lilly, Janssen, Merck, PLx Pharma, Pfizer, Sanofi, and The Medicines Company; b) Participation in review activities from CeloNova and St. Jude Medical. Institutional payments for grants from Amgen, AstraZeneca, Bayer, Biosensors, CeloNova, CSL Behring, Daiichi-Sankyo, Eisai, Eli-Lilly, Gilead, Janssen, Matsutani Chemical Industry Co., Merck, Novartis, Osprey Medical, and Renal Guard Solutions; in addition, DA is recipient of a funding from the Scott R. MacKenzie Foundation. Spartan Bioscience provided the genotyping platforms and kits for testing at the University of Florida Health, Jacksonville.
Footnotes
Author Contributions
P.E.E., J.S., S.T., K.W., D.J.A., A.L.B., J.C.C., J.D.D., F.F., N.A.L., K.A.M., R.P.K., A.O.O., J.F.P., J.P., V.M.P., F.R., S.A.S., T.C.S., G.A.S., R.A.W., L.H.C., and C.R.L. wrote the manuscript; P.E.E., J.S., S.T., K.W., D.J.A., A.L.B., J.C.C., J.D.D., F.F., L.J.B.J., J.A.J., N.A.L., R.P.K., A.O.O., J.F.P., J.P., N.P., V.M.P., F.R., S.A.S., T.C.S., M.R.V., G.A.S., R.A.W., L.H.C., and C.R.L. designed the research; P.E.E., J.S., S.T., K.W., D.J.A., A.L.B., J.C.C., J.D.D., F.F., L.J.B.J., J.A.J., N.A.L., R.P.K., A.O.O., J.F.P., J.P., N.P., V.M.P., F.R., S.A.S., T.C.S., M.R.V., G.A.S., R.A.W., L.H.C., and C.R.L. performed the research; P.E.E., J.S., S.T., D.J.A., A.L.B., J.C.C., J.D.D., F.F., N.A.L., A.O.O., J.F.P., J.P., V.M.P., T.C.S., R.A.W., L.H.C., and C.R.L. analyzed the data.
References
- 1.Cavallari LH, Beitelshees AL, Blake KV, Dressler LG, Duarte JD, Elsey A, et al. The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real-World Setting. Clin Transl Sci. 2017;10(3):143–146. doi: 10.1111/cts.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine GN, Bates ER, Blankenship JC, Bailey SR, Bittl JA, Cercek B, et al. 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation. 2011;124(23):e574–651. doi: 10.1161/CIR.0b013e31823ba622. [DOI] [PubMed] [Google Scholar]
- 3.Wallentin L, Becker RC, Budaj A, Cannon CP, Emanuelsson H, Held C, et al. Ticagrelor versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2009;361(11):1045–1057. doi: 10.1056/NEJMoa0904327. [DOI] [PubMed] [Google Scholar]
- 4.Wiviott SD, Braunwald E, McCabe CH, Montalescot G, Ruzyllo W, Gottlieb S, et al. Prasugrel versus clopidogrel in patients with acute coronary syndromes. N Engl J Med. 2007;357(20):2001–2015. doi: 10.1056/NEJMoa0706482. [DOI] [PubMed] [Google Scholar]
- 5.Fan W, Plent S, Prats J, Deliargyris EN. Trends in P2Y12 Inhibitor Use in Patients Referred for Invasive Evaluation of Coronary Artery Disease in Contemporary US Practice. Am J Cardiol. 2016;117(9):1439–1443. doi: 10.1016/j.amjcard.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, 2nd, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5(12):2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 7.Varenhorst C, James S, Erlinge D, Brandt JT, Braun OO, Man M, et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J. 2009;30(14):1744–1752. doi: 10.1093/eurheartj/ehp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon JY, Franchi F, Rollini F, Rivas Rios JR, Kureti M, Cavallari LH, et al. Role of Genetic Testing in Patients undergoing Percutaneous Coronary Intervention. Expert Rev Clin Pharmacol. 2017 doi: 10.1080/17512433.2017.1353909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudle KE, Dunnenberger HM, Freimuth RR, Peterson JF, Burlison JD, Whirl-Carrillo M, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC) Genet Med. 2017;19(2):215–223. doi: 10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scott SA, Sangkuhl K, Stein CM, Hulot JS, Mega JL, Roden DM, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther. 2013;94(3):317–323. doi: 10.1038/clpt.2013.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuldiner AR, O’Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302(8):849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 13.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome P450 genetic polymorphisms and the response to prasugrel: relationship to pharmacokinetic, pharmacodynamic, and clinical outcomes. Circulation. 2009;119(19):2553–2560. doi: 10.1161/CIRCULATIONAHA.109.851949. [DOI] [PubMed] [Google Scholar]
- 14.Wallentin L, James S, Storey RF, Armstrong M, Barratt BJ, Horrow J, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376(9749):1320–1328. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 15.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol. 2010;56(4):321–341. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 16.Cavallari LH, Lee CR, Beitelshees AL, Cooper-DeHoff RM, Duarte JD, Voora D, et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc Interv. 2018;11(2):181–91. doi: 10.1016/j.jcin.2017.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nutescu EA, Drozda K, Bress AP, Galanter WL, Stevenson J, Stamos TD, et al. Feasibility of implementing a comprehensive warfarin pharmacogenetics service. Pharmacotherapy. 2013;33(11):1156–1164. doi: 10.1002/phar.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.ASHP statement on the pharmacist’s role in clinical pharmacogenomics. Am J Health Syst Pharm. 2015;72(7):579–581. doi: 10.2146/sp150003. [DOI] [PubMed] [Google Scholar]
- 20.Shuldiner AR, Palmer K, Pakyz RE, Alestock TD, Maloney KA, O’Neill C, et al. Implementation of pharmacogenetics: the University of Maryland Personalized Anti-platelet Pharmacogenetics Program. Am J Med Genet C Semin Med Genet. 2014;166C(1):76–84. doi: 10.1002/ajmg.c.31396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee JA, Lee CR, Reed BN, Plitt DC, Polasek MJ, Howell LA, et al. Implementation and evaluation of a CYP2C19 genotype-guided antiplatelet therapy algorithm in high-risk coronary artery disease patients. Pharmacogenomics. 2015;16(4):303–313. doi: 10.2217/pgs.14.180. [DOI] [PubMed] [Google Scholar]
- 22.Deiman BA, Tonino PA, Kouhestani K, Schrover CE, Scharnhorst V, Dekker LR, et al. Reduced number of cardiovascular events and increased cost-effectiveness by genotype-guided antiplatelet therapy in patients undergoing percutaneous coronary interventions in the Netherlands. Neth Heart J. 2016;24(10):589–599. doi: 10.1007/s12471-016-0873-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanchez-Ramos J, Davila-Fajardo CL, Toledo Frias P, Diaz Villamarin X, Martinez-Gonzalez LJ, Martinez Huertas S, et al. Results of genotype-guided antiplatelet therapy in patients who undergone percutaneous coronary intervention with stent. Int J Cardiol. 2016;225:289–295. doi: 10.1016/j.ijcard.2016.09.088. [DOI] [PubMed] [Google Scholar]
- 24.Harada S, Zhou Y, Duncan S, Armstead AR, Coshatt GM, Dillon C, et al. Precision Medicine at the University of Alabama at Birmingham: Laying the Foundational Processes Through Implementation of Genotype-Guided Antiplatelet Therapy. Clin Pharmacol Ther. 2017 doi: 10.1002/cpt.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavallari LH, Weitzel KW, Elsey AR, Liu X, Mosley SA, Smith DM, et al. Institutional profile: University of Florida Health Personalized Medicine Program. Pharmacogenomics. 2017;18(5):421–426. doi: 10.2217/pgs-2017-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet. 2014;166C(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limdi NA, Holmes AM, Skaar TC, Cavallari LH, johnson JA, Beitelshees AL, et al. Real world cost-effectiveness of CYP2C19 guided antiplatelet therapy in patients with acute coronary syndrome and percutaneous coronary intervention. Society of Medical Decision Making (Abstract #11046) 2017 doi: 10.1038/s41397-020-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott SA. Clinical pharmacogenomics: opportunities and challenges at point of care. Clin Pharmacol Ther. 2013;93(1):33–35. doi: 10.1038/clpt.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weitzel KW, Cavallari LH, Lesko LJ. Preemptive Panel-Based Pharmacogenetic Testing: The Time is Now. Pharm Res. 2017;34(8):1551–1555. doi: 10.1007/s11095-017-2163-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson JE, Rohrer Vitek CR, Bell EJ, McGree ME, Jacobson DJ, St Sauver JL, et al. Participant-perceived understanding and perspectives on pharmacogenomics: the Mayo Clinic RIGHT protocol (Right Drug, Right Dose, Right Time) Genet Med. 2017;19(7):819–825. doi: 10.1038/gim.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cavallari LH, Lee CR, Duarte JD, Nutescu EA, Weitzel KW, Stouffer GA, et al. Implementation of inpatient models of pharmacogenetics programs. Am J Health Syst Pharm. 2016;73(23):1944–1954. doi: 10.2146/ajhp150946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Owusu-Obeng A, Weitzel KW, Hatton RC, Staley BJ, Ashton J, Cooper-Dehoff RM, et al. Emerging roles for pharmacists in clinical implementation of pharmacogenomics. Pharmacotherapy. 2014;34(10):1102–1112. doi: 10.1002/phar.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crews KR, Cross SJ, McCormick JN, Baker DK, Molinelli AR, Mullins R, et al. Development and implementation of a pharmacist-managed clinical pharmacogenetics service. Am J Health Syst Pharm. 2011;68(2):143–150. doi: 10.2146/ajhp100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanek EJ, Sanders CL, Taber KA, Khalid M, Patel A, Verbrugge RR, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther. 2012;91(3):450–458. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 35.Tuteja S, Haynes K, Zayac C, Sprague JE, Bernhardt B, Pyeritz R. Community pharmacists’ attitudes towards clinical utility and ethical implications of pharmacogenetic testing. Per Med. 2013;10(8) doi: 10.2217/pme.13.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitzel KW, Aquilante CL, Johnson S, Kisor DF, Empey PE. Educational strategies to enable expansion of pharmacogenomics-based care. Am J Health Syst Pharm. 2016;73(23):1986–1998. doi: 10.2146/ajhp160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorich MJ, Rowland A, McKinnon RA, Wiese MD. CYP2C19 genotype has a greater effect on adverse cardiovascular outcomes following percutaneous coronary intervention and in Asian populations treated with clopidogrel: a meta-analysis. Circ Cardiovasc Genet. 2014;7(6):895–902. doi: 10.1161/CIRCGENETICS.114.000669. [DOI] [PubMed] [Google Scholar]
- 38.Mega JL, Simon T, Collet JP, Anderson JL, Antman EM, Bliden K, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304(16):1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sorich MJ, Vitry A, Ward MB, Horowitz JD, McKinnon RA. Prasugrel vs clopidogrel for cytochrome P450 2C19-genotyped subgroups: integration of the TRITON-TIMI 38 trial data. J Thromb Haemost. 2010;8(8):1678–1684. doi: 10.1111/j.1538-7836.2010.03923.x. [DOI] [PubMed] [Google Scholar]
- 40.Peterson JF, Field JR, Unertl KM, Schildcrout JS, Johnson DC, Shi Y, et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clin Pharmacol Ther. 2016;100(1):67–74. doi: 10.1002/cpt.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Testing metrics


