Abstract
Background:
Regarding hydatid cyst (cystic echinococcosis, CE) as a human public health problem in the West of Iran, molecular data related to the genotypes of Echinococcus granulosus in cattle and sheep in these regions are still insufficient. Here, we evaluated the genotypes of E. granulosus infecting sheep and cattle in western Iran.
Methods:
Totally, 36 hydatid cysts including 18 hydatid cysts of sheep and 18 hydatid cysts of cattle were collected from Khorramabad slaughterhouse (Lorestan Province), Western Iran between May to September 2014. Protoscoleces or germinal layers were collected from individual cysts, DNA was extracted, and genotyping was performed by sequencing and analyzing mitochondrial cytochrome c oxidase subunit 1 (cox1) gene.
Results:
In sequencing analysis, all of sheep isolates belonged to genotype G1 (sheep strain). Among cattle hydatid cyst isolates, 16/18 (88.9%) were belonged to genotype G1 and 2/18 (11.1%) were belonged to G3 genotype. The phylogenetic analysis showed two clusters; one of the clusters includes cattle G3 genotype and the other cluster represents sheep and cattle G1 genotype that were isolated.
Conclusion:
The common sheep strain/G1 is predominant genotype in the western part of Iran, followed by G3 genotype, circulating among the animal hosts in this region. Further studies covering a larger number of isolates might be necessary to see if there are other genotypes in the hydatid cyst population in this region of Iran.
Keywords: Cystic echinococcosis, Cox1, Sheep, Echinococcus granulosus
Introduction
Hydatid cyst (cystic echinococcosis, CE), a parasitic infection caused by the larval stage of dog tapeworm Echinococcus granulosus, has been identified as a major public health and economic problem in developing countries, including Iran (1). The dog is the definitive host, where adult tapeworms attached to the intestinal epithelium, and humans as well as domestic livestock can be the intermediate host, where the worms form hydatid cysts in various organs (2).
In CE, clinical manifestations are mostly related to the localization, size, and number of cysts. There seemed to be a relationship between the genotypes and the size of hydatid cysts, in which all the patients infected with G7 genotype showed smaller liver cysts than those infected with G1 genotype (3). Molecular epidemiological data have identified 10 distinct genotypes of E. granulosus (G1–G10): E. granulosus sensustricto (G1, G2, G3), Echinococcus equines (G4), E. ortleppi (G5), E. intermedius (G6, G7, G8, G10) (4). In Iran, to date, three genotypes have been found including G1, G3, and G6 genotypes in humans and animals in different regions (5). Sheep, buffalo, and camels that are among common livestock in Iran are considered as main reservoirs of genotypes G1, G3, and G6, respectively (6). Reviews on the incidence of E. granulosus have reported that G1 (common sheep strain) is the most predominant genotype in Iran (6) and also most parts of the world (4). Studies have demonstrated that the, cytochrome c oxidase subunit 1 (cox1) and NADH dehydrogenase 1 (nad1) genes are good molecular markers for investigating genetic variation in a number of isolates of E. granulosus from a range of hosts (i.e., cattle, sheep, and goat) in Iran (5, 7). Moreover, mitochondrial genomes have the ability to eliminate some limitations in Echinococcus taxonomy (8).
Recently, Parsa et al (9) have demonstrated three genotypes (G1 [75%], G2 [10%] and G3 [15%]) from the E. granulosusisolates of 71 stray dogs, from Lorestan province, western Iran using DNA sequencing of the partial mitochondrial cox1 and nad1 (9). However, no genotyping data are available about E. granulosus isolates of domestic livestock in this province using DNA sequencing of cox1 and nad1.
Therefore, the present study aimed molecularly identify and genotype hydatid cysts of some livestock (sheep, and cattle) in Lorestan Province by sequencing and analyzing mitochondrial cox1 gene, to speculate on possible transmission patterns of this cestode by homology analysis, and to understand the phylogeny of genotypes of E. granulosus by constructing neighbor-joining trees.
Materials and Methods
Ethics statement
This study was approved by the Committee on the Ethics of Animal and Human Experiments of the Lorestan University of Medical Science, Khorramabad, Iran (Permit Number: 89/6).
Collection of hydatid cyst samples
A total of 36 hydatid cysts including 18 hydatid cysts of sheep and 18 hydatid cysts of cattle were collected from Khorramabad slaughterhouse (Lorestan Province), Western Iran between May to September 2014 and carried to the Parasitology Laboratory at the, Lorestan University of Medical Sciences, (Khorramabad, Iran).
Microscopic examination of protoscoleces
To confirm whether or not the protoscoleces exist in hydatid cysts, their contents were subjected to the microscopic examination. Protoscoleces and/or germinal layers were collected from cysts and washed five times with phosphate-buffered saline (PBS). The obtained pellets were fixed in 70 % ethanol and stored at 4 °C until genomic DNA isolation.
Extraction of genome DNA
To extract the genomic DNA (gDNA) of hydatid cyst samples, protoscoleces and/or germinal layers from each individual cyst were washed five times in sterile-distilled water by centrifugation to remove the ethanol and gDNA of each sample was extracted by High Pure PCR Template preparation kit (Roche, Mannheim, Germany) according to the manufacturer’s instructions. To ensure the accuracy of DNA extraction, concentration of the extracted DNA samples were analyzed by NanoDrop. The extracted DNAs were stored at −20°C until required for PCR.
Mitochondrial PCR amplification
The mitochondrial cox1 gene was amplified using specific primer sets JB3/JB4.5 (10). PCR was performed in a 25 μl final volume containing 1 X PCR buffer, 2.5 mM MgCl2, 0.2 mM of each deoxynucleotide triphosphate (dNTP), 0.2 μM of each primer and 1.5 μM Ampli-Taq Polymerase. Two primers, JB3 (forward), 5′-TTT TTT GGG CAT CCT GAG GTT TAT-3′ and JB4.5 (reverse), 5′-TAA AGA AAG AAC ATA ATG AAA ATG-3′ (10), were used to amplify a 450 bp fragment of cox1 gene under the following conditions: initial denaturation 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30s, annealing at 55°C for 30s and elongation for 45s at 72°C. Final extension was performed at 72°C for 10 min, and then the product of PCR was electrophoresed on 1.5% agar gel. All PCR amplifications were run with negative control (non-template water control).
Sequencing analysis
All PCR products were subjected to automated sequencing by the Illumina Genome Analysis System, employing the same primers used in the primary PCR. The electropherogram of each sequence was checked by eye, and the sequences were compared with each other using the software BioEdit. Accuracy of the sequencing data was confirmed by sequencing in both directions. Nucleotide sequences obtained in the present study were subjected to BLAST searches (http://www.ncbi.nlm.nih.gov/blast/), and were then aligned and analyzed with each other and E. granulosus reference sequences downloaded from GenBank using Clustal X 1.83.
Phylogenetic analysis
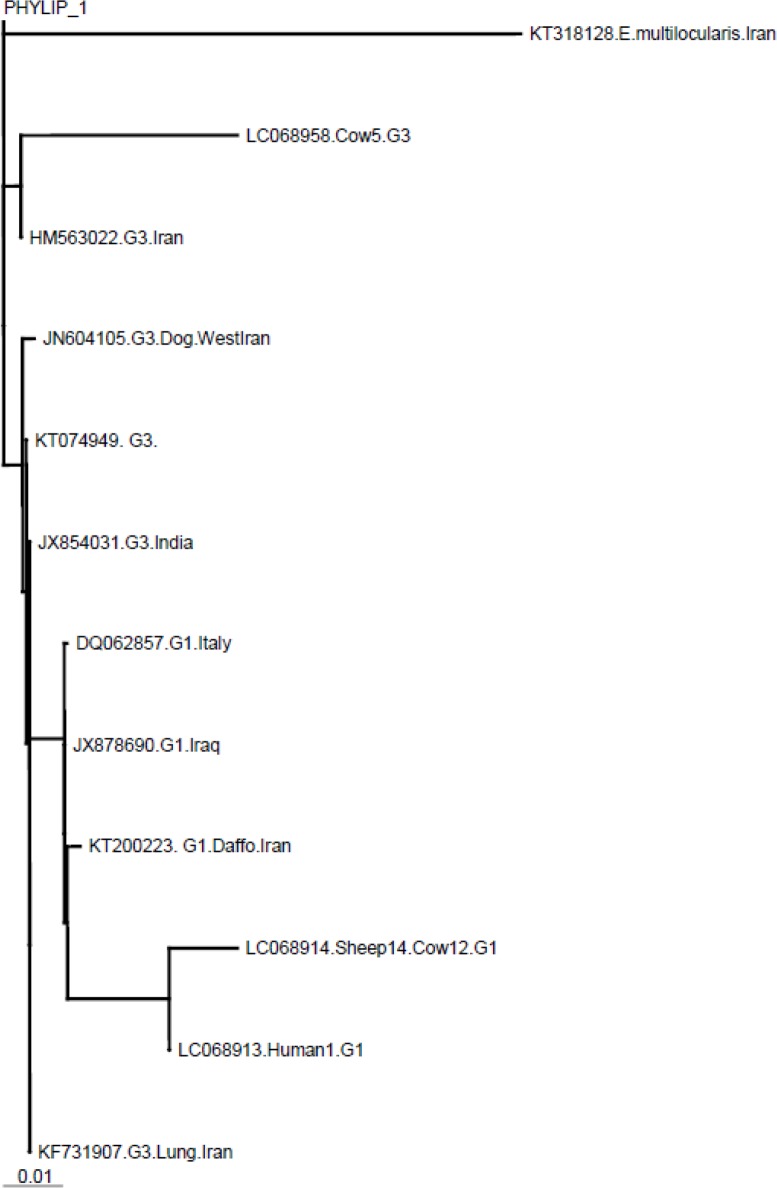
Sequences were aligned by means of the CLUSTALW software package (www.ebi.ac.uk/clustalw) and analyzed using the Neighbor–Joining (NJ) method provided in the MUST software package. The phylogenetic tree was run using sequences obtained in this study as well as reference sequences available for E. granulosus G1(accession number, KT200223.1) and G3 (accession number, HM563022.1) genotypes as shown in Fig. 1. Some of G1 (DQ062857.1, JX878690.1) and G3 (KT074949.1, JN604105.1, KF731907, JX854031.1) genotypes in the gene bank was used for comparison. E. multilocularis COX1 (KT318128.1) was applied in the model as anout-group.
Fig. 1:
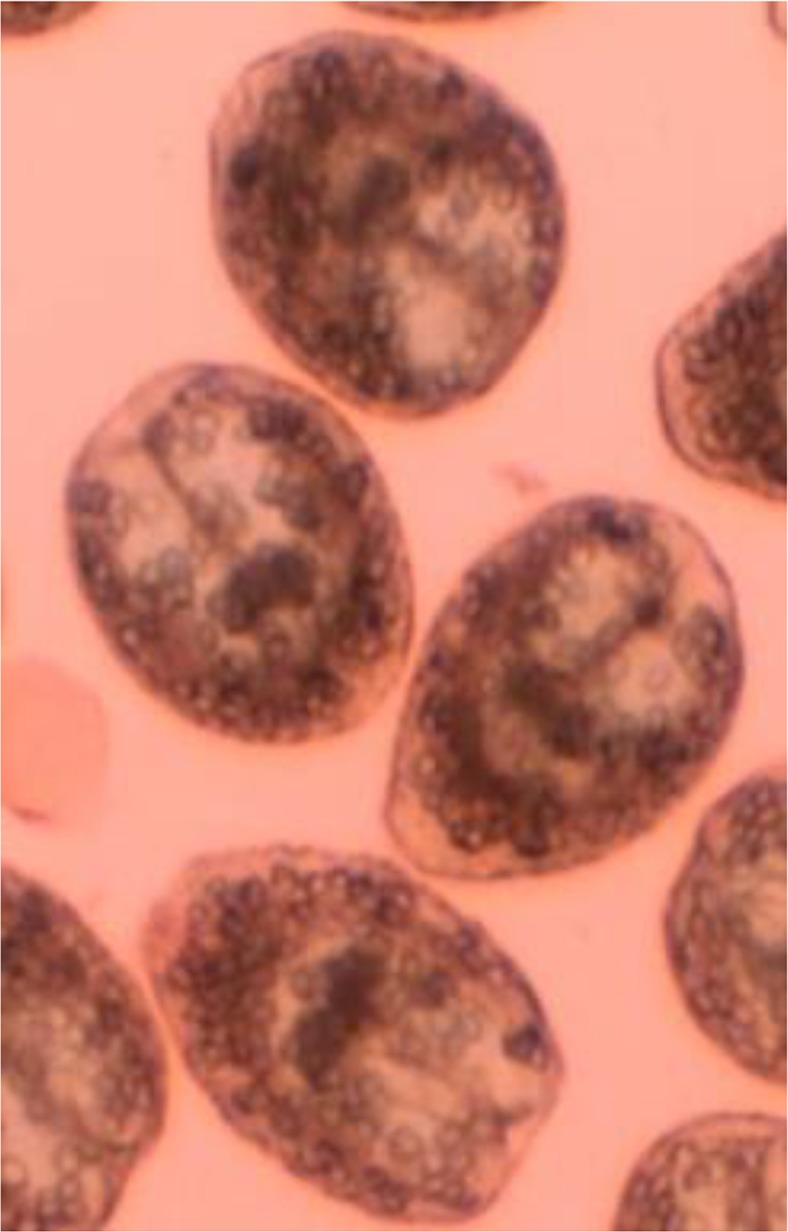
Live (colorless) protoscoleces of hydatid cysts with 0.1% eosin
Results
Totally, 36 hydatid cysts isolates from domestic animals from Lorestan Province were tested for the molecular analysis. All these samples were analyzed using mitochondrial cox1 primers. For all of isolates, fragment of about 450 bp were successfully PCR-amplified within cox1 gene. In sequencing analysis, the alignments of the sequences determined in this study with those of know genotypes of E. granulosus demonstrated all of sheep isolates belonged to genotype G1 (sheep strain). Among cattle hydatid cyst isolates, 16/18 (88.9%) were belonged to genotype G1 and 2/18 (11.1%) were belonged to G3 genotype. All the hydatid cysts belonged to G1 were confirmed to be fertile (with protoscoleces) by microscopy (Fig.1); whereas both hydatid cysts of belonged to G3 were observed to be fertile. Representative nucleotide sequences obtained in this study were deposited in the GenBank database under the accession numbers LC068914.1 and LC068958.1 for G1 genotype (for both sheep and cattle) and G3 genotype (cattle) respectively.
The phylogenetic analysis showed two clusters; one of the clusters includes cattle G3 genotype and the other cluster represents sheep and cattle G1 genotype that were isolated during in this study (Fig. 2).
Fig. 2:
Molecular phylogenetic tree of 3 E. granulosus isolates of sheep, cattle, and human along with reference isolates based on CO1 gene sequence. Accession numbers of KT200223 and HM563022 represent reference sequences of E. granulosus genotypes G1 and G3, respectively. Some of G1 (DQ062857, JX878690) and G3 (KT074949, JN604105, KF731907, JX854031) genotypes was used for comparison. E. multilocularis KT318128 was used as out group sequence data
Discussion
Lorestan Province due to favorable weather conditions and the spread of livestock is one of the prone provinces in Iran for prevalence of parasites (11–16). Cystic echinococcosis is a globally distributed parasitic infection of humans and livestock. At present, the World Health Organization (WHO) included CE in a subgroup of selected Neglected Tropical Diseases (NTDs) to be addressed within its 2008–2015 strategic plans for control of NTDs (1). In the present study, 36 hydatid cysts including 18 hydatid cysts of sheep and 18 hydatid cysts of cattle were tested for the molecular analysis. In this study, 34 (94.4%) and 2 (5.6%) of isolates were identified as G1 and G3 genotype of E. granulosus, respectively. The results presented here demonstrated that G1 was the dominant genotype of CE in some livestock animals such as sheep and cattle in Western Iran. The obtained findings revealed that all the hydatid cysts of isolated from sheep and 16/18 (88.8%) of cattle isolates were belonged to genotype G1. The G1 genotype can develop fertile cysts in sheep, but it can also infect goats, cattle, camel, pigs, and humans. We found only two isolates (3.6%) belonged to genotype G3. This isolates belonged to the cattle samples (2/18, 11.1%). The presence of G3 genotype producing fertile cysts in cattle, suggests the correlation between G3 genotype and producing fertile cysts in cattle and its high host specificity.
There are several studies that showed G1 genotype of E. granulosus is the predominant genotype in animals and humans around the world. For example, Utuk et al. (17) conducted a study in Turkey on 208 isolates (179sheep, 19 cattle, seven goats, one camel, one dog, and a single human sample) and detected only the G1 genotype in all of their isolates. Reviews have reported a similar result with different G1 to G3 proportions: for example, 95.5 % G1 vs. 4.5 % G3 in 112 sheep and cattle in Turkey (18); 78.75 % G1 vs. 12.5 % G3 in 80 cattle and water buffaloes in Italy (19); 93.3 % G1 vs. 6.7 % G3 in 30 ovine, bovine, and humans in Tunisia (20); 77.8 % G1 vs. 11.1 % G3 in 18 humans and dogs in southern Brazil (21); and 73.7 % G1 vs. 13.2 % G3 in 38 different intermediate hosts in southeast of Iran (22). Unlikely, Pednekar et al. (23) have demonstrated G3 in 63 % as the predominant genotype, whereas the G1 genotype was found only in six (13 %) isolates from 46 domestic livestock in India.
In line with our findings several investigations have been conducted on genotyping of E. granulosus in Iran. For example, dominant strain of E. granulosus in Chaharmahal-va –Bakhtyari Province, Central Iran, was G1 (24). Other study conducted in Isfahan Province, central Iran, on hydatid cysts with origination of cattle, sheep, goat, camel and human, showed G1 genotype as the dominant strain (25). Among 86 isolates E. granulosus isolated from humans and domestic animals from northwest of Iran (Zanjan Province) using the mitochondrial cox1 gene, 82 (95.35 %) were determined as G1 genotype, and the remaining four (4.65%), were identified as G3 genotype (26). All samples isolated in definitive and intermediate hosts of the E. granulosus belonged to G1 genotype (sheep strain) (27). In another study, three distinct genotypes (G1, G3, and G6) were reported from human, sheep, cattle, goats, and camels in Kerman Province, southeast of Iran (22). Ninety three isolated hydatid cysts from slaughtered livestock of Yasuj City, were G1 genotype (28). Moreover, in Ardabil Province, G1 was the dominant strain and for the first time in Iran G3 genotype in 2 human isolates was reported (29).
Based on the previous investigation three genotypes (G1, G2 and G3) were identified from the E. granulosus isolates of stray dogs, from Lorestan province using DNA sequencing of the partial mitochondrial cox1 and nad1 genes, whereas sheep appear to be a principal animal intermediate host for E. granulosus in this region. In other studies on human and domestic herbivores of this province, the results showed G1 as the dominant strain indicating the main intermediate host and biological maintenance in the nature are sheep and goat (30, 31). In Lorestan Province, the livestock (sheep and cattle) industry is a major economic component. Once these domestic animals are infected with E. granulosus, the habit of feeding raw offal of animals to dogs will easily lead to E. granulosus infection in dogs. Thus, this parasite can complete its life cycle in our area.
Conclusion
The common sheep strain/G1 is predominant genotype in the western part of Iran, followed by G3 genotype, circulating among the animal hosts in this region. The presence of dominant sheep strain (G1) in sheep and cattle of Lorestan Province shows that these animals probably infected of the same origin. This is because of the popularity of traditional live stock in the area that cattle and sheep use from one pasture. Further studies covering a larger number of isolates might be necessary to see if there are other genotypes in the hydatid cyst population in this region of the country.
Acknowledgements
This study was supported by Lorestan University of Medical Sciences, Khorramabad, Iran.
Footnotes
Declaration of Interest
The authors report no conflicts of interest.
References
- 1.World Health Organization (WHO) informal working group on echinococcosis. Bull WHO. 1996; 74:231–42. [PMC free article] [PubMed] [Google Scholar]
- 2.Mahmoudvand H, Kheirandish F, Dezaki ES, Shamsaddini S, Harandi MF. Chemical composition, efficacy and safety of Pistacia vera (var. Fandoghi) to inactivate protoscoleces during hydatid cyst surgery. Biomed Pharmacother. 2016; 82: 393–8. [DOI] [PubMed] [Google Scholar]
- 3.Schneider R, Gollackner B, Schindl M, Tucek G, Auer H. Echinococcus canadensis G7 (pig strain): an underestimated cause of cystic echinococcosis in Austria. Am J Trop Med Hyg. 2010; 82(5):871–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson RC. The taxonomy, phylogeny and transmission of Echinococcus. Exp Parasitol. 2008; 119(4):439–46. [DOI] [PubMed] [Google Scholar]
- 5.Rostami Nejad M, Taghipour N, Nochi Z, Mojarad EN, Mohebbi SR, Harandi MF, Zali MR. Molecular identification of animal isolates of Echinococcus granulosus from Iran using four mitochondrial genes. J Helminthol. 2012; 86(4):485–92. [DOI] [PubMed] [Google Scholar]
- 6.Sharbatkhori M, Mirhendi H, Jex AR, Pangasa A, Campbell BE, Kia EB, Eshraghian MR, Harandi MF, Gasser RB. Genetic categorization of Echinococcus granulosus from humans and herbivorous hosts in Iran using an integrated mutation scanning-phylogenetic approach. Electrophoresis. 2009; 30(15):2648–55. [DOI] [PubMed] [Google Scholar]
- 7.Sharbatkhori M, Fasihi Harandi M, Mirhendi H, Hajialilo E, Kia EB. Sequence analysis of cox1 and nad1 genes in Echinococcus granulosus G3 genotype in camels (Camelus dromedarius) from central Iran. Parasitol Res. 2011; 108(3):521–7. [DOI] [PubMed] [Google Scholar]
- 8.Rostami Nejad M, Nazemalhosseini Mojarad E, Fasihi Harandi M. Echinococcosis: based on molecular studies in Iran. Gastroenterology Hepatology Bed Bench. 2010; 3: 169–76. [Google Scholar]
- 9.Parsa F, Fasihi Harandi M, Rostami S, Sharbatkhori M. Genotyping Echinococcus granulosus from dogs from Western Iran. Exp Parasitol. 2012; 132(2):308–12. [DOI] [PubMed] [Google Scholar]
- 10.Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol Biochem Parasitol. 1992; 54(2):165–73. [DOI] [PubMed] [Google Scholar]
- 11.Kheirandish F, Sharafi AC, Kazemi B, Bandehpour M, Tarahi Mj, Khamesipour A. First molecular identification of Leishmania species in a new endemic area of cutaneous leishmaniasis in Lorestan, Iran. Asian Pac J Trop Med. 2013;6(9):713–7. [DOI] [PubMed] [Google Scholar]
- 12.Kheirandish F, Chegeni Sharafi A, Kazemi B, Mohebali M, Sarlak A, Tarahi MJ, Holakouee K, Hajaran H. Identification of Leishmania species using PCR assay on Giemsa-stained slides prepared from cutaneous leishmaniasis patients. Iran J Parasitol. 2013;8(3):382–8. [PMC free article] [PubMed] [Google Scholar]
- 13.Kheirandish F, Badparva E, Haghighi A, Nazemalhosseini Mojarad E, Kazemi B. Differential diagnosis of Entamoeba spp. in gastrointestinal disorder patients in Khorramabad, Iran. African J Microbiol Res. 2011; 5(18): 2863–6. [Google Scholar]
- 14.Kheirandish F, Tarahi MJ, Ezatpour B. Prevalence of intestinal parasites among food handlers in West of Iran. Rev Inst Med Trop Sao Paulo. 2014;56(2):111–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badparva E, Kheirandish F, Ebrahimzade F. Prevalence of intestinal parasites in Lorestan Province, West of Iran. Asian Pac J Trop Dis. 2014; 4: 930–4. [Google Scholar]
- 16.Kheirandish F, Tarahi M, Haghighi A, Nazemalhosseini-Mojarad E, Kheirandish M. Prevalence of intestinal parasites in bakery workers in Khorramabad, Lorestan Iran. Iran J Parasitol. 2011; 6(4): 76–83. [PMC free article] [PubMed] [Google Scholar]
- 17.Utuk AE, Simsek S, Koroglu E, McManus DP. Molecular genetic characterization of different isolates of Echinococcus granulosus in east and southeast regions of Turkey. Acta Trop. 2008; 107(2):192–4. [DOI] [PubMed] [Google Scholar]
- 18.Vural G, Baca AU, Gauci CG, Bagci O, Gicik Y, Lightowlers MW. Variability in the Echinococcus granulosus cytochrome C oxidase 1 mitochondrial gene sequence from livestock in Turkey and a reappraisal of the G1–3 genotype cluster. Vet Parasitol. 2008; 154(3–4):347–50. [DOI] [PubMed] [Google Scholar]
- 19.Casulli A, Manfredi MT, La Rosa G, et al. Echinococcus ortleppi and E. granulosus G1, G2 and G3 genotypes in Italian bovines. Vet Parasitol. 2008. August 1;155(1–2):168–72. [DOI] [PubMed] [Google Scholar]
- 20.M’rad S, Oudni-M’rad M, Filisetti D, et al. Molecular identification of Echinococcus granulosus in Tunisia: first record of the Buffalo strain (G3) in human and bovine in the country. Open Vet Sci J. 2010; 4:27–30. [Google Scholar]
- 21.de la Rue ML, Takano K, Brochado JF, Costa CV, et al. Infection of humans and animals with Echinococcus granulosus (G1 and G3 strains) and E. ortleppi in Southern Brazil. Vet Parasitol. 2011; 177(1–2):97–103. [DOI] [PubMed] [Google Scholar]
- 22.Hajialilo E, Harandi MF, Sharbatkhori M, Mirhendi H, Rostami S. Genetic characterization of Echinococcus granulosus in camels, cattle and sheep fromthe south-east of Iran indicates the presence of the G3 genotype. J Helminthol. 2012; 86(3):263–70. [DOI] [PubMed] [Google Scholar]
- 23.Pednekar RP, Gatne ML, Thompson RC, Traub RJ. Molecular and morphological characterisation of Echinococcus from food producing animals in India. Vet Parasitol. 2009; 165(1–2):58–65. [DOI] [PubMed] [Google Scholar]
- 24.Yoosefi H, Hashemzadeh M, Aliyari Z. Molecular study of hydatid cyst (sheep strain) in Chaahrmohal va Bakhteyari by PCR-RFLP. J Shahrkord Uni. 2007; 9(2):28–33. [Google Scholar]
- 25.Shahnazi M, Hejazi H, Salehi M, Andalib AR. Molecular characterization of human and animal Echinococcus granulosus isolates in Isfahan, Iran. Acta Trop. 2011; 117(1):47–50. [DOI] [PubMed] [Google Scholar]
- 26.Farhadi M, Fazaeli A, Haniloo A. Genetic characterization of livestock and human hydatid cyst isolates from northwest Iran, using the mitochondrial cox1 gene sequence. Parasitol Res. 2015; 114(12):4363–70. [DOI] [PubMed] [Google Scholar]
- 27.Yakhchali M, Mardani K. Study on strain variation of Echinococcus granulosus in domestic life cycle by amplification of nad-1 gene by PCR-RFLP. Iran J Vet. 2011;7(1):63–8. [Google Scholar]
- 28.Sadri A, Moshfe A, Doosti A. Characterization of isolated hydatid cyst from slaughtered livestock in Yasuj industrial slaughterhouse by PCR-RFLP. J Yasuj Uni. 2012; 17(3):143–52. [Google Scholar]
- 29.Pezeshki A, Akhlaghi L, Sharbatkhori M, Razmjou E, Oormazdi H, Mohebali M, Meamar AR. Genotyping of Echinococcus granulosus from domestic animals and humans from Ardabil Province, northwest Iran. J Helminthol. 2012;87(4):387–91. [DOI] [PubMed] [Google Scholar]
- 30.Parsa F, Haghpanah B, Pestechian N, Salehi M. Molecular epidemiology of Echinococcus granulosus strains in domestic herbivores of Lorestan, Iran. Jundishapur J Microbiol. 2011; 4(2): 123–30. [Google Scholar]
- 31.Kheirandish F, Mahmoudvand H, Ahmadinejad M, Karimi Rouzbahani A. Genetic characterization of human-derived hydatid cysts of Echinococcus granulosus in Lorestan Province, Western Iran. Tropical Biomed. 2017; 34(4): 863–9. [PubMed] [Google Scholar]