Abstract
Objective
To better understand the role of B cells, potential mechanisms for their aberrant activation, and the production of autoantibodies in the pathogenesis of Sjögren’s Syndrome (SS), we explored selection pressures and N-glycosylation acquired by somatic mutation (acN-glyc) in the immunoglobulin (Ig) variable regions (V-regions) of antibody secreting cells (ASCs) isolated from the minor salivary glands of SS patients and non-SS controls with sicca symptoms.
Methods
We report a novel method to produce and characterize recombinant monoclonal antibodies (mAbs) from SS patient and control labial salivary gland single-cell sorted ASC infiltrates that can be utilized to concurrently probe any other expressed genes. V-regions were amplified by RT-PCR, sequenced, and analyzed for incidence of N-glycosylation and selection pressure, then expressed as the native mAbs, or mutant mAbs lacking the acN-glyc for specificity testing. Protein modeling was used to demonstrate how even acN-glycs outside of the complementarity-determining region (CDR) could participate in, or inhibit, antigen binding.
Results
V-region sequence analyses revealed clonal expansions and evidence for secondary light chain editing and allelic inclusion not previously reported in SS. We found increased acN-glycs in the sequences from SS patients and that acN-glycs were associated with increased replacement mutations and lowered selection pressure. We also identified a clonal set of polyreactive mAbs with differential FWR1 acN-glycs and demonstrated that removal of the acN-glyc could nearly abolish binding to the autoantigens.
Conclusion
Our findings support an alternative mechanism involving V-region N-glycosylation for the selection and proliferation of some autoreactive B cells in SS patients.
Sjögren’s syndrome (SS) is a systemic, chronic autoimmune disease that is characterized by lymphocytic infiltration of the exocrine glands, inflammation, tissue damage, and secretory dysfunction. The lacrimal and salivary glands are typically affected, resulting in dry eyes (keratoconjuntivitis sicca) and dry mouth (xerosomia), but patients can also present with extraglandular complications, or overlapping autoimmune diseases. (1–5). In addition, SS patients have an increased risk of progression to various non-Hodgkin lymphomas (NHL) resulting in significant morbidity (6).
There is evidence for chronic immune cell stimulation by bacteria in the development of NHLs not associated with SS (7–9). This mechanism was demonstrated, where Pseudomonas aeruginosa and Burkholderia cenocepacia lectins bound to and activated B cells via B cell receptor variable region (V-region) glycans (10). In SS NHL there is also evidence for a bacterial etiology with the regression of a parotid mucosa-associated lymphoid tissue (MALT) lymphoma after the clearing of an H. pylori infection (11).
It is known that dysregulation of both innate and adaptive immunity contributes to the etiology of SS and its complications; however, the pathophysiology of SS as well as Sjögren’s-associated lymphoma is largely unknown. B cells play a role in the pathogenesis of SS as evidenced by the presence of autoantigen-specific memory B cells (12, 13) and the incidence of autoantibodies to Ro/SSA (Ro52 and Ro60) and La/SSB (14). Much evidence supports antigen-driven production of autoantibodies within the salivary glands (13, 15, 16). Ig V-region sequence analysis enables the identification of clonally expanded cells, which is strong evidence for antigen-driven B cell activation and proliferation. Antigen-driven activation can also be determined empirically by selection pressure analyses of V-region sequences where the observed frequency of non-synonymous (replacement) mutations is compared to their expected frequency in a state of no selection. When the frequency of replacement mutations is greater than expected, the Ig is considered to have undergone positive selection, and when the frequency is less than expected, negative selection is indicated. Typical antigen-driven activation results in positive selection in the CDR regions, which directly interact with the antigen, and negative selection in the framework regions (FWRs), which are important for structural integrity. Selective pressure patterns contrary to this model indicate non-specific activation.
Somatic hypermutations (SHMs) leading to amino acid replacements can also give rise to post-translational modifications by the introduction of N-linked glycosylation (N-glyc) motifs. This results in SHM-acquired N-glycs (acN-glyc) of the antibody at these sites, and may have implications in immune responses or disease states. Ig V-region acN-glycs have been reported in SS parotid B cells (17) and are strongly correlated to follicular lymphoma (18, 19), a disease increased 4-fold in SS patients (20). Single Ig V-region acN-glycs introduced by SHM have been demonstrated to strengthen (21), weaken, or abolish binding for self or foreign antigens (22). Conversely, bacterial or innate immune system lectins can bind V-region acN-glycs, causing activation of B cells in an antigen-independent fashion (10, 23). Therefore, analyses of acN-glyc motifs may give clues to antibody-antigen interactions, tolerance mechanisms, and non-specific modes of B-cell activation that may drive proliferation of B cells in SS.
We hypothesized that acN-glyc motifs in the V-regions of IgG ASCs isolated from the labial salivary glands of SS patients and non-SS patients with dry mouth/eye symptoms (sicca) controls may provide opportunities for antigen-independent proliferation of autoreactive B cells and antibody production in the salivary glands of SS patients. Our findings support this hypothesis, suggesting an alternative means for B cell selection and proliferation of some autoreactive B cells seen in SS patients.
Patients and Methods
Human Subject Sample Collection and Evaluation
Studies were approved by the Oklahoma Medical Research Foundation (OMRF) and University of Oklahoma Health Sciences Center Institutional Review Boards. Samples and data were obtained from subjects following written, informed consent. Participants were evaluated in the OMRF Sjögren’s Research Clinic (OSRC) as reported (24). Labial salivary glands were collected for single-cell sorting and mAb production as described (16). All 14 participants had symptomatic dry eyes and mouth without a current or historic diagnosis of NHL; 8 met American/European Consensus Group (AECG) primary SS classification criteria (2) and 1 also met the American College of Rheumatology criteria for SLE (25, 26). Six subjects that did not meet the SS classification criteria (DNMC) served as sicca controls. Hematoxylin and eosin (H&E)-stained salivary gland sections were obtained from the OSRC repository, and two independent evaluations were made for the presence of ectopic germinal center structures by both an oral pathologist and a hematopathologist. Participant demographics, classification criteria, clinical/extraglandular manifestations, and EULAR Sjögren’s syndrome disease activity index (ESSDAI) scores are summarized in Table 1. SS patients were evaluated for the predictive risk of developing NHL as described in (27) (Table S1).
Table 1.
Participant demographics, classification criteria, and extraglandular manifestations.
| SS (n=8) | DNMC (n=6) | |
|---|---|---|
| Demographics | ||
| Mean Age, S.D. (range) | 44.23, ± 10.48 (30-58) | 50, ± 11.97 (36-72) |
| Sex (Female/Male) | 8/0 | 5/1 |
| Race | Black (1), White (5), Wh/N.A. (2) | Black (1), White (4), Wh/N.A. (1) |
| Classification Criteria | ||
| Lissamine Green (No. Pos/Total) | 8/8 | 0/6 |
| Schirmer’s Test (No. Pos/Total) | 3/8 | 0/6 |
| WUSF (No. Pos/Total) | 3/8 | 2/6 |
| Biopsy Interpretation (No.) | FLS (8) | NSCS (5), FLS (1) |
| Mean Focus Score, S.D. (Range) | 4.41 ±3.39 (1.8 – 12) | 0.33 ± 0.82 (0 – 2) |
| Serum ANA (No. Pos/Total) | 8/8 | 5/6 |
| Serum anti-Ro/SSA (No. Pos/Total) | 6/8 | 1/6 |
| Serum anti-La/SSB (No. Pos/Total) | 3/8 | 0/8 |
| Mean ACR/EULAR Score, S.D. | 7.0 ± 2.07 | 1.33 ± 1.37 |
| Clinical/Extraglandular | ||
| Mean ESSDAI Score, S.D. | 1.625 ±0.92 | N/A |
| Hyper IgG (No. Pos/Total) | 5/8 | 1/6 |
| Hypo IgG (No. Pos/Total) | 0/8 | 1/6 |
| Hyper IgM (No. Pos/Total) | 1/8 | 0/6 |
| Low C3 (No. Pos/Total) | 0/8 | 0/6 |
| Low C4 (No. Pos/Total) | 1/8 | 0/6 |
| Low CH50 (No. Pos/Total) | 1/8 | 1/6 |
| Salivary Gland Enlargement - Current or Hx (No. Pos/Total) | 2/8 | 3/6 |
| Arthritis/Arthralgias (No. Pos/Total) | 2/8 | 2/6 |
| Synovitis (No. Pos/Total) | 1/8 | 1/6 |
| Raynaud’s Syndrome (No. Pos/Total) | 4/8 | 3/6 |
Abbreviations: ACR, American College of Rheumatology; ANA, anti-nuclear antibodies; DNMC, does not meet criteria (control); ESSDAI, EULAR Sjögren’s syndrome disease activity index; EULAR, European League Against Rheumatism; FLS, focal lymphocytic sialadenitis; Hx, history; Neg, negative; NSCS, non-specific chronic sialadenitis; Pos, positive; S.D., standard deviation; Wh/N.A., White/Native American; WUSF, whole unstimulated salivary flow.
Production of Recombinant mAbs
Single cell suspensions were stained for fluorescent activated cell sorting (FACS) as previously described (16). The mAbs were produced for subjects SLE/SS, pSS-1 and pSS-2 using IgG and kappa gene primers, as previously described (28). For the remaining subjects the protocol was modified as described herein (supplementary materials). Variable (V), diversity (D) and joining (J) domains, FWRs and CDRs along with the germline sequences were identified using the International Immunogenetics Information System database (IMGT/V-Quest).
To make the mutant mAbs clones used in this study, V-genes cloned into expression vectors were subjected to site directed mutagenesis using the Q5 site-directed mutagenesis kit (New England Biolabs, Ipswich, MA, USA) using non-overlapping primers designed using the NEBaseChanger online tool (v1.2.6). The mutated V-genes were sequenced, amplified, and expressed as above.
Ig Analyses for Antigen-Driven Selection
BASELINe (Bayesian estimation of Ag-driven selection) can be used to detect and quantify selection in individual or multiple sequences based on mutational patterns normalized to germline sequences and provides a visual representation of differences in selective pressure (29). Clonally related sequences were identified by manual examination of the genes called by IMGT/V-Quest and confirmed using the ClonalRelate program (30). To eliminate potential bias, we retained only the first randomly listed antibody in each of the clonal groups identified by ClonalRelate for the BASELINe analysis. The remaining productive heavy and light chain V-region sequences were then grouped (i.e. by subject classification (SS or DNMC), then by IgG allele (i.e. IGHV1-18*01, etc.)), and analyzed using BASELINe Version 1.3.
Ig Heavy and Light Chain N-Glycosylation Motif Analysis
Potential acN-glyc sites in the heavy and light chain V-region amino acid sequences were identified using the NetNGlyc web interface. N-glycosylation motifs were defined as previously described (31). To confirm that all identified acN-glyc sites were introduced by SHM, sequences were compared to their germline counterparts. The translated sequences of clonally related V-regions were analyzed using the online Clustal Omega multiple sequence alignment program (32). Clonal sequences were not removed because we found each could yield unique glycosylation patterns. We excluded any possible N-glyc sites with a potential of ≤ 0.5, or jury agreement of < 5 of 9 (33).
Glycoprotein and Specificity Analyses of mAbs
1.5 μg of mAbs and bovine IgG control (Equitech-Bio, Inc., Kerrville, TX, USA) were incubated at 37°C overnight with or without PNGase F (New England Biolabs), then electrophoresed in a 4-12% SDS-PAGE gel (GenScript, Piscataway, NJ, USA). For the Coomassie blue staining, the gel was fixed in a destaining solution of 50% methanol and 10% acetic acid for one hour at RT, incubated for one hour with Coomassie Brilliant Blue (BioRad, Hercules, CA, USA), then washed and destained with the destaining solution.
For lectin- and immune-blots, samples were loaded into a 4-12% SDS-PAGE gel (GenScript) and electrophoresed prior to transfer onto a cellulose membrane. For Concanavalin A (ConA) staining the membrane was blocked with a Tris buffered saline solution containing 0.05% Tween 20 (TBS-T) and 5% (w/v) BSA for 1h at RT, then incubated with ConA-biotin (0.2 μg/ml) (Vector Labs, Burlingame, CA, USA) in TBS-T with 1% BSA for 1 h at RT, followed by incubation with Streptavidin-HRP (0.2 μg/ml) (Vector) in TBS-T containing 1% BSA for 1h at RT. For hIgG staining, the membrane was incubated with peroxidase-labeled Goat anti-human IgG (Fc) antibody (KPL) at 0.2 μg/ml in TBS-T with 5% BSA for 1h at RT, then washed with TBS-T. Signals were detected using chemiluminescence horseradish peroxidase (HRP) substrate (SuperSignal® West Pico Chemiluminescent Substrate, Thermo Scientific, Waltham, MA, USA).
Nuclear Antigen Screening ELISAs were performed on all mAbs as previously described (16).
Immunoglobulin Protein Modeling
To visualize the location of the acN-glycs in the clonal set of mAbs (in Figure 3, A), we utilized the crystal structure of human IgG1 molecule (PDB Accession number 3EYQ, 2.4 Å resolution) as a template for the HHPRED modeling server (34). The individual somatic mutations from mAb clone 2-E04k were inserted where appropriate and a representative Fab framework 3 complex-type biantennary N-glycan identified in another published study (35) was modeled at the acN-glyc site using MAIN software (36). The model graphics were created using PyMOL (The PyMOL Molecular Graphics System, Version 1.8.2, Schrödinger, LLC).
Figure 3.
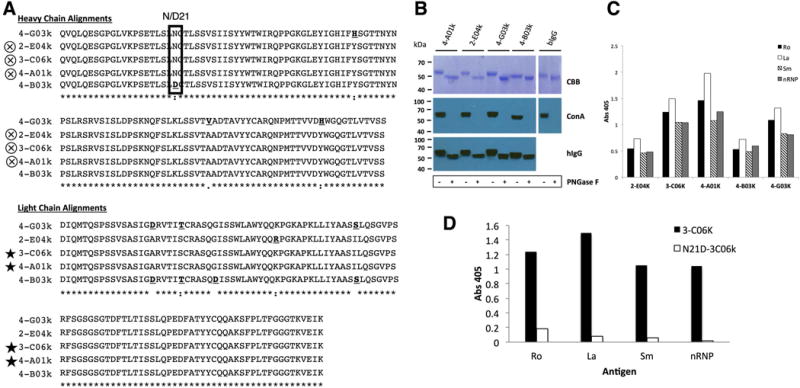
Clonal mAb N-glycosylation analysis and antigen-binding by ELISA. A) Alignment of heavy and light chain amino acids from clonally related mAbs from pSS-1 revealing differences that may affect antigen binding. Bold underlined amino acids are different from consensus residues. Box indicates site of acquired N-glycosylation motif (N-C-T, position 21) in 4 of the 5 clones. Symbols: Cross in circle = identical translated heavy chain V-region sequences; Star = identical translated light chain V-region sequences, “*” (asterisk) indicates fully conserved residues; “:” (colon) indicates conservation between groups with strongly similar properties; “.” (period) indicates conservation between groups with weakly similar properties. B) Mobility shift analysis of mAb heavy chains. After PNGase F digest, followed by Coomassie Blue staining a more modest shift is seen in 4-B03k and Bovine IgG (known to have 1 Fc region N-glyc) as compared to 4-A01k, 2-E04k and 4-G03k. Concanavalin A staining indicates the complete removal of glycans by PNGase F. C) Clonal mAbs binding of Ro, La, Sm, and nRNP by ELISA. D) Antigen ELISA shows loss of antigen binding by mutant N21D-306k (acN-glyc -) as compared to its glycosylated counterpart, 3-C06K (acN-glyc+).
Statistical Analyses
Fisher’s exact tests were performed using GraphPad QuickCalcs website. Prism v7.0b software was used for the F test to compare variances as well as all parametric t-tests and non-parametric Mann-Whitney tests, which were each preceded by D’Agostino & Pearson omnibus normality tests.
Results
Participant demographics and clinical characteristics
Evaluation of the H&E-stained sections from the salivary gland biopsies revealed that all SS patients and DNMC controls had varying levels of chronic inflammation with lymphocytic infiltration (Table 1). In addition, four of the SS patients (SS/SLE, pSS-1, pSS-2, and pSS-5) had probable ectopic germinal center structures, including various associating cells such as plasma cells, follicular dendritic cells and antigen-processing/presenting tingible body macrophages. We found no significant difference between the mean ages (44 ± 10.48 vs. 50 ± 11.97 years, respectively; p=0.35), or variances of ages (p=0.72) of the SS patient and DNMC control groups. We found that 5 of the 8 SS patients had an increased probability (39.9%) for future development of NHL based on the independent risk variables defined by a previously validated clinical tool (27) (Table S1).
Immunoglobulin heavy and light chain sequence analyses revealed clonal expansions, secondary light chain editing and allelic inclusion
We cloned and sequenced a total of 122 productive IgG heavy and kappa or lambda light chain pairs from ASCs from both groups. Of these, 100 pairs were from the SS patients (SS/SLE, n=4; pSS-1, n=18; pSS-2, n=23; pSS-3, n=11, pSS-4, n=36; pSS-5, n=1; pSS-6, n=5; pSS-7, n=2), and 22 pairs from the DNMC controls (DNMC-1, n=8; DNMC-2, n=6; DNMC-3, n=2; DNMC-4, n=1; DNMC-5, n=4; DNMC-6, n=1). We identified several clonally related mAbs (Table 2). Of the SS patients, pSS-1 had a single 5-member expansion; pSS-2 had a total of 5, 2-member expansions; and pSS-4 had a total of 5 expansions. There was one clonal pair identified in the DNMC-2 sequences.
Table 2.
Clonal expansions, light chain editing and allelic inclusion in 3 SS patients and one DNMC control.
| Subject | Clone ID | Status | VH-Gene | DH-Gene | JH-Gene | Light Chain V-Gene | Light Chain J-Gene |
|---|---|---|---|---|---|---|---|
| pSS-1 | 2-E04k | Clone 1§ | 4-59 | 4-23 | 4 | Vκ1-12 | Jκ4 |
| pSS-1 | 3-C06k | Clone 1§ | 4-59 | 4-23 | 4 | Vκ1-12 | Jκ4 |
| pSS-1 | 4-A01k | Clone 1§ | 4-59 | 4-23 | 4 | Vκ1-12 | Jκ4 |
| pSS-1 | 4-B03k | Clone 1 | 4-59 | 4-23 | 4 | Vκ1-12 | Jκ4 |
| pSS-1 | 4-G03k | Clone 1§ | 4-59 | 4-23 | 4 | Vκ1-12 | Jκ4 |
|
| |||||||
| pSS-2 | 1-B03K | Clone 1 | 2-5 | 3-22 | 4 | Vκ1-39 | Jκ2 |
| pSS-2 | 3-E04K | Clone 1 | 2-5 | 3-22 | 4 | Vκ1-39 | Jκ2 |
| pSS-2 | 1-C01K | Clone 2 | 1-69 | 2-2 | 5 | Vκ1-39 | Jκ2 |
| pSS-2 | 1-D03K | Clone 2 | 1-69 | 2-2 | 5 | Vκ1-39 | Jκ2 |
| pSS-2 | 1-F01K | Clone 3 | 3-7 | 2-21 | 4 | Vκ3-11 | Jκ5 |
| pSS-2 | 3-E01K | Clone 3 | 3-7 | 2-21 | 4 | Vκ3-11 | Jκ5 |
| pSS-2 | 1-G02K | Clone 4 - LC replacement | 3-33 | 3-9 | 4 | Vκ3-20 | Jκ4 |
| pSS-2 | 1-C06K | Clone 4 - LC replacement | 3-33 | 3-9 | 4 | Vκ3-15 | Jκ2 |
| pSS-2 | 2-D06K | Clone 5 - LC replacement | 3-33 | 3-9 | 1 | Vκ3-15 | Jκ3 |
| pSS-2 | 3-D06K | Clone 5 - LC replacement | 3-33 | 3-9 | 1 | Vκ4-1 | Jκ1 |
|
| |||||||
| pSS-4 | 1-G01K | Clone 1 | 5-10-1 | 3-3 | 5 | Vκ3-20 | Jκ5 |
| pss-4 | 6-D06K | Clone 1 | 5-10-1 | 3-3 | 5 | Vκ3-20 | Jκ5 |
| pss-4 | 1-C02L | Clone 2 - LC replacement | 4-34 | 3-22 | 4 | Vλ3-25 | Jλ2 |
| pss-4 | 2-A06K | Clone 2 - LC replacement | 4-34 | 3-22 | 4 | Vκ3-20 | Jκ1 |
| pss-4 | 1-F03K | Clone 3 - LC replacement | 3-23 | 2-21 | 4 | Vκ1-5 | Jκ1 |
| pss-4 | 4-C03K | Clone 3 - LC replacement | 3-23 | 2-21 | 4 | Vκ3-15 | Jκ2 |
| pss-4 | 6-C06K | Clone 3 - LC replacement | 3-23 | 2-21 | 4 | Vκ4-1 | Jκ5 |
| pss-4 | 6-E01K | Allelic inclusion | 3-21 | 6-19 | 6 | Vκ1-39 | Jκ1 |
| pss-4 | 6-E01L | Allelic inclusion | 3-21 | 6-19 | 6 | Vλ2-11 | Jλ2 |
| pss-4 | 6-E04K | Allelic inclusion | 3-53 | 6-13 | 5 | Vκ1-5 | Jκ1 |
| pss-4 | 6-E04L | Allelic inclusion | 3-53 | 6-13 | 5 | Vλ2-14 | Jλ2 |
|
| |||||||
| DNMC-2 | 1-F06K | Clone 1 - LC replacement | 3-15 | 6-19 | 4 | Vκ1-9 | Jκ2 |
| DNMC-2 | 1-G04K | Clone 1 - LC replacement | 3-15 | 6-19 | 4 | Vκ1-12 | Jκ1 |
clone has heavy chain N-glycosylation acquired by somatic mutation (acN-glyc) described in Figure 5.
We found evidence of secondary light chain editing in the sequences from subjects, pSS-2, pSS-4 and DNMC-2, indicated by clonally related heavy chains with differential light chain usage (clonal w/LC replacement; Table 2). In addition, allelic inclusion, or one heavy chain with two distinct light chains within the same cell, was identified in two instances from the ASCs from pSS-4 (6-E01, 6-E04; Table 2). In each case, both a kappa and a lambda light chain sequence were amplified from the same cell.
Whether analyzed with or without clonal sequences, the length of the heavy chain CDR3 was greater in the SS patients than in the DNMC sequences (with clones: 17 vs. 15.5 amino acids, p=0.047, and without clones: 17 vs. 15 amino acids, p=0.048). There were no differences between patients and controls in the lengths of the light chain CDR3s (11.00 vs. 11.00, with or without clones) or in the number of amino acid replacements in the heavy or light chains.
Differential acquisition of V-region N-glycoslyation motifs in SS and DNMC
All of the N-glyc motifs identified in the heavy and light chain V-regions of patients and controls were not germline encoded and therefore, had been acquired via SHM (acN-glyc). We identified acN-glycs in sequences from 6 of 8 SS patients (pSS/SLE, pSS-1, pSS-2, pSS-3, pSS-5, and pSS-6) and 2 of 6 DNMC controls (DNMC-3 and DNMC-5). The acquisition of Fab N-glycosylation by SHM is a process that occurs in germinal centers. Therefore, we looked for correlations between incidence of acN-glycs and the presence of germinal centers in the glands. We found that in each of the 4 subjects identified with germinal center-like structures all had V-region acN-glycs, but not all subjects with V-region acN-glycs had germinal center-like structures in the glands examined. The incidence of acN-glyc in a heavy/light chain pair was more than doubled in SS sequences (29%, or 28/97) as compared to controls (9%, or 2/22); however, these differences only trended towards statistical significance (p=0.061).
We found a trend for increased acN-glycs in the SS FWRs compared to those from DNMC controls (22%, or 21/97 and 5%, or 1/22, respectively, p = 0.072). In the SS patients, half (n=11) of the FWR acN-glycs were located in the FWR1 of the heavy and light chains. There were no FWR1 acN-glycs in the DNMC sequences. We found no instances of acN-glycs in the CDR3 regions of heavy or light chains and no differences in CDR1/CDR2 acN-glycs between SS patients or DNMCs (11/97 and 3/22, respectively; p = 0.722).
acN-glycs are associated with increased SHM and amino acid replacements
We compared the heavy and light chain sequences that had acN-glycs (n = 31) to those without acN-glycs, (n = 91) without regard to disease classification (Figure 1, A-C). Heavy chains with acN-glycs had significantly more SHMs than those without acN-glycs (22.45 ± 1.047 vs 17.27 ± 0.8383; p = 0.001). The SHMs in heavy chains with acN-glycs also resulted in replacement mutations more often than in those without acN-glycs (16.65 ± 0.86 vs 12.88 ± 0.62, p = 0.002). For light chains there were trends for more SHMs and replacement mutations in those with acN-glycs than those without (SHM; 13 vs 10, p = 0.096 and replacement mutations; 9 vs 7, p = 0.146). Both heavy and light chains with acN-glycs had significantly more amino acid replacements than did those without acN-glycs (heavy chains, 13.26 ± 0.599, vs 10.46 ± 0.462, p = 0.002; light chains 8.910 ± 0.843 vs 6.980 ± 0.416, p = 0.028).
Figure 1.
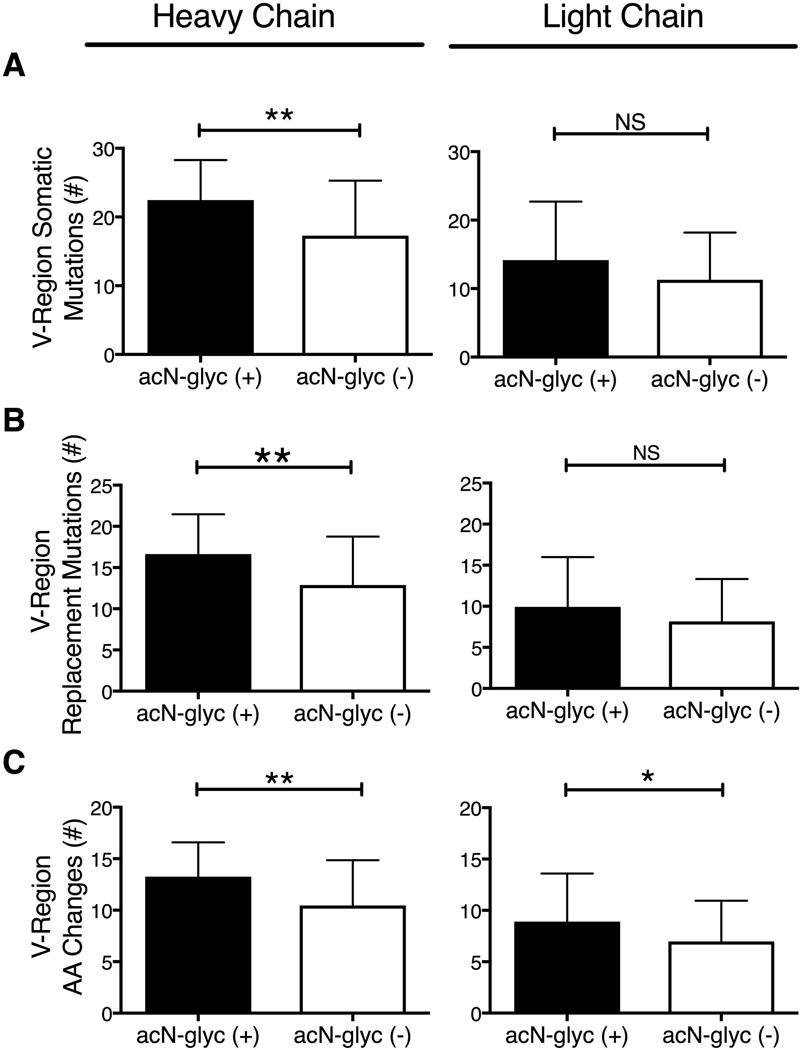
Somatic hypermutations, replacement mutations and amino acid changes in sequences with and without acN-glycs. A – C) Sequence analyses of V-regions from SS patient and DNMC control minor salivary gland ASCs with (black columns, n = 31) and without (white columns, n = 91) acN-glycs. A) Number of heavy and light chain V-region somatic hypermutations. B) Number of heavy and light chain replacement mutations. C) Number of heavy and light chain V-region amino acid changes.
Selection pressure analyses in SS and DNMC V-regions
We performed a BASELINe Focused binomial test (29, 37, 38) on all of the unique sequences from SS and DNMCs and found overall positive selection in the CDRs and negative selection in the FWRs of the heavy chains from each group (CDRs: SS p=0.009, DNMC p=0.05 and FWRs: SS p=2.99×10−7, DNMC p=0.02). We found no significant selection pressure in the light chain CDRs of either SS or DNMC sequences (p=0.19 and p=0.39, respectively). However, both SS and DNMC light chain sequences had negative selection in the FWRs (p=2.85×10−14 and 4.46×10−8, respectively). When we compared all of SS sequences with all of the DNMC sequences, we found no significant differences in selection pressures in either the heavy or light chain CDRs or FWRs between the two groups (Figure 2-, A, heavy chains and B, light chains).
Figure 2.
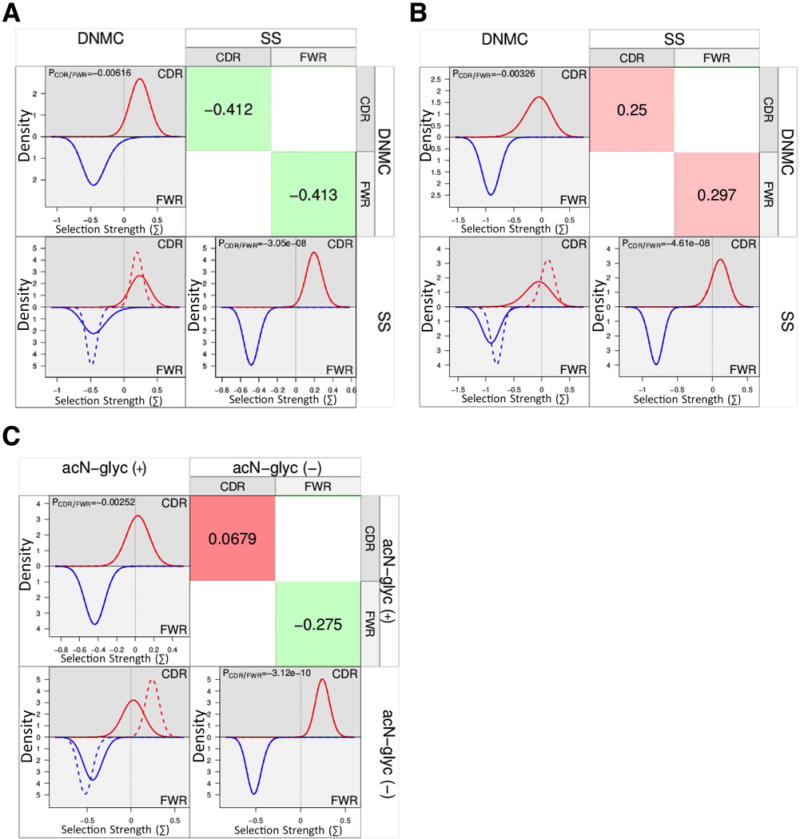
BASELINe comparison of selection pressure differences between groups. The histograms provide a visualization of selection pressures for the CDRs (top half of plots, red lines) and the FWRs (bottom half of plots, blue lines) and an overlay (lower left quadrant) showing the differences between the compared groups, where the solid lines represent the column variable (ie. DNMC in panel A) and the dashed lines represent the row variable (ie. SS in panel A). Statistically derived relative differences (binomial statistical test used by BASELINe) are shown (upper right quadrant) for each comparison, where red color and positive numbers are representative of positive selection, and green color and negative numbers are representative of negative selection. A) Comparison of selection pressures in heavy chains between SS and DNMC controls. B) Comparison of selection pressures in light chains between SS and DNMC controls. C) Comparison of selection pressures in heavy chains with acN-glycs vs those without acN-glycs.
Fab sequences with acN-glyc motifs have differential selection pressure patterns
Since we had found an increased frequency of acN-glycs in SS sequences and bacterial lectins have been reported to bind acN-glycs and facilitate non-specific B cell activation (10), we analyzed the effects of acN-glycs on selection pressure. We compared heavy chain sequences with acN-glycs to those without acN-glycs, without regard to disease classification. A BASELINe focused binomial test indicated differential CDR selection between the two groups - no selection in the CDRs from heavy chains with acN-glycs (p=0.41) vs. strong selection in the CDRs from heavy chains without acN-glycs (p=0.0007). All sequences, with or without acN-glycs had negative selection in the FWRs (p=3.91×10−5 and 5.6×10−8, respectively). When we compared the selection strengths of heavy chains with acN-glycs to those without acN-glycs, we found that heavy chains with acN-glycs have trends for less positive selection in their CDRs (p=0.0679) relative to the sequences without acN-glycs (Figure 2, C).
Clonally related mAbs with differential FW1 acN-glyc motifs
Our acN-glyc analysis also revealed a clonal set of differentially glycosylated Ig sequences (Table 2; pSS-1, clone 1). A multiple alignment of the heavy and light chain (Figure 3, A) amino acid sequences revealed that three of the clones’ heavy chain translated sequences were identical (2-E04k, 3-C06k, and 4-A01k). Clone 2-C06k and 4-A01k also shared an identical light chain sequence. Clone 4-B03k was the only one of the 5 clonal Igs that did not have the acN-glyc at position 21 of the heavy chain (D21) in FWR1. To confirm that the FWR1 (N21) N-glyc motifs in this clonal group could indeed be glycosylated at the protein level, the heavy and light chain V-regions of the 5 pairs were cloned into expression vectors and expressed as mAbs. Glycosylation studies were performed on mAbs 4-A01k, 2-E04k, 4-G03k and 4-B03k. Clone C06k was not analyzed, as it was identical in amino acid sequence to clone 4-A01k (Figure 3, A). After enzymatic removal of N-glycans with PNGase F and separation on SDS-PAGE gel, staining (CBB) showed there was a motility shift in the heavy chain of the Ig molecules indicating the loss of N-glycans (Figure 3, B). The shift for clone 4-B03k, lacking the FWR1 N-glycan motif, was moderate compared to the 3 others and comparable to that observed for bovine IgG (bIgG) known to only carry 1 N-glycan per heavy chain. The ConA staining, which binds mannose-containing N-glycans, confirmed the presence of N-glycans in the untreated sample and that they were quantitatively removed after treatment with PNGase F.
FWR1 N-glycoslyation affects mAb antigen binding
To determine if these clones were autoreactive, the mAbs were tested for binding Ro, La, Sm and nRNP by ELISA (Figure 3, C). All 5 clones had some degree of binding to all antigens with 3-C06k, 4-A01k, and 4-G03k having the highest levels of reactivity. We next determined the effects of N-glycosylation in the FWR1 on antigen binding. We removed the acN-glyc motif from the FWR1 of 3-C06k by replacing the asparagine (N) at position 21 with an aspartic acid (D) (N21D-3C06k). Conversely, we introduced an N-glyc motif to 4-B03k at the same position, converting the aspartic acid to an asparagine (D21N-4B03k). We expressed these mutant mAbs and retested them for binding to Ro, La, Sm, and nRNP. We found that N21D-3C06k had almost completely lost all binding to all 4 antigens after removal of the FWR1 N-glyc (Figure 3, D). However, D21N-4B03k did not gain binding any of the antigens after the addition of an N-glyc in FWR1, but instead lost binding to La, Sm, and nRNP.
Protein modeling reveals a potential mechanism for FW1 acN-glyc participation in antigen binding by expansion of antigen binding site
We used protein modeling of clone 2-E04k to visualize the location of the FWR1 N-glyc in relation to the CDR antigen-binding pocket (Figure 4, A - B). The heavy chain FWR1 acN-glyc oriented on the molecule exterior very near the CDR binding site. We replaced the light chain in the model with the other light chains in the clonal group and found no appreciable difference in the orientation of the acN-glyc (data not shown). To visualize how an N-glyc might affect or participate in antigen binding, we added and remodeled an FWR3 complex, biantennary N-glycan structure (4FQC) identified in another study (35) to our model (Figure 4, C) and found that the binding surface could effectively be expanded by the close proximity of the branched carbohydrate structure (Figure 4, D).
Figure 4.
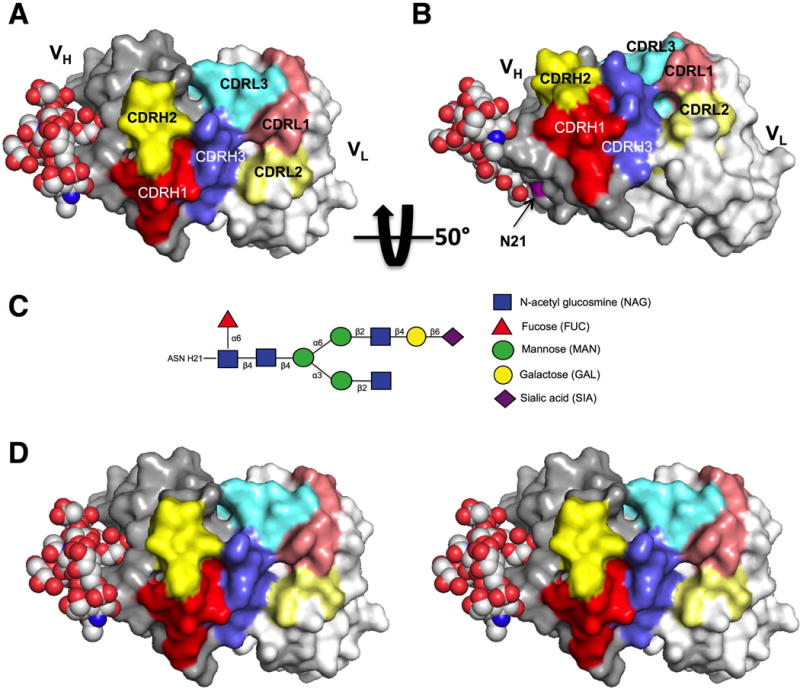
Protein model of 2-E04k mAb variable regions with complex-type FWR1 N-glycan. A) Mono image with heavy chain variable region (VH) oriented on the left and light chain variable region (VL) on the right. Heavy chain CDR region residues are indicated by red (CDR1), yellow (CDR2), and blue (CDR3). Light chain CDR region residues are indicated by muted colors, pink (CDR1), light yellow (CDR2) and light blue (CDR3). B) The same model rotated upward approximately 50° about the horizontal axis with the position of the heavy chain acN-glyc motif (N21) shown in magenta. C) Schematic of the representative complex-type N-glycan utilized in the model. D) Stereo image of model in panel A.
Discussion
The DNMC controls used in this study were not healthy subjects, but unlike healthy controls, they had sufficient ASC’s for isolation by single cell sorting and therefore, served as excellent sicca controls. Since the controls each had some degree of glandular inflammation (see Table 1), the differences we reported between SS and DNMCs in this study cannot be attributed to an inflammatory milieu or the lack thereof. It will be interesting to follow the DNMC controls longitudinally to determine if some progress to SS with time, or if they represent a distinct disease or subset not yet defined.
The ASCs from the first 3 subjects enrolled into this study were interrogated using a previously published RT-PCR protocol (28) that limited the probing of each single-sorted cell with multiplexed primers for only one heavy and one light chain isotype. The modification of our protocol for the remaining subjects to a two-step RT-PCR method enabled probing for multiple heavy and light chain isotypes. The utility of this method could also extend to any other expressed gene of interest in single-sorted cells from any B cell population. Our efforts revealed what appear to be rare cases of allelic inclusion in pSS-4, which would not have been identified with the previous method. We are uncertain if both light chains are translated and expressed with heavy chains in these cells, or if they remain as remnant mRNA from earlier selection events. B cells that express dual light chains on their surface have been described in healthy humans (39) and in various B cell lymphomas (40–42). Lymphoma is increased in SS, and it is possible that the dual-light chain ASC’s we have identified are precursors to neoplastic lymphoblasts. Interestingly, pSS-4 was one of the patients that we also identified as having an increased probability of lymphoma development (27)
Secondary light chain replacement, or receptor revision as it is called in the periphery, has been demonstrated in both mice and man (43, 44). Receptor revision has also been reported in patients with SLE (45) and in the cerebral spinal fluid infiltrating B cells from multiple sclerosis patients (46, 47), but to our knowledge has not been previously reported in SS. These findings are not likely a result of PCR contamination, as each light chain replacement was a unique sequence, not found paired with any other heavy chain in the study. Whether the light chain replacements we have identified represent a novel peripheral tolerance mechanism is unknown.
Our finding of somatically mutated, clonally related ASCs in three SS patients and one DNMC control, is evidence for antigen-driven proliferation within the labial salivary gland and agrees with our previous observations (16). The BASELINe analyses also support this. A recent study by Hamza, et al, found no evidence for positive selection in SS parotid B cell heavy chains using the BASELINe method (17), but this may be easily explained by differences in study designs. Theirs involved bulk RNA from parotid tissue B cells, instead of single-cell sorted labial salivary gland ASCs as in our study. They also probed only the IgG VH3-family genes, while we included primers for all VH-family genes. Alternatively, there may be micro-environmental differences between the parotid and labial salivary gland tissues, or differences in B cell subsets, since memory B cells accumulate in Sjögren’s parotid tissue (48).
The increased incidence of acN-glycs, particularly, those occurring in the FWRs, was interesting. Hamza’s group also reported increased FWR acN-glycs in SS patients. The increased SHM rates in Igs with acN-glycs suggest that acN-glycs may drive SHM by facilitating increased activation signals via bacterial lectins, independent of antigen. This is supported by our finding that heavy chains with acN-glycs do not indicate selection of the CDR antigen binding regions, suggesting an antigen-independent mechanism for their genesis. Alternatively, the likelihood of acquiring novel N-glycosylation motifs may increase as the rate of SHM increases. It was interesting that we found evidence of salivary gland germinal center structures in only 4 of the subjects with acN-glycs. This may be explained by the fact that these analyses were not performed in the same glands. Since the glands processed for single cell sorting and monoclonal antibody production/sequencing must be used in their entirety for the production of mAbs, the germinal center analyses were performed on other glands biopsied at the same clinic visit.
Sequences with acN-glycs have at least one replacement mutation leading to the acquisition of the N-glyc, so it is possible that we have introduced bias to this analysis by parsing them into acN-glyc(+) vs acN-glyc(-) groups. However, BASELINe predicts selection pressures by identifying replacement mutations that occur at higher rates than would be expected in a scenario of “no selection,” so we would expect any bias of this nature to have resulted in increased positive selection, but we observed the opposite, which supports our data and reinforces the idea of a non-specific activation of these cells. Since bacterial lectins have been demonstrated to bind IgG acN-glycs and trigger non-specific activation of the B cell (10), this is a possible mechanism for a break in tolerance leading to immunoglobulin autoreactivity, as well as the increased risk for lymphoma seen in SS patients.
After removing the FWR1 acN-glyc from clone, 3-C06k, binding to 4 common autoantigens was almost completely abolished indicating that the acquisition of an N-glyc via SHM can actually increase binding to the self-antigens that are commonly bound by mAbs produced from the labial salivary glands of SS patients (16). Similar findings have been reported for a polyreactive mAb after removal of a FWR3 N-glyc (49). The heavy chain is only partially responsible for antigen affinity and between the 5 clones there were 4 light chains with unique mutations, which could explain the lowered binding by both D21N-4B03k and 2-E04k. Oligosacharides are inherently flexible and individual sugar linkages can exist in various conformations (50). The N-glyc conformation in our model is one possible, but hypothetical conformation. In reality, the glycan at N21 probably freely explores the solvent space within its cone of rotation, and for it to add surface area to the CDR3, must be trapped in this conformation by stabilizing influences of the incoming antigen, or by antigen binding in those moments when the glycan stochastically occupies that space. Therefore our model provides visual evidence of how glycan structures originating outside of the CDRs could affect antigen binding, depending on their placement, structure and rotational conformations by extending the surface of, or interfering with the antigen-binding plane. This supports the possibility for antigen-independent, or co-dependent stimulation via lectin-glycan interactions, leading to B cell activation.
In summary, we have developed an improved method for the isolation, characterization and mAb production for rare B cells populations, which has provided data suggesting the possibility of peripheral tolerance mechanisms in SS labial ASCs not previously reported. Overall, we found evidence for antigen-driven positive selection in these cells. However, our finding of reduced positive selection in ASCs harboring Ig acN-glycs suggests potential alternative mechanisms for B cell selection, hyperactivation and the proliferation of some of the autoreactive cells frequently seen in SS.
Supplementary Material
Acknowledgments
We thank J. Andrew Duty for discussions in our RT-PCR protocol development, Christina Lawrence for the preparation of the salivary gland single-cell suspensions, Diana Hamilton and Jacob Bass of the OMRF Flow Cytometry Core for their assistance in cell sorting, and Steven H. Kleinstein for his consultations concerning our BASELINe analyses.
Financial Support
This work was supported by the NIH’s NIAMS (P30-AR053483 and P50-AR060804), the NIH/NIAID, Autoimmunity Centers of Excellence Grant (U19- AI082714), the Oklahoma Shared Clinical and Translational Resources Grant (U54-GM104938), the NIH/NIGMS Centers of Biomedical Research Excellence Grant (P30-GM103510) and the National Center for Functional Glycomics (FCFG) at Beth Israel Deaconess Medical Center, Harvard Medical School (P41-GM103694)
Footnotes
DR. ROBERT HAL SCOFIELD (Orcid ID : 0000-0003-1015-5850)
Websites Utilized in this Study.
NEBaseChanger online tool: nebasechanger.neb.com
GraphPad QuickCalcs: www.graphpad.com/quickcalcs/contingency1
ClonalRelate program: www.cse.unsw.edu.au/~ihmmune/ClonalRelate
iHMMune web interface: www.emi.unsw.edu.au/~ihmmune/
BASELINe Version 1.3: http://clip.med.yale.edu/selection
NetNGlyc: www.cbs.dtu.dk/services/NetNGlyc/
Clustal Omega multiple sequence alignment program for protein sequences: http://www.ebi.ac.uk/Tools/msa/clustalo/
References
- 1.Shiboski SC, Shiboski CH, Criswell L, Baer A, Challacombe S, Lanfranchi H, et al. American College of Rheumatology classification criteria for Sjogren’s syndrome: a data-driven, expert consensus approach in the Sjogren’s International Collaborative Clinical Alliance cohort. Arthritis Care Res (Hoboken) 2012;64(4):475–87. doi: 10.1002/acr.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjogren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61(6):554–8. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ramos-Casals M, Brito-Zeron P, Font J. The overlap of Sjogren’s syndrome with other systemic autoimmune diseases. Semin Arthritis Rheum. 2007;36(4):246–55. doi: 10.1016/j.semarthrit.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 4.Garcia-Carrasco M, Ramos-Casals M, Rosas J, Pallares L, Calvo-Alen J, Cervera R, et al. Primary Sjogren syndrome: clinical and immunologic disease patterns in a cohort of 400 patients. Medicine (Baltimore) 2002;81(4):270–80. doi: 10.1097/00005792-200207000-00003. [DOI] [PubMed] [Google Scholar]
- 5.Davidson BK, Kelly CA, Griffiths ID. Primary Sjogren’s syndrome in the North East of England: a long-term follow-up study. Rheumatology (Oxford) 1999;38(3):245–53. doi: 10.1093/rheumatology/38.3.245. [DOI] [PubMed] [Google Scholar]
- 6.Voulgarelis M, Dafni UG, Isenberg DA, Moutsopoulos HM. Malignant lymphoma in primary Sjogren’s syndrome: a multicenter, retrospective, clinical study by the European Concerted Action on Sjogren’s Syndrome. Arthritis Rheum. 1999;42(8):1765–72. doi: 10.1002/1529-0131(199908)42:8<1765::AID-ANR28>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 7.Wotherspoon AC. Gastric MALT lymphoma and Helicobacter pylori. Yale J Biol Med. 1996;69(1):61–8. [PMC free article] [PubMed] [Google Scholar]
- 8.Schollkopf C, Melbye M, Munksgaard L, Smedby KE, Rostgaard K, Glimelius B, et al. Borrelia infection and risk of non-Hodgkin lymphoma. Blood. 2008;111(12):5524–9. doi: 10.1182/blood-2007-08-109611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponzoni M, Ferreri AJ, Guidoboni M, Lettini AA, Cangi MG, Pasini E, et al. Chlamydia infection and lymphomas: association beyond ocular adnexal lymphomas highlighted by multiple detection methods. Clin Cancer Res. 2008;14(18):5794–800. doi: 10.1158/1078-0432.CCR-08-0676. [DOI] [PubMed] [Google Scholar]
- 10.Schneider D, Duhren-von Minden M, Alkhatib A, Setz C, van Bergen CA, Benkisser-Petersen M, et al. Lectins from opportunistic bacteria interact with acquired variable-region glycans of surface immunoglobulin in follicular lymphoma. Blood. 2015;125(21):3287–96. doi: 10.1182/blood-2014-11-609404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwai H, Nakamichi N, Nakae K, Konishi M, Inaba M, Hoshino S, et al. Parotid mucosa-associated lymphoid tissue lymphoma regression after Helicobacter pylori eradication. Laryngoscope. 2009;119(8):1491–4. doi: 10.1002/lary.20258. [DOI] [PubMed] [Google Scholar]
- 12.Le Pottier L, Devauchelle V, Fautrel A, Daridon C, Saraux A, Youinou P, et al. Ectopic germinal centers are rare in Sjogren’s syndrome salivary glands and do not exclude autoreactive B cells. J Immunol. 2009;182(6):3540–7. doi: 10.4049/jimmunol.0803588. [DOI] [PubMed] [Google Scholar]
- 13.Salomonsson S, Jonsson MV, Skarstein K, Brokstad KA, Hjelmstrom P, Wahren-Herlenius M, et al. Cellular basis of ectopic germinal center formation and autoantibody production in the target organ of patients with Sjogren’s syndrome. Arthritis Rheum. 2003;48(11):3187–201. doi: 10.1002/art.11311. [DOI] [PubMed] [Google Scholar]
- 14.Ben-Chetrit E, Fischel R, Rubinow A. Anti-SSA/Ro and anti-SSB/La antibodies in serum and saliva of patients with Sjogren’s syndrome. Clin Rheumatol. 1993;12(4):471–4. doi: 10.1007/BF02231773. [DOI] [PubMed] [Google Scholar]
- 15.Jonsson MV, Skarstein K, Jonsson R, Brun JG. Serological implications of germinal center-like structures in primary Sjogren’s syndrome. J Rheumatol. 2007;34(10):2044–9. [PubMed] [Google Scholar]
- 16.Maier-Moore JS, Koelsch KA, Smith K, Lessard CJ, Radfar L, Lewis D, et al. Antibody-secreting cell specificity in labial salivary glands reflects the clinical presentation and serology in patients with Sjogren’s syndrome. Arthritis Rheumatol. 2014;66(12):3445–56. doi: 10.1002/art.38872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamza N, Hershberg U, Kallenberg CG, Vissink A, Spijkervet FK, Bootsma H, et al. Ig gene analysis reveals altered selective pressures on Ig-producing cells in parotid glands of primary Sjogren’s syndrome patients. J Immunol. 2015;194(2):514–21. doi: 10.4049/jimmunol.1302644. [DOI] [PubMed] [Google Scholar]
- 18.Zhu D, McCarthy H, Ottensmeier CH, Johnson P, Hamblin TJ, Stevenson FK. Acquisition of potential N-glycosylation sites in the immunoglobulin variable region by somatic mutation is a distinctive feature of follicular lymphoma. Blood. 2002;99(7):2562–8. doi: 10.1182/blood.v99.7.2562. [DOI] [PubMed] [Google Scholar]
- 19.Radcliffe CM, Arnold JN, Suter DM, Wormald MR, Harvey DJ, Royle L, et al. Human follicular lymphoma cells contain oligomannose glycans in the antigen-binding site of the B-cell receptor. Journal of Biological Chemistry. 2007;282(10):7405–15. doi: 10.1074/jbc.M602690200. [DOI] [PubMed] [Google Scholar]
- 20.Ekstrom Smedby K, Vajdic CM, Falster M, Engels EA, Martinez-Maza O, Turner J, et al. Autoimmune disorders and risk of non-Hodgkin lymphoma subtypes: a pooled analysis within the InterLymph Consortium. Blood. 2008;111(8):4029–38. doi: 10.1182/blood-2007-10-119974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallick SC, Kabat EA, Morrison SL. Glycosylation of a VH residue of a monoclonal antibody against alpha (1----6) dextran increases its affinity for antigen. J Exp Med. 1988;168(3):1099–109. doi: 10.1084/jem.168.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabouri Z, Schofield P, Horikawa K, Spierings E, Kipling D, Randall KL, et al. Redemption of autoantibodies on anergic B cells by variable-region glycosylation and mutation away from self-reactivity. Proc Natl Acad Sci U S A. 2014;111(25):E2567–75. doi: 10.1073/pnas.1406974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coelho V, Krysov S, Ghaemmaghami AM, Emara M, Potter KN, Johnson P, et al. Glycosylation of surface Ig creates a functional bridge between human follicular lymphoma and microenvironmental lectins. Proc Natl Acad Sci U S A. 2010;107(43):18587–92. doi: 10.1073/pnas.1009388107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen A, Ice JA, Li H, Grundahl K, Kelly JA, Radfar L, et al. Comparison of the American-European Consensus Group Sjogren’s syndrome classification criteria to newly proposed American College of Rheumatology criteria in a large, carefully characterised sicca cohort. Ann Rheum Dis. 2014;73(1):31–8. doi: 10.1136/annrheumdis-2013-203845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 26.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 27.Fragkioudaki S, Mavragani CP, Moutsopoulos HM. Predicting the risk for lymphoma development in Sjogren syndrome: An easy tool for clinical use. Medicine (Baltimore) 2016;95(25):e3766. doi: 10.1097/MD.0000000000003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith K, Garman L, Wrammert J, Zheng NY, Capra JD, Ahmed R, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nature protocols. 2009;4(3):372–84. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uduman M, Yaari G, Hershberg U, Stern JA, Shlomchik MJ, Kleinstein SH. Detecting selection in immunoglobulin sequences. Nucleic Acids Res. 2011;39:W499–504. doi: 10.1093/nar/gkr413. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen Z, Collins AM, Wang Y, Gaeta BA. Clustering-based identification of clonally-related immunoglobulin gene sequence sets. Immunome research. 2010;6(Suppl 1):S4. doi: 10.1186/1745-7580-6-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kasturi L, Chen H, Shakin-Eshleman SH. Regulation of N-linked core glycosylation: use of a site-directed mutagenesis approach to identify Asn-Xaa-Ser/Thr sequons that are poor oligosaccharide acceptors. Biochem J. 1997;323(Pt 2):415–9. doi: 10.1042/bj3230415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balonova L, Hernychova L, Mann BF, Link M, Bilkova Z, Novotny MV, et al. Multimethodological approach to identification of glycoproteins from the proteome of Francisella tularensis, an intracellular microorganism. Journal of proteome research. 2010;9(4):1995–2005. doi: 10.1021/pr9011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–8. doi: 10.1093/nar/gki408. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A. 2012;109(47):E3268–77. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turk D. MAIN software for density averaging, model building, structure refinement and validation. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 8):1342–57. doi: 10.1107/S0907444913008408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaari G, Vander Heiden JA, Uduman M, Gadala-Maria D, Gupta N, Stern JN, et al. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Frontiers in immunology. 2013;4:358. doi: 10.3389/fimmu.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaari G, Uduman M, Kleinstein SH. Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 2012;40(17):e134. doi: 10.1093/nar/gks457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Giachino C, Padovan E, Lanzavecchia A. kappa+lambda+ dual receptor B cells are present in the human peripheral repertoire. J Exp Med. 1995;181(3):1245–50. doi: 10.1084/jem.181.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.del Senno L, Gandini D, Gambari R, Lanza F, Tomasi P, Castoldi G. Monoclonal origin of B cells producing k, lambda and k lambda immunoglobulin light chains in a patient with chronic lymphocytic leukemia. Leukemia research. 1987;11(12):1093–8. doi: 10.1016/0145-2126(87)90162-7. [DOI] [PubMed] [Google Scholar]
- 41.Matsuo Y, Nakamura S, Ariyasu T, Terao R, Imajyo K, Tsubota T, et al. Four subclones with distinct immunoglobulin light chain phenotypes. (kappa+lambda+, kappa+, lambda+ and kappa-lambda-) from acute leukemia. Leukemia. 1996;10(4):700–6. [PubMed] [Google Scholar]
- 42.Peltomaki P, Bianchi NO, Knuutila S, Teerenhovi L, Elonen E, Leskinen R, et al. Immunoglobulin kappa and lambda light chain dual genotype rearrangement in a patient with kappa-secreting B-CLL. Eur J Cancer Clin Oncol. 1988;24(7):1233–8. doi: 10.1016/0277-5379(88)90133-2. [DOI] [PubMed] [Google Scholar]
- 43.Wang YH, Diamond B. B cell receptor revision diminishes the autoreactive B cell response after antigen activation in mice. J Clin Invest. 2008;118(8):2896–907. doi: 10.1172/JCI35618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Wildt RM, Hoet RM, van Venrooij WJ, Tomlinson IM, Winter G. Analysis of heavy and light chain pairings indicates that receptor editing shapes the human antibody repertoire. J Mol Biol. 1999;285(3):895–901. doi: 10.1006/jmbi.1998.2396. [DOI] [PubMed] [Google Scholar]
- 45.Dorner T, Foster SJ, Farner NL, Lipsky PE. Immunoglobulin kappa chain receptor editing in systemic lupus erythematosus. J Clin Invest. 1998;102(4):688–94. doi: 10.1172/JCI3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambracht-Washington D, O’Connor KC, Cameron EM, Jowdry A, Ward ES, Frohman E, et al. Antigen specificity of clonally expanded and receptor edited cerebrospinal fluid B cells from patients with relapsing remitting MS. J Neuroimmunol. 2007;186(1-2):164–76. doi: 10.1016/j.jneuroim.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monson NL, Brezinschek HP, Brezinschek RI, Mobley A, Vaughan GK, Frohman EM, et al. Receptor revision and atypical mutational characteristics in clonally expanded B cells from the cerebrospinal fluid of recently diagnosed multiple sclerosis patients. J Neuroimmunol. 2005;158(1-2):170–81. doi: 10.1016/j.jneuroim.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 48.Hansen A, Odendahl M, Reiter K, Jacobi AM, Feist E, Scholze J, et al. Diminished peripheral blood memory B cells and accumulation of memory B cells in the salivary glands of patients with Sjogren’s syndrome. Arthritis Rheum. 2002;46(8):2160–71. doi: 10.1002/art.10445. [DOI] [PubMed] [Google Scholar]
- 49.Leibiger H, Hansen A, Schoenherr G, Seifert M, Wustner D, Stigler R, et al. Glycosylation analysis of a polyreactive human monoclonal IgG antibody derived from a human-mouse heterohybridoma. Mol Immunol. 1995;32(8):595–602. doi: 10.1016/0161-5890(95)00009-4. [DOI] [PubMed] [Google Scholar]
- 50.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol. 2010;6(10):713–23. doi: 10.1038/nchembio.437. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


