Abstract
NOMe-seq (nucleosome occupancy and methylome sequencing) identifies nucleosome- depleted regions that correspond to promoters, enhancers, and insulators. The NOMe-seq method is based on the treatment of chromatin with the M.CviPI methyltransferase, which methylates GpC dinucleotides that are not protected by nucleosomes or other proteins that are tightly bound to the chromatin (GpCm does not occur in the human genome and therefore there is no endogenous background of GpCm). Following bisulfite treatment of the M.CviPI-methylated chromatin (which converts unmethylated Cs to Ts and thus allows the distinction of GpC from GpCm) and subsequent genomic sequencing, nucleosome-depleted regions can be ascertained on a genome-wide scale. The bisulfite treatment also allows the distinction of CpG from CmpG (most endogenous methylation occurs at CpG dinucleotides) and thus the endogenous methylation status of the genome can also be obtained in the same sequencing reaction. Importantly, open chromatin is expected to have high levels of GpCm but low levels of CmpG; thus, each of the two separate methylation analyses serve as independent (but opposite) measures which provide matching chromatin designations for each regulatory element.
Keywords: NOMe-seq, nucleosome-depleted regions, enhancers, promoters, insulators, open chromatin, DNA methylation1
NOMe-seq has advantages over ChIP-seq for identification of regulatory elements because it is not reliant upon knowing the exact modifications on the surrounding nucleosomes. Also, NOMe-seq has advantages over DHS (DNase hypersensitive site)-seq, FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements)-seq, and ATAC (Assay for Transposase-Accessible Chromatin)-seq because it also gives positioning information for several nucleosomes on either side of each open regulatory element. Here, we provide a detailed protocol for NOMe-seq that begins with the isolation of chromatin, followed by methylation of GpCs with M.CviPI and treatment with bisulfite, and ending with the creation of next generation sequencing libraries. We also include sequencing QC analysis metrics and bioinformatics steps that can be used to identify nucleosome-depleted regions throughout the genome.
1. INTRODUCTION
Regulatory elements such as promoters, enhancers, and insulators are regions of open chromatin that are created and maintained by the binding of site-specific transcription factors (TFs) and their associated protein complexes. These genomic landing platforms are delineated by nucleosome-depleted regions (NDRs), flanked on either side by a series of phased nucleosomes. At promoters and enhancers, the flanking nucleosomes can harbor one or more modifications, such as acetylation of lysine 27 on histone H3 (H3K27ac) at enhancers or methylation of lysine 4 on histone H3 (H3K4me3) at promoters [1–5], that provide additional information about the specific functional state of a particular NDR. These histone modifications are created by the recruitment of histone-modifying enzymes (e.g. acetylases and methylases) to the NDR via interaction with site-specific transcription factors bound to the DNA [6,7]. Insulators, on the other hand, are characterized by the presence of site-specific DNA binding components of the cohesin complex, such as CTCF and RAD21, often in the absence of marks associated with active enhancers or promoters [8].
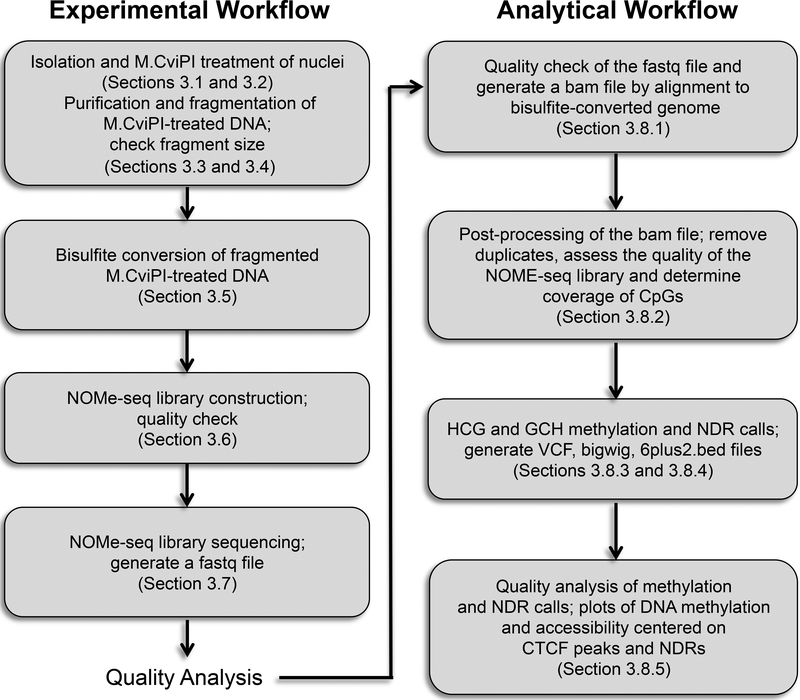
NOMe-seq (nucleosome occupancy and methylome sequencing) identifies NDRs that correspond to promoters, enhancers, and insulators; see Figure 1 [9]. The NOMe-seq method is based on the treatment of chromatin with the M.CviPI methyltransferase. This enzyme, which is isolated from Chlorella virus, methylates Cs in the context of GpC dinucleotides. GpCm does not occur in the human genome (the vast majority of DNA methylation in the human genome is at CpG dinucleotides, not GpC dinucleotides) and therefore there is no endogenous background of GpCm. The enzyme can only methylate GpC dinucleotides that are accessible in the context of chromatin, i.e. not protected by nucleosomes or other proteins that are tightly bound to the chromatin. Following bisulfite treatment of the M.CviPI-methylated chromatin (which converts unmethylated Cs to Ts and thus allows the distinction of GpC from GpCm) and subsequent genomic sequencing, the status of GpC-containing regions can be ascertained on a genomewide scale. Using this method, NDRs are defined as regions having increased GpCm methylation over background (i.e. they were in open regions and thus were methylated by the M.CviPI enzyme) that are at least 140 bp in length. The bisulfite treatment also allows the distinction of CpG from CmpG and thus the endogenous methylation status of the genome can also be obtained in the same sequencing reaction. It is important to note that in contrast to the induced GpCm which represents nucleosome-free, open chromatin that is available for TF binding, the endogenous CmpG represents nucleosome-bound chromatin that is not available for TF binding. We note that GCG trinucleotides cannot be used to distinguish between enforced GpC methylation and endogenous CpG methylation (on the other strand); therefore, in the analysis of NOMe-seq datasets GCH (H=A, C, or T) trinucleotides are selected and analyzed for nucleosome positioning whereas HCG trinucleotides are selected and analyzed for endogenous DNA methylation. As reported earlier, GCG trinucleotides are not frequent in the genome and are almost always within 20 bp of a GCH [9], thus allowing an NDR containing a GCG to be identified by nearby GCH sequences. Importantly, open chromatin is expected to have high levels of GpCm but low levels of CmpG; thus, each of the two separate methylation analyses serve as independent (but opposite) measures which should provide matching chromatin designations (open vs. closed).
Figure 1. Schematic overview of NOMe-seq.
Left: Experimental workflow, representing Sections 3.1–3.7 in the Methods. Right: Analytical workflow, representing Sections 3.8.1–3.8.5 in the Methods.
Although ChIP-seq performed using antibodies to specifically modified histones can also be used to identify regulatory elements [10], NOMe-seq has advantages over ChIP-seq because it is not reliant upon knowing the exact modifications on the surrounding nucleosomes. NOMe-seq may also provide information not easily gained from ChIP-seq. As noted above, regulatory regions identified using NOMe-seq should have high levels of GpCm or CmpG, but not high levels of both types of methylation. However, previous analyses using NOMe-seq have found that a small number of regions of the genome have been identified as having both types of methylation in the same cell population [9]. It has been suggested that these regions represent allelic differences, with one allele having an active regulatory element (high GpCm) but the other allele being in a closed state (high CmpG). In support of this hypothesis, Kelly et al. [9] previously showed that doubly-identified regions (i.e. NDRs identified as having high GpCm and high CmpG) are enriched for known imprinted promoters. Thus, NOMe-seq can help to identify new allele-specific regulatory elements without the need for a SNP to be within the element (as is the case for analysis of allele-specific ChIP-seq). Of course, sequencing depth is important in such analyses because high coverage of the examples of “nucleosome-depleted” and “DNA methylated” reads in the same region is needed to be certain that the regions are doubly marked.
NOMe-seq has similarities to other techniques used to detect regions of open chromatin such as DHS (DNase hypersensitive site)-seq [11] and FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements)-seq [12], both of which rely on the physical separation of nucleosome- free vs. nucleosome-bound DNA, or ATAC (Assay for Transposase-Accessible Chromatin)-seq [13,14] which identifies regions of open chromatin using transposon integration. However, because DNA breakage (sonication or treatment with DNase) or transposon integration is not used in NOMe-seq, there is no bias towards open chromatin and there may be fewer false positive identified regions. Two other advantages of NOMe-seq are that, unlike the other methods, it also gives positioning information of several nucleosomes on either side of each open regulatory element and it provides information concerning the endogenous methylation state of every CpG dinucleotide in the genome.
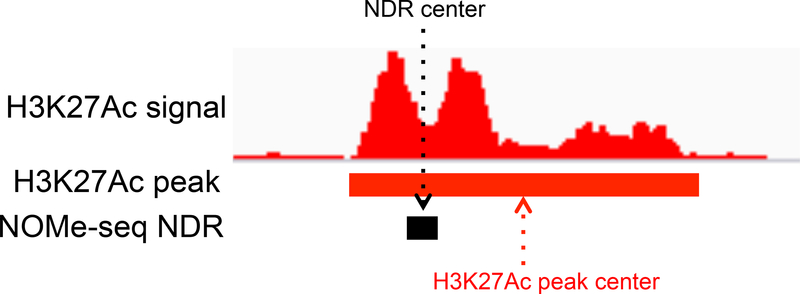
It is also important to consider the size of the regulatory element identified by the different techniques. For example, the average width of the set of H3K27ac peaks is quite large and it is not reasonable to simply define the center of a H3K27ac-covered area as the functional (i.e. the TF binding platform) region. On the other hand, the NDRs called by NOME-seq are smaller in width, corresponding to inter-nucleosomal regions, and therefore more closely match the region containing TF binding sites (Figure 2). The ability to refine the functional compartment within open chromatin domains to a small region can have considerable influence on the quality of downstream analyses, such as motif finding and interpretation of non-coding variants identified by GWAS. It is also important to precisely delineate the functional compartment of an open regulatory region when using DNA methylation status to link activity of an element to gene expression. For example, DNA methylation levels may be high throughout a large H3K27ac peak, only showing a small hypomethylated region that corresponds to the NDR; averaging methylation levels over a large region may obscure the presence of a differentially active enhancer when comparing different tissues types or disease states.
Figure 2. Refinement of a regulatory element using NOMe-seq.
Shown is a nucleosome- depleted region (NDR) flanked by nucleosomes harboring the histone modification H3K27ac; the centers of the NDR and the region covered by H3K27ac are also indicated.
To date, NOMe-seq has been performed in IMR90 lung cells and glioblastoma cells [9], normal (PREC) and cancer (PC3) prostate cells, normal (HMEC) and cancer (MCF7) breast cells [15,16], and HCT116 and DKO colon cancer cells [17]. However, due to technology improvements, our current protocol has changed as compared to that used in those initial studies. Here, we provide a detailed protocol for NOMe-seq which differs from that used in previous studies in several important steps, such as the order in which the DNA is treated with bisulfite in the library protocol, which can have a considerable influence in the yield of DNA in the resultant library.
2. MATERIALS
2.1 Isolation of Nuclei
1X Dulbecco's phosphate-buffered saline (DPBS): sterile, no calcium, no magnesium
Trypsin or dispase (if needed for your cell type)
Trypan Blue and hemocytometer
Lysis Buffer: 10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA, 0.5% NP-40
Wash Buffer: 10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1 mM EDTA
2.2 Treatment of Nuclei with M.CviPI
10X GpC Buffer (New England Biolabs)
32 mM S-adenosylhomocysteine (SAM) (New England Biolabs)
50 U/μL M.CviPI (New England Biolabs)
1 M Sucrose
Nuclease-free water
Stop Buffer: 20 mM Tris pH 7.4, 600 mM NaCl, 1% SDS, 10 mM EDTA
2.3 Isolation of M.CviPI-treated DNA
5 M NaCl
Proteinase K (Promega)
1:1 Phenol:Chloroform
100% Ethanol
TE buffer: 10 mM Tris pH 8, 10 mM EDTA pH 8
Nanodrop spectrophotometer
2.4 Fragmentation of M.CviPI-treated DNA
Covaris sonicator (S220, formerly S2)
Covaris MicroTUBE AFA Pre-slit Snap-Cap 6x16mm
Nanodrop Spectrophotometer
DNA High Sensitivity Kit (Agilent) for use with Agilent 2100 Bioanalyzer
2.5 Bisulfite Conversion of M.CviPI-treated DNA
EZ DNA Methylation Kit #D5001 (Zymo Research)
2.6 NOMe-seq Library Construction
Accel-NGS Methyl-Seq DNA Library Kit for Illumina Platforms (Swift Biosciences #30024)
Methyl-Seq Set A Indexing Kit (Swift Biosciences)
SPRIselect Magnetic Beads (Beckman Coulter)
Qubit dsDNA HS (High Sensitivity) Assay Kit (Thermo Fisher Scientific)
DNA High Sensitivity Kit (Agilent) for use with Agilent 2100 Bioanalyzer
3. METHODS
3.1 Isolation of Nuclei (Note: stopping points throughout the experimental protocol are indicated with a stop sign symbol.  )
)
Treat adherent cells with trypsin or dispase or collect suspension cells and place into a pre-chilled 15 mL tube; see Note 1. Centrifuge at 250 g at 4°C for 5 minutes.
Place cells on ice or at 4°C for the remaining steps in Section 3.1
Remove the media and wash cells with 10 mL ice-cold sterile PBS.
Remove 10 μL of the cell suspension and combine with 10 μL of trypan blue in a 1.5 mL tube; mix well.
Pipette 10 μL of the cell/trypan blue mixture onto the hemocytometer. Count the number of intact cells (i.e. cells that are not blue) in each of the four quadrants. Take the average of these four counts, multiply by a dilution factor of 2 and multiply by 10,000 to get the number of cells per milliliter.
Transfer a volume equivalent to 1 million cells into a new 15 mL conical vial. Centrifuge at 250 g at 4°C for 5 minutes, remove PBS wash, and save the pellet.
Resuspend the pelleted cells in 1 mL ice-cold Lysis Buffer and let sit undisturbed on ice for 5–10 minutes to lyse the cells.
Check a small aliquot of cells under the microscope using trypan blue and a hemocytometer in the same way as used for counting the cells. The majority of the cells should have blue nuclei, indicating that the cell membrane has been ruptured but the nuclei are intact; see Note 2.
After confirming that most cells (but not most nuclei) are lysed, centrifuge the cells for 5 minutes at 750 g in 4°C and discard the supernatant, taking care not to disturb the nuclear pellet.
Using a P1000 pipetman, gently resuspend the nuclei in 1 mL ice-cold wash buffer. Centrifuge for 5 minutes at 750 g in 4°C, discard supernatant, and immediately proceed to M.CviPI treatment of the pelleted nuclei.
3.2 Treatment of Nuclei with M. CviPI to Methylate Accessible GpCs
Prepare at least 378 μL of 1X GpC Buffer (it is recommended that you start with 4 tubes of 250,000 cells and 94.5 μL is needed per 250,000 cells) by diluting the stock 10X GpC buffer in nuclease-free water.
Using a P1000 pipetman, resuspend the nuclei obtained from 1 million cells in 378 μL of 1X GpC buffer to obtain a final concentration of 250,000 nuclei per 94.5 μL; keep nuclei on ice.
- In 4 pre-chilled 1.7 mL microcentrifuge tubes, prepare 4 reaction mixtures containing the following components in the order listed (see Note 3):
1 M Sucrose 45.0 μL 10x GpC Buffer 5.0μL Nuclei (250,000) 94.5μL 32 mM SAM 1.5μL 50 U/ μl M.CviPI 4.0μ L ---------------------------- Total 150.0 μL/tube - Incubate for 7.5 minutes at 37°C, then boost the reaction by adding the following:
32 mM SAM 1.5μL 50 U/lI M.CviPI 2.0μL/tube ---------------------- Total 3.5μL/tube Incubate for an additional 7.5 minutes at 37°C, then stop the reaction by adding 153.5 μL of the Stop Buffer.
3.3 Purification of M.CviPI-Treated DNA
Add 200 μg/rnl of Proteinase K (3 μL of 20 mg/mL Proteinase K) to each of the 4 reaction mixtures and incubate for 16 hours at 55°C to inactivate the M.CviPI enzyme and digest proteins present in the treated nuclei preparations.
Purify the DNA in the 4 reaction mixtures using a standard phenol/chloroform extraction method, removing the aqueous layer to a new 1.7 mL tube; note that phase-lock gel can be used to assist the separation of the aqueous and organic phases. Add 2.5X volumes (775 μL) of 100% ethanol to each tube containing the aqueous layer and incubate at - 20°C for overnight or at −80°C for 1–2 hours; see Note 4.

Pellet the DNA by centrifuging at a maximum speed in a micro centrifuge for 15 minutes. Carefully remove the ethanol and add 300 μL of ice cold 70% ethanol to the pellet.
Pellet the DNA again by centrifuging at a maximum speed in a micro centrifuge for 15 minutes. Remove the ethanol and allow the pellet to air dry (~ 20 minutes).
Resuspend the DNA pellet in 20 μl of nuclease-free water or TE buffer.
Quantify the DNA and combine the treated DNA from the 4 tubes into a single 1.7 mL tube. In general, a quantity of 100 ng/μL from the starting 1 million cells (~8 μg total) is expected. Store long term at −20°C.

3.4 Fragmentation of M.CviPI-treated DNA
Dilute M.CviPI-treated DNA to a total volume of 130 μl and transfer into one 6X16mm microTUBE, taking care to avoid air bubbles; see Note 5
Perform sonication using the Covaris system, producing 150 bp fragments; see Note 6.
Ethanol precipitate the sonicated DNA by adding 2.5 volumes of ice-cold 100% ethanol; incubate at −20°C for overnight or at −80°C for 1–2 hours.
Pellet the DNA by centrifuging at a maximum speed in a micro centrifuge for 15 minutes. Remove the ethanol and allow the pellet to air dry (~ 20 minutes).
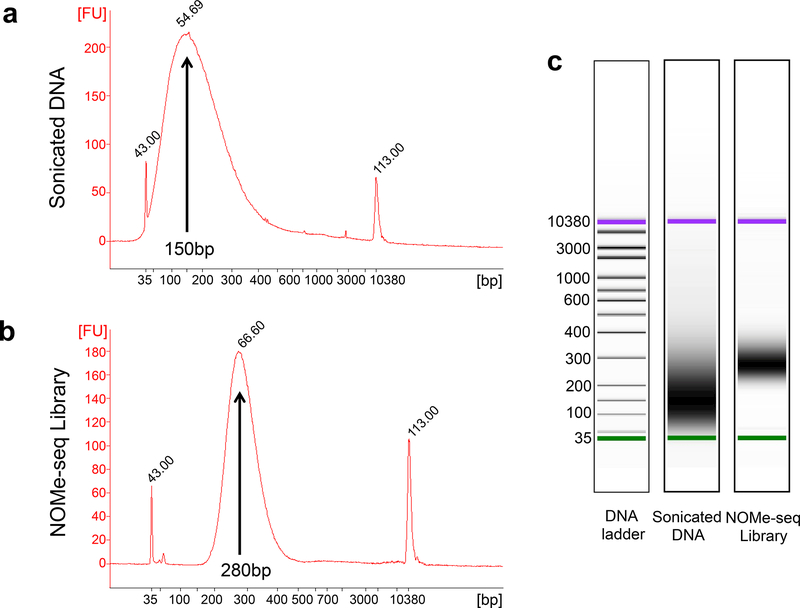
Resuspend the DNA pellet in 15 μL of nuclease free water. Quantify DNA using a nanodrop spectrophotometer. Check the fragment size using an Agilent Bioanalyzer with a DNA High Sensitivity chip (Figure 3). Store long term at −20°C; see Note 6.

Figure 3. Size distribution analysis of a NOMe-seq sample and library.
Shown is a Bioanalyzer trace, obtained using an Agilent 2100 Bioanalyzer instrument and an Agilent High Sensitivity DNA chip, of the DNA after M.CviPI treatment and fragmentation using a Covaris S220 sonicator (a) and of the resultant NOMe-seq library (b). The leftmost and rightmost peaks (labeled 43 and 113) are size markers of 35 bp and 10380 bp, respectively. The average length of the fragmented DNA is calculated to be 150 bp whereas the average length of the library fragments is calculated to be 280 bp. (c) For comparison to the Bioanalyzer traces, the gel images of the fragmented DNA and the NOMe-seq library are also shown.
3.5 Bisulfite Treatment of M.CviPI-methylated DNA to Convert All Unmethylated Cs to Ts
Use the EZ DNA Methylation kit from Zymo Research to convert unmethylated Cs in up to 1ug of M.CviPI-treated and fragmented DNA.
Add 5 μL of M-Dilution Buffer to the DNA and adjust total volume to 50 μL with water. Mix the sample by flicking or pipetting up and down.
Incubate the sample at 37°C for 15 minutes.
After the above incubation, add 100 μL of the prepared CT Conversion Reagent to the sample and mix.
Incubate the sample in a thermocycler at (95°C for 30 seconds, 50°C for 60 minutes) for 16 cycles, then hold at 4°C.
Add 400μL of M-Binding Buffer to a Zymo-Spin IC Column and place the column in the provided collection tube.
Transfer the sample being held at 4°C to the column containing the M-Binding Buffer. Mix by inverting the column in the collection tube several times.
Centrifuge at full speed for 30 seconds. Discard flow through.
Add 100 μL of M-Wash Buffer to the column. Centrifuge at full speed for 30 seconds.
Add 200 μL of M-Desulphonation Buffer to the column and let stand at room temperature for 15–20 minutes. After incubation, centrifuge at full speed for 30 seconds.
Add 200 μL of M-Wash Buffer to the column. Centrifuge at full speed for 30 seconds. Add an additional 200 μL of M-Wash Buffer and centrifuge for an additional 30 seconds.
Place the column into a 1.7mL microcentrifuge tube. Add 20 μL of nuclease-free water to the column matrix. Centrifuge for at full speed for 30 seconds to collect the DNA solution. Bisulfite-converted DNA is stable at −20°C for up to a year.

3.6 NOMe-seq Library Construction
Use the bisulfite-converted DNA isolated in the previous step to generate a library using the Accel-NGS Methyl-Seq DNA Library Kit for lllumina Platforms; see Note 7. The basic steps in the library preparation include an adaptase step (end repair, tailing of 3’ ends, and ligation of the first truncated sequencing adapter in a single step), extension, ligation of the second truncated adapter, and indexing PCR. A detailed protocol is provided with the kit; however, we note that we generally use the entire amount of converted DNA with 7–10 PCR cycles and that for all steps involving SPRl (Solid Phase Reversible Immobilization) select beads, the volumes indicated for a 165bp insert size should be used. Importantly, this kit should be purchased along with indexing reagents to barcode your library allowing for the pooling of multiple libraries; see Note 8.

Measure the concentration of the library using the Qubit DNA HS assay kit.
Check the library size using an Agilent Bioanalyzer with a DNA High Sensitivity chip (Figure 3); see Note 6.

3.7 Sequencing a NOMe-seq Library
NOMe-seq libraries can be sequenced either using single-end or paired-end methods at standard read lengths using lllumina sequencers (e.g. Hi-Seq or NextSeq machines) (see http://www.illumina.com/systems/sequencing.html for details). To check the quality of a NOME- seq library, a low pass run should be performed; see Note 8. After determining that the library is of high quality (see Section 3.8), a minimum of 200 million reads should be obtained, which corresponds to ~5x coverage of all methylated loci in the human genome.
3.8 Quality analysis of a NOMe-seq Library
Genome alignment. After obtaining fastq files from the Illumina sequencer, the quality of the fastq files is examined using software tools such as FastQC (http://www.bioinformatics.babraham.ac.uk/proiects/fastqc/). The fastq files must be aligned to a bisulfite-converted genome, which can be done using bisulfite sequencing mapping programs such as BSMAP [18], BWA-METH (https://github.com/brentp/bwa-meth). Bismark [19], or BS- SEEKER [20,21]. The aligned file produced from the fastq file using the bisulfite sequencing mapping programs should be saved as a bam file format; see Note 9.
-
Post-processing of bam files. The bam file generated from Section 3.8.1 must be post- processed for further analyses, such as DNA methylation and NDR calling. To remove duplicate reads caused by PCR from a bam file, the MarkDuplicates function of Picard (http://picard.sourceforge.net) should be used. If multiple sequencing lanes for a given sample were obtained, the bam files from each lane can be combined. Multiple bam files can be merged by using the MergeSamFiles function of Picard or the merge function of SAMTOOLS [22]. When there are multiple bam files, it is important that each read from multiple sequencing lanes has the proper read group in order to remove duplicates and keep track of the data. By using the AddOrReplaceReadGroups function of Picard, read groups of each read can be added or replaced. The final bam file should be sorted using the sort function of SAMTOOLS and indexed, producing a bai file which contains indices of the bam file required for access to arbitrary genomic coordinates
To assess the quality of the bam file, the flagstat function of SAMTOOLS or the CollectAlignmentSummaryMetrics function of Picard can be used. The output file of the flagstat function will list the total number of mapped reads (which includes QC-passed reads and QC- failed reads), the number of duplicates reads, the number of mapped reads, and the number of correctly paired reads if the library was sequenced using a paired-end method. Similarly, using the Picard CollectAlignmentSummary function, statistics such as total reads, aligned reads and percent of aligned pairs can be measured. The coverage of CpGs vs coverage of random regions of the genome can be calculated using the BamToElementEnrichment script from ECWorkflows (https://github.com/uec). This value is critical to assess the quality of the NOMe- seq library. It has been observed that CpG islands can often be poorly represented in bisulfite- converted libraries. Because CpG islands are enriched in promoter regions, it is critical that these regions of the genome be adequately represented in the libraries; see Note 10.
Methylation calling. To identify the methylation status of CpG sites (in all HCG trinucleotides) and GpC sites (in all GCH trinucleotides) from the bam file, the Bis-SNP [23] program can be used. The BisulfiteGenotyper function of the Bis-SNP pipeline takes a bam file and generates a VCF file, which contains detailed information about the SNPs in the analyzed genome and provides DNA methylation information. The Vcf2bed6plus2 script in the Bis-SNP pipeline converts vcf files to a 6plus2.bed format file which contains information about each CpG or GpC site, including the chromosome start and end position, status indicating if a SNP or a reference CG is present, a score showing the methylation level (0–1000), the strand orientation, the methylation level (0–100%), and the number of CT reads covered at each locus. The Vcf2wig script in the Bis-SNP pipeline converts vcf files to wiggle files such as bedGraph and bigwig files, which can be used to visualize the DNA methylation levels across the genome by using browsers such as the UCSC genome browser [22], IGV [24], or IGB [25] or to make plots and heatmaps showing the DNA methylation density at regions of interest using the Bis-tools (https://github.com/dnaase/Bis-tools). The MethylSummarizeList.txt file generated from the Bis- SNP pipeline contains statistics of the methylation calling, such as visited bases, callable bases, confidently called bases, and average good reads coverage in all visited and callable loci.
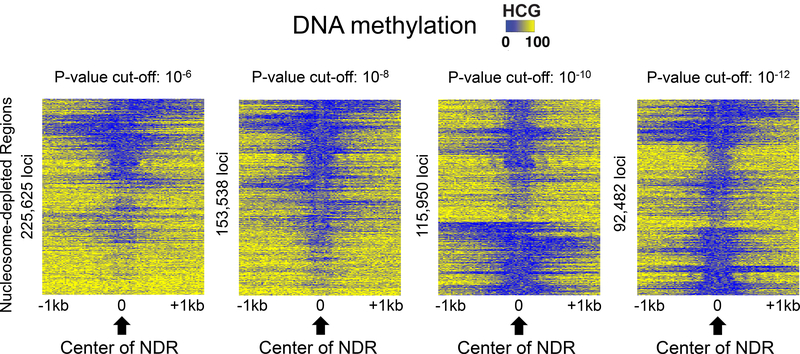
Calling NDRs. For identification of NDRs, the findNDRs function in the aaRon R package can be used (see https://github.com/astatham/aaRon for details). To use the aaRon R package, a GCH.6plus2.bed file, which contains methylation calls from the Bis-SNP program (see section 3.8.3), should be transformed to a tsv file, which contains the number of CT reads as methylation levels, multiplying total number of CT reads by the methylation level at each GpC site. For the findNDRs function, different p-value cut-offs and window sizes can be used; see Note 11. Although the number of NDRs will differ for each NOMe-seq library and for each p- value cut-off, a standard number of NDRs for further analyses of human genomes is 70,000100,000.
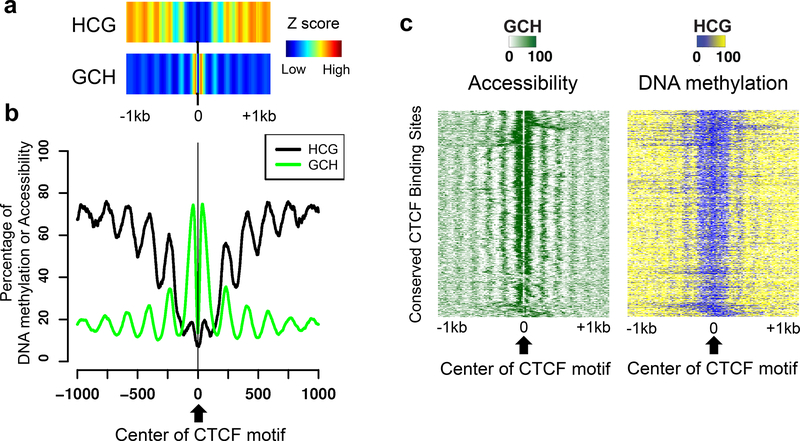
Quality analysis of methylation and NDR calls. To determine the quality of the DNA methylation data, the HCG and GCH methylation levels can be visualized at the center of conserved motif-containing CTCF peaks; see Note 12. A high quality NOMe-seq library with proper DNA methylation calls will show phasing of HCG and GCH signals; see Figure 4. To determine the quality of the NDR calls, the HCG and GCH methylation levels can be visualized at the center of the called NDRs. As discussed above, the NDRs should have high GCH signals but low HCG signals at their centers; see Figure 5. By generating a heatmap that can visualize methylation signals at each NDR locus, one can remove false positive NDRs and decide p- value cut-offs for NDR calls; see Figure 6.
Figure 4. Quality analysis of NOMe-seq data using CTCF peaks.
A set of 3,216 CTCF peaks that were commonly identified in 58 different human cell types and that have a CTCF motif were used to compare endogenous DNA methylation (HCG) and accessibility (GCH). In each panel, data is shown for a 2 kb region, centered on the CTCF motifs within the CTCF peaks; the genomic locations of the set of 3,216 CTCF sites are provided in Supplementary Table S1. (a) The density (Z scores) of HCG methylation and GCH methylation, centered on the CTCF motif, is shown for all common CTCF peaks. (b) The average methylation levels of HCG (endogenous DNA methylation) and the average methylation of GCH (accessibility) are shown for all common CTCF peaks. (c) A heatmap representing the percentage of GCH methylation (left) and endogenous HCG methylation (right) is shown for all common CTCF peaks. The heatmap was made by first clustering the GCH values at the CTCF peaks, then plotting both the GCH and HCG values in the same order.
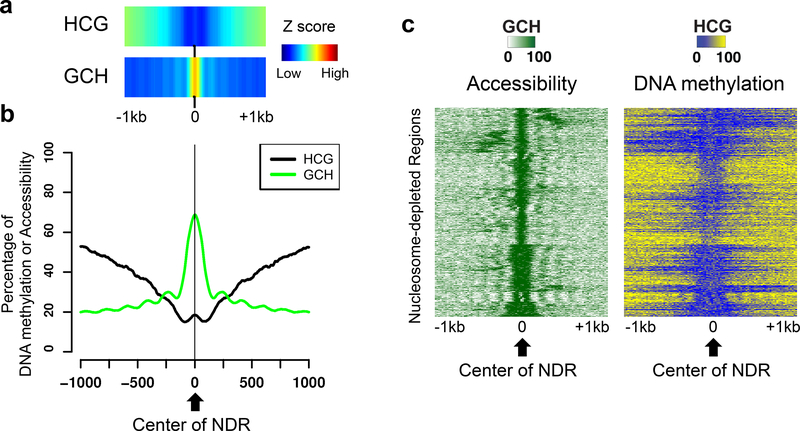
Figure 5. Quality analysis of called NDRs.
Data is shown for a 2 kb region centered on 92,482 called NDRs (P-value cut-off = 10−12) for a NOMe-seq dataset. (a) The density (Z scores) of HCG methylation (endogenous DNA methylation) and GCH methylation (accessibility), centered on the NDRs, is shown. (b) The average methylation levels of HCG (endogenous DNA methylation) and the average methylation of GCH (accessibility) are shown for all NDRs. (c) A heatmap representing the percentage of GCH methylation (left) and endogenous HCG methylation (right) is shown for all NDRs. The heatmap was made by first clustering the GCH values at the NDRs, then plotting both the GCH and HCG values in the same order.
Figure 6. Comparison of DNA methylation levels at NDRs identified using different P- value cut-offs.
Shown are heatmaps indicating the percentage of endogenous methylation at HCG sites for a 2 kb region centered on NDRs selected using different P-value cut-offs. The heatmaps were made by first clustering the GCH values at each NDR, then plotting the HCG values in the same order.
3.9. Example analyses of a NOMe-seq library
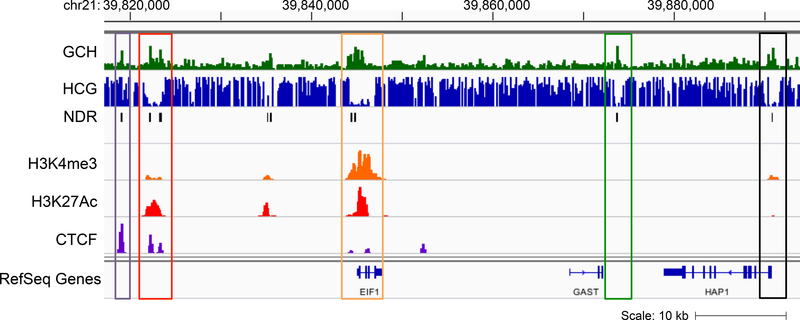
We generated a NOME-seq library using CNON (Cultured Neuronal cells derived from Olfactory Neuroepithelium) cells [26] from patient sample 45 as part of the PsychENCODE project (https://www.synapse.org/#iSynapse:syn4921369/wiki/235539) [27] using 100bp paired-end sequencing with the Illumina Hi-Seq 2500. To compare NDRs to regulatory elements defined by the H3K4me3 promoter mark, the H3K27ac enhancer mark, and CTCF binding sites, we also generated ChIP-seq libraries with proper antibodies using our previously published protocols for histones and site-specific DNA binding factors [10,28]. See Supplementary File 1 for specific cell culture and ChIP-seq protocols for CNON cells; we recommend using MACS2 [29] to call the peaks (see also https://github.com/taoliu/MACS/). When we overlapped NDRs with H3K4me3 peaks, H3K27ac peaks and CTCF peaks from CNON cells we found that about 80% of the NDRs were in these regulatory elements. However, we also identified NDRs which are distal from transcription start sites and do not have a significant H3K27ac or CTCF signal (Figure 7).
Figure 7. Examples of NDRs at regulatory elements.
Shown is a genome browser screen shot (hg19) of a region from chr21q22.2 with tracks representing accessibility (GCH), endogenous DNA methylation (HCG), called NDRs, and H3K4me3, H3K27ac and CTCF ChIP- seq data; all data is from CNON cells. The purple box highlights an NDR classified as an insulator (a genomic region bound by CTCF that does not have the histone modifications found at promoters or enhancers), the red box highlights an NDR representing an enhancer (a distal genomic region marked by H3K27ac), the orange box highlights an NDR in the promoter of the EIF1 gene, the green box highlights an NDR that lacks promoter, enhancer, and insulator marks, and the black box highlights an NDR at the promoter of the HAP1 gene, which is not actively transcribed in these cells (as shown by the small H3K4me3 signal).
Supplementary Material
4. NOTES
A protocol for growing and processing CNON cells (used to obtain the example NOMe- seq dataset), including the dispase treatment used to collect the cells needed for nuclei isolation, is provided as Supplementary Information. It is recommended that exponentially growing cells be used for these experiments. However, if it is necessary to use tissue samples, then methods such as those used to perform native ChIP (no crosslinking) from tissues should be employed [30,31]. In addition, other protocols suggest that crosslinked cells can be used as the starting material for NOMe-seq (http://www.activemotif.com/documents/1847.pdf).
The time needed to lyse the cells varies based on cell type, so it is recommended to optimize the cell lysis condition for each cell line prior to beginning the NOMe-seq protocol. If intact cells (i.e. cells that do not have blue nuclei) remain after the initial cell lysis treatment, you will need to extend the time in Lysis Buffer, monitoring progress using trypan blue staining. It is critical that the nuclei remain intact throughout the subsequent wash and reaction steps. Therefore, the shortest amount of time needed to lyse the majority of the cells should be used.
The standard concentration of M.CviPI from NewEngland BioLabs is 4 U/μl. However, we recommend that a special, high concentration order of 50 U/μl be purchased from the company; otherwise, the reaction volumes must be adjusted accordingly if the low concentration enzyme is used.
| 1 M Sucrose | 45.0 μL |
| 10x GpC Buffer | 5.0μL |
| Nuclei (250,000) | 94.5μL |
| 32 mM SAM | 1.5μL |
| 50 U/ μl M.CviPI | 4.0μ L(200 units) |
| ---------------------------- | |
| Total | 150.0 μL/tube |
| 32 mM SAM | 1.5μL |
| 50 U/lI M.CviPI | 2.0μL (100 units) |
| ---------------------- | |
| Total | 3.5μL/tube |
Incubate for 7.5 minutes at 37°C, then stop by adding 153.5 μL of Stop Buffer.
| 1 M Sucrose | 15.0 μL |
| 10x GpC Buffer | 17.0μL |
| Nuclei (250,000) | 282.0μL |
| 32 mM SAM | 1.5μL |
| 50 U/ μl M.CviPI | 50.0μ L(200 units) |
| ---------------------------- | |
| Total | 500.0 μL/tube |
| 32 mM SAM | 1.5μL |
| 50 U/lI M.CviPI | 25.0μL (100 units) |
| ---------------------- | |
| Total | 26.5μL/tube |
Incubate for 7.5 minutes at 37°C, then stop by adding 526.5 μL of Stop Buffer.
In addition to the phenol chloroform extraction method, other methods of isolating human genomic DNA may be used, such as the column-based genomic DNA isolation kit from Zymo (Genomic DNA Clean & Concentrator −25).
If using the Covaris S220 sonicator, no more than 10 μg of DNA should be fragmented at a time; if you obtained more than 10 μg of DNA from the treated cells, you should dilute to 100 ng/ul and only use 8–10 μg per sonication tube.
Sonication must be optimized for each cell type to produce 100–200 bp fragments. If using a Covaris S220 sonicator, it is recommended that you start by using a 10% duty cycle, an intensity setting of 5, and 200 cycles per burst for 6 minutes. If the fragments in the resultant sonicated fragments or library are far from the appropriate size when examined on the Bioanalyzer (Sections 3.4.5 or 3.6.3) it is recommended that the sonication step be optimized and the protocol repeated prior to proceeding with library preparation or sequencing.
Although previous NOMe-seq studies [17,16,9] have used methods in which adaptors are ligated to the fragmented DNA prior to bisulfite treatment, these methods result in considerable losses of DNA. We recommend using the Accel-NGS Methyl-Seq DNA Library Kit because it enables the preparation of high complexity next-generation sequencing libraries after bisulfite conversion of the DNA. Importantly, this kit is compatible with single-stranded DNA, making it a good choice for use with DNA fragments damaged and denatured by bisulfite conversion. This single-strand compatibility also overcomes the library loss associated with methylated adapter ligation prior to bisulfite conversion.
It is highly recommended that NOMe-seq libraries be barcoded so that they can be pooled with other libraries prior to sequencing. It is recommended that a low-pass sequencing run be performed (~10 million reads) for each NOMe-seq library to assess various quality metrics before proceeding to sequence a library at a high depth. It is important to note that, once a library has passed quality assessment, sequencing data from multiple NOMe-seq libraries from the same cells can be combined to increase the genomic coverage.
A bisulfite-converted genome is low in complexity, due to the conversion of the vast majority of Cs to Ts, which results in a lower percentage of alignment of sequenced fragments to the genome than obtained from standard genomic libraries. However, the use of paired-end sequencing methods can improve the alignment and therefore this method is preferable rather than single-end sequencing for NOMe-seq libraries; if single-end sequencing is used, longer reads (at least 100 bp) can help increase the alignment percentage. Increasing the number of allowed mismatch parameters in the bisulfite sequencing mapping programs may also improve alignment of reads to the bisulfite genome.
A bias for or against CpG islands could be due to size selection of the NOMe-seq library. Smaller library fragments tend to be enriched for CpG islands. Therefore, if the size of library is too small, CpG islands will be represented but perhaps other regulatory elements, such as distal enhancers, may be lost. On the other hand, if the size of library is too big, there will be low coverage of CpG islands, which will negatively affect the ability to identify NDRs in promoter regions. A ratio of CpG vs random coverage close to 1 is desired; if the ratio is lower than 0.5 the coverage may be biased toward non-CpG islands and if the ratio is larger than 1, the coverage may be biased in favor of CpG islands. Therefore, it is important that the library size be optimized (see Note 6).
We restrict window size to 140bp for NDRs, which provides a more precise region of the inter-nucleosomal region of open chromatin that can be used for motif analyses.
The CTCF protein binds with high affinity to a specific DNA motif, which contains a CpG dinucleotide, which helps to visualize DNA methylation calls. Binding of CTCF is not compatible with high levels of endogenous DNA methylation and therefore the CmpG levels should be very low at these sites. Conversely, because CTCF binds in regions of open chromatin, the levels of GpCm should be high at the sites. To assist investigators who do not have CTCF ChIP-seq data for their particular cell type in which NOMe-seq is being performed, we have generated a file of conserved, motif-containing CTCF sites, which are distal from transcription start sites and commonly found across 114 CTCF ChIP-seq samples from 58 different cell types [1]. This file, which includes 3,216 genomic coordinates of CTCF sites (hg19), can be used for quality analysis of any human NOMe-seq library (Supplementary Table S1).
5. REFERENCES
- 1.ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489 (7414):57–74. doi:10.1038/Nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470 (7333):279–283. doi:nature09692 [pii] 10.1038/nature09692 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roadmap Epigenomics Consortium (2015) Integrative analysis of 111 reference human epigenomes. Nature 19:317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heintzman ND, Ren B (2009) Finding distal regulatory elements in the human genome. Current opinion in genetics & development 19 (6):541–549. doi:10.1016/j.gde.2009.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nature genetics 39 (3):311–318. doi:10.1038/ng1966 [DOI] [PubMed] [Google Scholar]
- 6.DesJarlais R, Tummino PJ (2016) Role of Histone-Modifying Enzymes and Their Complexes in Regulation of Chromatin Biology. Biochemistry 55 (11):1584–1599. doi:10.1021/acs.biochem.5b01210 [DOI] [PubMed] [Google Scholar]
- 7.Allis CD, Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nature reviews Genetics 17 (8):487–500. doi:10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- 8.Merkenschlager M, Nora EP (2016) CTCF and Cohesin in Genome Folding and Transcriptional Gene Regulation. Annual review of genomics and human genetics 17:17–43. doi:10.1146/annurev-genom-083115-022339 [DOI] [PubMed] [Google Scholar]
- 9.Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA (2012) Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome research 22 (12):2497–2506. doi:10.1101/gr.143008.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Geen H, Echipare L, Farnham PJ (2011) Using ChIP-Seq Technology to Generate High- Resolution Profiles of Histone Modifications. Methods in molecular biology 791:265–286. doi:10.1007/978-1−61779-316-5_20 [doi]] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford Ge, Stamatoyannopoulos JA(2012) The accessible chromatin landscape of the human genome. Nature 489 (7414):75–82. doi:10.1038/nature11232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD (2007) FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome research 17 (6):877–885. doi:10.1101/gr.5533506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buenrostro JD, Giresi PG, Zaba Lc, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods 10 (12):1213–1218. doi:10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ (2015) ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109:21 29 21–29. doi:10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taberlay PC, Statham AL, Kelly TK, Clark SJ, Jones PA (2014) Reconfiguration of nucleosome-depleted regions at distal regulatory elements accompanies DNA methylation of enhancers and insulators in cancer. Genome research 24 (9):1421–1432. doi:10.1101/gr.163485.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Statham AL, Taberlay PC, Kelly TK, Jones PA, Clark SJ (2015) Genome-wide nucleosome occupancy and DNA methylation profiling of four human cell lines. Genom Data 3:94–96. doi:10.1016/j.gdata.2014.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lay FD, Liu Y, Kelly TK, Witt H, Farnham PJ, Jones PA, Berman BP (2015) The role of DNA methylation in directing the functional organization of the cancer epigenome. Genome research 25 (4):467–477. doi:10.1101/gr.183368.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xi Y, Li W (2009) BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics 10:232. doi:10.1186/1471-2105-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27 (11):1571–1572. doi:10.1093/bioinformatics/btr167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo W, Fiziev P, Yan W, Cokus S, Sun X, Zhang MQ, Chen PY, Pellegrini M (2013) BS- Seeker2: a versatile aligning pipeline for bisulfite sequencing data. BMC genomics 14:774. doi:10.1186/1471−2164-14-774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen PY, Cokus SJ, Pellegrini M (2010) BS Seeker: precise mapping for bisulfite sequencing. BMC Bioinformatics 11:203. doi:10.1186/1471-2105-11−203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 (16):2078–2079. doi:10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Siegmund KD, Laird PW, Berman BP (2012) Bis-SNP: combined DNA methylation and SNP calling for Bisulfite-seq data. Genome biology 13 (7):R61. doi:10.1186/gb-2012-13-7-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29 (1):24–26. doi:10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicol JW, Helt GA, Blanchard SG Jr., Raja A, Loraine AE (2009) The Integrated Genome Browser: free software for distribution and exploration of genome-scale datasets. Bioinformatics 25 (20):2730–2731. doi:10.1093/bioinformatics/btp472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evgrafov OV, Wrobel BB, Kang X, Simpson G, Malaspina D, Knowles JA (2011) Olfactory neuroepithelium-derived neural progenitor cells as a model system for investigating the molecular mechanisms of neuropsychiatric disorders. Psychiatr Genet 21 (5):217–228. doi:10.1097/YPG.0b013e328341a2f0 [DOI] [PubMed] [Google Scholar]
- 27.ThePsychEncodeConsortium, Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, Jaffe AE, Pinto D, Dracheva S, Geschwind DH, Mill J, Nairn AC, Abyzov A, Pochareddy S, Prabhakar S, Weissman S, Sullivan PF, State MW, Weng Z, Peters MA, White KP, Gerstein MB, Amiri A, Armoskus C, Ashley-Koch AE, Bae T, Beckel-Mitchener A, Berman BP, Coetzee GA, Coppola G, Francoeur N, Fromer M, Gao R, Grennan K, Herstein J, Kavanagh DH Ivanov NA, Jiang Y, Kitchen RR, Kozlenkov A, Kundakovic M, Li M, Li Z, Liu S, Mangravite LM, Mattei E, Markenscoff-Papadimitriou E, Navarro FC, North N, Omberg L, Panchision D, Parikshak N, Poschmann J, Price AJ, Purcaro M, Reddy TE, Roussos P, Schreiner S, Scuderi S, Sebra R, Shibata M, Shieh AW, Skarica M, Sun W, Swarup V, Thomas A, Tsuji J, van Bakel H, Wang D, Wang Y, Wang K, Werling DM, Willsey AJ, Witt H, Won H, Wong CC, Wray GA, Wu EY, Xu X, Yao L, Senthil G, Lehner T, Sklar P, Sestan N (2015) The PsychENCODE project. Nature neuroscience 18 (12):1707– 1712. doi:10.1038/nn.4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Geen H, Frietze S, Farnham PJ (2010) Using ChIP-seq technology to identify targets of zinc finger transcription factors. Methods in molecular biology 649:437–455. doi:10.1007/978-1-60761-753-2_27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome biology 9 (9):R137. doi:10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Neill LP, Turner BM (2003) Immunoprecipitation of native chromatin: NChIP. Methods 31:76–82 [DOI] [PubMed] [Google Scholar]
- 31.Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC (2015) An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nature communications 6:6033. doi:10.1038/ncomms7033 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.