Abstract
Objective
An unconventional population of CD4+SLAMF7+ cytotoxic TEM cells (CD4+CTLs) has been linked causally to IgG4-related disease (IgG4-RD). Glucocorticoids represent the first line therapeutic approach in patients with IgG4-RD but their mechanism of action in this specific condition remains unknown. Here we describe the impact of glucocorticoids on CD4+CTLs in IgG4-RD.
Methods
CD8α, granzyme A, perforin, and SLAMF7 expression within the effector/memory compartment of CD45RO (TEM) and CD45RA (TEMRA) CD4+ T cells was quantified by flow cytometry in 18 active IgG4-RD patients at baseline and after 6 months of glucocorticoid treatment. Eighteen healthy subjects were studied as controls. Next-generation sequencing of the T-cell receptor α and β chain gene was performed on circulating CD4+CTLs in patients with IgG4-RD before and after treatment, and in affected tissues.
Results
Circulating CD4+ TEM and TEMRA cells were not expanded in IgG4-RD patients compared to healthy controls. CD4+SLAMF7+ TEM cells (but not TEMRA cells) were significantly increased among IgG4-RD patients. Within CD4+SLAMF7+ TEM cells, CD8α− but not CD8αlow cells were elevated in IgG4-RD patients. The same dominant clones of CD8α−CD4+SLAMF7+ TEM cells found in the peripheral blood were also identified in affected tissue. Both CD8α− and CD8αlow CD4+SLAMF7+ TEM cells expressed cytolytic molecules. Clonally expanded CD8α− but not CD8αlow CD4+SLAMF7+ TEM cells decreased following glucocorticoid-induced disease remission.
Conclusions
A subset of CD8α−CD4+SLAMF7+ cytotoxic TEM cells is oligoclonally expanded in patients with active IgG4-RD. This population contracts following glucocorticoid-induced remission. Further characterization of this cell population may provide prognostic information and targets for therapeutic intervention.
Keywords: IgG4, IgG4-related disease, CD4 cytotoxic T cells, glucocorticoids, cytomegalovirus
INTRODUCTION
IgG4-related disease (IgG4-RD) is an increasingly recognized immune-mediated condition that includes a variety of clinical manifestations previously regarded as unique, single-organ disorders unrelated to each other. The clinical manifestations of IgG4-RD include autoimmune pancreatitis, retroperitoneal fibrosis, chronic sialadenitis, and hypertrophic pachymeningitis, among others [1,2]. IgG4-RD is also commonly referred to as a fibro-inflammatory disorder characterized by tumefactive lesions, frequent elevation of serum IgG4 levels, and tissue fibrosis [1]. Glucocorticoids represent the treatment of choice to induce IgG4-RD remission but their effect on the cells orchestrating the disease remains unknown [1]. Likewise, the pro-fibrotic mechanisms driving IgG4-RD remain poorly elucidated.
Recently, the accepted view of IgG4-RD as a Th2-mediated disorder has been questioned by the identification of an unusual population of clonally-expanded “CD4+ cytotoxic T cells” (CD4+CTLs) that accumulate within affected tissues [3–5]. In contrast to the lack of correlation with Th2 lymphocytes, in fact, CD4+ CTLs appear linked to IgG4-RD activity because they are increased in the peripheral blood of subjects with active disease and decrease following rituximab-induced remission despite their lack of CD20 expression [3–8]. They express CD8αα homodimers in addition to CD4 and share several features with CD8+ CTLs (which express CD8 αβ heterodimers as well as CD8 αα homodimers), including the production of cytolytic molecules [4,9]. Moreover, CD4+ CTLs in patients with IgG4-RD secrete pro-fibrotic cytokines (such as IL-1β, INF-γ, and TGF-β) and express signaling lymphocytic activation molecule family member 7 (SLAMF7), a surface antigen implicated in homotypic interactions with activated B cells and disease immunopathogenesis [4,10]. Thus, because CD4+ CTLs have the potential both to stimulate fibroblast activation and to interact with antigen presenting B cells, these cells may play a central role in the pathogenesis of IgG4-RD [1].
In order to better clarify the mechanisms of action of glucocorticoids in IgG4-RD and the pathogenic relevance of CD4+ CTLs, we herein aim to describe the effects of corticosteroid treatment on CD4+ CTLs. Glucocorticoids, in fact, offer a unique angle to observe variations of putatively pathogenic T lymphocytes because they have been shown to not drastically perturb B and T cell counts in the peripheral blood [11,12]. Rituximab, on the other hand, reduces the circulating levels of activated CD4+ T lymphocytes in a global manner by abrogating the survival and differentiation signals provided by naïve B cells [13,14]. In the present work, we also identify a population of CD8α-negative CD4+SLAMF7+ effector/memory T (TEM) cells - corresponding to a subset of CD4+ CTLs - that are oligoclonally expanded in the peripheral blood of patients with active IgG4-RD and contract with disease remission.
PATIENTS AND METHODS
Patients
Eighteen consecutive patients with active untreated IgG4-RD referred to the San Raffaele Scientific Institute were included in this observational prospective study [15]. IgG4-RD was diagnosed according to the “Consensus Statement on the Pathology of IgG4-RD” and to the “Comprehensive diagnostic criteria for IgG4-RD” [16,17]. Patients with isolated pancreatic involvement who did not undergo pancreatic biopsy were diagnosed with “definite” IgG4-RD according to the “International consensus diagnostic criteria for autoimmune pancreatitis” [18]. All patients were initially treated with prednisone (0.6-1 mg/kg) for one month; prednisone was gradually tapered and withdrawn after six months in accordance with international guidelines [14]. Blood for serological, immunological studies, and clinical correlations was drawn at baseline and 6 months after the institution of glucocorticoid treatment. Eighteen healthy age- and sex-matched subjects were used as controls. All patients gave written informed consent for the analyses performed. The study was conducted according to the Declaration of Helsinki and approved by the Ethical Committee of the San Raffaele Scientific Institute.
Disease activity
IgG4-RD activity was assessed by means of the IgG4-RD responder index (RI). Active disease was defined by an IgG4-RD RI ≥ 3. Disease remission was defined by an IgG4-RD RI < 3. A reduction of the IgG4-RD RI of ≥ 2 points still with a total score ≥ 3 was considered a partial response to glucocorticoid treatment [19].
Flow cytometry
Peripheral blood mononuclear cells (PBMCs) were isolated from IgG4-RD subjects and healthy controls by Ficoll density-gradient centrifugation (GE Healthcare), re-suspended in fetal bovine serum containing 10% dimethyl sulfoxide and cryopreserved. For T cells surface staining, cells were stained at 4°C for 30 minutes in staining buffer containing optimized concentrations of the following fluorochrome-conjugated anti-human antibodies (purchased from Biolegend, San Diego, CA, unless otherwise specified): CD4-PE (clone RPA-T4, BD Biosciences, San Jose, CA), CD4-PE/Cy7 (clone OKT4), CD8α-FITC (clone HIT8a, BD Biosciences), CD8α-PE (clone SK1), CD27-APCCy7 (clone 0323), CD27-BV510 (clone MT271), CD45RO-Pacific Blue (clone UCHLa), CD45RA-BV605 (clone Hl100, BD Biosciences), CD62L-PECy7 (clone DREG-56), CD319/SLAMF7-APC (clone 162.1), CD319/SLAMF7-Alexa Fluor 647 (clone 235614, BD Biosciences). Viable cells were identified among those negative for the 7-aminoactinomycin D Viability Staining Solution (Biolegend). For intracellular staining of granzyme A- Pacific Blue (clone CB9d) and perforin-FITC (clone dG9) on T cells, cells were fixed and permeabilized with the BD Cytofix/Cytoperm kit (BD Biosciences, San Jose, CA), according to the manufacturer’s instructions and then stained in permeabilization buffer at 4°C for 30 minutes. CD45RO+ TEM cells were defined as CD4+CD45RO+CD27lowCD62Llow cells. CD45RA+ effector/memory T cells (TEMRA) were identified as CD4+CD45RA+CD27lowCD62Llow cells (Supplementary Figure). In order to identify CD4+ CTLs we assessed granzyme A and perforin expression among SLAMF7+ cells within the TEM and the TEMRA compartments. Flow cytometry was performed on a BD LSR II (BD Biosciences, San Jose, CA) and the FCS files were analyzed using FlowJo software (version 10). For plasmablasts staining, ten colour flow cytometry (Navios cytometer - Beckman-Coulter, Brea, CA) was readily performed on EDTA whole blood fresh samples using a lyse-no-wash technique (ammonium chloride) and the following panel of directly conjugated antibodies: CD3-FITC, CD56-PE, CD4-ECD, CD138-PC5.5, CD27-PC7, CD20-APC, CD19-A700, CD38-A750, CD8-PB, CD45-KO (Beckman-Coulter, Brea, CA). Plasmablasts were identified within the lymphocyte gate as CD19+CD20−CD27+CD38+bright cells.
Cell sorting and immunofluorescence
CD4+CD45RO+CD27lowCD62LlowCD319/SLAMF7+CD8− were sorted using a Beckman Coulter MoFlo™ XDP instrument (Beckman-Coulter, Brea, CA) from two patients with IgG4-RD at the time of diagnosis (#1 and #5) and after 6 months of glucocorticoids (#1). Cells were cultured overnight in DMEM 10% FBS, glutamine and antibiotics on lab-Tek® Chamber Slides (ThermoFisher, Waltham, MA) previously coated by polylysine (Sigma-Aldrich, MO USA). Cells were fixed in 4% paraformaldehyde for 10 minutes at room temperature followed by incubation for 1 hour with PBS containing 0.1% Triton X100, 10% serum, and 3% BSA (blocking solution). For granzyme staining, cells were incubated with mouse anti-human granzyme A monoclonal antibody (Chemicon, Temecula, CA) overnight at 4°C in PBS 10% BSA followed by a secondary goat anti-mouse Alexafluor-488 conjugated antibody (Invitrogen, Waltham, MA) and Hoechst (for nuclear staining). Unrelated mouse IgG antibody was used in parallel as isotype control. Images were captured using a Perkin Elmer Ultraview ESR Laser Scanning Confocal microscope (Perkin Elmer, MA USA). Linear adjustments of the images were done using Adobe Photoshop CS4.
Next Generation Sequencing
Total RNA was extracted from FFPE tissues of patient #1 (pancreas) and #5 (lacrimal gland and lungs) (Pathology Unit of San Raffaele Institute) with Maxwell 16 LEV RNA FFPE kit (Promega, Madison, WI), according to the manufacturer protocol. RNA extraction from sorted CD8α−CD4+SLAMF7+ CTLs was performed with PicoPure RNA Isolation Kit (ThermoFisher, Waltham, MA). RNA concentration was assessed by Qubit 2.0 Fluorometer (ThermoFisher, Waltham, MA) using Qubit RNA Assay kit (Thermofisher, Waltham, MA). The RNA integrity (RNA Integrity Number, RIN) was evaluated on Agilent 2100 Bioanalyzer with RNA 6000 Nano kit (Agilent Technologies, Santa Clara, CA). High throughput sequencing of the TCR α and β chain repertoire was performed by using a modified RACE PCR protocol [20,21]. TCR-specific cDNA was reverse transcribed using the SmartScribe enzyme (Takara Bio, Mountain View, CA) in the presence of biotinylated primers specific for the constant α and β chain genes and of a barcoded (5bp) template switching primer. Enrichment of the TCR-specific biotinylated cDNA molecules was obtained using streptavidin magnetic beads (Dynabeads, Life Technologies, Woburn, MA). Upon magnetic capture, TCR-specific cDNA was washed and resuspended in 10 μl water. 1 μl cDNA and 1 μl first PCR product were used as input material for the first and second exponential PCR, respectively. In the first exponential PCR, primers specific for the constant α and β chain gene and for the template switching sequence were used. In the second exponential PCR, fusion-primers harboring Illumina MiSeq sequencing adaptors were added (Illumina, San Diego, CA). In particular, a barcoded (10 bp) fusion primer specific for the constant TCR genes was used to tag each individual sample. PCR amplicons were purified using the AmPure beads (Beckman Coulter, Brea, CA). Library quantification was performed by Qubit 2.0 Fluorometer (ThermoFisher, Waltham, MA) using Qubit dsDNA HS kit (ThermoFisher, Waltham, MA) and the fragment length distribution was assessed on Agilent 2100 Bioanalyzer with High Sensitivity DNA kit (Agilent Technologies, Santa Clara, CA). Libraries were sequenced in paired-end mode (300 bp Read 1 + 150 bp Read 2) on a MiSeq platform using MiSeq v3 600 cycles reagents (Illumina, San Diego, CA). After demultiplexing of the sequencing results, analysis of the complementaruty determining region 3 (CDR3) clonotypes was carried out using the MiTCR software [22]. To determine the richness of the TCR repertoire (represented by its total number of species, e.g. CDR3 nucleotidic sequences), we calculated the Shannon Diversity Index as follows: SA = −Σi (ni/N) lg (ni/N), where i is an index that is chosen between 1 and the number of species s, ni is the number of sequencing reads in specie i and N is the total number of reads [23].
Statistical analysis
Statistical analysis was performed with Graphpad Prism 6.0 software (La Jolla, CA, USA). The populations were analyzed for normality distribution with D’Agostino and Pearson test. Comparisons between the different populations were performed by mean of paired and unpaired Mann Whitney and Wilcoxon correction tests. Correlation studies were performed by mean of Pearson or Spearman correlation tests. Linear regression analyses were performed and expressed by mean of R2 values and p values. Results are presented as mean ± range unless otherwise specified. P values < 0.05 were considered statistically significant.
RESULTS
Clinical, serological, and immunological features of the IgG4-RD patients’ cohort
Eighteen patients (11 male, 7 female) with a mean age of 64 years (range, 47-83 years) were included in the study (Table 1). All patients had “definite” IgG4-RD based on the 2011 Consensus Statement on the pathology of IgG4-RD [16]. The pancreas was the most commonly affected organ (10 cases), followed by lymph nodes (six cases), the biliary tree (three cases), the aorta (three cases), and lacrimal glands (two cases). Parotid glands, lung, and submandibular glands were involved in one case each. Seven patients (40%) had two or more organs involved by IgG4-RD. All patients had active IgG4-RD as defined by a mean IgG4-RD RI of 8 (range 6-15; normal < 3). The mean levels of serum IgG4 and circulating plasmablasts were 423 mg/dL (range 156-1.360 mg/dL; normal < 135 mg/dL) and 5.781 cells/mL (range 140-40.840 cells/mL; normal < 653 cells/mL), respectively. Serum IgG4 levels were elevated in all patients. Circulating plasmablasts were elevated in 16 patients (89%).
Table 1.
Clinical, serological, and immunological features of the patients cohort
| Patient # | Age (yrs) | Sex | TEM (cells/mL) | SLAMF7+ TEM (cells/mL) | CD8α−SLAMF7+ TEM (cells/mL) | Plamablasts (cells/mL) | Serum IgG4 (mg/dL) | Organ involvement | IgG4-RD RI |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 83 | F | 226.389 | 188.840 | 146.877 | 9.000 | 1.360 | Pancreas - Biliary tree | 12 |
| 2 | 66 | M | 40.299 | 17.071 | 14.459 | 7.140 | 485 | Biliary tree - Parotid glands – Lymph nodes | 15 |
| 3 | 67 | M | 25.778 | 14.595 | 13.962 | 940 | 720 | Pancreas | 6 |
| 4 | 70 | M | 25.037 | 9.430 | 9.176 | 5.400 | 343 | Pancreas - Lymph nodes | 9 |
| 5 | 72 | M | 46.035 | 31.540 | 29.221 | 250 | 368 | Lacrimanl glands - Lungs | 6 |
| 6 | 70 | M | 239.643 | 131.204 | 129.987 | 3.560 | 221 | Pancreas – Lymph nodes | 9 |
| 7 | 47 | F | 90.251 | 45.535 | 44.635 | 140 | 177 | Pancreas - Lacrimal glands | 9 |
| 8 | 73 | F | 30.749 | 15.619 | 15.558 | 7.000 | 534 | Pancreas – Lymph nodes | 12 |
| 9 | 53 | M | 141.512 | 72.899 | 68.959 | 9.300 | 973 | Lymph nodes | 6 |
| 10 | 63 | M | 101.729 | 48.667 | 45.213 | 810 | 156 | Submandibular glands | 6 |
| 11 | 65 | M | 48.234 | 24.250 | 23.819 | 4.000 | 308 | Pancreas | 6 |
| 12 | 71 | F | 84.439 | 46.744 | 37.063 | 1.000 | 420 | Aorta | 6 |
| 13 | 51 | F | 45.877 | 26.225 | 23.796 | 6.000 | 178 | Aorta | 6 |
| 14 | 53 | F | 38.232 | 16.677 | 15.891 | 1.030 | 173 | Aorta | 6 |
| 15 | 66 | M | 48.115 | 26.576 | 26.040 | 1.760 | 362 | Pancreas | 6 |
| 16 | 75 | M | 93.084 | 47.096 | 38.084 | 2.150 | 314 | Biliary tree | 6 |
| 17 | 51 | F | 18.462 | 9.974 | 9.688 | 40.840 | 253 | Pancreas | 15 |
| 18 | 71 | M | 201.073 | 161.133 | 156.111 | 3.740 | 282 | Pancreas – Lymph nodes | 6 |
Abbreviations: IgG4-related disease responder index (IgG4-RD RI). Normal values: plasmablasts 0-653[7]; serum IgG4 < 135mg/dL; IgG4-RD RI 0-3.
CD4+ TEM and TEMRA cells are not expanded in patients with newly diagnosed, active, treatment-naïve IgG4-RD
In patients with IgG4-RD, CD4+ CTLs have been identified following the observation of expanded circulating CD4+CD45RO+CD27lowCD62Llow TEM cells during active disease [4]. In order to study CD4+ CTLs, we therefore first performed flow cytometry analysis on CD27lowCD62Llow cells within the CD4+CD45RO+ memory T cell compartment. Circulating CD4+ TEM cells were not increased in our cohort of active, untreated IgG4-RD patients compared to age- and sex-matched healthy individuals. In particular, the mean absolute counts of circulating CD4+ TEM cells in IgG4-RD patients and healthy subjects were 85.830 cells/mL (range 18.462-239.643 cells/mL) and 82.385 cells/mL (range 32.832-194.000 cells/mL), respectively (p>0.05). CD4+ TEM cells in IgG4-RD patients and controls represented on average 20% (range 10.4-64.7%) and 18% (range 10.8-46.2%) of the total CD4+CD45RO+ memory T cells, respectively (p>0.05) (Figure 1.A–B). Absolute counts of CD4+ TEM cells did not correlate with the number of organs involved by IgG4-RD, IgG4-RD RI, serum IgG4 levels, nor circulating plasmablasts (data not shown).
Figure 1. CD4+ TEM and TEMRA cells are not expanded in patients with active untreated IgG4-RD.
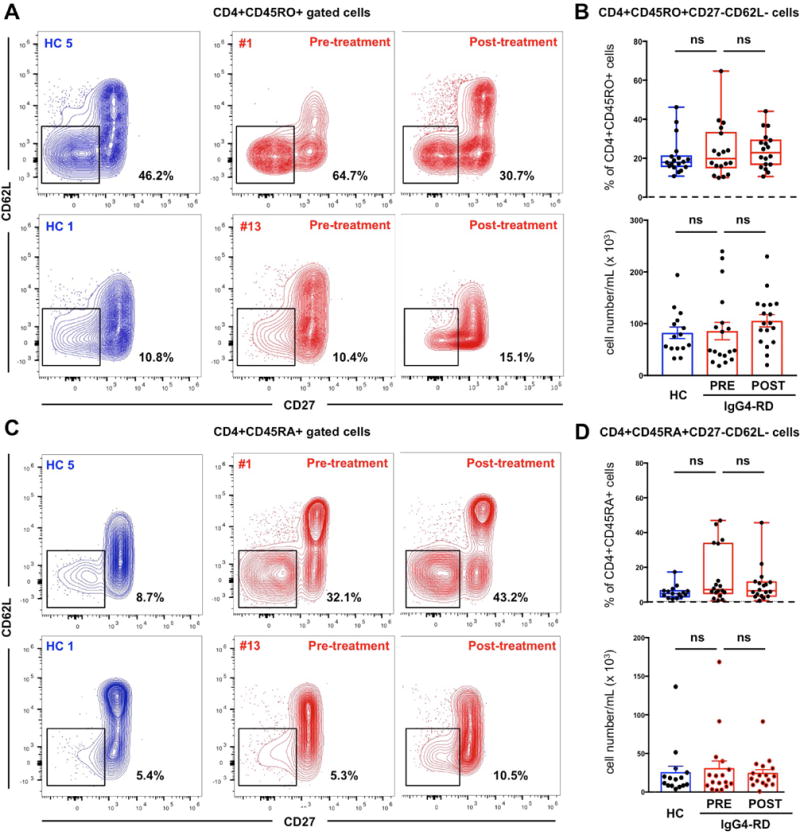
Contour plots showing different frequencies of CD4+CD45RO+CD27lowCD62Llow TEM cells (A) and CD4+CD45RA+CD27lowCD62Llow TEMRA cells (C) in the peripheral blood of two representative healthy controls (HC 1 and HC 5) and two representative patients with IgG4-RD (#1 and #13) before and after glucocorticoid treatment. Box plots of CD4+CD45RO+CD27lowCD62Llow TEM cells (B) and CD4+CD45RA+CD27lowCD62Llow TEMRA cells (D) in the peripheral blood of healthy controls and patients with IgG4-RD displaying mean values and range before and after glucocorticoid treatment. P values are based on the Mann-Whitney test.
CD4+ CTLs were described originally in the setting of chronic viral infections as CD27lowCD62Llow CD4+CD45RA+ effector/memory (TEMRA) cells that acquire a cytolytic gene program after down-regulation of the transcription factor ThPOK (T-helper-inducing POZ/Kruppel-like factor) and up-regulation of Eomesodermin and Runx3 (Runt-related transcription factor 3) [24,25]. We therefore also examined the expansion of CD27lowCD62Llow cells within the CD4+CD45RA+ (TEMRA) compartment by flow cytometry but did not identify significant differences between IgG4-RD patients and healthy subjects (p > 0.05) (Figure 1.C–D).
CD8α−CD4+SLAMF7+ TEM cells are expanded in the peripheral blood of patients with active untreated IgG4-RD
CD4+ CTLs in IgG4-RD have been described as SLAMF7+ expressing cells within the CD4+ TEM compartment [4]. In order to assess the frequency of circulating CD4+ CTLs in patients with active untreated IgG4-RD, we analysed TEM cells for the expression of SLAMF7. The fraction of circulating CD4+SLAMF7+ TEM cells over total TEM cells in IgG4-RD patients was significantly increased compared to healthy individuals (mean 54.1% (range 35.9-83.4%) and 40.5% (range 20.7-66.8%), respectively) (p < 0.01). Absolute counts of circulating CD4+SLAMF7+ TEM cells in IgG4-RD patients were increased compared to healthy individuals but not to a significant extent (mean 51.893 cells/mL (range 9.430-188.840 cells/mL) and 26.570 cells/mL (range 9.740-53.670 cells/mL), respectively) (p = 0.07) (Figure 2.A and C).
Figure 2. CD4+CD8α−SLAMF7+ TEM cells are expanded in patients with active untreated IgG4-RD and decrease following glucocorticoid treatment.
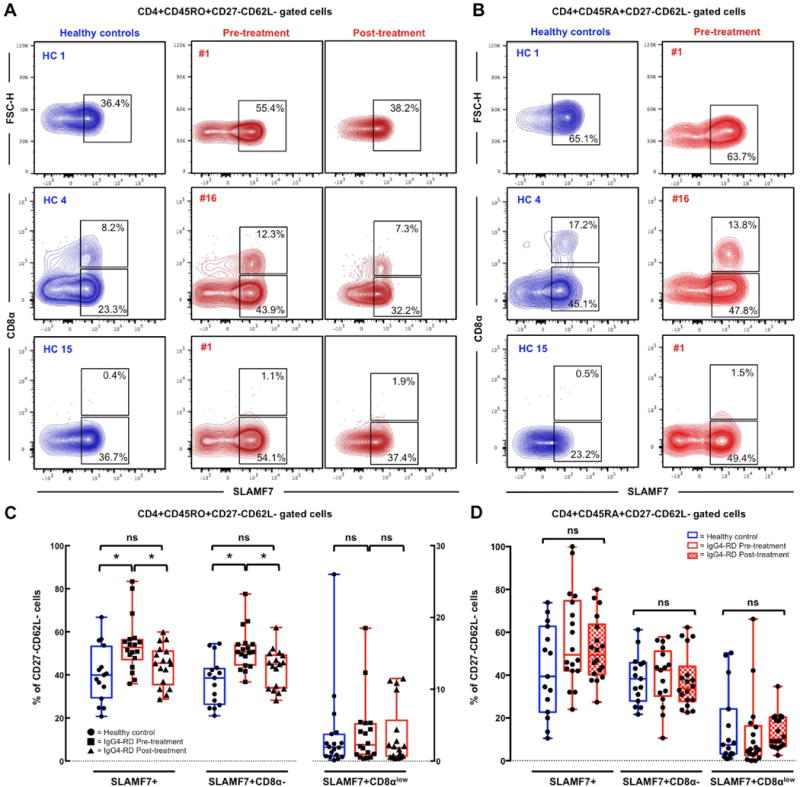
Contour plots showing different frequencies of CD8αlow and CD8α− SLAMF7+CD4+ TEM cells (A) and TEMRA cells (B) in the peripheral blood of representative healthy controls (HC 1, 4, and 15) and patients with IgG4-RD (#1 and 16) before and after glucocorticoid treatment. Box plots of CD8αlow and CD8α− SLAMF7+CD4+ TEM cells (C) and TEMRA cells (D) in the peripheral blood of healthy controls and patients with IgG4-RD displaying mean values and range. P values are based on the Mann-Whitney test. Asterisks indicate a p value < 0.05.
Because CD4+ CTLs have been also shown to express CD8α, we further assessed circulating CD4+SLAMF7+ TEM cells for the expression of this surface marker [24,25]. CD8α negative but not CD8αlowCD4+SLAMF7+ TEM cells were significantly expanded in patients with active IgG4-RD compared to healthy controls, both in absolute numbers and in percentage of TEM cells. In particular, the mean values of circulating CD8α−CD4+SLAMF7+ TEM cells in patients with active IgG4-RD and healthy subjects were 47.141 cells/mL (range 9.176-146.877 cells/mL) and 23.872 cells/mL (range 9.430-42.420 cells/mL), respectively (p = 0.05). The mean fraction of circulating TEM cells that had a CD8α−CD4+SLAMF7+ phenotype was 51.8% (range 37.6-77.6%) in active IgG4-RD patients and 37.1% (range 21.0-53.7%) in healthy controls, (p = 0.0008). The mean value of circulating CD8αlowCD4+SLAMF7+ TEM cells in patients with active IgG4-RD and healthy subjects was 4.478 cells/mL (range 200-41.960 cells/mL) and 2.989 cells/mL (range 160-20.890 cells/mL), respectively (p > 0.05). Circulating CD8αlowCD4+SLAMF7+ TEM cells in patients with active IgG4-RD and healthy subjects represented on average 3.9% (range 0.4-18.5%) and 3.7% (range 0.1-26%) of TEM cells, respectively (p > 0.05) (Figure 2.A and C).
Great variability in the frequency of circulating CD8αlowCD4+SLAMF7+ TEM cells was observed in both IgG4-RD patients and healthy individuals. Indeed, while circulating CD8α−CD4+SLAMF7+ TEM cells were uniformly expanded in IgG4-RD patients compared to controls, circulating CD8αlowCD4+SLAMF7+ TEM cells were present in 10/18 (56%) of both IgG4-RD patients and healthy subjects (Figure 2). Absolute numbers of total CD4+SLAMF7+ TEM cells and CD8α− CD4+SLAMF7+ TEM cells did not correlate with the number of organs involved by IgG4-RD, IgG4-RD RI, serum IgG4 levels, or with circulating plasmablasts (data not shown). In addition, the absolute number as well as the percentage of SLAMF7+ cells within the TEMRA compartment were increased compared to controls but not to a significant extent (Figure 2.B–D). The circulating levels of CD8αlow and CD8α− CD4+SLAMF7+ TEMRA cells did not differ between IgG4-RD patients and healthy subjects (Figure 2.B–D). Of note, CD8αlowCD4+SLAMF7+ TEMRA cells were absent in the same IgG4-RD patients and healthy controls lacking CD8αlowCD4+SLAMF7+ TEM cells (Figure 2.B–D).
CD8α−CD4+SLAMF7+ TEM cells in IgG4-RD are oligoclonally expanded, bear a cytolytic phenotype, and the same clones found in the blood also infiltrate affected organs
In order to compare the cytotoxic phenotype of the expanded CD8α−CD4+SLAMF7+ TEM cells and of CD8αlowCD4+SLAMF7+ TEM cells in patients with IgG4-RD, we assessed the expression of granzyme A and perforin by flow cytometry on both cell populations. Co-expression of the two cytolytic molecules was detected in half of the CD8α−CD4+SLAMF7+ TEM cells (average 51.7%, range 37.1-62.0%) and in almost all CD8αlowCD4+SLAMF7+ TEM cells (Figure 3.A). Granzyme A expression on sorted CD8α−CD4+SLAMF7+ TEM cells was also confirmed by immunofluorescence (Figure 3.B). Of note, the expression levels of granzyme A and perforin in both CD8αlow and CD8α− CD4+SLAMF7+ TEM cells were comparable to that of professional cytotoxic CD8+ T cells (Figure 3.C). Conversely, the expression level of CD8α on CD8αlowCD4+SLAMF7+ TEM cells was comparable in IgG4-RD patients and healthy individuals, but significantly lower than that of professional CD8+ T cells (Figure 3.D).
Figure 3. Expression of cytolytic molecules on CD8α− and CD8αlow CD4+SLAMF7+ TEM cells.
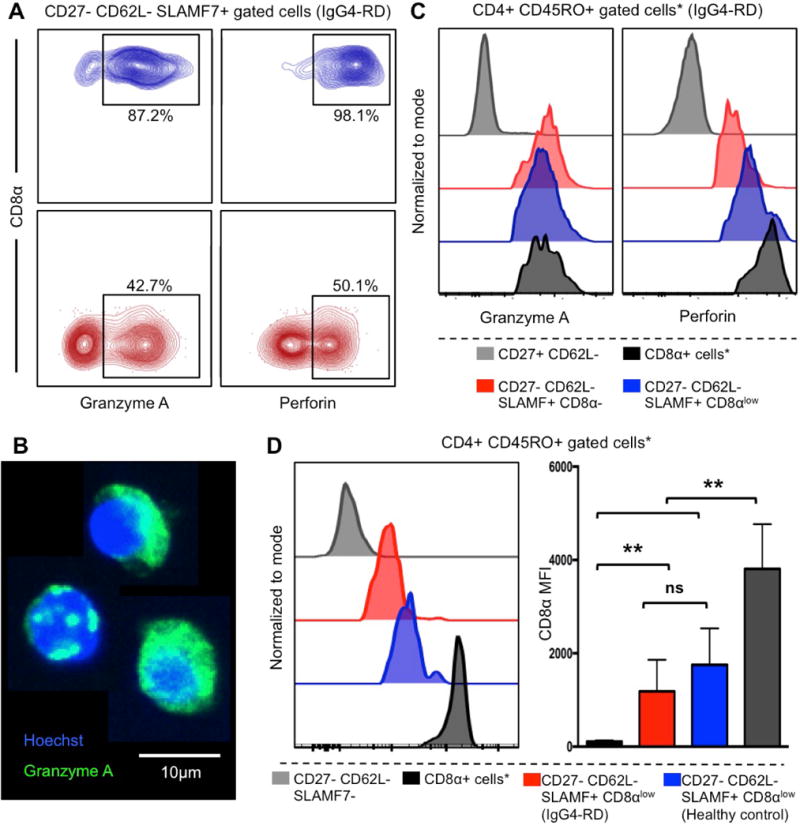
(A) Contour plots showing different frequencies of granzyme A and perforin positive CD8α− and CD8αlow CD4+ TEM SLAMF7+ CTLs in a patient with IgG4-RD representative of n=10 independent experiments. (B) Immunofluorescence on bulk sorted CD8α− CD4+ TEM SLAMF7+ CTLs showing granzyme A production. (C) Histograms showing the expression of granzyme A and perforin on CD8α− and CD8αlow CD4+ TEM SLAMF7+ CTLs in in a patient with IgG4-RD representative of n=10 independent experiments. *Gated on CD8+ T lymphocytes. (D) Expression level of CD8α on CD8αlowCD4+SLAMF7+ CTLs in a representative patient with IgG4-RD and in a representative healthy control (histograms), and in the study cohort presented as Median Fluorescence Intensity (MFI) (* gated on CD8+ T lymphocytes; ** p < 0.01).
To gain clues about CD8α−CD4+CTLs involvement in IgG4-RD pathogenesis, we profiled Patient’s #1 and #5 TCR α and β chain repertoire in the peripheral blood and looked for shared clones in tissue biopsies obtained before treatment at the time of diagnosis. Comprehensive analysis of the TCR α and β chain nucleotide sequences showed an oligoclonal TCRαβ repertoire of circulating CD8α−CD4+SLAMF7+ TEM cells and CDR3 clonotypes that were identical to those sequenced within the tissue in each patient (Figure 4.A–B).
Figure 4. Clinical improvement induced by glucocorticoid treatment correlates with contraction of clonally expanded circulating CD8α−CD4+SLAMF7+ TEM cells.
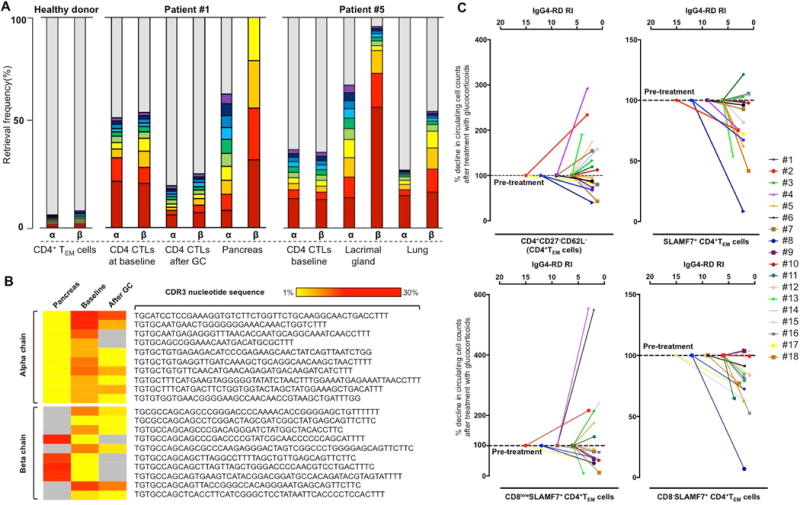
(A) Retrieval frequency of CDR3 amino acid sequences of TCRα and β chain from circulating CD8α-CD4+SLAMF7+ TEM cells before and after treatment and in the pancreas of Patient #1, and in the blood, lacrimal gland, and lung tissues of Patient #5 show an oligoclonal pattern on Next-Generation Sequencing analysis. Circulating CD4+ TEM cells from a healthy donor are polyclonal. Red bars indicate the most frequently encountered sequences; violet bars represent the 10th most frequently encountered sequences; grey bars represent overall remaining CDR3 sequences. The higher the grey bar, the more polyclonal is the sample. Patient #1 also shows an increased polyclonality of the TCRα and β chain repertoire after treatment. (B) Ten most frequent shared CDR3 nucleotide sequences of the TCR α and β chain between circulating CD8α-CD4+SLAMF7+ TEM cells and pancreatic tissue of Patient #1. The frequency of clones expanded at baseline decreases after glucocorticoid treatment together with clinical improvement. Gray panels indicate missing nucleotide sequences. (C) Percentage decrease of circulating CD4+ TEM cells, total SLAMF7+CD4+CTLs, CD8αlowCD4+SLAMF7+CTLs, and CD8α-CD4+SLAMF7+CTLs after six months of glucocorticoids (normalized to pre-treatment levels) is plotted against the IgG4-RD RI.
Clonally expanded CD8α−CD4+SLAMF7+ TEM cells contracts following glucocorticoid induced disease remission
Six months after the institution of glucocorticoid treatment, we performed a second flow cytometry analysis to assess the effects of glucocorticoids on CD4+ TEM cells and CD4+SLAMF7+ TEM cells. At that time-point, the mean IgG4-RD RI value was 2 (range 1-4), consistently with a clinical improvement in all patients. Fifteen patients were in remission and three had achieved a partial response.
The mean value of circulating CD4+ TEM cells was 105.672 cells/mL (range 38.501-229.949 cells/mL), representing on average 23.9% of total CD4+CD45RO+ memory T cells (range 10.6-44.1%), and did not differ from pre-treatment level (paired p > 0.05) (Figure 1.A–B). Following clinical improvement, circulating CD4+SLAMF7+ TEM cells decreased to a mean value of 46.585 cells/mL (range 2.304-104.390 cells/mL) and to an average of 45.5% of total CD4+ TEM (range 28.7-59.9%) (paired p < 0.05 compared to baseline values) (Figure 2.C and 3.A). The mean percentage decline of circulating SLAMF7 expressing CD4+ TEM cells over pre-treatment level was 19%.
Among CD4+SLAMF7+ TEM cells, the counts of CD8α-negative lymphocytes decreased selectively and reached levels comparable to healthy individuals following clinical improvement (Figure 3.C). In particular, circulating CD8α−CD4+SLAMF7+ CTLs decreased to an average of 42.8% of total CD4+ TEM cells (range 28.2-50.4%) (paired p = 0.001 compared to baseline value). The mean percentage decline of circulating CD8α−CD4+SLAMF7+ CTLs compared to pre-treatment level was 28%. CD8αlowCD4+SLAMF7+ TEM cells were not affected by glucocorticoid treatment (Figure 2.C and 3.C). The reduction of CD4+SLAMF7+ TEM cells after glucocorticoids, therefore, was mainly due to the selective decrease of the CD8α negative subset of CD4+ SLAMF7+ TEM cells (Figure 4.C). In a single patient in which the analysis was performed, the TCRαβ chain repertoire of CD8α−CD4+SLAMF7+ CTLs showed a decrease in the frequency of the initially expanded clones corresponding with an increase in the repertoire diversity (Figure 4.A–B). No significant effect of glucocorticoids were observed on the TEMRA compartment.
DISCUSSION
The concept of “cytotoxicity associated with CD4+ T-helper lymphocytes” has steadily emerged in recent years, in contrast with the traditional view of CTLs originating only from major histocompatibility complex (MHC) class I-restricted CD8+ T lymphocytes. CD4+ CTLs, which represent uncommon subsets of highly differentiated CD4+ TEM and CD4+ TEMRA lymphocytes that retain the ability of killing target cells in a MHC class II-dependent manner, arise in response to repeated antigen stimulation [24,25]. In particular, CD4+ CTLs have been extensively characterized in the setting of chronic viral infections such as cytomegalovirus, dengue, HIV, hepatitis B and hepatitis C viruses, where they likely complement CD8+ and natural killer T cell immune responses [26–28]. These cells appear to be of particular importance and superior to CD8+ cytotoxic T cells in maintaining viral latency [31]. CD4+ CTLs have been also identified in autoimmune diseases such as rheumatoid arthritis, inflammatory bowel disease, and systemic sclerosis, indicating that they might contribute to chronic inflammation and fibrosis, as well [24,25,29]. More recently, we contributed to the identification of CD4+ CTLs in the increasingly recognized fibro-inflammatory condition IgG4-RD, and further characterized them as SLAMF7 expressing cells within the CD4+ TEM compartment [4]. Indeed, SLAMF7+CD4+ TEM cells exhibited distinctive features of CD8+ CTLs, such as (i) expression of the surface antigen CD8α; (ii) expression of the transcription factors Eomes and Runx3; (iii) secretion of the cytolytic molecules granzyme A and perforin; and (iv) in vitro cytotoxic activity [4].
In the present work, we describe for the first time the effects of glucocorticoids on CD4+ CTLs, and demonstrate that the expression of CD8α identifies at least two distinct subsets of CD4+SLAMF7+ CTLs within the TEM and TEMRA compartments of patients with IgG4-RD. CD8α-negative but not CD8αlow CDCD4+SLAMF7+ TEM CTLs correlate with IgG4-RD activity as the former, in contrast to the latter, are increased in the peripheral blood of patients with active untreated disease, express TCR clones also found in affected lesions, and appear to be selectively modulated with disease remission after glucocorticoids treatment. Indeed, the finding of common CDR3 sequences expressed by dominant circulating CD8α−CD4+SLAMF7+ CTLs clones and tissue infiltrating lymphocytes strongly supports a pathophysiological relevance of these cells in IgG4-RD because it indicates that antigen specific cytotoxic T-cells activated in secondary lymphoid organs are directly present at the site of active disease. In this sense, CD8α−CD4+SLAMF7+ CTLs might represent the intralesional SLAMF7+ granzyme A+CD4+ T cells that have been previously shown to infiltrate IgG4-RD tissues, although further studies are warranted to address this issue [5]. Definitive confirmation of this finding would require TCR repertoire analysis on CD8α−CD4+SLAMF7+ CTLs sorted directly from fresh IgG4-RD biopsies.
The response of CD8α−CD4+SLAMF7+ TEM cells to glucocorticoids also represents an important finding above and beyond the previously reported effect of rituximab on CD4+SLAMF7+ CTLs in patients with IgG4-RD [4]. By depleting CD20+ naïve B cells, in fact, rituximab is presumed to deprive activated CD4+ T lymphocytes of B cell-derived survival signals, thus limiting the expansion of both pathogenic and bystander T cell clones [11,12]. Accordingly, the decrease of CD4+SLAMF7+ CTLs counts that we previously observed in IgG4-RD patients treated with rituximab might have been a general consequence of B cell depletion on activated CD4+ T cells. Conversely, glucocorticoids have minimal effects in the long-term on circulating CD19+ and CD20+ B cells in IgG4-RD patients, offering a different perspective to identify alterations of putatively pathogenic cells eventually associated with disease remission [30]. Therefore, the selective decrease of CD8α−CD4+SLAMF7+ but not of CD8αlowCD4+SLAMF7+ CTLs induced by glucocorticoid treatment further strengthens the pathogenic link between CD8α−CD4+SLAMF7+ CTLs and IgG4-RD, and unveils a potentially relevant mechanism behind the efficacy of glucocorticoids in patients with this condition. The reduction of the frequency of expanded CD8α−CD4+SLAMF7+ T cell clones that we observed in a single patient after glucocorticoids also supports a contraction of antigen specific T lymphocytes but deserves additional confirmation on a larger number of patients.
On the other hand, CD8αlowCD4+CTLs represented a minor population within the expanded CD4+SLAMF7+ TEM cell pool in IgG4-RD patients, and could also be found in healthy individuals, arguing against a causal role in IgG4-RD inflammation and fibrosis. In particular, while circulating CD8α−CD4+SLAMF7+ TEM cells appeared uniformly expanded only in patients with active IgG4-RD, CD8αlowCD4+SLAMF7+ T cells were detectable in 50 to 60% of both patients and controls. These results, therefore, suggest that CD8αlowCD4+CTLs may not be linked to active IgG4-RD per se but may reflect an underlying chronic disease or pre-existing immune responses to common chronic viral infections (e.g. cytomegalovirus, hepatitis B or hepatitis C viruses), as it occurs in otherwise healthy subjects. Activation assays with viral peptides would be necessary to confirm this hypothesis and to address the antigen specificity of CD8αlow compared to CD8α−CD4+SLAMF7+ TEM cells.
In conclusion, we report for the first time that glucocorticoid-induced remission in patients with IgG4-RD is associated with a decrease of circulating CD4+CTLs, a recently identified potentially pathogenic population of T lymphocytes. Moreover, we herein identify a CD8α− subpopulation of CD4+SLAMF7+ CTLs that better associates with IgG4-RD as it is selectively depleted by glucocorticoids. Additional studies are needed to address the biological relationship between CD8α−CD4+SLAMF7+ TEM and TEMRA cells in IgG4-RD, to functionally characterize them, and to understand the pathogenic relevance of concomitant chronic viral infections in patients with IgG4-RD.
Supplementary Material
Supplementary Figure. Gating strategies for the identification of “CD4+ T effector memory” and “CD4+ T effector memory CD45RA+“ cells.
Acknowledgments
This work was supported by a “Fondazione Italiana per la Ricerca sull’Artrite (FIRA Onlus) (2014)” award to EDT, by a “TRIDEO 2014” award to EDT from the “Italian Association for Cancer Research (AIRC)/Cariplo Foundation”, and by an Autoimmune Center of Excellence U19 AI 110495 award to SP from the National Institutes of Health. EDT received support from the “Collegio Ghislieri” (Pavia, Italy). The authors are grateful to Dr. Raffaella Milani (Hematology and Bone Marrow Transplant Unit - IRCCS San Raffaele Scientific Institute, Milan, Italy) and to Dr. Monica Romanò (FRACTAL - Flow cytometry Resource Advanced Cytometry Technical Applications Laboratory - IRCCS San Raffaele Scientific Institute, Milan, Italy) for helpful discussion, and to Dr. Antonella Monno (Division of Immunology, Transplantation and Infectious Disease, IRCCS San Raffaele Scientific Institute, Milan, Italy) for acquisition of immunofluorescence pictures.
Footnotes
DISCLOSURE - The authors have not received any financial support or other benefits from commercial sources for the work reported on in the manuscript, or any other financial interests that could create a potential conflict of interest or the appearance of a conflict of interest with regard to the work.
References
- 1.Della-Torre E, Lanzillotta M, Doglioni C. Immunology of IgG4-related disease. Clin Exp Immunol. 2015;181:191–206. doi: 10.1111/cei.12641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu LX, Della-Torre E, Stone JH, Clark SW. IgG4-related hypertrophic pachymeningitis: clinical features, diagnostic criteria, and treatment. JAMA Neurol. 2014;71:785–93. doi: 10.1001/jamaneurol.2014.243. [DOI] [PubMed] [Google Scholar]
- 3.Mattoo H, Della-Torre E, Mahajan VS, Stone JH, Pillai S. Circulating Th2 memory cells in IgG4-related disease are restricted to a defined subset of subjects with atopy. Allergy. 2014;69:399–402. doi: 10.1111/all.12342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattoo H, Mahajan VS, Maehara T, Deshpande V, Della-Torre E, Wallace ZS, et al. Clonal expansion of CD4(+) cytotoxic T lymphocytes in patients with IgG4-related disease. J Allergy Clin Immunol. 2016;138:825–38. doi: 10.1016/j.jaci.2015.12.1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maehara T, Mattoo H, Ohta M, Mahajan VS, Moriyama M, Yamauchi M, et al. Lesional CD4+ IFN-γ+ cytotoxic T lymphocytes in IgG4-related dacryoadenitis and sialoadenitis. Ann Rheum Dis. 2017;76:377–385. doi: 10.1136/annrheumdis-2016-209139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Della-Torre E, Feeney E, Deshpande V, Mattoo H, Mahajan V, Kulikova M, et al. B-cell depletion attenuates serological biomarkers of fibrosis and myofibroblast activation in IgG4-related disease. Ann Rheum Dis. 2015;74:2236–43. doi: 10.1136/annrheumdis-2014-205799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallace ZS, Mattoo H, Carruthers M, Mahajan VS, Della Torre E, Lee H, et al. Plasmablasts as a biomarker for IgG4-related disease, independent of serum IgG4 concentrations. Ann Rheum Dis. 2015;74:190–5. doi: 10.1136/annrheumdis-2014-205233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mattoo H, Mahajan VS, Della-Torre E, Sekigami Y, Carruthers M, Wallace ZS, et al. De novo oligoclonal expansions of circulating plasmablasts in active and relapsing IgG4-related disease. J Allergy Clin Immunol. 2014;134:69–87. doi: 10.1016/j.jaci.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Detre C, Keszei M, Romero X, Tsokos GC, Terhorst C. SLAM family receptors and the SLAM-associated protein (SAP) modulate T cell functions. Semin Immunopathol. 2010;32:157–71. doi: 10.1007/s00281-009-0193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JK, Mathew SO, Vaidya SV, Kumaresan PR, Mathew PA. CS1 (CRACC, CD319) induces proliferation and autocrine cytokine expression on human B lymphocytes. J Immunol. 2007;179:4672–8. doi: 10.4049/jimmunol.179.7.4672. [DOI] [PubMed] [Google Scholar]
- 11.Olnes MJ, Kotliarov Y, Biancotto A, Cheung F, Chen J, Shi R, et al. CHI Consortium Effects of Systemically Administered Hydrocortisone on the Human Immunome. Sci Rep. 2016;6:23002. doi: 10.1038/srep23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khosroshahi A, Wallace ZS, Crowe JL, Akamizu T, Azumi A, Carruthers MN, et al. International Consensus Guidance Statement on the Management and Treatment of IgG4-Related Disease. Arthritis Rheumatol. 2015;67:1688–99. doi: 10.1002/art.39132. [DOI] [PubMed] [Google Scholar]
- 13.Lavielle M, Mulleman D, Goupille P, Bahuaud C, Sung HC, Watier H, et al. Repeated decrease of CD4+ T-cell counts in patients with rheumatoid arthritis over multiple cycles of rituximab treatment. Arthritis Res Ther. 2016;18:253. doi: 10.1186/s13075-016-1152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford A, Macleod M, Schumacher T, Corlett L, Gray D. Primary T cell expansion and differentiation in vivo requires antigen presentation by B cells. J Immunol. 2006;176:3498–506. doi: 10.4049/jimmunol.176.6.3498. [DOI] [PubMed] [Google Scholar]
- 15.Campochiaro C, Ramirez GA, Bozzolo EP, Lanzillotta M, Berti A, Baldissera E, et al. IgG4-related disease in Italy: clinical features and outcomes of a large cohort of patients. Scand J Rheumatol. 2016;45:135–45. doi: 10.3109/03009742.2015.1055796. [DOI] [PubMed] [Google Scholar]
- 16.Deshpande V, Zen Y, Chan JK, Yi EE, Sato Y, Yoshino T, et al. Consensus statement on the pathology of IgG4-related disease. Mod Pathol. 2012;25:1181–92. doi: 10.1038/modpathol.2012.72. [DOI] [PubMed] [Google Scholar]
- 17.Umehara H, Okazaki K, Masaki Y, Kawano M, Yamamoto M, Saeki T, et al. Comprehensive diagnostic criteria for IgG4-related disease (IgG4-RD), 2011. Mod Rheumatol. 2012;22:21–30. doi: 10.1007/s10165-011-0571-z. [DOI] [PubMed] [Google Scholar]
- 18.Shimosegawa T, Chari ST, Frulloni L, Kamisawa T, Kawa S, Mino-Kenudson M, et al. International consensus diagnostic criteria for autoimmune pancreatitis: guidelines of the International Association of Pancreatology. Pancreas. 2011;40:352–8. doi: 10.1097/MPA.0b013e3182142fd2. [DOI] [PubMed] [Google Scholar]
- 19.Carruthers MN, Stone JH, Deshpande V, Khosroshahi A. Development of an IgG4-RD Responder Index. Int J Rheumatol. 2012;2012:259408. doi: 10.1155/2012/259408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bolotin DA, Mamedov IZ, Britanova OV, Zvyagin IV, Shagin D, Ustyugova SV, et al. Next generation sequencing for TCR repertoire profiling: platform-specific features and correction algorithms. Eur J Immunol. 2012;42:3073–83. doi: 10.1002/eji.201242517. [DOI] [PubMed] [Google Scholar]
- 21.Ruggiero E, Nicolay JP, Fronza R, Arens A, Paruzynski A, Nowrouzi A, et al. High-resolution analysis of the human T-cell receptor repertoire. Nat Commun. 2015;6:8081. doi: 10.1038/ncomms9081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolotin DA, Shugay M, Mamedov IZ, Putintseva EV, Turchaninova MA, Zvyagin IV, et al. MiTCR: software for T-cell receptor sequencing data analysis. Nat Methods. 2013;10:813–4. doi: 10.1038/nmeth.2555. [DOI] [PubMed] [Google Scholar]
- 23.Shannon CE. The mathematical theory of communication. MD Comput. 1963;14:306–17. [PubMed] [Google Scholar]
- 24.Tian Y, Sette A, Weiskopf D. Cytotoxic CD4 T Cells: Differentiation, Function, and Application to Dengue Virus Infection. Front Immunol. 2016;7:531. doi: 10.3389/fimmu.2016.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oja AE, Vieira Braga FA, Remmerswaal EB, Kragten NA, Hertoghs KM, Zuo J, et al. The Transcription Factor Hobit Identifies Human Cytotoxic CD4+ T Cells. Front Immunol. 2017;8:325. doi: 10.3389/fimmu.2017.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zloza A, Schenkel JM, Tenorio AR, Martinson JA, Jeziorczak PM, Al-Harthi L. Potent HIV-specific responses are enriched in a unique subset of CD8+ T cells that coexpresses CD4 on its surface. Blood. 114:3841–53. doi: 10.1182/blood-2009-02-202481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nascimbeni M, Pol S, Saunier B. Distinct CD4+ CD8+ double-positive T cells in the blood and liver of patients during chronic hepatitis B and C. PLoS One. 2011;6:e20145. doi: 10.1371/journal.pone.0020145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nascimbeni M, Shin EC, Chiriboga L, Kleiner DE, Rehermann B. Peripheral CD4(+)CD8(+) T cells are differentiated effector memory cells with antiviral functions. Blood. 2004;104:478–86. doi: 10.1182/blood-2003-12-4395. [DOI] [PubMed] [Google Scholar]
- 29.Parel Y, Aurrand-Lions M, Scheja A, Dayer JM, Roosnek E, Chizzolini C. Presence of CD4+CD8+ double-positive T cells with very high interleukin-4 production potential in lesional skin of patients with systemic sclerosis. Arthritis Rheum. 2007;56:3459–67. doi: 10.1002/art.22927. [DOI] [PubMed] [Google Scholar]
- 30.Lanzillotta M, Della-Torre E, Milani R, Bozzolo EP, Bozzalla-Cassione E, Rovati L, et al. Effects of Glucocorticoids on B-cell Subpopulations in patients with IgG4-Related Disease. Submitted. [PubMed] [Google Scholar]
- 31.Mason G, Jackson S, Okecha G, Poole E, Sissons JG, Sinclair J, et al. Human Cytomegalovirus Latency-Associated Proteins Elicit Immune-Suppressive IL-10 Producing CD4+ T Cells. PLOS, Pathog. 9(10):e1003635. doi: 10.1371/journal.ppat.1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure. Gating strategies for the identification of “CD4+ T effector memory” and “CD4+ T effector memory CD45RA+“ cells.


