Dear Editor,
Heterochromatin, the highly-condensed nucleosome arrays in chromosomes, plays an important role in regulating multiple cellular events including transcriptional silencing and chromosome segregation (Allis and Jenuwein, 2016). Aberrant heterochromatin formation is linked to tumor progression, as well as other severe physiological disorders (Hahn et al., 2010). However, the mechanism by which factors contribute to heterochromatin formation remains elusive.
In Saccharomyces cerevisiae, heterochromatin forms at two hidden mating-type loci (HM loci, namely HMR and HML), telomeres and rRNA-encoding DNA (rDNA) region (Grunstein and Gasser, 2013). Besides the four silent information regulators, Sir1p–Sir4p, it was reported that histone chaperones are also required for heterochromatin formation. For instance, H3–H4 chaperones chromatin assembly factor 1 (CAF-1), anti-silencing function protein 1 (Asf1p), histone transcription regulator 1 (Hir1p), and regulator of Ty1 transposition protein 106 (Rtt106p) are important for heterochromatin silencing (Enomoto and Berman, 1998; Huang et al., 2005; Huang et al., 2007; Sharp et al., 2001). Genetic studies revealed two parallel pathways among these histone chaperones: Asf1p, Hir1p, and Rtt106p seem to mediate a genetically distinguishable silencing pathway from CAF-1. Interestingly, cells deficient in both pathways are only partially defective in heterochromatin silencing, which suggests other factors might also be involved (Huang et al., 2007).
Histone chaperone complex FACT (facilitates chromatin transcription/transaction), consisting of two essential subunits, Spt16p and Pob3p (SSRP1 in humans), plays a critical role in transcription, replication, and other chromatin based processes in eukaryotes (Formosa, 2012). While both Spt16p and Pob3p are essential in S. cerevisiae, Pob3p is dispensable in Schizosaccharomyces pombe, and contributes to heterochromatin silencing (Lejeune et al., 2007). However, whether FACT also has a role in heterochromatin silencing in S. cerevisiae remains to be determined. Here, we report that Spt16p functions redundantly with CAF-1 and Rtt106p in heterochromatin silencing and is required for heterochromatin formation at both mating-type loci and telomeres in budding yeast.
To identify other histone chaperones that might function in heterochromatin silencing, we performed a small-scale candidate gene screen using a GFP-based reporter gene silencing assay (Fig. 1A). The expression of GFP gene inserted at the silent HMR locus (hmr::GFP) was used to monitor the degree of silencing by quantifying the percentage of yeast cells expressing GFP (Fig. 1A). Yeast cells harboring double deletion of CAC1 (the large subunit of histone chaperone CAF-1) and RTT106 was made as a query strain (Fig. 1B). Consistent with prior studies, 47.5% of cac1Δrtt106Δ cells lost GFP silencing while only 0.4% wild-type cells expressed GFP, likely from the background. As a positive control, sir3Δ almost completely abolishes cells’ ability in heterochromatin silencing (99.8%). Interestingly, spt16-m, a newly identified Spt16p mutant allele in our group, dramatically exacerbates the phenotype of cac1Δrtt106Δ (77.3%) (Fig. 1C). The spt16-m allele contains two amino acids substitution (K692AR693A) in its tandem pleckstrin homology (PH) domain and displays a subtle defect in transcription initiation but a substantial defect in DNA replication-coupled nucleosome assembly (Yang et al., 2016). Thus, this result indicates that Spt16p may also be involved in heterochromatin silencing.
Figure 1.
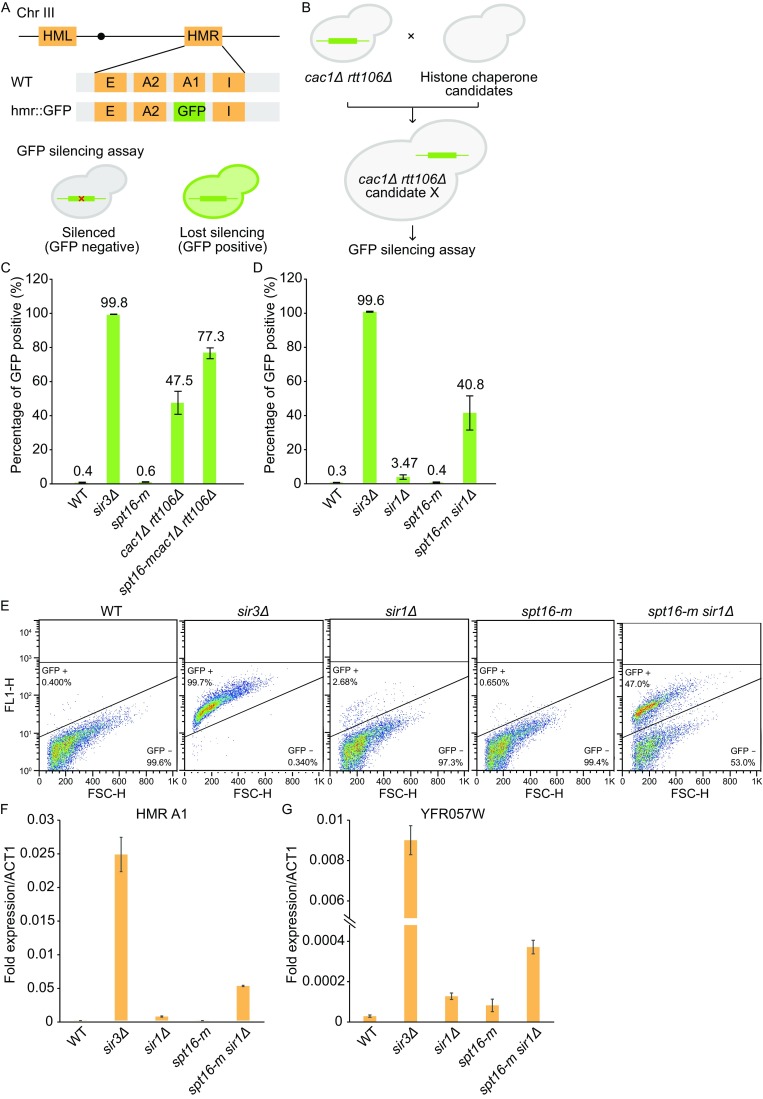
Histone chaperone Spt16p is required for heterochromatin silencing in S. cerevisiae. (A) A scheme for the GFP silencing assay. Top: A diagram of yeast chromosome III (Chr III) in either wild-type (WT) strain or strain used for GFP silencing assay (hmr::GFP). In WT cells, HMR contains a silenced copy of mating-type gene including HMR A1 (A1) and HMR A2 (A2), which are surrounded by two silencers, silencer E (E) and silencer I (I). In hmr::GFP cells, A1 gene is replaced with a gene encoding GFP as a reporter for silencing assay. Bottom: Cells lost transcriptional silencing will express GFP and become GFP positive during flow cytometer analysis (FACS). (B) Screening strategy for searching histone chaperone that functions in parallel with CAF-1 and Rtt106p in heterochromatin silencing at HMR locus. A series of yeast strains containing defects in different histone chaperones were crossed with cac1Δrtt106Δ hmr::GFP query strain to make triple mutant strains, which were then analyzed with GFP silencing assay. (C) Silencing at HMR locus is reduced more dramatically in cells deficient in spt16-m cac1Δrtt106Δ triple mutant cells than in cac1Δrtt106Δ double mutant cells. Expression of the GFP gene at the HMR locus in each strain was measured by FACS. The percentage of GFP expressing cells was calculated based on the average of three independent experiments. Error bars: standard deviations (SDs) calculated from three biological repeats. (D) Silencing at HMR locus is reduced more dramatically in spt16-m sir1Δ double mutant cells than either of the spt16-m or sir1Δ single mutant cells. Error bars: SDs calculated from three biological repeats. (E) One representative FACS analysis in Fig. 1D. (F) Quantification of A1 gene expression at the silenced HMR locus. The relative expression levels were analyzed by RT-qPCR and normalized to ACT1 gene. Error bars: SDs calculated from three technical repeats. (G) Quantification of YFR057W expression at the right arm of chromosome VI (VI-R) near telomeric region. The relative expression levels were analyzed by RT-qPCR and normalized to ACT1 gene. Error bars: SDs calculated from three technical repeats
In yeast, Sir1p is involved in the establishment of heterochromatin silencing at the HM loci (Rusche et al., 2003). To dissect the role of Spt16p in heterochromatin silencing in yeast, we first examined the role of Spt16p at the HMR locus in combination with sir1Δ (Fig. 1D and 1E). Consistent with the published results, sir1Δ causes a mild silencing defect (3.47%). GFP silencing appears to be normal in spt16-m cells (0.4%). However, spt16-m strongly exacerbates the silencing defect of sir1Δ (40.8%) (Fig. 1D). Similar synergistic behavior between SPT16 and SIR1 was also observed in the background of cac1Δ rtt106Δ (Fig. S1). These results suggest that Spt16p cooperates with Sir1p, CAF-1, and Rtt106 in heterochromatin silencing at the HMR locus.
To quantify the transcriptional activity at the HMR locus, we used reverse transcription quantitative PCR (RT-qPCR) to analyze the expression level of A1 gene that resides within HMR region (Fig. 1F). Consistent with the previous observations, A1 gene is silenced in wild-type cells, but is highly transcribed in sir3Δ strain. The expression level of A1 in spt16-m sir1Δ double mutant cells is dramatically higher than that in either spt16-m or sir1Δ single mutant cells. Moreover, we also analyzed the expression level of a gene YFR057W near the telomere of chromosome VI (Fig. 1G). Although no aberrant expression was detected in spt16-m strain, the transcription level has been apparently elevated in spt16-m sir1Δ strain, indicating a potential role of Spt16p and Sir1p in telomere silencing. While Sir1p is dispensable for telomeric silencing at the modified telomere (Aparicio et al., 1991), it has also been suggested that SIR1 contributes to silencing at natural telomeres which contains intact subtelomeric repeat sequences (Pryde and Louis, 1999). Together, our data suggested that the cooperation between Spt16p and Sir1p might be required for efficient silencing at both HM loci and telomeres.
In yeast, Sir2p is a nicotinamide adenine dinucleotide (NAD) dependent histone deacetylase, which deacetylates nearby histones to facilitate binding of Sir3p and Sir4p. Subsequently, more Sir2p is recruited through direct interaction with Sir3p and Sir4p to spread Sir proteins (Grunstein and Gasser, 2013). A reduction of Sir2p and Sir3p proteins over heterochromatin region was reported in cac1Δ rtt106Δ strain (Huang et al., 2007). To examine whether Spt16 affects the binding of Sir2–4p to different silent regions, we performed chromatin immunoprecipitation (ChIP) assay using antibodies against Sir2–4p (Fig. 2A–D). As reported, we found that all the Sir proteins are highly enriched at heterochromatin region rather than euchromatin region in wild-type cells, while sir3Δ completely abolishes the Sir proteins binding over the heterochromatin region. Moreover, while spt16-m mutation led to a mildly reduction of Sir2–4p binding at heterochromatin region, this mutation in combination with cac1Δ rtt106Δ led to a dramatic reduction of Sir2–4p binding comparing with either single or double mutant cells (Fig. 2A). This data suggests that Spt16p might function in parallel with CAF-1 and Rtt106p in regulating Sir proteins’ occupancy over heterochromatin regions. Furthermore, we found that the levels of Sir proteins drop dramatically in spt16-m sir1Δ strain at both HMR locus and telomere region (Fig. 2B and 2C). This dramatic defect is not due to alteration in Spt16p expression or chromatin occupancy in the mutant cells (Fig. S2). To further characterize the defect of heterochromatin formation in spt16-m mutant cells, we also analyzed the distribution of all three Sir proteins across HMR locus (Fig. 2D) and the histone modifications at HMR locus (Fig. 2E). Silencers, including E and I elements, are genomic regions where heterochromatin formation initiates (Grunstein and Gasser, 2013). In agreement with the silencer function of HMR-E and HMR-I that recruits Sir proteins and promotes formation of heterochromatin inwards toward the mating-type genes, an “M” shape distribution of Sir proteins was detected in wild-type cells. Compared with wild-type cells, the amount of Sir proteins is comparable in sir1Δ or spt16-m cells and the similar “M” pattern was observed, indicating that the silencer function is largely maintained in either single mutant strains. By contrast, the M pattern is lost in spt16-m sir1Δ strain (Fig. 2D). Acetylation of H4 lysine 16 (H4K16Ac) is low at heterochromatin region to facilitate the Sir proteins binding but is high at euchromatin regions adjacent to heterochromatin to restrict heterochromatin spreading into euchromatin (Grunstein and Gasser, 2013). We found that the level of H4K16Ac at the heterochromatin region is more dramatically enhanced in spt16-m sir1Δ double mutant cells compared to either single mutant alone (Fig. 2E, left panel). No obvious alterations was observed in H3K56Ac, an acetylation mark of newly synthesized H3 (Li et al., 2008), which was analyzed by ChIP assays side by side (Fig. 2E, right panel). Together, these data further supports the idea that Spt16p may cooperate with Sir1p and function in regulating the binding of Sir2–4p binding to heterochromatic region.
Figure 2.
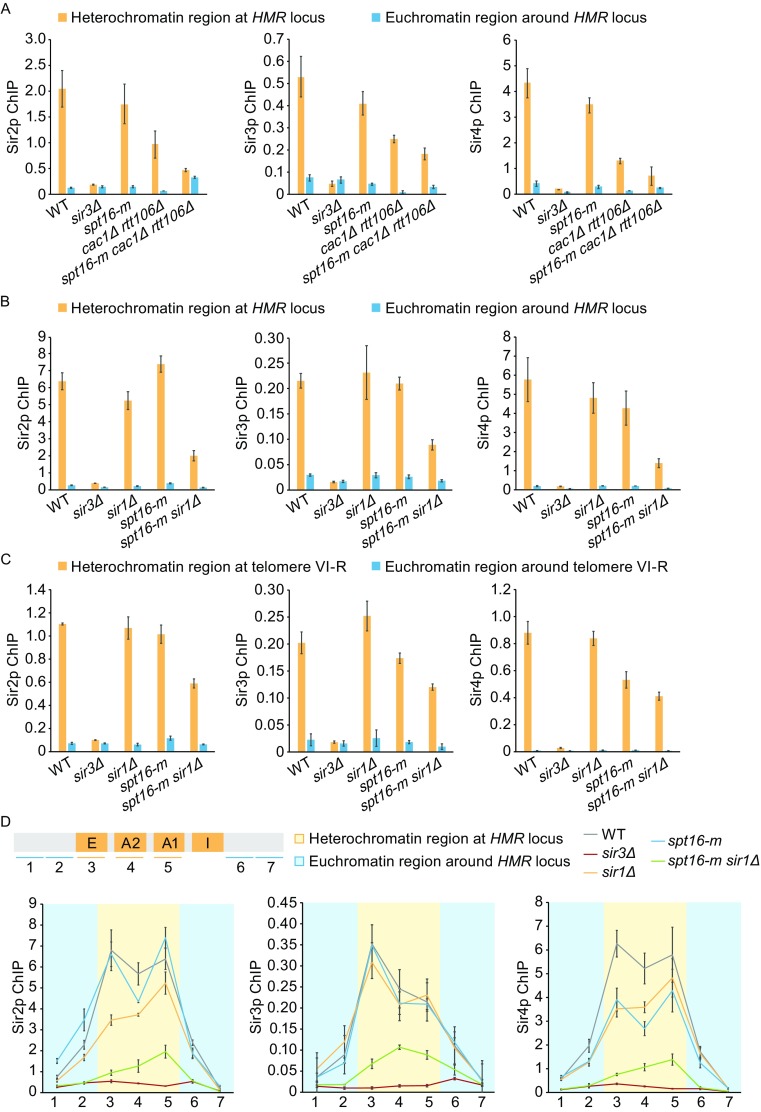
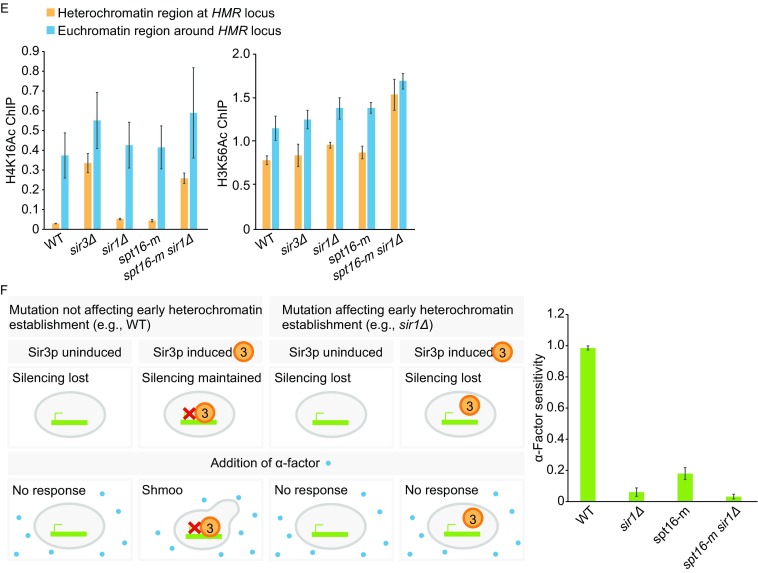
Spt16p is required for Sir2-4p binding to heterochromatin region and Spt16p cooperates with Sir1p during the establishment of heterochromatin silencing. (A) Sir2-4p binding at HMR locus is dramatically reduced in spt16-m cac1Δ rtt106Δ triple mutant cells. ChIP assays were performed using yeast strains of the indicated genotype and antibodies against Sir2p, Sir3p, and Sir4p, respectively. The error bars here represent SDs calculated from three technical repeats. The data are presented as the ratio of Sir proteins ChIP signal over input signal. sir3Δ strain was used as a control. (B and C) The association of Sir proteins to both HMR locus (B) and Chr VI-R telomeric region (C) is dramatically reduced in spt16-m sir1Δ double mutant cells. Sir2-4p ChIP was performed as described in (A). (D) The distribution pattern of Sir proteins at HMR locus has been changed in spt16-m sir1Δ double mutant cells. Top: A schematic diagram of HMR locus indicating the location of the primers used in this study. Bottom: The same ChIP assay was performed as described in (A) to map the distribution of Sir proteins across HMR locus in the indicated strains. (E) The level of H4K16Ac is increased at the heterochromatin region in spt16-m sir1Δ double mutant cells. ChIP assays were performed with antibodies against H4K16Ac and H3K56Ac. The data are presented as the ratio of H4K16Ac or H3K56Ac ChIP signal over input signal, with error bars indicating the SD for three technical repeats. (F) Spt16p has a role in the establishment of heterochromatin silencing. Left: A schematic diagram of the inducible heterochromatin establishment assay. There are two different mating types in S. cerevisiae: MATa and MATα. Selected strains for this assay are all MATa type cells, which normally only express a-specific genes and inhibit the expression of α-specific genes because of silencing at both HM loci. MATa cells will respond to the opposite mating pheromone, α-factor, by growing a protrusion called shmoo due to its distinct shape. In the absence of Sir3p (Sir3p uninduced), heterochromatin silencing is lost and such cells fail to respond to α-factor due to the expression of both a-specific and α-specific genes. This defect could be rescued by the induction of Sir3p if there are no other mutations except sir3Δ. In cells lacking Sir1p, a protein involved in heterochromatin establishment, establishment of silencing is affected. Green bar: HM loci; green arrow: the expression of α-specific genes; red cross: the inhibition of α-specific genes; orange circle with number 3: the presence of Sir3p; blue dot: α-factor. Right: The percentage of shmoo cells after Sir3p induction was counted (n = 3 with at least 200 cells for each genotype). Error bars: SDs calculated from three individual experiments
As Sir1p is known to be required for establishment of heterochromatin (Pillus and Rine, 1989), we speculate that Spt16p may also have a role in this stage. To test this, we used a Sir3p-induction system in sir3Δ cells to monitor the establishment of heterochromatin at silenced mating-type region (Fig. 2F). Before induction, no Sir3p proteins are present and heterochromatin silencing is completely abolished. Therefore, MATa cells will not respond to the mating pheromone α-factor due to lost silencing at yeast silent mating-type locus. Upon induction of Sir3p expression, heterochromatin is re-established in these cells and as a result, the cells will respond to α-factor and form shmoos. Deletion of SIR1 leads to reduced number of shmoos after Sir3p induction. Notably, comparing with wild-type cells, spt16-m also exhibits an apparent defect in shmoo formation after induction of Sir3p. Moreover, spt16-m exacerbates the defect of shmoo formation of sir1Δ (Fig. 2F). Thus, we conclude that Spt16p has a role in the establishment of heterochromatin silencing and it may function cooperatively with Sir1p during this process.
In summary, we show that histone chaperone FACT is required for the heterochromatin silencing in S. cerevisiae via the study of its subunit Spt16p. We find that the spt16-m mutant aggravates the HMR silencing defect in cells lacking both CAC1 and RTT106 genes. Moreover, Spt16p functions in parallel with CAF-1 and Rtt106p in regulating Sir proteins’ occupancy. Therefore, besides functioning with CAF-1 and Rtt106p during nucleosome assembly (Yang et al., 2016), we speculate that Spt16p also cooperates with CAF-1 and Rtt106 in heterochromatin formation. Additionally, we find that Spt16p likely functions cooperatively with Sir1p in the establishment of heterochromatin silencing at both telomeres and cryptic mating-type loci. Pob3p has been previously reported to be important for heterochromatin silencing in S. pombe (Lejeune et al., 2007). Our data suggests that Spt16p might also play a crucial role during heterochromatin silencing in S. cerevisiae, a highly divergent eukaryotic species from S. pombe. Therefore, it is likely that FACT’s role in heterochromatin silencing is conserved among eukaryotes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Footnotes
This work was supported by Grants from the National Science Foundation for Excellent Young Scholars of China (31322017) and the National Natural Science Foundation of China (Grant Nos. 31370767 and 31671332). Q.L. is a scholar from the Chinese 1000 Plan-the young talents group.
This article does not contain any studies with human or animal subjects performed by the any of the authors. Xiaowei Yan, Jiayi Yang, Jiawei Xu, Jianxun Feng, and Qing Li declare that they have no conflict of interest.
Footnotes
Xiaowei Yan, Jiayi Yang, and Jiawei Xu are co-first authors.
Electronic supplementary material
The online version of this article (doi:10.1007/s13238-017-0485-4) contains supplementary material, which is available to authorized users.
Contributor Information
Jianxun Feng, Email: fengjx@pku.edu.cn.
Qing Li, Email: li.qing@pku.edu.cn.
References
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Enomoto S, Berman J. Chromatin assembly factor I contributes to the maintenance, but not the re-establishment, of silencing at the yeast silent mating loci. Genes Dev. 1998;12:219–232. doi: 10.1101/gad.12.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2012;1819:247–255. doi: 10.1016/j.bbagrm.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M, Gasser SM. Epigenetics in Saccharomyces cerevisiae. Cold Spring Harb Perspect Biol. 2013;5:a017491. doi: 10.1101/cshperspect.a017491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Dambacher S, Schotta G. Heterochromatin dysregulation in human diseases. J Appl Physiol. 2010;1985:232–242. doi: 10.1152/japplphysiol.00053.2010. [DOI] [PubMed] [Google Scholar]
- Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z. Rtt106p is a histone chaperone involved in heterochromatin-mediated silencing. Proc Natl Acad Sci USA. 2005;102:13410–13415. doi: 10.1073/pnas.0506176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z. A novel role for histone chaperones CAF-1 and Rtt106p in heterochromatin silencing. EMBO J. 2007;26:2274–2283. doi: 10.1038/sj.emboj.7601670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z. Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell. 2008;134:244–255. doi: 10.1016/j.cell.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillus L, Rine J. Epigenetic inheritance of transcriptional states in S. cerevisiae. Cell. 1989;59:637–647. doi: 10.1016/0092-8674(89)90009-3. [DOI] [PubMed] [Google Scholar]
- Pryde FE, Louis EJ. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/S0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang X, Feng J, Leng H, Li S, Xiao J, Liu S, Xu Z, Xu J, Li D, et al. The histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 2016;14:1128–1141. doi: 10.1016/j.celrep.2015.12.096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


