Abstract
Background
Plastome mutants are ideal resources for elucidating the functions of plastid genes. Numerous studies have been conducted for the function of plastid genes in barley and tobacco; however, related information is limited in Chinese cabbage.
Results
A chlorophyll-deficient mutant of Chinese cabbage that was derived by ethyl methanesulfonate treatment on isolated microspores showed uniformly pale green inner leaves and slow growth compared with that shown by the wild type “Fukuda 50′ (‘FT’). Genetic analysis revealed that cdm was cytoplasmically inherited. Physiological and ultrastructural analyses of cdm showed impaired photosynthesis and abnormal chloroplast development. Utilizing next generation sequencing, the complete plastomes of cdm and ‘FT’ were respectively re-mapped to the reference genome of Chinese cabbage, and an A-to-C base substitution with a mutation ratio higher than 99% was detected. The missense mutation of plastid ribosomal protein S4 led to valine substitution for glycine at residue 193. The expression level of rps4 was analyzed using quantitative real-time PCR and found lower in than in ‘FT’. RNA gel-blot assays showed that the abundance of mature 23S rRNA, 16S rRNA, 5S rRNA, and 4.5S rRNA significantly decreased and that the processing of 23S, 16S rRNA, and 4.5S rRNA was seriously impaired, affecting the ribosomal function in cdm.
Conclusions
These findings indicated that cdm was a plastome mutant and that chlorophyll deficiency might be due to an A-to-C base substitution of the plastome-encoded rps4 that impaired the rRNA processing and affected the ribosomal function.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1353-y) contains supplementary material, which is available to authorized users.
Keywords: Chinese cabbage, Maternal inheritance, Plastome mutant, Plastid ribosomal protein, rRNA processing
Background
Mutant lines are ideal resources for discovering novel gene functions and deciphering unknown mechanisms [1, 2]. In higher plants, chlorophyll-deficient mutants are considered important for gaining a better insight into the mechanism of photosynthesis, chlorophyll synthesis, development and differentiation of chloroplast structure, gene functional identification, and nucleo-cytoplasmic interactions [3, 4].
Chlorophyll-deficient mutants can be generally divided into three types: mutants controlled by nuclear genes, mutants controlled by cytoplasmic genes, and mutants controlled by nuclear-cytoplasmic interactions [5]. Most chlorophyll-deficient mutants in different species, such as Arabidopsis thaliana [6], rice [7], barley [8], soybean [9], carrot [10], and cabbage [11], are controlled by nuclear genes involved in plastid development and pigment biosynthesis that transferred from the plastids to the nucleus during evolution [12]. Only a small number of chlorophyll-deficient mutants are cytoplasmic mutants [13].
Chlorophyll-deficient mutants have been used as a genetic tool for the initial study of cytoplasmic inheritance [14, 15]. The existence of extranuclear DNA and non-Mendelian inheritance was first reported in leaf color mutants of Mirabilis jalapa and Pelargonium zonale [14–16]. Ris and Plaut [17] as well as Nass and Nass [18] reported that plastids and mitochondria have their own DNA and independent genetic systems [19]. Cytoplasmic mutations are inherited in a non-Mendelian fashion, which includes three basic modes: maternal inheritance, paternal inheritance, and bi-parental inheritance [20]. Of these, maternal inheritance is the main mode in angiosperms. In Brassica campestris, plastids and mitochondria are maternally inherited, whereas in Brassica napus, mitochondria are not strictly maternally inherited [21, 22].
A limited number of cytoplasmic inherited chlorophyll-deficient mutants with a non-chromosomal stripe phenotype are mitochondrial mutants generally caused by rearranging [23, 24]. Most cytoplasmic-inherited chlorophyll-deficient mutants are plastome mutants that occur spontaneously [25, 26] or induced by artificial mutagenesis, plastome mutator alleles [27–29], or transformation [30–32]. Previous studies showed that the rate of spontaneous plastome mutations causing chlorophyll deficiency varies between 0.006 and 0.3% in different plant species [33]. Spontaneous plastome mutations include all mutation types, including indels and point mutations. Artificial mutagenesis used to generate plastome mutants is either physical mutagenesis [34] or chemical mutagenesis [35]. N-nitroso-N-methyl-urea (NMU), methyl-nitro-nitrosoguanidine (MNNG), 5-bromo-2′-deoxyuridine (BrdU), and 9-aminoacridine hydrochloride (9AA) have been widely used as chemical mutagens to induce plastome mutations [36–38]. Ethyl methanesulfonate (EMS) mainly induces nuclear gene mutations, but also plastome mutations [39]. Nuclear genes can induce plastome mutations by plastome mutator alleles in frequencies much higher than those of spontaneous mutations [40–42].
Plastome mutants are classified into three categories: those with mutations in genetic system genes, in photosynthesis-related genes, and other genes and conserved reading frames [43]. Numerous plastome mutants with mutations in genetic system genes showed chlorophyll deficiency [13, 42, 44], antibiotic resistance [45, 46], misshapen leaves, or low-temperature tolerance [30].
Genetic system genes, including 62 plastome-encoded genes, 30 tRNAs, 4 rRNAs, and 21 ribosomal protein genes, constitute the largest group in the plastome, which is associated with plastid gene expression [47, 48]. Plastid ribosomal proteins are essential components of the plastid ribosome composed by a 50S subunit and a 30S subunit. The former subunit comprises of 23S rRNA, 5S rRNA, 4.5S rRNA, and 33 plastid ribosomal proteins, whereas the latter subunit comprises of 16S rRNA and 24 chloroplast ribosomal proteins. Additionally, 12 proteins of the 30S subunit and nine proteins of the 50S subunit are encoded by plastid genes [31]. Some plastid ribosomal proteins are essential for plastid translation, plant growth, and plant development, whereas others are non-essential [30, 44, 49]. Plastid ribosomal proteins encoded by plastid genes have been widely studied in tobacco, whereas those encoded by nuclear genes are well studied in A. thaliana. The rps2, rps4, rpl18, and rpl20 knockout mutants in tobacco show misshapen leaves, revealing the essential role of plastid proteins in leaf development [30, 44], whereas the rps15 knockout mutant shows chlorophyll deficiency [31]. In addition, the prps1, rps5, prsp17, prpl11, and prpl24 mutants in A. thaliana show chlorophyll deficiency and reduced growth, and of these, prps17 and prpl24 show rRNA process impairment [50–52]. Therefore, plastid ribosomal proteins play essential roles in plastid translation, plant growth, and plant development. In contrast to simple knock-out analysis of plastid transformation, point mutations or indels in plastome mutants are valuable for elucidating plastid functions [33]. The identification of plastome mutants by the phenotype or using conventional genetic methods is considered unreliable, or it may be due to a mitochondrial mutations secondarily resulting in plastid malfunction. Next-generation sequencing (NGS) provides a relatively reliable and rapid method to directly identify plastome mutants at the DNA level.
In this study, a maternal inherited mutation causing chlorophyll deficiency was identified in Chinese cabbage. The complete plastomes of cdm and Fukuda 50 (wild type; ‘FT’) were respectively re-mapped to the cpDNA reference genome of Chinese cabbage [53] using Illumina NGS, and a missense mutation was found in the ribosomal protein S4 (RPS4). The accumulation and processing of plastid rRNAs, including 23S, 16S, 5S, and 4.5S rRNAs, were found to be aberrant in cdm compared with those in ‘FT’. Our results suggested that RPS4 is associated with chloroplast rRNA processing and chlorophyll deficiency in cdm.
Methods
Plant material
In this study, a doubled haploid line rived from the Chinese cabbage variety ‘FT’ was used as a donor for isolated microspore culturing to create a mutant library. In August 2014, ‘FT’ seeds of a doubled haploid line derived from the Chinese cabbage variety Fukuda 50 (‘FT’) were vernalized at 4 °C for 20 d after accelerating germination and then, sown in an aperture disk on a seedbed. Seedlings were re-potted in late September, and the unopened flower buds were used for isolated microspore culturing.
EMS mutagenesis on isolated microspores
Procedures of microspore isolation, purification, and culture were performed as described previously [54, 55]. Isolated microspore culturing was conducted at the Liaoning Key Laboratory of Genetics and Breeding of Cruciferous Vegetable Crops, Shenyang Agricultural University, China. EMS mutagenesis was performed as described by Huang et al. [56]. In brief, EMS dissolved in B5 medium [57] at a concentration of 0.12% (v/v) was filter-sterilized through a 0.22-μm filter membrane. Isolated microspores were washed by B5 medium and centrifuged twice at 120×g for 3 min. The precipitate acquired after the first centrifugation was suspended in B5 medium containing EMS (0.12%, v/v) for 10 min. The microspores were re-suspended and cultured in NLN-13 medium [58], incubated at 33 °C for 24 h, and transferred to 25 °C for more than 7 d in the dark. Regenerated plants from cotyledonous embryoids were transferred to Murashige and Skoog (1962) medium [59] and cultured at 25 °C under a 16-h photoperiod for subculture and root induction. The remaining regenerated plants were transplanted in pots after rooting for 20 d.
Quantification of chlorophyll
Total chlorophyll was extracted as described by Inskeep and Bloom [60]. In brief, fresh inner leaves (0.1 g) from cdm and ‘FT’ were respectively selected at the seedling stage (4-week-old) and the flowering stage (8-week-old) and submerged in 80% acetone for 24 h in the dark. The absorbance of the supernatants was recorded at 645 nm and 663 nm using the DU 800 UV/Vis Spectrophotometer (Beckman Coulter, Brea, CA, USA).
Characterization of chloroplast and mitochondrial ultrastructure
The chloroplast and mitochondrial ultra-structures were characterized as described by Lichtenthaler et al. [61] and Huang et al. [56] Fresh inner leaves of cdm and ‘FT’ were selected at the seedling stage, cut into small pieces (approximately 1 mm2), and fixed in 4% (v/v) glutaraldehyde in a 0.1 M phosphate buffer solution (PBS, pH 7.3) at 4 °C for 7 d. The fixed samples were post-fixed in 1% (w/v) aqueous osmium tetroxide for 4 h and rinsed 3 times in PBS for 5 min each time. The samples were gradually dehydrated with various concentrations of ethanol, and then, impregnated and embedded in Epon812. Ultrathin sections were made using the LKB2088 ultramicrotome (LKB Company, Saffle, Sweden) and double stained with uranyl acetate and lead citrate. Finally, the samples were observed under the H-7650 transmission electron microscope (TEM; Hitachi, Tokyo, Japan).
Measurement of fluorescence kinetic parameters
Ten 4-week-old cdm and ‘FT’ plants with consistent growth were selected for measuring fluorescence kinetic parameters by applying A Fluor Cam Portable ChI/GFP Luminoscope (Handy GFPCam; Eco Tech, Beijing, China) after 20 min of dark adaption using leaf clips. The optional maximal photochemical efficiency of PS II (Fv/Fm), the effective quantum yield of PSII (ФPSII), the photochemical quenching coefficient (QP), and the non-photochemical Chl fluorescence quenching (NQP) were calculated as described by Bilger and Bjorkman [62] and Maxwell and Johnson [63]. Each measurement was performed in triplicate.
Chloroplast DNA (cpDNA) isolation
At the seedling stage, well-grown cdm and ‘FT’ plants were placed at 4 °C for 48 h in the dark to eliminate chloroplast starch granules and reduce the plastid breakage. Fresh leaves collected from three plants were mixed, cleaned with double distilled water, dried on filter paper, and the main nerve was cut off. A 30-g sample was ground to less than 1 mm2 in liquid nitrogen. Total cpDNA was isolated using the column Plant chloroplast DNAout kit (TIANDZ, Tianjin, China), according to the manufacturer’s instructions with minor modifications. This experiment was performed on ice, and all vessels and solution were precooled.
Chloroplast genome re-sequencing by Illumina HiSeq
Purified cpDNA from cdm and ‘FT’ was used for constructing an NGS library using the NEBNext Ultra DNA Library Prep Kit for Illumina, according to the manufacturer’s instructions. In brief, 1 μg cpDNA was randomly fragmented to < 500-bp fragments by sonication and treated with End Prep Enzyme Mix, followed by adaptor additions to both ends. Fragments of approximately 410 bp with a 350-bp insert were purified using the AxyPrep Mag PCR Clean-up kit (Axygen, Corning, NY, USA) and then, amplified using P5 and P7 primers. Purified PCR products were validated by the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and quantified by the Qubit2.0 Fluorometer (Invitrogen, Carlsbad, CA, USA). DNA libraries were multiplexed, loaded on an Illumina HiSeq instrument, and sequenced by a 2 × 150 paired-end (PE) configuration with the HiSeq Control Software (HCS) + OLB + GAPipeline-1.6 (Illumina) reading sequence information.
Based on the reference cpDNA genome of Chinese cabbage (http://www.ncbi.nlm.nih.gov/genomes/GenomesGroup.cgi?opt=plastid&taxid=3398), cpDNA sequence reads were isolated from the raw sequence reads of mixed nucleus and mitochondrial DNA. The filtered cpDNA sequence reads from cdm and ‘FT’ were used for re-sequencing as described by [53].
Sequence analysis
Based on gene annotation information [64] and nucleotide sequences, PCR primers were designed to verify the mutation site of the candidate gene. Total cpDNA extracted from three cdm and plants, respectively, was used as a template. PCR products were purified by a Gel Extraction Kit (Omega Bio-tek, Norcross, GA, USA), introduced into the pGEM®-T Easy Vector (Promega, Madison, WI, USA), and transformed into the JM109 competent cell (Takara, Beijing, China). The recombinant plasmids were sequenced by the Institute of Beijing Genomics, China, and the sequences were aligned using DNAMAN (Lynnon, San Ramon, CA, USA).
RNA isolation and qRT-PCR
Fresh inner leaves collected from 4-week-old ‘FT’, cdm, ‘FT’ × cdm and cdm × ‘FT’ plants were used for RNA isolation using TRIZOL (Invitrogen). First-strand cDNA was synthesized with random hexamer primers. A 10-fold dilution of cDNA was used for quantitative real time PCR (qRT-PCR) by the Bio-Rad IQ5 Real Time PCR System (Bio-Rad, Hercules, CA, USA) with gene-specific primer sets (Additional file 1: Table S1) and SYBR Green PCR master mix (Takara). The qRT-PCR thermal cycling conditions were as follows: denaturation at 95 °C for 3 min, followed by 40 cycles at 95 °C for 30 s, 60 °C for 30 s, and 70 °C for 30 s. The 2-△△Ct method was used for analyzing the relative gene expression levels as described by Livak et al. [65] and normalized using the house-keeping gene actin and the expression of responsive sample controls. All reactions were quantified in triplicate with three independent biological replicates.
Northern blot analysis
Total RNA was quantified by Infinite M200PRO (Tecan, Mannedorf, Switzerland). Then, 5-μg aliquots were denatured in RNA loading buffer with ethidium bromide for 15 min at 65 °C, separated in formaldehyde-containing 2% agarose gels, transferred onto Hybond N+ membranes (GE, Boston, MA, USA) by capillary blotting using standard protocols, and fixed by applying a crosslinker (UVP American, Upland, CA, USA). Digoxigenin-labeled plasmid DNA probes were amplified using the PCR DIG Probe Synthesis Kit (Roche, Basel, Switzerland) with gene-specific primers listed in Additional file 1: Table S1 [66], denatured at 100 °C for 5 min, and immerged into a hybridization solution, containing 50% (v/v) formamide, 5× saline sodium citrate (SSC), 50 mM phosphate buffer, 0.1% N-lauroylsarcosine sodium salt, 7% (m/v) sodium dodecyl sulfate (SDS), and 2% (m/v) blocking reagent (Roche). The membrane was pre-hybridized at 50 °C for 1 h, and then hybridized at 55 °C for 24 h after adding the specific probe in a hybridization oven (UVP American). The membrane was washed in 2× SSC containing 0.1% SDS at 25 °C, and subsequently, washed twice in 0.1× SSC, containing 0.1% SDS, at 68 °C. A 1.5% blocking reagent (Roche) in maleic buffer was applied for immunological detection as described by Huang et al. [67] and the membrane was detected and analyzed by Tano-5200 Chemi-luminescent Imaging System (Tanon Science Technology, China).
Results
Isolation of maternal -inherited chlorophyll-deficient mutant (cdm)
Microspore-derived regenerated plants (M0) were obtained from EMS mutagenesis on isolated microspores, and the variant plants were screened. All the plants were selfed to obtain M1 seeds. In M1 generation, all the new variant plants were also screened. A chlorophyll-deficient mutant (cdm) (Fig. 1a) was identified in the M0 generation and stably inherited in the progeny. Chlorophyll deficiency and reduced growth in cdm were more apparent in winter.
Fig. 1.
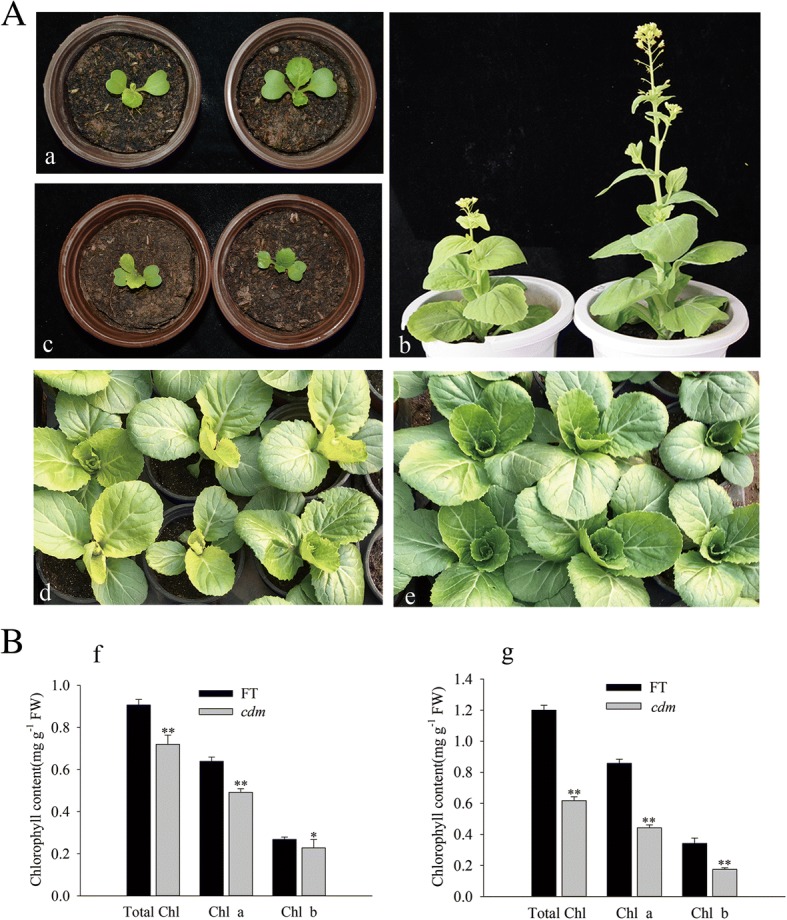
Identification of chlorophyll-deficient mutant (cdm) plants and their phenotypes at the seedling and mature stage. (Aa, d, e) (left) and wild-type (‘FT’) plants at the seedling stage; (Ab) cdm (left) and ‘FT’ plants at the mature stage; (Ac) F1 plants derived from a cross of cdm× genetically distant inbred line (WZ) (left) and WZ × cdm (B) Chlorophyll content of cdm and ‘FT’ plants grown for 7 d and 25 d under long-day conditions. Error bars indicate one standard deviation of the mean (n = 5). Asterisks (**) indicate statistically significant differences between cdm and ‘FT’ at P < 0.01. fw, fresh weight
For genetic analysis, cdm plants were reciprocally crossed with ‘FT’ plants. The 376 F1 individuals were different, but consistent with the characteristics of female parent (Table 1), suggesting that cdm was a maternal mutant. For further verification, cdm plants were reciprocally crossed with a genetically-distant inbred Chinese cabbage line ‘WZ’ and found that only the maternal parent could transmit the mutant phenotype (Fig. 1a; Table 1).
Table 1.
Genetic analysis of offspring derived from reciprocal crosses between chlorophyll-deficient mutant (cdm) and wild-type (‘FT’) or genetically-distant inbred line (WZ)
| Cross | Number | Leaf color | |
|---|---|---|---|
| Green | Yellow | ||
| cdm (selfed) | 213 | 0 | 213 |
| ‘FT’ × cdm | 174 | 174 | 0 |
| cdm × ‘FT’ | 202 | 0 | 202 |
| WZ × cdm | 91 | 91 | 0 |
| cdm× WZ | 87 | 0 | 87 |
Phenotypic characterization of cdm
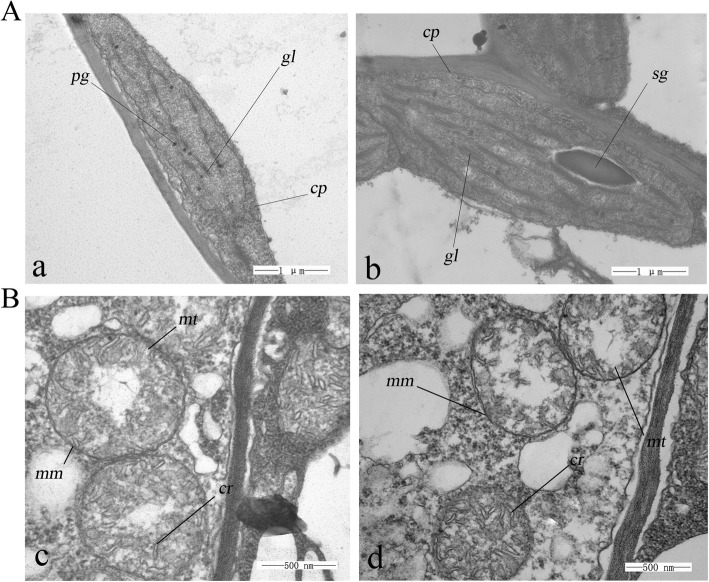
All Plants of cdm showed pale-yellow inner leaves and reduced growth. Consistently, the contents of total chlorophyll, chlorophyll a, and chlorophyll b were significantly lower in cdm at the seedling and mature stages compared with those in ‘FT’ (Fig. 1b). The results indicated that chloroplast development was severely impaired (Fig. 2a); however, no differences were observed in the mitochondria of cdm and ‘FT’ (Fig. 2b).
Fig. 2.
Chloroplast and mitochondrial ultrastructure of chlorophyll-deficient mutant (cdm) and wild-type (‘FT’) plants at the seedling stage (Chloroplast, × 30,000; Mitochondrion, × 60,000). (Aa and b) Chloroplasts of cdm and ‘FT’. (Bf and g) Mitochondria of cdm. cp, chloroplast; sg, starch grain; pg, plastoglobule; gl, grana lamella; mt, mitochondrion; mm, mitochondrial membrane; cr, mitochondrial crista. Scale bar, 1 μm. Bars = 1 μm (A) and 500 nm (B)
The PSII capacity of cdm was analyzed for various chlorophyll fluorescence parameters compared with that of ‘FT’. The results showed that Fv/Fm (Fig. 3a) and the electron transport rate (Fig. 3b) were lower in cdm than in ‘FT’, whereas qN (Fig. 3c) was higher in cdm than in ‘FT’, revealing PSII photo-inhibition and reduced photosynthetic efficiency in cdm.
Fig. 3.
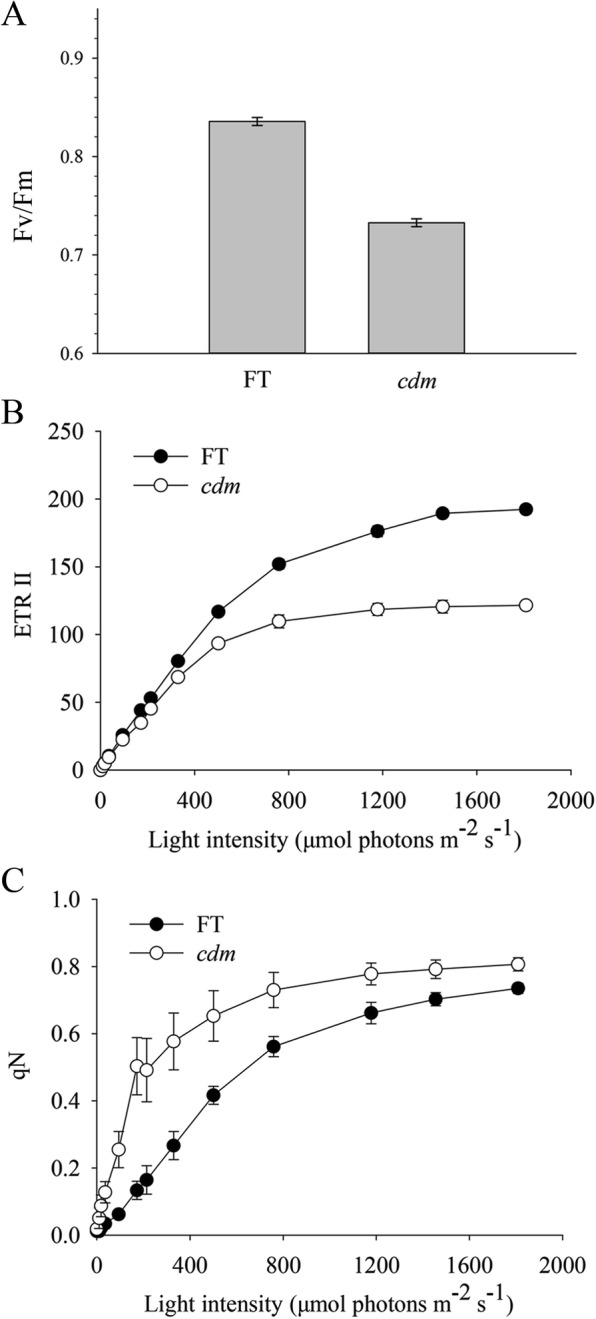
Photosynthetic activity in the leaves of chlorophyll-deficient mutant (cdm) and wild-type (‘FT’) plants. a Maximum quantum efficiency of PSII (Fv/Fm). b Light saturation curve of linear electron flux as calculated from the PSII yield (ETR II). c Non-photochemical quenching (NQP). Error bars indicate the SD (n = 4)
A missense mutation of chloroplast RPS4 in cdm
The chloroplast genome of cdm and ‘FT’ was re-sequenced and compared. Based on the inter-comparison of sequencing data and the reference genome, the frequency of A, T, C, and G on each site was recorded for the corresponding ratio in the chloroplast genome according to the sequence depth. All sites in the chloroplast genomes of cdm and ‘FT’ are presented in Additional file 2: Tables S2 and Additional file 3: Table S3. A site with a mutation ratio higher than 99% was detected in cdm after comparison of sequencing data (Table 2) and identified at nucleotide 44,398 of rps4, based on the reference genome and gene annotation as described by Wu et al. [64]. The A-to-C base substitution on this site was detected as a missense mutation that led to valine (Val) substitution for glycine (Gly) at residue 193, which is a highly conserved residue of RPS4 in plants. qRT-PCR was performed to detect any effects of the mutation on the expression of rps4 and showed that the transcript level of RPS4 in cdm was lower than that in ‘FT’ (Fig. 4).
Table 2.
Mutation sites in chlorophyll-deficient mutant (cdm) and wild-type (WT) compared with the reference genome
| Location in Ref genome | Region | Mutation type | Reference | FT | cdm |
|---|---|---|---|---|---|
| 265 | trnH-psbA spacer | SNP | A | T | T |
| 266 | trnH-psbA spacer | SNP | A | T | T |
| 267 | trnH-psbA spacer | SNP | A | T | T |
| 268 | trnH-psbA spacer | SNP | A | T | T |
| 20,164 | rpoC1 coding region | SNP | C | T | T |
| 44,398 | rps4 coding region | SNP | A | A | C |
| 66,083 | psaJ-rpl33 spacer | SNP | A | T | T |
| 66,084 | psaJ-rpl33 spacer | SNP | G | T | T |
| 66,085 | psaJ-rpl33 spacer | SNP | A | C | C |
| 66,086 | psaJ-rpl33 spacer | SNP | A | T | T |
| 116,457 | psaC coding region | SNP | G | A | A |
Fig. 4.
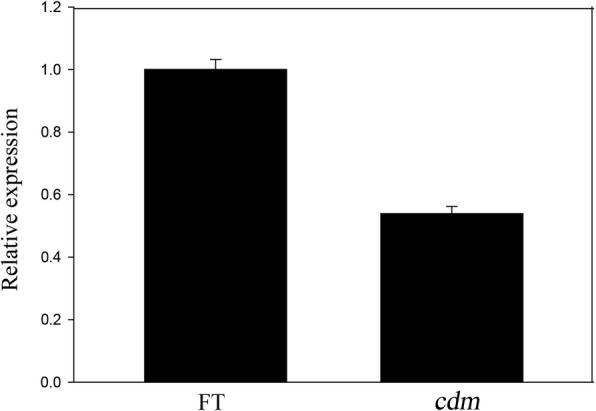
Expression of rps4 in chlorophyll-deficient mutant (cdm) and wild-type (‘FT’) plants detected by quantitative real-time PCR. Data are presented as means ± standard deviation of three biological replicates with three independent biological replicates
Verifying the mutation in rps4
The inner leaves of cdm were uniformly pale green without any dark green sections, suggesting that the mutation might be homoplasmic and non-segregating. This assumption was supported by the A-to-C point mutation ratio in rps4 that was higher than 99% and also by the sequence analysis of cpDNA from cdm and ‘FT’ which showed that the point mutation of rps4 was present in different cdm. Therefore, the point mutation of rps4 was homoplasmic in cdm.
RPS4 structure
RPS4 is a plastid ribosomal protein and a component of the plastid ribosome 30S small subunit. Its N-terminal region is highly conserved in higher plants (Fig. 5a). To study whether the amino acid substitution of RPS4 affected its secondary structure, the three-dimensional structure and the protein domain were predicted online at https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_sopma.html; https://swissmodel.expasy.org/; http://smart.embl-heidelberg.de/. The predicted secondary structure of RPS4 showed that the site of amino acid substitution was located at the extended strand in ‘FT’, whereas at the alpha helix in cdm (Fig. 5b). Thus, the predicted secondary structure of RPS4 in cdm was different from that in ‘FT’. However, the three-dimensional structure of RPS4 was the same in cdm and ‘FT’ (Fig. 5c). Zhang et al. [52] considered that the mutant site might not be analyzed by the prediction. RPS4 has two domains: the ribosomal protein S4 domain (3–88) and the S4 domain (89–153) (Fig. 5d). The ribosomal protein S4 domain binds to the ribosomal RNA and is composed of four helices that are discontinuous in sequence. The S4 domain probably mediates the binding to RNA. The amino acid substitution of RPS4 is not located in the two domains.
Fig. 5.
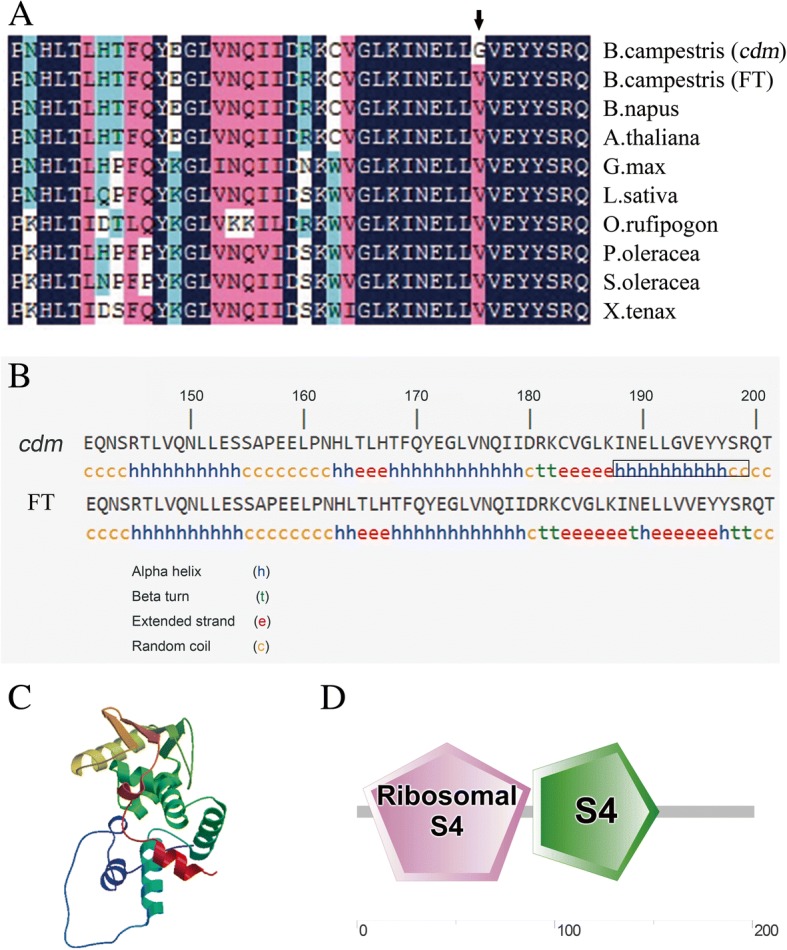
Partial sequence alignment and secondary structure prediction. a N-terminal domain sequence alignment of RPS4 in chlorophyll-deficient mutant (cdm) and wild-type (‘FT) of Brassica campestris ssp., Arabidopsis thaliana, Brassica napus, Glycine max, Lactuca sativa, Oryza rufipogon, Portulaca oleracea, Spinacia oleracea, and Cyanophora paradoxa (P23402). Vertical arrow indicates the substituted amino acid site. b Predicted secondary structure of the RPS4 N-terminal domain. Black box indicates sites altered secondary structure. c Predicted three-dimensional structure of RPS4. d Predicted protein domain of RPS4
Efficiency of chloroplast rRNA processing in cdm
To research whether the chloroplast rRNA processing was affected in cdm and cdm × ‘FT’, we investigated the pattern of total RNA by using denatured agarose gels electrophoresis. The results showed that the plastid rRNAs were significantly reduced in cdm and cdm × ‘FT’, compared with those in ‘FT’ and ‘FT’ × cdm (Fig. 6a, Additional file 4: Figure S1).
Fig. 6.
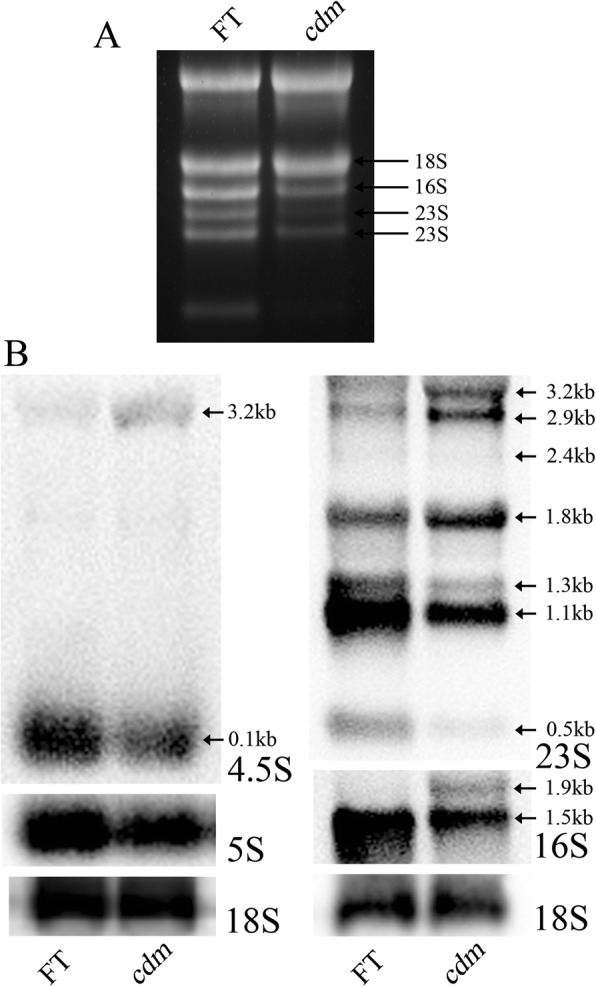
Chloroplast rRNA accumulation and processing in chlorophyll-deficient mutant (cdm) and wild-type (‘FT’) plants. a rRNA accumulation pattern in ethidium bromide-stained gel. b Northern blot analysis of chloroplast 23S, 16S, 5S, and 4.5S rRNAs and cytoplasmic 18S rRNA. Equal loading controls are shown in Additional file 5: Figure S2
The plastid 70S ribosome is composed of a 50S subunit and a 30S subunit. The former is comprised by 23S rRNA, 5S rRNA, and 4.5S rRNA, 33 chloroplast ribosomal proteins, whereas the latter by 16S rRNA and 24 chloroplast ribosomal proteins. Chloroplast rrn operon is transcribed to generate large rRNA precursors, which form mature rRNAs after processing. For a more detailed analysis of the rRNA accumulation pattern of cdm, northern blot experiments were performed using gene-specific probes (Additional file 1: Table S1). The results showed that the abundance of mature 23S rRNA, 16S rRNA, 5S rRNA, and 4.5S rRNA was decreased in cdm compared with that in ‘FT’, whereas the precursors of 23S (3.2 kb, 2.9 kb, and 2.5 kb) rRNA, 16S (1.9 kb) rRNA, and 4.5S rRNA were over-accumulated in cdm (Fig. 6b). However, cytosolic 18S rRNA that used as a control showed similar levels in cdm and ‘FT’, indicating that cytosolic rRNA was not affected in cdm plants. Therefore, the mutation in rps4 possibly impaired rRNA processing in cdm.
Discussion
In the present study, we identified a chlorophyll-deficient mutant derived by EMS treatment on isolated microspores. The mutation showed non-Mendelian inheritance, indicating that it was located either in the chloroplast or in the mitochondrial genome. TEM studies showed that the ultrastructure of mitochondria was the same in cdm and ‘FT’, whereas that of plastids was abnormal in cdm. The plastome includes a conserved set of genes most of which are directly involved in photosynthesis or plastid translation [47, 49]. Thus, the mutation was likely in the plastome. The plastomes genomes of cdm and ‘FT’ were re-mapped to the reference genome of Chinese cabbage, revealing an A-to-C base substitution at nucleotide 44,398 of rps4 with a mutation ratio higher than 99% that led to Val substitution for Gly in RPS4, which altered its secondary structure.
In our study, cdm showed uniformly pale green leaves and a slow growth rate. Almost all the chlorophyll-deficient cytoplasmic mutants are variegated [33], containing aberrant and normal plastids in distinct tissues of an individual plant, indicating heteroplasmy. However, all the inner leaves of cdm had a homogeneous pale-green color and extremely rare plant uniformity, suggesting homoplasmy and non-segregation. NGS showed that the mutation ratio of A-to-C base substitution was higher than 99% in rps4, verifying that the mutation was homoplasmic across all plastids. The pale green leaves of cdm plants were uniform, suggesting that the uniform mutant phenotype was due to a homoplasmic mutation. These results indicated that rps4 was homoplasmic mutation, and might be responsible for the uniform chlorophyll deficiency of cdm plants.
The mechanism of EMS mutagenesis is based on the alkylation of guanine (G) residues, which primarily induces C-to-T changes and ultimately leads to an amino acid change or deletion [68]. The A-to-C base substitution in the present study did not correspond to a known mutagenesis mechanism, and thus, the homoplasmic mutation in rps4 was assumed to be induced by a plastome mutator allele generated by EMS mutagenesis on isolated microspores. Plastome mutator alleles have been widely studied in A. thaliana [69], barley [27, 70], and the genus Oenothera [40, 71]. The cytoplasmic line 3 (CL3) in barley that selected from a barley plastid mutator genotype displays a homogeneous light green phenotype [27]. The plastome mutator can induce various plastome mutations in a homozygous recessive condition [33]. Since EMS treatment on isolated microspores can rapidly generate homozygous mutants, we speculated that a homozygous recessive nuclear mutation was generated, which was a plastome mutator that induced the point mutation in rps4.
Ribosomal proteins are essential components of the plastid ribosome and play different roles in rRNA processing and abundance. Mutations in plastid ribosomal proteins result in a range of developmental phenotypes, of which, some impair rRNA processing. Previous studies demonstrated that a mutation in rps5, prps17, and prpl24 impairs rRNA processing in A. thaliana [51, 72]. Plastome-encoded ribosomal proteins, as well as their function, are highly conserved. For instance, the rpl33 mutants in E. coli showed low-temperature tolerance [73, 74], whereas the rpl33 mutants in tobacco showed decreased viability and growth under chilling stress [30]. RPS4 is required early in the assembly process, since it directly binds to the 16S rRNA in Escherichia coli [75] and is essential in tobacco [30]; thus, these studies support our assumption that mutation in rps4 might affect the chloroplast ribosomal assembly process and impair plastid rRNA processing in cdm. By running denaturing agarose gels, we found that abundance of plastid rRNAs in cdm and cdm × ‘FT’ was decreased significantly than those in ‘FT’ and ‘FT’ × cdm. These results indicated that the impaired rRNA processing was maternal inherited. Northern blot data revealed a reduced abundance of mature 23S rRNA, 16S rRNA, and 4.5S rRNA, but over-accumulation of 23S rRNA, 16S rRNA, and 4.5S rRNA precursors in cdm. The missense mutation in RPS4 was the cause of aberrant rRNA processing in cdm that might be due to the reduced transcript level of RPS4 or the Val to Gly mutation in RPS4.
rRNA is stable only when integrated into ribosomal subunits; thus, rRNA abundance can serve as an indicator of the chloroplast ribosome content [76]. The ribosome is primarily composed of rRNA; thus, the aberrant processing of rRNA can affect ribosome function and consequently, plastid translation [47, 48, 77], which is indispensable in plant growth, development, and photosynthesis. Overall, our data suggested that the missense mutation in RPS4 might be the cause of aberrant rRNA processing, which affected plastid translation and resulted in chlorophyll deficiency and reduced plant growth.
Conclusions
We first reported the identification of a homoplasmic plastome mutant in Chinese cabbage this study. The A-to-C point in the plastome-encoded rps4 was associated with Chlorophyll deficiency and slow growth mutation in Chinese cabbage that impaired the rRNA processing and affected the ribosomal function and plastid translation.
Additional files
Table S1. Primers used in this study. (XLSX 11 kb)
Table S2. Mutation sites in the plastome of wild type ‘FT’ compared with the reference genome. (FREQ 10682 kb)
Table S3. Mutation sites in the plastome of cdm compared with the reference genome. (XLSX 9748 kb)
Figure S1. rRNA accumulation pattern of cdm, ‘FT’, ‘FT’ × cdm and cdm × ‘FT’ in ethidium bromide-stainde gel. (TIF 21348 kb)
Figure S2. Loading controls for RNA gel blotting. (TIF 16387 kb)
Acknowledgements
We thank Jinwen Liu and Junxiang Zhang for data interpretation and technical assistance.
Funding
This study was supported by the National Natural Science Foundation of China (No.31672144). The funding played roles in the design of the study and collection, analysis, and interpretation of data.
Availability of data and materials
All data are fully available without restriction.
Abbreviations
- 9AA
9-aminoacridine hydrochloride
- BrdU
5-bromo-2′-deoxyuridine
- EMS
Ethyl methanesulfonate
- Fv/Fm
optional maximal photochemical efficiency of PS II
- MNNG
methyl-nitro-nitrosoguanidine
- NGS
Next-generation sequencing
- NMU
N-nitroso-N-methyl-urea
- RPS4
chloroplast ribosomal protein S4
- SSC
saline sodium citrate
- ФPSII
-
effective quantum yield of PSII, the photochemical quenching coefficient (QP), and qN
non-photochemical Chl fluorescence quenching
Authors’ contributions
XT, YW and FH conceived the study and designed the experiments. XT drafted the manuscript. XT, DF conducted the experiments, ZL, SH, and YZ performed data analysis, YZ provided guidance in phenotype investigation assays, and SH in genetic analysis. HF, YW, YZ, SH and YZ participated in revised the manuscript. All authors read and approved the final manuscript.
Authors’ information
Xiaoyan Tang and Yiheng Wang are postgraduates from College of Horticulture, Shenyang Agricultural University, Shenyang City, Liaoning Province, People’s Republic of China. Yun Zhang and Shengnan Huang are lecturers at Shenyang Agricultural University, Shenyang City, Liaoning Province, People’s Republic of China. Zhiyong Liu, associate professor, Shenyang Agricultural University, Shenyang City, Liaoning Province, People’s Republic of China. Danli Fei, an undergraduate from College of Horticulture, Shenyang Agricultural University, Shenyang City, Liaoning Province, People’s Republic of China. Hui Feng, professor, College of Horticulture, Shenyang Agricultural University, Shenyang City, Liaoning Province, People’s Republic of China.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1353-y) contains supplementary material, which is available to authorized users.
Contributor Information
Xiaoyan Tang, Email: yitangxy@126.com.
Yiheng Wang, Email: sywyhyx@126.com.
Yun Zhang, Email: zhangyun-511@163.com.
Shengnan Huang, Email: hsn870806@163.com.
Zhiyong Liu, Email: lzyky99@163.com.
Danli Fei, Email: danli.fei@hotmail.com.
Hui Feng, Email: fenghuiaaa@263.net.
References
- 1.Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, Sundaresan V, Leung H. Rice mutant resources for gene discovery. Plant Mol Biol. 2004;54:325–334. doi: 10.1023/B:PLAN.0000036368.74758.66. [DOI] [PubMed] [Google Scholar]
- 2.Krishnan A, Guiderdoni E, An G, Hsing YI, Han CD, Lee MC, Yu SM, Upadhyaya N, Ramachandran S, Zhang Q, Sundaresan V, Hirochika H, Leung H, Pereira A. Mutant resources in rice for functional genomics of the grasses. Plant Physiol. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leon P, Arroyo A, Mackenzie S. Nuclear control of plastid and mitochondrial development in higher plants. Annu Rev Plant Biol. 1998;49(1):453–480. doi: 10.1146/annurev.arplant.49.1.453. [DOI] [PubMed] [Google Scholar]
- 4.Liu YY, Wang X, Zhang CZ, Yang L, Gao DL, Chen HM, Huang SW. A CsYcf54 variant conferring light green coloration in cucumber. Euphytica. 2016;208(3):509–517. doi: 10.1007/s10681-015-1592-z. [DOI] [Google Scholar]
- 5.Glick RE, Sears BB. Genetically programmed chloroplast dedifferentiation as a consequence of plastome–genome incompatibility in Oenothera. Plant Physiol. 1994;106:367–373. doi: 10.1104/pp.106.1.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frick G, Su Q, Apel K, Armstrong GA. An Arabidopsis porB porC double mutant lacking light-dependent NADPH: protochlorophyllide oxidoreductases B and C is highly chlorophyll-deficient and developmentally arrested. Plant J. 2003;35(2):141–153. doi: 10.1046/j.1365-313X.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu W, Fu Y, Hu G, Si H, Zhu L, Wu C, Sun Z. Identification and fine mapping of a thermo-sensitive chlorophyll deficient mutant in rice (Oryza sativa L.) Planta. 2007;226(3):785–795. doi: 10.1007/s00425-007-0525-z. [DOI] [PubMed] [Google Scholar]
- 8.Qin DD, Dong J, Xu FC, Guo GG, Ge ST, Xu Q, Xu YX, Li MF. Characterization and fine mapping of a novel barley stage greenrevertible albino gene (HvSGRA) by bulked segregant analysis based on SSR assay and specific length amplified fragment sequencing. BMC Genomics. 2015;16(1):838. doi: 10.1186/s12864-015-2015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell BW, Mani D, Curtin SJ, Slattery RA, Michno JM, Ort DR, Schaus PJ, Palmer RG, Orf JH, Stupar RM. Identical substitutions in magnesium chelatase paralogs result in chlorophyll-deficient soybean mutants. G3: Genes, Genomes, Genetics. 2015;5:123–131. doi: 10.1534/g3.114.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nothnagel T, Straka P. Inheritance and mapping of a yellow leaf mutant of carrot (Daucus carota) Plant Breed. 2003;122(4):339–342. doi: 10.1046/j.1439-0523.2003.00884.x. [DOI] [Google Scholar]
- 11.Liu XP, Yang C, Han FQ, Fang ZY, Yang LM. Genetics and fine mapping of a yellow-green leaf gene (ygl-1) in cabbage (Brassica oleracea var. capitata L) Molecular Breeding. 2016;36(6):1–8. doi: 10.1007/s11032-016-0509-4. [DOI] [Google Scholar]
- 12.Hess WR, Börner T. Organellar RNA polymerases of higher plants. Int Rev Cytol. 1999;190:1–59. doi: 10.1016/S0074-7696(08)62145-2. [DOI] [PubMed] [Google Scholar]
- 13.Colombo N, Emanuel C, Lainez V, Maldonado S, Prina AR, Börner T. The barley plastome mutant CL2 affects expression of nuclear and chloroplast housekeeping genes in a cell-age dependent manner. Mol Gen Genomics. 2008;279:403–414. doi: 10.1007/s00438-008-0321-x. [DOI] [PubMed] [Google Scholar]
- 14.Correns C. Vererbungsversuche mit blass (gelb) grünen und buntblättrigen Sippen bei Mirabilis Jalapa, Urtica pilulifera und Lunaria annua. Z Indukt Abstamm Vererbungs. 1909;1:291–329. [Google Scholar]
- 15.Baur E. Das Wesen und die Erblichkeitsverhältnisse der Varietates albomarginatae hort.vonPelargonium zonale. Mol Gen Genet. 1909;1(1):330–351. doi: 10.1007/BF01990603. [DOI] [Google Scholar]
- 16.Kirk JTO, Tilney-Bassett RAE. The plastids; their chemistry, structure, growth and inheritance. 1967. [Google Scholar]
- 17.Ris H, Plaut W. Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas. J Cell Biol. 1962;13(3):383. doi: 10.1083/jcb.13.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nass MK, Nass S. Intramitochondrial fibers with DNA characteristics. J Cell Biol. 1963;19:593–611. doi: 10.1083/jcb.19.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chloroplasts BL. J Cell Biol. 1981;91:256–270. doi: 10.1083/jcb.91.3.256s. [DOI] [Google Scholar]
- 20.Hagemann R. The sexual inheritance of plant organelles. In: Daniell H, Chase C, editors. Molecular biology and biotechnology of plant organelles. Heidelberg: Springer; 2004. pp. 93–113. [Google Scholar]
- 21.Reboud X, Zeyl C. Organelle inheritance in plants. Heredity. 1994;72:132–140. doi: 10.1038/hdy.1994.19. [DOI] [Google Scholar]
- 22.Bock R. Structure, function, and inheritance of plastid genomes. In: Bock R, editor. Cell and molecular biology of plastids. Berlin Heidelberg: Springer; 2007. pp. 29–63. [Google Scholar]
- 23.Newton KJ, Knudsen C, Gabay-Laughnan S, Laughnan JR. An abnormal growth mutant in maize has a defective mitochondrial cytochrome oxidase gene. Plant Cell. 1990;2:107–113. doi: 10.1105/tpc.2.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton KJ. Analysis of cytoplasmically inherited mutants. In: Freeling M, Walbot V, editors. The maize handbook. New York: Springer; 1994. pp. 413–417. [Google Scholar]
- 25.Gordon KHJ, Hildebrandt JW, Bohnert HJ, Herrmann RG, Schmitt JM. Analysis of the plastid DNA in an Oenothera plastome mutant deficient in ribulose bisphosphate carboxylase. Theor Appl Genet. 1980;57:203–207. doi: 10.1007/BF00264671. [DOI] [PubMed] [Google Scholar]
- 26.Bonnema AB, Castillo C, Reiter N, Cunningham M, Adams HP, O'Connell M. Molecular and ultrastructural analysis of a nonchromosomal variegated mutant (tomato mitochondrial mutants that cause abnormal leaf development) Plant Physiol. 1995;109:385–392. doi: 10.1104/pp.109.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prina AR. Mutator-induced cytoplasmic mutants in barley: genetic evidence of activation of a putative chloroplast transposon. J Hered. 1996;87:385–389. doi: 10.1093/oxfordjournals.jhered.a023020. [DOI] [Google Scholar]
- 28.Prina AR, Landau AM, Colombo N, Jaureguialzo M, Arias MC, Rios RD. Pacheco MG. Genetically unstable mutants as novel sources of genetic variability: the chloroplast mutator genotype in barley as a tool for exploring the plastid genome. In: Shu QY, editor. Induced plant mutations in the genomics era. Rome: Food and agriculture Organization of the United Nations; 2009. pp. 227–228. [Google Scholar]
- 29.Prina AR, Arias MC, Lainez V, Landau A, Maldonado S. A cytoplasmically inherited mutant controlling early chloroplast development in barley seedlings. Theor Appl Genet. 2003;107:1410–1418. doi: 10.1007/s00122-003-1391-0. [DOI] [PubMed] [Google Scholar]
- 30.Rogalski M, Schottler MA, Thiele W, Schulze WX, Bock R. Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell. 2008;20:2221–2237. doi: 10.1105/tpc.108.060392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleischmann TT, Scharff LB, Alkatib S, Hasdorf S, Schottler MA, Bock R. Nonessential plastid-encoded ribosomal proteins in tobacco: a developmental role for plastid translation and implications for reductive genome evolution. Plant Cell. 2011;23:3137–3155. doi: 10.1105/tpc.111.088906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Larosa V, Remacle C. Transformation of the mitochondrial genome. Int J Dev Biol. 2013;57:659–665. doi: 10.1387/ijdb.130230cr. [DOI] [PubMed] [Google Scholar]
- 33.Greiner S. Plastome mutants of higher plants. In: Bock R, Knoop V, editors. Genomics of chloroplasts and mitochondria. Netherlands: Springer; 2012. pp. 237–266. [Google Scholar]
- 34.Draper CK, Hays JB. Replication of chloroplast, mitochondrial and nuclear DNA during growth of unirradiated and UVB-irradiated Arabidopsis leaves. Plant J. 2000;23:255–265. doi: 10.1046/j.1365-313x.2000.00776.x. [DOI] [PubMed] [Google Scholar]
- 35.Azhagiri AK, Maliga P. Exceptional paternal inheritance of plastids in Arabidopsis suggests that low–frequency leakage of plastids via pollen may be universal in plants. Plant J. 2007;52:817–823. doi: 10.1111/j.1365-313X.2007.03278.x. [DOI] [PubMed] [Google Scholar]
- 36.Hagemann R. Induction of plastome mutations by nitro-urea-compounds. In: Edelman M, Hallick RB, Chua N-H, editors. Methods in chloroplast molecular biology. New York: Elsevier; 1982. pp. 119–127. [Google Scholar]
- 37.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 38.GuhaMajumdar M, Baldwin S, Sears BB. Chloroplast mutations induced by 9-aminoacridine hydrochloride are independent of the plastome mutator in Oenothera. Theor Appl Genet. 2004;108:543–549. doi: 10.1007/s00122-003-1454-2. [DOI] [PubMed] [Google Scholar]
- 39.Shikanai T, Foyer CH, Dulieu H, Parry MA, Yokota A. A point mutation in the gene encoding the rubisco large subunit interferes with holoenzyme assembly. Plant Mol Biol. 1996;31:399–403. doi: 10.1007/BF00021801. [DOI] [PubMed] [Google Scholar]
- 40.Chang TL, Stoike LL, Zarka D, Schewe G, Chiu WL, Jarrell DC, Sears BB. Characterization of primary lesions caused by the plastome mutator of Oenothera. Curr Genet. 1996;30(6):522–530. doi: 10.1007/s002940050165. [DOI] [PubMed] [Google Scholar]
- 41.Herrmann RG, Westhoff P, Link G. Biogenesis of plastids in higher plants. In: Herrmann RG, editor. Cell organelles. Vienna: Springer; 1992. pp. 275–349. [Google Scholar]
- 42.Landau A, Paleo AD, Civitillo R, Jaureguialzo M, Prina AR. Two infA gene mutations independently originated from a Mutator genotype in barley. J Hered. 2007:272–6. [DOI] [PubMed]
- 43.Kahlau S, Aspinall S, Gray JC, Bock R. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J Mol Evol. 2006;63:194–207. doi: 10.1007/s00239-005-0254-5. [DOI] [PubMed] [Google Scholar]
- 44.Rogalski M, Ruf S, Bock R. Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 2006;34(16):4537–4545. doi: 10.1093/nar/gkl634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kavanagh TA, O’Driscoll KM, McCabe PF, Dix PJ. Mutations conferring lincomycin, spectinomycin, and streptomycin resistance in Solanum nigrum are located in three different chloroplast genes. Mol Gen Genet. 1994;242:675–680. doi: 10.1007/BF00283422. [DOI] [PubMed] [Google Scholar]
- 46.Yeh KC, KY T, Sun SW, Wu MC, Lin TY, Chen CC. Point mutations in the chloroplast 16s rRNA gene confer streptomycin resistance in Nicotiana plumbaginifolia. Curr Genet. 1994;26:132–135. doi: 10.1007/BF00313800. [DOI] [PubMed] [Google Scholar]
- 47.Wicke S, Schneeweiss GM, Müller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011;76(3–5):273–297. doi: 10.1007/s11103-011-9762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tiller N, Bock R. The translational apparatus of plastids and its role in plant development. Mol Plant. 2014;7:1105–1120. doi: 10.1093/mp/ssu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahlert D, Ruf S, Bock R. Plastid protein synthesis is required for plant development in tobacco. Proc Natl Acad Sci. 2003;100:15730–15735. doi: 10.1073/pnas.2533668100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pesaresi P, Varotto C, Meurer J, Jahns P, Salamini F, Leister D. Knock-out of the plastid ribosomal protein L11 in Arabidopsis: effects on mRNA translation and photosynthesis. Plant J. 2001;27(3):179–189. doi: 10.1046/j.1365-313x.2001.01076.x. [DOI] [PubMed] [Google Scholar]
- 51.Romani I, Tadini L, Rossi F, Masiero S, Pribil M, Jahns P, Kater M, Leister D, Pesaresi P. Versatile roles of Arabidopsis plastid ribosomal proteins in plant growth and development. Plant J. 2012;72:922–934. doi: 10.1111/tpj.12000. [DOI] [PubMed] [Google Scholar]
- 52.Zhang JX, Yuan H, Yang Y, Fish T, Lyi SM, Thannhauser TW, Zhang LG, Li L. Plastid ribosomal protein S5 is involved in photosynthesis, plant development, and cold stress tolerance in Arabidopsis. J Exp Bot. 2016;67:2731–2744. doi: 10.1093/jxb/erw106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang JB, Tang M, Li HT, Zhang ZR, Li DZ. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 2013;13(1):84. doi: 10.1186/1471-2148-13-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Genyi L, Muqiang G, Xiushan Z. Isolate microspore culture of Chinese cabbage (Brassica campestris ssp. pekinensis) Acta Hortic Sinica. 1993;20:167–170. [Google Scholar]
- 55.Huang SN, Liu ZY, Li DY, Yao RP, Meng Q, Feng H. Screening of Chinese cabbage mutants produced by 60Co γ-ray mutagenesis of isolated microspore cultures. Plant Breed. 2014;133(4):480–488. doi: 10.1111/pbr.12166. [DOI] [Google Scholar]
- 56.Huang SN, Liu ZY, Yao RP, Li DY, Zhang T, LI X, Hou L, Wang YH, Tang XY, Feng H. Candidate gene prediction for a petal degeneration mutant, pdm, of the Chinese cabbage (Brassica campestris ssp. pekinensis) by using fine mapping and transcriptome analysis. Mol Breed. 2016;36(3):1–10. [Google Scholar]
- 57.Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res. 1968;50(1):151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- 58.Lichter R. Induction of haploid plants from isolated pollen of Brassica napus. Z Pflanzenphysiol. 1982;105:427–434. doi: 10.1016/S0044-328X(82)80040-8. [DOI] [Google Scholar]
- 59.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15(3):473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x. [DOI] [Google Scholar]
- 60.Inskeep WP, Bloom PR. Extinction coefficients of chlorophyll a and b in N, N-dimethylformamide and 80% acetone. Plant Physiol. 1985;77:483–485. doi: 10.1104/pp.77.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lichtenthaler HK, Kuhn G, Prenzel U, Meier D. Chlorophyll-protein levels and degree of thylakoid stacking in radish chloroplasts from high-light, low-light and bentazon-treated plants. Physiol Plant. 1982;56(2):183–188. doi: 10.1111/j.1399-3054.1982.tb00322.x. [DOI] [Google Scholar]
- 62.Bilger W, Björkman O. Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis. Photosynth Res. 1990;25(3):173–185. doi: 10.1007/BF00033159. [DOI] [PubMed] [Google Scholar]
- 63.Maxwell K, Johnson GN. Chlorophyll fluorescence—a practical guide. J Exp Bot. 2000;51(345):659–668. doi: 10.1093/jexbot/51.345.659. [DOI] [PubMed] [Google Scholar]
- 64.Wu J, Liu B, Cheng F, Ramchiary N, Choi SR, Lim YP, Wang XW. Sequencing of chloroplast genome using whole cellular DNA and Solexa sequencing technology. The Brassica Genome. 2012:s 23. [DOI] [PMC free article] [PubMed]
- 65.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 66.Liu JW, Zhou WB, Liu GF, Yang CP, Sun Y, Wu WJ, Cao SQ, Wang C, Hai GH, Wang ZF, Bock R, Huang JR, Cheng YX. The conserved endoribonuclease YbeY is required for chloroplast ribosomal RNA processing in Arabidopsis. Plant Physiol. 2016;168:205–221. doi: 10.1104/pp.114.255000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang JR, Takano T, Akita S. Expression of ɑ-expansin genes in young seedlings of rice (Oryza sativa L.) Planta. 2000;211:76–473. doi: 10.1007/s004250000311. [DOI] [PubMed] [Google Scholar]
- 68.Maple J, Møller SG. Mutagenesis in Arabidopsis. Circadian Rhythms: Methods and Protocols. 2007:197–206. [DOI] [PubMed]
- 69.Redei GP, Plurad SB. Hereditary structural alterations of plastids induced by a nuclear mutator gene in Arabidopsis. Protoplasma. 1973;77(2):361–380. doi: 10.1007/BF01276769. [DOI] [Google Scholar]
- 70.Prina AR. A mutator nuclear gene inducing a wide spectrum of cytoplasmically inherited chlorophyll deficiences in barley. Theor Appl Genet. 1992;85(2):245–251. doi: 10.1007/BF00222866. [DOI] [PubMed] [Google Scholar]
- 71.Stoike LL, Sears BB. Plastome mutator–induced alterations arise in Oenothera chloroplast DNA through template slippage. Genetics. 1998;149(1):347–353. doi: 10.1093/genetics/149.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tiller N, Weingartner M, Thiele W, Maximova E, Schottler MA, Bock R. The plastid-specific ribosomal proteins of Arabidopsis thaliana can be divided into non-essential proteins and genuine ribosomal proteins. Plant J. 2012;69:302–316. doi: 10.1111/j.1365-313X.2011.04791.x. [DOI] [PubMed] [Google Scholar]
- 73.Maguire BA, Wild DG. The roles of proteins L28 and L33 in the assembly and function of Escherichia coli ribosomes in vivo. Mol Microbiol. 1997;23(2):237–245. doi: 10.1046/j.1365-2958.1997.2131578.x. [DOI] [PubMed] [Google Scholar]
- 74.Dabbs ER. Mutants lacking individual ribosomal proteins as a tool to investigate ribosomal properties. Biochimie. 1991;73:639–645. doi: 10.1016/0300-9084(91)90043-Z. [DOI] [PubMed] [Google Scholar]
- 75.Kaczanowska M, Rydén-Aulin M. Ribosome biogenesis and the translation process in Escherichia coli. Microbiol Mol Biol Rev. 2007;71(3):477–494. doi: 10.1128/MMBR.00013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walter M, Piepenburg K, Schöttler MA, Petersen K, Kahlau S, Tiller N, Bock R. Knockout of the plastid RNase E leads to defective RNA processing and chloroplast ribosome deficiency. Plant J. 2010;64(5):851–863. doi: 10.1111/j.1365-313X.2010.04377.x. [DOI] [PubMed] [Google Scholar]
- 77.Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in this study. (XLSX 11 kb)
Table S2. Mutation sites in the plastome of wild type ‘FT’ compared with the reference genome. (FREQ 10682 kb)
Table S3. Mutation sites in the plastome of cdm compared with the reference genome. (XLSX 9748 kb)
Figure S1. rRNA accumulation pattern of cdm, ‘FT’, ‘FT’ × cdm and cdm × ‘FT’ in ethidium bromide-stainde gel. (TIF 21348 kb)
Figure S2. Loading controls for RNA gel blotting. (TIF 16387 kb)
Data Availability Statement
All data are fully available without restriction.