Abstract
The microRNA (miR)-30 family has been reported to be aberrantly expressed in several types of cancer. However, its contributions to lung cancer remain to be fully elucidated. Myocyte enhancer factor 2D (MEF2D), an oncogene in liver cancer, has been shown to be aberrantly expressed in lung cancer. In the present study, it was found that MEF2D and miR-30a were inversely correlated in lung cancer samples. Using an online database, it was predicted that miR-30a targeted the 3′ untranslated region (UTR) of MEF2D mRNA. The activity of luciferase with MEF2D 3′UTR was suppressed by transfecting cells with miR-30a mimics. The results of western blot analysis showed that the miR-30a mimics also suppressed the MEF2D protein. The miR-30a mimics were able to reduce the growth and colonies of lung cancer cells by suppressing MEF2D. The results of FACS and western blot assays showed that the apoptotic rate was reduced by transfection with the miR-30a mimics. Collectively, the aberrant expression of miR-30a in lung cancer promoted the expression of MEF2D protein. miR-30a inhibited the growth and colony formation of the lung cancer cells by promoting apoptosis.
Keywords: lung cancer, microRNA-30a, myocyte enhancer factor 2D
Introduction
The microRNA (miR)-30 family contains five members and six mature miRs, namely miR-30a, miR-30b, miR-30c-1, mIR-30c-2, miR-30d and miR-30e (1). The majority of previous studies have shown that miR-30 family members act as tumor suppressors in various types of cancer (2–4). In addition, miR-30 family members are frequently found to be downregulated in these types of tumor tissue. Several studies have revealed that miR-30d and miR-30a are antimetastatic factors in tumors (5,6), and interfere with the epithelial-mesenchymal transition (EMT) (7). Certain miR-30 family members have been identified to abrogate chemoresistance and promote cancer cell apoptosis (7–12). In a previous study on lung cancer, Chan et al established the potential role of the miR-30 family in the modulation of the phosphoinositide 3-kinase-seven in absentia homolog 2 interaction in non-small cell lung cancer (NSCLC) (13). In other investigations, miR-30b and miR-30c were found to inhibit sarcoma viral oncogene homolog, which affected gefitinib-induced apoptosis and the EMT of NSCLC cells (14). miR-30a was also found to be negatively associated with the expression of N-cadherin, a mesenchymal marker (2), and miR-30d was demonstrated to inhibit tumor growth and invasion in NSCLC (15). However, the possible role of miR-30a in lung cancer remains to be elucidated. In the present study, it was found that miR-30a was underexpressed in lung cancer tissue, compared with that in non-cancerous tissue. Furthermore, the expression of miR-30a was inversely correlated with the mRNA and protein expression levels of myocyte enhancer factor 2D (MEF2D) in these tissue samples. The data showed that miR-30a suppressed lung cancer cell proliferation via directly targeting the 3′untranslated region (UTR) of MEF2D mRNA, inducing tumor cell apoptosis.
Materials and methods
Tissues and cell culture
The present study involved the collection of eight NSCLC specimens with the approval of the Ethical Committee of Qingdao Municipal Hospital (Qingdao, China) and each patient provided written informed consent. All procedures performed in investigations involving human participants were in accordance with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The tumor specimens were homogenized in tissue homogenization buffer immediately following resection and stored at −80°C until use. Patient information is available in Table I.
Table I.
Patient information.
| No. | Gender | Age | Dates of collection | Method of collection |
|---|---|---|---|---|
| 1 | Male | 42 | 2015.4.5 | Puncture surgery |
| 2 | Male | 44 | 2015.4.12 | Puncture surgery |
| 3 | Male | 57 | 2015.4.19 | Puncture surgery |
| 4 | Male | 60 | 2015.4.22 | Puncture surgery |
| 5 | Male | 38 | 2015.4.25 | Puncture surgery |
| 6 | Male | 51 | 2015.5.30 | Puncture surgery |
| 7 | Male | 59 | 2015.6.4 | Puncture surgery |
| 8 | Male | 65 | 2015.6.5 | Puncture surgery |
The A549 human lung cancer cell line (American Type Culture Collection, Manassas, VA, USA) was cultured in Dulbecco's modifed Eagle's medium (DMEM; Invitrogen; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA, Carlsbad, CA, USA) and penicillin (100 U/ml) RPMI 1640 at 37°C with 5% CO2.
Immunohistochemistry and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
The formalin-fixed, paraffin-embedded tissue sections underwent immunohistochemical staining using the streptavidin-biotin-peroxidase complex method. For antigen retrieval, ultrathin sections (5 µm) were heated in 0.5 M Tris-HCl (pH 9.0), for 1–2 h at 95°C. Rabbit anti-human MEF2D antibody (cat. no. HPA004807; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was used to detect the expression of MEF2D. Primary antibody was diluted to 1:500 and incubated at 4°C overnight. The slides were counterstained using hematoxylin. The images were captured using a Leica Aperio VERSA system (Leica Microsystems, Inc., Buffalo Grove, IL, USA), at magnification, ×200. The 3′UTR of MEF2D mRNA was predicted to be targeted by miR-30a in the TargetScan database (www.targetscan.org). The processing of all cell lines and tissue samples for RNA extraction were performed using an mRNA and miRNA isolation kit [RNeasy Mini Kit (50), cat. no. 74104; Qiagen China Co., Ltd., Shanghai, China]. According to the manufacturer's protocol The stem-loop RT-qPCR primers were as follows: miR-30a, RT primer 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACGCTGCA-3′, forward primer 5′-CGACGACTTTCA-GTCGGATGTT-3′ and reverse primer 5′-GTGCAGGGTCCGAGGT-3′; U6, RT and reverse primer 5′-GTGCA-GGGTCCGAGGT-3′ and forward primer, 5′-CTCGCTTCGGCAGCACA-3′ as previously described (2). The method of quantification was the 2−ΔΔCq method (16).
Transfection and vectors
The A549 cells were plated in 6-well plates (6×105 cells/well) and 100 nm of miR-30a mimics or negative control of miR mimics were transfected into the A549 cells using Lipofectamine™ RNAiMAX transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The above mimics were purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China).
Cell proliferation assay
At 24 h post-transfection, the medium was replaced with 100 µl fresh medium, and 20 µl of CellTiter 96 AQueous One solution (Promega Corporation, Madison, WI, USA; cat. no. G3582). The plate was incubated at 37°C for 1 h, and 60 µl of medium in each well was transferred into an ELISA 96-well plate, following which the absorbance at 490 nm (450–540 nm) was recorded with a 96-well plate reader.
Colony formation assay
In a glass bottle, 4% agar was melted in a microwave and maintained in a 56°C water bath. The bottom layer was prepared from 5 ml of 10% FBS DMEM containing 0.75% agar. The top layer was prepared from 3 ml of 10% FBS DMEM-containing 3×104 cells and 0.36% agar. The dish was cultured in a 37°C incubator for 15 days.
Analysis of apoptosis
The cells were collected and washed twice with PBS. Each pellet was resuspended in PBS (400 µl). Subsequently, 100 µl of incubation buffer was added to the cells with 2 µl of Annexin (1 mg/ml) and 2 µl of propidium iodide (PI; 1 mg/ml). The cells were then subjected to flow cytometry (BD Accuri™ C6; BD Biosciences, Franklin Lakes, NJ, USA).
Western blot analysis
Western blot analyses were performed as described by Ma et al (17). Total protein was extracted and quantified using Beyotime protein systems (pre-chilled lysis buffer, cat. no. P0013; bicinchoninic acid assay, cat. no. P0012; Beyotime Institute of Biotechnology, Haimen, China) according to the manufacturer's protocol. Proteins were separated using 10% SDS-PAGE. Gels were transferred to nitrocellulose membranes, which were blocked in 5% fetal bovine serum (FBS; Sigma-Aldrich; Merck KGaA) in Tris-buffered saline with Tween-20 for 1 h. The membrane was subsequently incubated with MEF2D antibody (1:1,000; cat. no. 610774, BD Biosciences) at 4°C, overnight. The horseradish peroxidase-conjugated secondary antibody (1:3,000; cat. no. 7074/7076; Cell Signaling Technology, Inc., Danvers, MA, USA) was incubated with the membranes at room temperature for 1 h. Chemiluminescent horseradish peroxidase substrate was purchased from Merck KGaA (cat. no. WBKLS0500). Quantification of blots bands was performed using ImageJ software (version 1.8.0; National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
The data obtained for RT-qPCR analysis, cell growth rate and colony formation are expressed as the mean ± standard deviation, and were compared at a given time point using one-way analysis of variance followed by Fisher's LSD test. Analysis was performed using SPSS software (version 19.0.1; IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a statistically significant difference.
Results
MEF2D is overexpressed in lung cancer
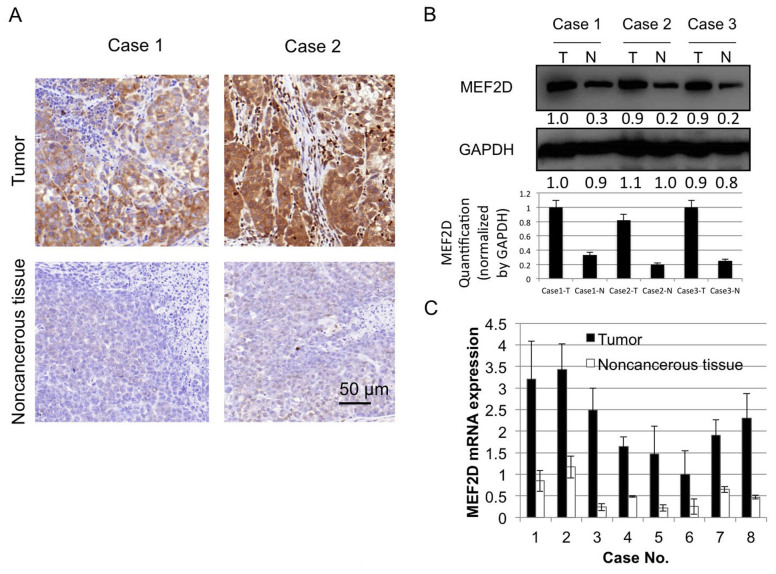
MEF2D has been identified as an oncogene in liver cancer (17), however, the expression level of MEF2D in lung cancer has not to been elucidated. To investigate whether MEF2D is aberrantly expressed in lung cancer tissue, immunohistochemical staining for MEF2D was performed in the present study to evaluate differential staining in cancer tissue, vs. non-cancerous tissue. Eight cancer tissue samples were stained for the expression of MEF2D, of which six were strongly positive and two were moderately positive. By contrast, the paired non-cancerous tissues were negative or weakly positive (Fig. 1A). The western blot and RT-qPCR analyses of the protein and mRNA expression levels of MEF2D in paired tumor and non-cancerous tissues showed similar results (Fig. 1B and C).
Figure 1.
MEF2D is expressed at a high level in lung cancer tissue samples. (A) Immunohistochemical analysis of the expression of MEF2D was performed in tumor and paired non-cancerous tissues. (B) Western blot analysis of the protein expression of MEF2D in lung cancer and paired non-cancerous tissue. Quantification of western blots is shown below corresponding bands. (C) Reverse transcription-quantitative polymerase chain reaction analysis of mRNA levels of MEF2D in paired patient cancer and normal tissues. MEF2D, myocyte enhancer factor 2D; T, tumor; N, non-cancerous.
Expression of MEF2D is regulated by miR-30a in lung cancer
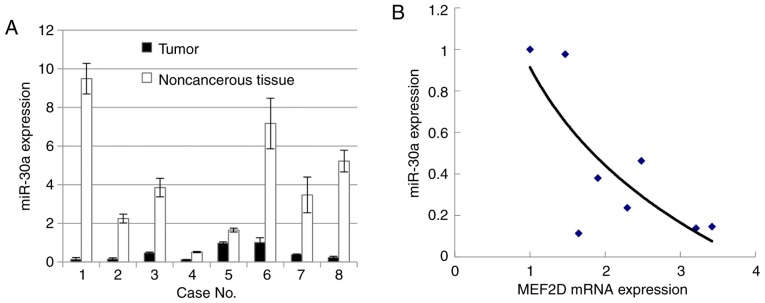
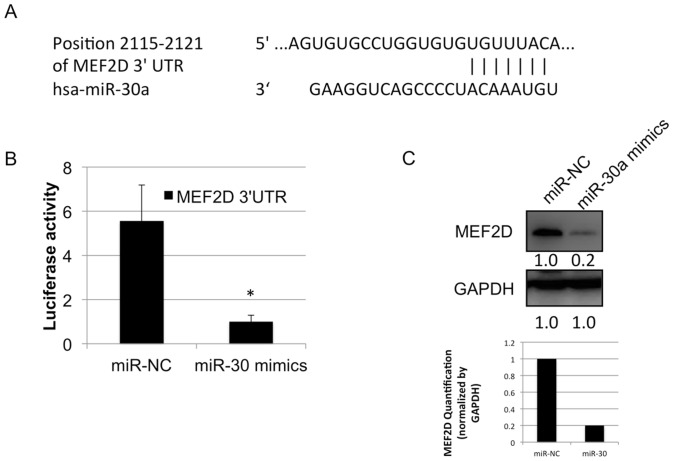
miR-30c was previously reported to be poorly expressed in lung cancer as a marker of EMT (18). To investigate whether miR-30a is underexpressed in cancer tissues, vs. non-cancerous tissues, RT-qPCR analysis was used to evaluate the level of miR-30a in cancer tissues and the paired non-cancerous tissues. The results showed that miR-30a was underexpressed in the lung cancer samples, compared with that in the paired non-cancerous samples. Subsequently, a correlation curve was established between the mRNA level of MEF2D and expression level of miR-30a (Fig. 2A). The data showed that the mRNA level of MEF2D was negatively correlated with that of miR-30a (Fig. 2B). The 3′UTR of MEF2D mRNA was predicted to be targeted by miR-30a in the TargetScan database (www.targetscan.org) (Fig. 3A). To investigate whether miR-30a directly targets the MEF2D 3′ UTR, a luciferase reporter vector was constructed which carried the MEF2D 3′UTR containing the specific seed sequence of miR-30a. This vector was transfected with or without miR-30a mimics into the A549 cell line. The results showed that the luciferase activity was significantly inhibited by transfection with the miR-30a mimics, compared with that in cells transfected with the miR-negative control (Fig. 3B). The protein level of MEF2D was markedly downregulated following the transfection of miR-30a mimics in the A549 cell line (Fig. 3C). These data indicated that miR-30a regulated the protein expression of MEF2D in the human lung cancer cell line.
Figure 2.
miR-30a is underexpressed in lung cancer samples. (A) Reverse transcription-quantitative polymerase chain reaction analysis of the expression of miR-30a was performed in tumor tissues and paired non-cancerous tissues. (B) Correlation curve between mRNA expression level of MEF2D and expression level of miR-30a. MEF2D, myocyte enhancer factor 2D; miR, microRNA.
Figure 3.
miR-30a regulates the expression of MEF2D in lung cancer. (A) An miR-30a binding site was predicted to exist in the 3′UTR of MEF2D mRNA. (B) miR-30a mimics and a luciferase vector with the MEF2D 3′UTR were co-transfected into A549 cells and the luciferase activity was detected. (C) Western blot analysis of the protein expression of MEF2D was performed in A549 cells following transfection with miR-30a mimics and miR-NC. Quantified results of the western blot analysis are shown below the corresponding bands. MEF2D, myocyte enhancer factor 2D; miR, microRNA; NC, negative control; 3′UTR, 3′untranslated region. *P<0.05 vs. NC.
Low expression of miR-30a promotes the growth of lung cancer cells
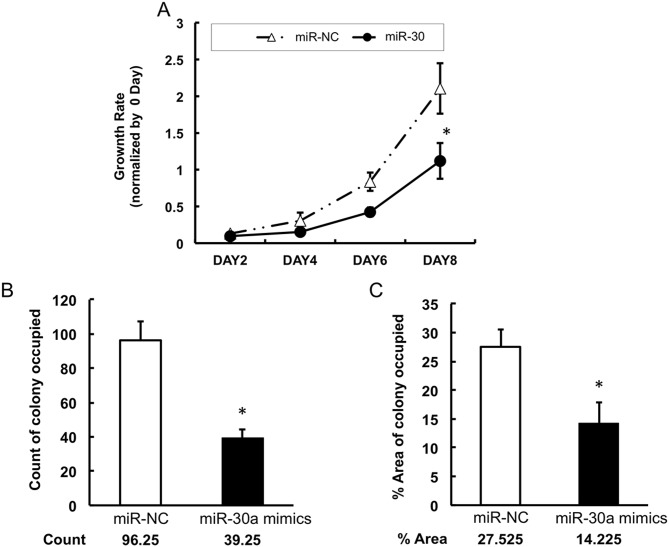
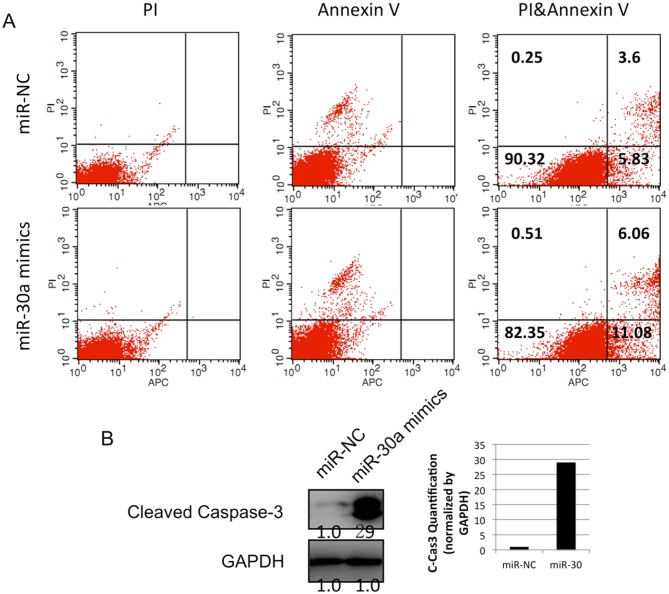
To investigate whether miR-30a can inhibit lung cancer cell growth, the present study measured the proliferation of A549 cells following transfection with miR-30a mimics or negative control mimics. It was found that the proliferation of A549 cells was significantly suppressed by the restoration of miR-30a from day 6, compared with that in the negative control group (Fig. 4A). Similarly, the colony formation assays showed that the miR-30a-transfected group had fewer colonies of A549 cells, compared with the control group (Fig. 4B and 4C). To determine the mechanisms by which miR-30a reduced the proliferation of lung cancer cells, the apoptosis of the A549 cell line was examined using an Annexin V/PI kit and flow cytometry. The data showed that, following transfection with miR-30a mimics, the percentage of Annexin V+/PI− cells in the population was increased from 5.83 to 11.08% in the lung cancer cells (Fig. 5A). This result revealed that miR-30a induced the apoptosis of lung cancer cells. In addition, the level of cleaved caspase 3 was detected using western blot analysis. The protein level of cleaved caspase 3 was increased following the infection of miR-30a mimics (Fig. 5B). Together, these data indicated that the overexpression of miR-30a in lung cancer cells induced cancer cell apoptosis by stimulating the apoptotic pathway involving caspase 3.
Figure 4.
miR-30a suppresses the proliferation and colony formation of A549 cells. (A) Growth rates of A549 cells transfected with miR-30a mimics were determined. (B) Numbers of colonies and (C) percentage area of formed colonies were measured in the cells transfected with miR-30a mimics. MEF2D, myocyte enhancer factor 2D; miR, microRNA; NC, negative control. *P<0.05 vs. NC.
Figure 5.
miR-30a promotes the apoptosis of A549 cells. (A) Annexin V/PI staining analysis was performed on the A549 cells using FACS. (B) Western blot analysis of cleaved caspase-3 was performed in the miR-30a-mimics-transfected A549 cells. Quantified results of the western blot analysis are shown below the corresponding bands. MEF2D, myocyte enhancer factor 2D; miR, microRNA; NC, negative control; PI, propidium iodide.
Discussion
MEF2D is a member of a family of four myocyte enhancer factor transcription factors, (MEF2A, 2B, 2C and 2D), with an important role in muscle differentiation (19,20). The transcription factor MEF2D promotes the survival of various types of cells and functions as an oncogene in liver cancer (17,21,22).
Zhao et al reported that oleanolic acid (OA) suppressed the proliferation of lung carcinoma cells (23). Furthermore, their data revealed that OA induced the expression of miR-122. Cyclin G1 and MEF2D, two putative miR-122 targets, were found to be downregulated by OA treatment. However, the MEF2D expression profile in lung cancer and its regulatory mechanism remained to be elucidated.
In the present study, the mRNA and protein levels of MEF2D were measured in lung cancer using immunohistochemistry, western blot analysis and RT-qPCR analysis (Fig. 1A-C). The data showed that MEF2D was overexpressed in the lung cancer tissues, compared with the non-cancerous tissues. Of note, it was observed that miR-30a was downregulated in lung cancer samples, compared with control samples (Fig. 2A). In addition, the levels of MEF2D mRNA and miR-30a were inversely correlated (Fig. 2B).
To investigate whether miR-30a regulated the expression of MEF2D in lung cancer, mimics of miR-30a and its comparative control were synthesized and transfected into A549 lung cancer cells. The data showed that the increased expression of miR-30a suppressed the protein level of MEF2D (Fig. 3C). These results led to the investigation of whether miR-30a regulated MEF2D by directly targeting its 3′UTR. The present study analyzed miR-30a and MEF2D mRNA 3′UTR sequences using the online miRNA database, TargetScan. It was found that the targeted sequence sites in the 3′UTR of MEF2D by miR-30a were conserved among species, which indicated that these sites may be functional for physiological processes. In addition, a luciferase reporter vector was constructed, which carried specific miR-30a seed sequences within the MEF2D mRNA 3′UTR. The results showed that the activity of luciferase was markedly inhibited by transfection with miR-30a mimics in the lung cancer cell line (Fig. 3B). Therefore, it was hypothesized that the suppression of MEF2D was responsible, at least in part, for the antitumor effect of miR-30a. In accordance, Song et al (24) demonstrated that the expression of MEF2D is decreased by another miR (miR-218), in lung carcinoma. One possible explanation is that the same miR may have different targets and the same mRNA may be targeted by different miRs in diverse cell types (22,24).
In the present study, the data also showed that the upregulation of miR-30a inhibited the proliferation of lung cancer cells by inducing apoptosis. In addition, an increase in the level of cleaved-caspase 3 was detected, which is a critical executioner of apoptosis. Taken together, the results indicated that the upregulation of miR-30a induced the apoptosis of lung cancer cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
NL performed functional assays. YW collected specimens. XL performed data analyses and wrote the manuscript. All authors participated in discussions and interpretation of the data and results.
Ethics approval and consent to participate
The present study involved the collection of eight NSCLC specimens with the approval of the Ethical Committee of Qingdao Municipal Hospital (Qingdao, China) and each patient provided written informed consent.
Consent for publication
Each patient provided written informed consent for publication.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumarswamy R, Mudduluru G, Ceppi P, Muppala S, Kozlowski M, Niklinski J, Papotti M, Allgayer H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int J Cancer. 2012;130:2044–2053. doi: 10.1002/ijc.26218. [DOI] [PubMed] [Google Scholar]
- 3.Liu Z, Tu K, Liu Q. Effects of microRNA-30a on migration, invasion and prognosis of hepatocellular carcinoma. FEBS Lett. 2014;588:3089–3097. doi: 10.1016/j.febslet.2014.06.037. [DOI] [PubMed] [Google Scholar]
- 4.Sousa JF, Nam KT, Petersen CP, Lee HJ, Yang HK, Kim WH, Goldenring JR. miR-30-HNF4γ and miR-194-NR2F2 regulatory networks contribute to the upregulation of metaplasia markers in the stomach. Gut. 2016;65:914–924. doi: 10.1136/gutjnl-2014-308759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu K, Guo L, Guo Y, Zhou B, Li T, Yang H, Yin R, Xi T. AEG-1 3′-untranslated region functions as a ceRNA in inducing epithelial-mesenchymal transition of human non-small cell lung cancer by regulating miR-30a activity. Eur J Cell Biol. 2015;94:22–31. doi: 10.1016/j.ejcb.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 6.Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2010;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- 7.Kao CJ, Martiniez A, Shi XB, Yang J, Evans CP, Dobi A, deVere White RW, Kung HJ. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene. 2014;33:2495–2503. doi: 10.1038/onc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershkovitz-Rokah O, Modai S, Pasmanik-Chor M, Toren A, Shomron N, Raanani P, Shpilberg O, Granot G. MiR-30e induces apoptosis and sensitizes K562 cells to imatinib treatment via regulation of the BCR-ABL protein. Cancer Lett. 2015;356:597–605. doi: 10.1016/j.canlet.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Zou Z, Wu L, Ding H, Wang Y, Zhang Y, Chen X, Chen X, Zhang CY, Zhang Q, Zen K. MicroRNA-30a sensitizes tumor cells to cis-platinum via suppressing beclin 1-mediated autophagy. J Biol Chem. 2012;287:4148–4156. doi: 10.1074/jbc.M111.307405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ouzounova M, Vuong T, Ancey PB, Ferrand M, Durand G, Le-Calvez Kelm F, Croce C, Matar C, Herceg Z, Hernandez-Vargas H. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genomics. 2013;14:139. doi: 10.1186/1471-2164-14-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heinzelmann J, Unrein A, Wickmann U, Baumgart S, Stapf M, Szendroi A, Grimm MO, Gajda MR, Wunderlich H, Junker K. MicroRNAs with prognostic potential for metastasis in clear cell renal cell carcinoma: A comparison of primary tumors and distant metastases. Ann Surg Oncol. 2014;21:1046–1054. doi: 10.1245/s10434-013-3361-3. [DOI] [PubMed] [Google Scholar]
- 12.Fuster O, Llop M, Dolz S, García P, Such E, Ibáñez M, Luna I, Gómez I, López M, Cervera J, et al. Adverse prognostic value of MYBL2 overexpression and association with microRNA-30 family in acute myeloid leukemia patients. Leuk Res. 2013;37:1690–1696. doi: 10.1016/j.leukres.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Chan LW, Wang F, Meng F, Wang L, Wong SC, Au JS, Yang S, Cho WC. MiR-30 family potentially targeting PI3K-SIAH2 predicted interaction network represents a novel putative theranostic panel in non-small cell lung cancer. Front Genet. 2017;8:8. doi: 10.3389/fgene.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang L, Liu L, Huang S, Zhao Y, He X. MicroRNA-30d-5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer. Cancer Lett. 2015;362:208–217. doi: 10.1016/j.canlet.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Ma L, Liu J, Liu L, Duan G, Wang Q, Xu Y, Xia F, Shan J, Shen J, Yang Z, et al. Overexpression of the transcription factor MEF2D in hepatocellular carcinoma sustains malignant character by suppressing G2-M transition genes. Cancer Res. 2014;74:1452–1462. doi: 10.1158/0008-5472.CAN-13-2171. [DOI] [PubMed] [Google Scholar]
- 18.Zhong Z, Xia Y, Wang P, Liu B, Chen Y. Low expression of microRNA-30c promotes invasion by inducing epithelial mesenchymal transition in non-small cell lung cancer. Mol Med Rep. 2014;10:2575–2579. doi: 10.3892/mmr.2014.2494. [DOI] [PubMed] [Google Scholar]
- 19.Molkentin JD, Olson EN. Combinatorial control of muscle development by basic helix-loop-helix and MADS-box transcription factors. Proc Natl Acad Sci USA. 1996;93:9366–9373. doi: 10.1073/pnas.93.18.9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinsey TA, Zhang CL, Olson EN. MEF2: A calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/S0968-0004(01)02031-X. [DOI] [PubMed] [Google Scholar]
- 21.Hong X, Hong XY, Li T, He CY. Pokemon and MEF2D co-operationally promote invasion of hepatocellular carcinoma. Tumour Biol. 2015;36:9885–9893. doi: 10.1007/s13277-015-3744-0. [DOI] [PubMed] [Google Scholar]
- 22.Su L, Luo Y, Yang Z, Yang J, Yao C, Cheng F, Shan J, Chen J, Li F, Liu L, et al. MEF2D transduces microenvironment stimuli to ZEB1 to promote epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2016;76:5054–5067. doi: 10.1158/0008-5472.CAN-16-0246. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Liu M, Li D. Oleanolic acid suppresses the proliferation of lung carcinoma cells by miR-122/Cyclin G1/MEF2D axis. Mol Cell Biochem. 2015;400:1–7. doi: 10.1007/s11010-014-2228-7. [DOI] [PubMed] [Google Scholar]
- 24.Song L, Li D, Zhao Y, Gu Y, Zhao D, Li X, Bai X, Sun Y, Zhang X, Sun H, et al. miR-218 suppressed the growth of lung carcinoma by reducing MEF2D expression. Tumour Biol. 2016;37:2891–900. doi: 10.1007/s13277-015-4038-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.