Abstract
A previous study revealed that DEP domain containing 1 (DEPDC1) is involved in the carcinogenesis of bladder cancer via forming a complex with zinc finger protein 224 (ZNF224) to suppress A20 expression, resulting in the activation of the nuclear factor (NF)-κB signaling pathway; however, the role of DEPDC1 in liver cancer remains unclear. Hep G2 cells were treated with 11R-DEP: 611–628, a peptide capable of disrupting the DEPDC1-ZNF224 complex. Cell proliferation was examined using an MTT assay and apoptosis was analyzed via detection of the apoptotic marker caspase-3 using western blot analysis. A20 expression was examined via reverse transcription-quantitative polymerase chain reaction and NF-κB subcellular localization was determined via immunofluorescence staining. microRNA (miR)-130a was overexpressed in HepG2 cells and its effects on proliferation and apoptosis were examined. The results demonstrated that 11R-DEP: 611–628 (3 µM) and miR-130a inhibited cell proliferation and promoted apoptosis in HepG2 cells by activating A20 expression, which blocks the nuclear transportation of NF-κB. In addition, the results demonstrated that the 11R-DEP: 611–628 (3 µM) treatment resulted in downregulation of DEPDC1 expression, indicating that DEPDC1 expression is regulated by the DEPDC1-ZNF224 complex. In conclusion, the data indicated that DEPDC1 suppresses apoptosis to promote cell proliferation through the NF-κB signaling pathway in HepG2 cells and that DEPDC1 is a potential target for the treatment of liver cancer.
Keywords: DEP domain containing 1, liver cancer, apoptosis, nuclear factor-κB, microRNA-130a
Introduction
Primary liver cancer is one of the leading causes of lethal malignancy worldwide, with an increasing rate of incidence (1). In the last few decades, a number of medical approaches, including improvements in patient stratification and introduction of novel therapies such as sorafenib (2), were applied in clinical management; however, liver cancer remains a leading life-threatening disease worldwide (3). Therefore, it is necessary to discover effective molecule-targeting drugs against liver cancer.
DEP domain containing 1 (DEPDC1), a highly conserved protein normally expressed only in the testes, was first reported as being upregulated in bladder cancer cells (4). Subsequently, the ectopic expression of DEPDC1 has also been detected in other cancer types, including breast cancer (5), lung adenocarcinoma (6) and hepatocellular carcinomas (7). A study of the bladder cancer UM-UC-3 cell line elucidated the mechanism underlying carcinogenesis caused by DEPDC1; it interacts with zinc finger protein 224 (ZNF224) to form a complex, which acts as a transcription inhibitor of A20, a negative regulator of the nuclear factor (NF)-κB pathway (8). Notably, a peptide from the DEPDC1 protein sequence 611–628 (11R-DEP: 611–628) has the ability to disrupt the DEPDC1-ZNF224 complex efficiently in the bladder cancer UM-UC-3 cell line and thereby induces apoptosis in vitro and in vivo, inhibits cell proliferation and suppresses tumor growth (8). These data indicated that this peptide is a potential therapeutic candidate for different types of cancer with DEPDC1 expression. A recent study determined that microRNA (miR)-130a acts as a tumor suppressor in prostate cancer by targeting DEPDC1 and SEC23B (9); however, the role of miR-130a in liver cancer remains unknown.
In the present study, the role of DEPDC1 in liver cancer was investigated and the efficacy of peptide 11R-DEP: 611–628 and miR-130a in HepG2 cells was evaluated.
Materials and methods
Antibodies and peptides
The anti-DEPDC1 antibody (cat. no. LS-C167364; dilution 1:500) was purchased from LifeSpan BioSciences, Inc. (Seattle, WA, USA.). The caspase-3 (cat. no. 9665S; dilution 1:1,000), cleaved-caspase-3 (cat. no. 9661; dilution 1:300) and NF-κB antibodies (cat. no. 8242S; dilution 1:75) were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). The β-actin antibody (cat. no. TA-09; dilution 1:1,000) was purchased from OriGene Technologies, Inc. (Beijing, China). The Goat Anti-Rabbit IgG DyLight® 488 antibody (cat. no. A32731; dilution 1:500) was purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). 11R-DEP: 611–628 peptide and the scramble peptide were synthesized by Shanghai Top-Peptide Biotechnology Co., Ltd., (Shanghai, China) according to the sequences previously reported (8).
Cell culture and transfection
HepG2 (cat. no. KCB 200507YJ) and A549 (cat. no. KCB 200434YJ) cells were purchased from the Kunming Cell Bank of the Chinese Academy of Sciences (Kunming, Yunnan, People's Republic of China. http://www.kmcellbank.com/). Bel-7402 (cat. no. TcHu10), SK-Hep-1 (cat. no. TcHu109) and SMMC-7721 (cat. no. TcHu52) cells were obtained from the Cell Resource Center, Shanghai Institute of Life Sciences, Chinese Academy of Sciences (Shanghai, China). The cells were maintained in Dulbecco's modified Eagle's medium (cat. no. 11965-092; Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS; cat. no. 10437-028; Gibco; Thermo Fisher Scientific, Inc.) in a humidified atmosphere at 37°C containing 5% CO2 and 95% air. HepG2 cells were transiently transfected miR-130a (1.5 µg) plasmid and empty vector GV514 (1.5 µg) (purchased from Shanghai GenePharma Co., Ltd., Shanghai, China) using the Lipofectamine® 3000 reagent (cat. no. L3000015; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocols. After 72 h, the cells were harvested for cell counting and an examination of the cell morphology and apoptosis was conducted.
Western blot analysis
HepG2, A549, Bel-7402, SK-Hep-1 and SMMC-7721 cells were lysed with RIPA lysis buffer (150 mM NaCl; 50 M Tris-HCl; pH, 7.5; 1% Triton X-100; 0.1% sodium deoxycholate; and 0.1% SDS) containing 0.1% phenylmethane sulfonyl fluoride. Protein concentration of cell lysates was measured by BCA kit (Beyotime Institute of Biotechnology, Shanghai, China). Equal protein lysate samples (30 µg) were separated by 10% SDS-PAGE and then transferred onto polyvinylidene fluoride membrane, which was blocked in 5% nonfat milk at room temperature for 2 h, followed by incubation with primary antibodies at 4°C overnight. The membrane was then incubated with secondary antibody (horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG; catalog no. G-21234, dilution 1:5,000) or goat anti-mouse IgG (catalog no. G-21040, dilution 1:5,000) antibodies (Pierce; Thermo Fisher Scientific, Inc.) at room temperature for 1 h. Blots were developed using an enhanced chemiluminescence kit (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). The densitometric analysis of bands was performed using FluorChem SA (6.0.0) (Informer Technologies, Inc, Chicago, IL, USA).
Cell proliferation assay
The MTT dissolved in 0.1 M PBS at a concentration of 5 mg/ml was purchased from Beijing Solarbio Science & Technology Co., Ltd. The HepG2 cells were seeded onto 96-well plates in medium containing 2% FBS in the presence of 11R-DEP: 611–628 peptide or scramble peptide at 0, 0.5, 1, 2 and 3 µM. The medium was replaced with medium containing same concentration of peptides every 24 h for 4 days, and then the proliferation of the cells was detected. A total of 10 µl MTT was added into each well and then incubated at 37°C for 4 h. Finally, the DMEM culture medium supplemented with 2% FBS was discarded and 150 µl dimethyl sulfoxide was added into each well to dissolve the crystals completely. Absorbance was measured with a microplate reader at 490 nm.
Immunofluorescence staining
HepG2 cells were fixed with 4% paraformaldehyde for 30 min at room temperature following treatment with 11R-DEP: 611–628 peptide or scramble peptide at 1, 6 and 12 h. Following this, the cells were permeabilized with a 5% blocking solution (0.1 M PBS; pH, 7.4 containing 5% bovine serum albumin; and 0.3% Triton X-100) for 2 h at room temperature. The sections were subsequently incubated with the rabbit anti-NF-κB antibody overnight at 4°C in a humid chamber. Following three washes with PBS, the cells were incubated with Goat Anti-Rabbit IgG DyLight 488 antibody for 2 h at room temperature. The cell nuclei were counterstained with DAPI for 10 min at room temperature. Images of the cells were captured with a fluorescence microscope (IX71; Olympus Corporation, Tokyo, Japan) at a magnification of ×200.
Terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling (TUNEL) staining
The HepG2 cells (8×104/well) were seeded into wells of the chamber slides one day prior to the experiments. Cells were treated with 3 µM 11R-DEP: 611–628 peptide for 3 h at 37°C. Cells were fixed in freshly diluted 1% paraformaldehyde in PBS, pH 7.4 for 10 min at room temperature. TUNEL staining was performed with the Apoptosis Detection kit (cat. no. S7100; EMD Millipore, Billerica, MA, USA) according to the manufacturer's protocols. The nuclei were counterstained in 0.1% hematoxylin for 1 min at room temperature, and then rinsed with running tap water for 5 min. The slides were mounted with mounting medium Permount (cat. no. SP15-500, Thermo Fisher Scientific, Inc.), and a total of 10 randomly selected fields of view were observed under a fluorescent microscope (magnification, ×200).
Flow cytometric analysis
The HepG2 Cells (2.5×105) were transfected (the transfection protocol was performed as previously described) with the miR-130a expression plasmid or a control plasmid. After 72 h, the cells were harvested and washed with PBS twice. The cells then were stained with the FITC Annexin V/Dead Cell kit (Merck KGaA, Darmstadt, Germany) for 20 min at room temperature in the dark, according to the manufacturer's protocols. Finally, cells were analyzed using a flow cytometer (Muse® Cell Analyzer, MuseSoft_V1.5.0.0; Merck Millipore, Darmstadt, Germany).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the HepG2 cells using the TRIzol® reagent (Takara Bio Inc., Otsu, Japan). For RT-qPCR, the first-strand cDNA was reverse transcribed from 2 µg total RNA using the Reverse Transcription kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's protocol. The following primers were used: A20 forward, 5′-CGTCCAGGTTCCAGAACACCATTC-3′ and reverse, 5′-TGCGCTGGCTCGATCTCAGTTG-3′; DEPDC1 forward, 5′-GAGGTCACTGATGATACATAC-3′ and reverse, 5′-TGCAGTCTGTAAGTAAGAGG-3′; and GAPDH forward, 5′-CAGGAGGCATTGCTGATGAT-3′ and reverse, 5′-GAAGGCTGGGGCTCATTT-3′. To analyze miR-130a expression, the cDNA was amplified according to the manufacturer's protocol using the Mir-X miRNA First Strand Synthesis kit (#638313, Takara Bio Inc.). qRT-PCR was conducted according to the manufacturer's instructions of the Mir-X miRNA qRT-PCR SYBR kit (#638314, Takara Bio Inc.). The U6 (forward, 5′-GGAACGATACAGAGAAGATTAGC-3′ and reverse, 5′-TGGAACGCTTCACGAATTTGCG-3′) and miR-130a special primer (forward, 5′-GGCCAGAGCTCTTTTCACAT-3′ and reverse, 5′-CGGCCAATGCCCTTTTAACAT-3′) were purchased from Takara Bio Inc. The relative level of gene expression was quantified using the 2−∆∆Cq method (10), normalized to GAPDH or U6, and expressed as the fold induction of the control. PCR conditions were as followed: 95°C for 10 sec; 40 cycles: 95°C for 5 sec, and 60°C for 20 sec.
Statistical analysis
The values are expressed as the mean ± standard deviation. Comparisons between two groups were conducted using the Student's t-test and comparisons among multiple groups were performed using one-way analysis of variance, followed by the Student-Newman-Keuls post hoc test. SPSS 13.0 software (SPSS, Chicago, IL, USA) and GraphPad Prism 5.0 (GraphPad Software, Inc., La Jolla, CA, USA) software were utilized for statistical analyses. All experiments were performed at least three independent times, and representative data are presented. P<0.05 was considered to indicate a statistically significant difference.
Results
DEPDC1 is expressed in HepG2 cells
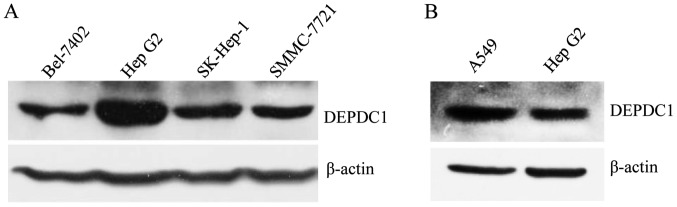
To understand the role of DEPDC1 in liver cancer, DEPDC1 expression at a protein level in HepG2 cells and other liver cancer cell lines was examined using a western blotting assay. To the best of our knowledge, DEPDC1 expression has not been detected in liver cancer cell lines in any published report to date; therefore, the A549 cell line was used as a positive control due to the role of DEPDC1 in A549 having been recently reported and due to the fact that samples were available (11). The results demonstrated that DEPDC1 is expressed in all of the liver cancer cell lines used, and its protein levels in HepG2 cells are the highest among the examined liver cancer cell lines (Fig. 1A), but lower than the A549 lung cancer cells (Fig. 1B); therefore, the HepG2 cell line was used to conduct further experiments.
Figure 1.
DEPDC1 was expressed in HepG2 cells. DEPDC1 protein expression levels were detected by western blotting. (A) The results demonstrated that HepG2 cells exhibited the highest DEPDC1 expression levels among the liver cancer cell lines used. (B) The DEPDC1 expression levels in the HepG2 cells were lower, compared with the A549 lung cancer cells. DEPDC1, DEP domain containing 1.
11R-DEP: 611–628 treatment inhibits HepG2 cell proliferation
A study reported that DEPDC1 has the ability to promote cell proliferation by forming a complex with ZNF224, which inhibits associated gene expression in the bladder cancer UM-UC-3 cell line (8). It was hypothesized that DEPDC1 functions in the same manner in HepG2 cells, and this hypothesis was investigated by treating HepG2 cells with 11R-DEP: 611–628, which is capable of disrupting the DEPDC1-ZNF224 complex in the bladder cancer UM-UC-3 cell line (8), and in the lung cancer A549 cell line (11). Cell proliferation was then analyzed using an MTT assay. Compared with the scramble peptide (Fig. 2A), 11R-DEP: 611–628 (3 µM) significantly inhibited HepG2 cell proliferation from day 1–4 (P<0.05) (Fig. 2B); therefore, 3 µM was selected as the optimal concentration for further experiments. Since the 11R-DEP: 611–628 peptide was designed to disrupt the DEPDC1-ZNF224 complex (8), this peptide inhibited the proliferation of HepG2 cells, indicating that the DEPDC1-ZNF224 complex serves a key role in promoting HepG2 cell proliferation. To summarize, DEPDC1 promotes HepG2 cell proliferation via interacting with ZNF224.
Figure 2.
11R-DEP: 611–628 inhibited HepG2 cell proliferation. Cell proliferation was analyzed using an MTT assay. (A) The scramble peptide had no effect on cell proliferation. (B) The 11R-DEP: 611–628 peptide (3 µM) suppressed cell proliferation. *P<0.05, indicating significant difference between 0 and 3 µM treatment at 1 and 4 days.
11R-DEP: 611–628 treatment induces the apoptosis of HepG2 cells
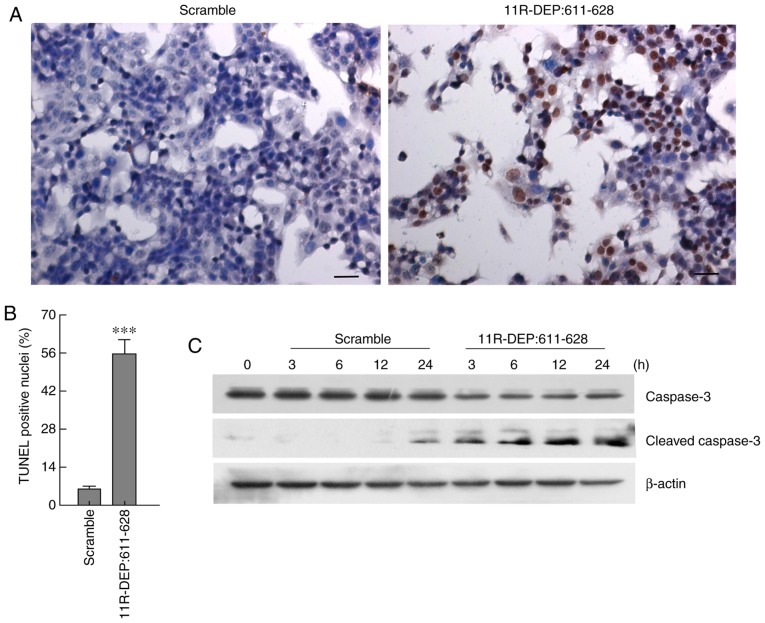
To further reveal how DEPDC1 affects cell proliferation, HepG2 cells were treated with 3 µM 11R-DEP: 611–628 peptide for 3, 6, 12, and 24 h; following this, apoptosis was analyzed by TUNEL staining, and the apoptotic marker caspase-3 protein, which is cleaved when apoptosis occurs, was detected via western blotting (12). The results demonstrated that treatment with 11R-DEP: 611–628 causes a significant increase (P<0.001) in TUNEL positive cells (Fig. 3A and B), a decrease in caspase-3 protein levels and an increase in the cleaved caspase-3 protein level, compared with the scramble peptide (Fig. 3C). These results demonstrated that 11R-DEP: 611–628 treatment enhances apoptosis, indicating that DEPDC1 inhibits apoptosis in HepG2 cells.
Figure 3.
11R-DEP: 611–628 induced the apoptosis of HepG2 cells. The TUNEL staining results indicated that the 11R-DEP: 611–628 (3 µM) peptide treatment caused a significant increase in the apoptosis of HepG2 cells. (A) Representative images of TUNEL staining. (B) Quantitative analysis results of TUNEL staining. ***P<0.001. (C) The western blot analysis results demonstrated that the caspase-3 levels in cells treated with the 11R-DEP: 611–628 (3 µM) peptide were notably decreased, while the cleaved caspase-3 levels were markedly increased, compared with those in the cells treated with the scramble peptide. TUNEL, terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling.
11R-DEP: 611–628 treatment results in the blocking of NF-κB nuclear translocation in HepG2 cells
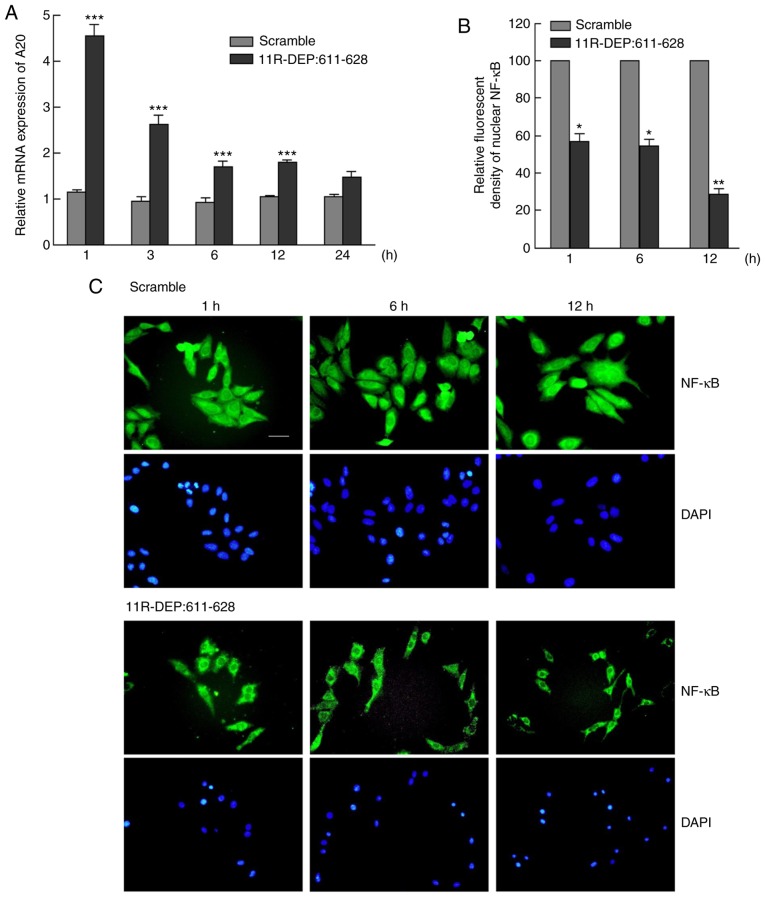
A previous study indicated that DEPDC1 functions through regulating A20 expression to inhibit NF-κB function in bladder cancer cell lines (8), and thus the A20 mRNA level should be increased following the disruption of the DEPDC1-ZNF224 complex with 11R-DEP: 611–628 (8). Therefore, it was investigated whether DEPDC1 serves the same role in HepG2 cells. The A20 mRNA expression levels were examined by RT-qPCR and the results demonstrated a significant increase (P<0.05) (Fig. 4A) between 1 and 24 h following treatment with the 11R-DEP: 611–628 peptide, compared with the scramble peptide. A20 protein expression levels were detected with commercial antibodies, but the protein level was not obtained due to poor antibody quality (data no shown). However, A20 mRNA increase following 11R-DEP: 611–628 treatment should be sufficient to support the theory that A20 expression is regulated by the DEPDC1-ZNF224 complex. In addition, the nuclear NF-κB expression was significantly decreased (P<0.05) in 11R-DEP: 611–628 peptide-treated cells, compared with scramble peptide-treated cells (Fig. 4B and C). These data indicated that the 11R-DEP: 611–628 peptide treatment induces A20 expression, which blocks NF-κB (p65) nuclear translocation.
Figure 4.
The 11R-DEP: 611–628 peptide treatment upregulated A20 expression and blocked NF-κB nuclear translocation. (A) A20 mRNA expression was determined by reverse transcription-quantitative polymerase chain reaction. The results demonstrated that, compared with the control, A20 mRNA levels reached >4-fold after 1 h of the 11R-DEP: 611–628 peptide treatment and then decreased gradually, but remained greater than the control group (n=3, ***P<0.0001). (B) The relative fluorescent density of nuclear NF-κB shown in panel C. (C) NF-κB was detected by immunofluorescent staining following the 11R-DEP: 611–628 peptide (3 µM) treatment for 1, 6, and 12 h. The representative images are displayed at ×200 magnification. *P<0.05, **P<0.01. NF-κB, nuclear factor-κB.
11R-DEP: 611–628 treatment downregulates DEPDC1 protein expression
DEPDC1 is upregulated in a variety of cancer types such as breast (5), lung cancer (6) and bladder cancer (8); however, the mechanism underlying the regulation of DEPDC1 expression in cancer cells remains unknown. It was hypothesized that DEPDC1 expression may be regulated by the DEPDC1-ZNF224 complex. To investigate this theory, HepG2 cells were treated with 11R-DEP: 611–628 peptide and then the DEPDC1 protein expression levels were examined. A notable decrease in DEPDC1 protein expression was observed in cells treated with 11R-DEP: 611–628 peptide (Fig. 5) compared with those in cells treated with the scramble peptide, indicating that DEPDC1 expression is regulated by the DEPDC1-ZNF224 complex.
Figure 5.
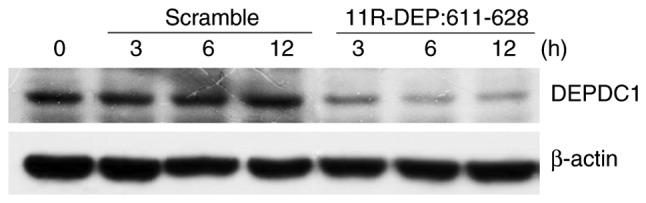
The 11R-DEP: 611–628 peptide treatment downregulated DEPDC1 protein levels. Western blot analysis results demonstrated that the 11R-DEP: 611–628 peptide treatment resulted in a decrease in DEPDC1 protein levels. DEPDC1, DEP domain containing 1.
miR-130a modulates proliferation and apoptosis by targeting DEPDC1 in HepG2 cells
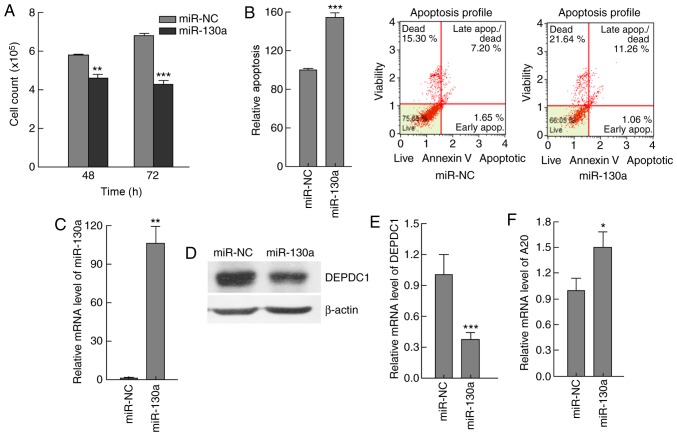
It was reported that the overexpression of miR-130a induces apoptosis and inhibits proliferation in prostate cancer cells by targeting DEPDC1 and SEC23B, and demonstrated that miR-130a inhibits DEPDC1 MRE1-directed luciferase activity (9). Based on these data, miR-130a was used to investigate the role of DEPDC1 in HepG2 cells. HepG2 cells were transfected with miR-130a expressing plasmids, and the effects of miR-130a expression were examined. The results demonstrated that, compared with the control group, the total cell number was decreased, whilst the number of apoptotic cells was increased in the miR-130a expression group (Fig. 6A-C). DEPDC1 expression at the protein and mRNA level was significantly reduced (P<0.0005) (Fig. 6D and E), and A20 expression was significantly increased (P<0.05) at 72 h post-transfection, compared with the control group (Fig. 6F). These results indicated that miR-130a may regulate HepG2 cell apoptosis by targeting DEPDC1.
Figure 6.
miR-130a modulated apoptosis by downregulating DEPDC1 expression in HepG2 cells. HepG2 cells were transfected with miR-130a expressing plasmids or NC plasmids. (A) Cells were counted at 48 and 72 h, and the results indicated that miR-130a expression resulted in a significant decrease in the cell number, compared with miR-NC. (B) Apoptosis was analyzed by flow cytometry, and the results demonstrated that miR-130a expression enhanced apoptosis, compared with miR-NC. (C) miR-130a expression in cells transfected with miR-130a expressing plasmids or NC plasmids was examined by RT-qPCR, and the results demonstrated that miR-130a expression increased ~100-fold in cells transfected with miR-130a expressing plasmids, compared with miR-NC. (D) The DEPDC1 protein levels were significantly reduced 72 h post-transfection as demonstrated by western blot analysis data, compared with miR-NC. (E) The DEPDC1 mRNA levels were significantly reduced 72 h post-transfection as demonstrated by RT-qPCR data, compared with miR-NC. (F) A20 expression levels were increased significantly 72 h post-transfection, compared with miR-NC. These results indicated that miR-130a has the ability to regulate HepG2 apoptosis by targeting DEPDC1. The data are presented as the mean ± standard deviation. *P<0.05, **P<0.01, ***P<0.001. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; NC, control; miR-130a, microRNA 130a; DEPDC1, DEP domain containing 1.
Discussion
It was previously reported that DEPDC1 is highly expressed in bladder cancer cells, and downregulating its expression with siRNA significantly suppressed the proliferation of bladder cancer cells (4). DEPDC1 interacts with ZNF224 to form a complex and inhibits the expression of A20, resulting in an increase in nuclear NF-κB and an inhibition of apoptosis in bladder cancer cells (8).
In the present study, the role of DEPDC1 was first investigated in HepG2 cells. It was demonstrated that interfering with DEPDC1-ZNF224 complex formation using the 11R-DEP: 611–628 peptide resulted in decreased cell proliferation and enhanced apoptosis. A comprehensive review determined that NF-κB was constitutively elevated in a variety of tumor types, including hematological and solid tumors (13). Continuously activated NF-κB signaling results in the upregulation of anti-apoptosis and pro-proliferation genes, which is notably correlated with tumorigenesis and tumor progression (14–16). In the present study, it was determined that the DEPDC1-ZNF224 complex suppresses A20 expression and thus activates the NF-κB signaling pathway, eventually resulting in the inhibition of apoptosis in HepG2 cells (17). To the best of our knowledge, the present study was the first to study the role of DEPDC1 in HepG2 cells, and to demonstrate the efficacy of 11R-DEP: 611–628 and miR-130a in HepG2 cells; these data may ultimately benefit patients with liver cancer. Performing further experiments in other liver cancer cell lines will provide further evidence regarding the role of DEPDC1 in liver cancer and the potential clinical application of 11R-DEP: 611–628 and miR-130a. Notably, the results demonstrated that the 11R-DEP: 611–628 peptide treatment resulted in a decrease in DEPDC1 protein, indicating that the DEPDC1-ZNF224 complex serves a role in regulating DEPDC1 protein levels; however, the underlying mechanisms require further elucidation in the future. The experiments analyzing the characteristics of the DEPDC1 promoter may be beneficial to elucidate the mechanisms underlying the regulation of DEPDC1 expression by the DEPDC1-ZNF224 complex.
miRNAs, a class of endogenous 22-nucleotide non-coding RNAs, inhibit gene expression by imperfectly pairing with the 3′-untranslated region of the target genes (18). It was reported that miRNAs, including miR-9, miR-21 and miR-224, regulate NF-κB activity by targeting the family members of NF-κB or its upstream signaling molecules (19–22). miR-130a suppresses cell proliferation and induces apoptosis by targeting DEPDC1 and SEC23B in prostate cancer cells (9). The present study indicated that miR-130a regulates apoptosis through the NF-κB signaling pathway in HepG2 cells by targeting DEPDC1, although luciferase assay and rescue experiments are required to further confirm the association between miR-130a and DEPDC1; therefore, miR-130a may be a potential therapeutic factor for liver cancer with DEPDC1 expression.
The limitation of the present study is that only the HepG2 cell line was used to conduct the experiments. The HepG2 cell line was originally identified as a hepatocellular carcinoma cell line, but was later confirmed to be a hepatoblastoma-derived cell line (23); however, the misidentification of HepG2 does not affect the outcomes and conclusions of the present study.
In summary, the data demonstrated for the first time that 11R-DEP: 611–628 and miR-130a are capable of inhibiting proliferation by inducing apoptosis through the NF-κB signaling pathway in HepG2 cells; therefore, DEPDC1 could be a therapeutic target for the treatment of liver cancer with DEPDC1 expression.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the National Natural Science Foundation of China (grant no. 81560453), Natural Science Foundation of Guangxi (grant no. 2015GXNSFAA139178), the Guangxi Health and Family Planning Commission (grant no. S2015-34), the Lijiang Scholar Award and the ‘Sphingolipids and Related Diseases’ Program for Innovative Research Team of Guilin Medical University. G. H. was supported by the Hundred Talents Program of Guangxi.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
AL, QW and GFH performed the experiments. AL, JJ and GJH analyzed and interpreted the data. AL, QW, JJ and GJH wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Reig M, Sherman M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology. 2016;150:835–853. doi: 10.1053/j.gastro.2015.12.041. [DOI] [PubMed] [Google Scholar]
- 3.Chacko S, Samanta S. ‘Hepatocellular carcinoma: A life-threatening disease’. Biomed Pharmacother. 2016;84:1679–1688. doi: 10.1016/j.biopha.2016.10.078. [DOI] [PubMed] [Google Scholar]
- 4.Kanehira M, Harada Y, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y, Katagiri T. Involvement of upregulation of DEPDC1 (DEP domain containing 1) in bladder carcinogenesis. Oncogene. 2007;26:6448–6455. doi: 10.1038/sj.onc.1210466. [DOI] [PubMed] [Google Scholar]
- 5.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer. 2011;10:15. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72:100–111. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 7.Yuan SG, Liao WJ, Yang JJ, Huang GJ, Huang ZQ. DEP domain containing 1 is a novel diagnostic marker and prognostic predictor for hepatocellular carcinoma. Asian Pac J Cancer Prev. 2014;15:10917–10922. doi: 10.7314/APJCP.2014.15.24.10917. [DOI] [PubMed] [Google Scholar]
- 8.Harada Y, Kanehira M, Fujisawa Y, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y, Katagiri T. Cell-permeable peptide DEPDC1-ZNF224 interferes with transcriptional repression and oncogenicity in bladder cancer cells. Cancer Res. 2010;70:5829–5839. doi: 10.1158/0008-5472.CAN-10-0255. [DOI] [PubMed] [Google Scholar]
- 9.Ramalho-Carvalho J, Martins JB, Cekaite L, Sveen A, Torres-Ferreira J, Graça I, Costa-Pinheiro P, Eilertsen IA, et al. Epigenetic disruption of miR-130a promotes prostate cancer by targeting SEC23B and DEPDC1. Cancer Lett. 2017;385:150–159. doi: 10.1016/j.canlet.2016.10.028. [DOI] [PubMed] [Google Scholar]
- 10.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q, Li A, Jin J, Huang G. Targeted interfering DEP domain containing 1 protein induces apoptosis in A549 lung adenocarcinoma cells through the NF-κB signaling pathway. Onco Targets Ther. 2017;10:4443–4454. doi: 10.2147/OTT.S142244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf BB, Schuler M, Echeverri F, Green DR. Caspase-3 is the primary activator of apoptotic DNA fragmentation via DNA fragmentation factor-45/inhibitor of caspase-activated DNase inactivation. J Biol Chem. 1999;274:30651–30656. doi: 10.1074/jbc.274.43.30651. [DOI] [PubMed] [Google Scholar]
- 13.Pacifico F, Leonardi A. NF-kappaB in solid tumors. Biochem Pharmacol. 2006;72:1142–1152. doi: 10.1016/j.bcp.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 14.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 15.Kasibhatla S, Brunner T, Genestier L, Echeverri F, Mahboubi A, Green DR. DNA damaging agents induce expression of Fas ligand and subsequent apoptosis in T lymphocytes via the activation of NF-kappa B and AP-1. Mol Cell. 1998;1:543–551. doi: 10.1016/S1097-2765(00)80054-4. [DOI] [PubMed] [Google Scholar]
- 16.Meteoglu I, Erdogdu IH, Meydan N, Erkus M, Barutca S. NF-KappaB expression correlates with apoptosis and angiogenesis in clear cell renal cell carcinoma tissues. J Exp Clin Cancer Res. 2008;27:53. doi: 10.1186/1756-9966-27-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. Hep G2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 18.Gu X, Li A, Liu S, Lin L, Xu S, Zhang P, Li S, Li X, Tian B, Zhu X, Wang X. MicroRNA124 regulated neurite elongation by targeting OSBP. Mol Neurobiol. 2016;53:6388–6396. doi: 10.1007/s12035-015-9540-4. [DOI] [PubMed] [Google Scholar]
- 19.Bazzoni F, Rossato M, Fabbri M, Gaudiosi D, Mirolo M, Mori L, Tamassia N, Mantovani A, Cassatella MA, Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proc Natl Acad Sci USA. 2009;106:5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, Becker Buscaglia LE, Barker JR, Li Y. MicroRNAs in NF-kappaB signaling. J Mol Cell Biol. 2011;3:159–166. doi: 10.1093/jmcb/mjr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niu J, Shi Y, Tan G, Yang CH, Fan M, Pfeffer LM, Wu ZH. DNA damage induces NF-κB-dependent microRNA-21 up-regulation and promotes breast cancer cell invasion. J Biol Chem. 2012;287:21783–21795. doi: 10.1074/jbc.M112.355495. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Scisciani C, Vossio S, Guerrieri F, Schinzari V, de Iaco R, D'Onorio de Meo P, Cervello M, Montalto G, Pollicino T, Raimondo G, et al. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol. 2012;56:855–861. doi: 10.1016/j.jhep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 23.López-Terrada D, Cheung SW, Finegold MJ, Knowles BB. Hep G2 is a hepatoblastoma-derived cell line. Hum Pathol. 2009;40:1512–1515. doi: 10.1016/j.humpath.2009.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.