Abstract
Lipids are known to serve important roles in energy storage, membrane structure and signal transduction as well as in human cancers. In the present study, lipidomics was employed in order to identify plasma lipid markers for the early detection of lung cancer. Mass spectrometry was performed to profile 390 individual lipids in 44 plasma samples obtained from a training discovery cohort, which included 22 patients with squamous cell lung carcinoma (SqCC) and 22 high-risk individuals. An additional cohort that included 22 high-risk individuals and 22 patients with SqCC was further used for validation. During the training stage, a total of 20 distinct lipids that were significantly distributed between the high-risk and SqCC cases, were identified. A panel of 2 lipid markers (C18:2 cholesterol esters and sphingomyelin 22:0) were then further defined using the training accuracy values of 95.5% sensitivity, 90.9% specificity and 95.2% area under the receiver operating characteristic curve (AUC). The validation accuracy values applied for the additional cohort were 93.9% sensitivity, 92.9% specificity and 98.7% AUC. Thus, in the present study, 2 lipid markers that were able to discern SqCC patients from high-risk individuals with a high sensitivity, specificity and accuracy, were identified. These results may provide vital information for the development of a quick and safe blood test for the early diagnosis of SqCC.
Keywords: lipidomics, plasma lipids, squamous cell lung carcinoma, cancer screening, biomarkers
Introduction
Lung cancer is the leading cause of cancer-associated morbidity and mortality in the world (1). Lung cancer is a heterogeneous disease with a number of histological and molecular subtypes; it is usually classified according to the histological types associated with tumor behavior and prognosis (2). The majority of lung cancer types are known as non-small cell lung cancers (NSCLCs), which are carcinoma malignancies that arise from epithelial cells. The two most common NSCLC subtypes are adenocarcinoma and squamous cell lung cancer (SqCC). SqCC originates in the large airways in the central part of the lungs and is the most common histological subtype of lung cancer among European smokers (3,4). It is thought that the detection of lung cancer during the early stages could reduce the mortality rates by 10- to 50-fold (5). However, SqCC is often diagnosed at an advanced stage, by which point approximately two-thirds of all patients have metastatic tumors when diagnosed. Currently the low-dose computed tomography (LDCT) scan approach is used to as a non-invasive method for detecting tumors at early stages; however, this method has yielded conflicting results (6,7). Therefore, it is necessary to develop novel, minimally invasive methods, such as molecular biomarkers, for the early detection of lung cancer.
Lipids, which contain multiple different types of molecules, have a number of key biological functions in cellular energy storage, membrane structure and signaling processes. Within the human body, lipid levels are regulated, spatially and temporally, in various areas. Abnormal regulation of lipid metabolism contributes to the development of a variety of human diseases, including diabetes (8), Alzheimer's disease (9), hypertension (10) and many types of cancer (11–14). Abnormal lipid metabolism in lung cancer has also been demonstrated in a previous study, in which the distribution of lipids in 21 pairs of resected frozen NSCLCs and adjacent normal tissue samples were analyzed (14).
The involvement of lipid abnormalities in disease pathogenesis increases the potential of using lipids as biomarkers for various human diseases. However, due to technical limitations in lipid measurement, only a few studies have investigated this potential so far. Lipidomics is a relatively novel field that allows for the quantitative assessment of a range (hundreds) of fatty (lipids) species at one time; this analysis can be used to generate lipid profiles for the majority of pathophysiological states. Recently, lipidomics has been employed to study a number of diseases such as diabetes (15) and obesity (16), as well as some types of human cancers, including colon cancer (17), thyroid papillary cancer (18) and prostate cancer (19). Direct tissue matrix-assisted laser desorption/ionization (MALDI) mass spectrometry (MS) analysis has been applied previously to perform lipid analysis in resected frozen lung cancer tissue samples (14); however, large-scale studies on plasma lipid profiling have yet to be reported in lung cancer.
In a previous study, the authors of the present study evaluated the predictive power of this lipid panel in diagnosis of early stage NSCLC (20). In the present study, a lipid profiling study using tandem MS was performed in order to measure 390 distinct lipids in the plasma specimens obtained from patients with specifically in SqCC and high-risk controls. The aim of the present study was to develop a plasma lipid marker panel to use in high-risk patients as a companion test with LDCT-based methods for the screening and early detection of SqCC.
Patients and methods
Human research ethical statement
The present study was conducted in accordance with the Declaration of Helsinki. The Rush University Medical Center Institution Review Board (Chicago, IL, USA) approved the experimental protocol, including the human rights and protection aspects of the study, and all participants provided written informed consent.
Patient cohorts
In the present study, ~1,250 individuals were enrolled at the Lung Cancer Biorepository of Rush University Medical Center between January 2004 and December 2010. Participants were aged ≥50 years with: i) smoking histories of >30 pack-years or ii) current smokers or a former smokers who quit within the past 15 years. Of these patients, a sub-cohort of 44 patients with SqCC and 44 high-risk individuals were selected for this pilot study (Table I). Patients were then further divided into the following cohorts: i) Pathologically diagnosed SqCC patients (n=22); and ii) matched high-risk individuals (n=22) for the training stage; iii) independent SqCC patients (n=22); and iv) random high-risk controls (n=22) for the validation stage. The early-stage SqCC patient inclusion criteria included the following: Disease was confined to the chest without evidence of distant metastases; no preoperative chemo- or radiotherapy within 1 year of the present study's initial blood sampling; and a minimum of 2 years of clinical follow-up data available. High risk patients recruited to the present study were defined as individuals aged 55 to 75 years, had a smoking history of >30 years and quit smoking <15 years prior to randomization in the present study. All high-risk individuals were followed with annual LDCT and remained cancer-free for a minimum of 2 years during follow-up. At the training stage, SqCC cases and high-risk participants were matched in terms of race, sex, age and smoking status. The demographic information for these patients and controls is listed in Table I.
Table I.
Patient characteristics in all of the samples used in the training and validation stages.
| Training stage | Validation stage | |||
|---|---|---|---|---|
| Characteristic | SqCC (n=22) | High risk (n=22) | SqCC (n=22) | High risk (n=22) |
| Sex (n) | ||||
| Male | 11 | 11 | 15 | 16 |
| Female | 11 | 11 | 7 | 6 |
| Race (n) | ||||
| Caucasian | 18 | 21 | 14 | 19 |
| Noncaucasian | 4 | 1 | 8 | 3 |
| Age (years) | ||||
| Median | 62.95 | 64.14 | 56.75 | 73.44 |
| Range | 49–81 | 51–82 | 69–82 | 49–73 |
| Smoking history | ||||
| Median (years) | 59.17 | 47.04 | 52.90 | 32.28 |
| Nonsmoker (n) | 2 | 0 | 0 | 0 |
| Tumor stage (n) | ||||
| Stage I | 12 | − | 14 | − |
| Stage II | 4 | − | 1 | − |
| Stage III | 6 | − | 7 | − |
SqCC, squamous cell lung cancer.
Electrospray ionization-tandem mass spectrometry (ESI-MS) lipid profiling
In the present study, a total of 390 lipids in plasma samples were tested using an automated ESI-MS system at the Kansas State University Lipidomics Research Center (Manhattan, KS, USA), as previously described (21). Briefly, an aliquot of 3 µl plasma was used to identify plasma lipid species at the level of the head group plus the total acyl carbons:total double bonds in this assay; precise amounts of internal standards were obtained and quantified, as previously described (21). A series of spectra were generated from the sequential precursor and neutral loss scans of the extracts; each spectrum identified a group of lipid species containing a common head group fragment. In total, 13 lipid classes were measured: Phosphatidylcholine (PC), sphingomyelin (SM), lysoPC, phosphatidylethanolamine (PE), lysoPE, phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidic acid, cholesterol esters (CE), PS with one ether-linked (alkyl or alkenyl) chain, PC with one ether-linked (alkyl or alkenyl) chain, PE with one ether-linked (alkyl or alkenyl) chain and ceramide PE, as detailed previously (19). The background of each spectrum was removed, the data were normalized, and the customization script and Applied Biosystems Analyst software (version 1.5; Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were used to integrate the peak area. The data for the analyzed sample portion were then corrected and the sample volume was normalized to produce data with units of nmol/µl.
Statistical and bioinformatics analysis
In the present study, a Student's unpaired t-test was performed using SPSS 18 software (SPSS, Inc., Chicago, IL, USA) to compare the mean plasma concentrations of 390 lipid species between the SqCC patient and control groups following the log transformation of data. The GeneSpring 12.6 program (Agilent Technologies, Inc., Santa Clara, CA, USA) was used to perform Hierarchical Clustering Analysis (HCA), and the Weka version 3.73 software (the University of Waikato, Hamilton, New Zealand) (22) was used in bioinformatics analysis. The simple logistics classification algorithm and information gain algorithm in Weka 3.7 were used to rank the individual apparent lipid species and lipid class according to their predictive powers in patients with SqCC; 10-fold cross validation was employed to estimate the performance of a predictive model. The unpaired Student's t-test in SPSS 18 software was used to compare the mean plasma lipid concentrations in the controls and patients. The chi-squared test was used to compare the differences between sex and race in the patient and control groups. The differences in age between the groups was analyzed by a Mann Whitney U test. P<0.05 was considered to indicate a statistically significant difference.
Results
Lipid profiling of 390 lipid species in the training cohort
During the training stage, the plasma lipid profiles of 390 individual lipid species from 13 classes of phospholipids and CEs were identified using lipidomics in 44 plasma samples, which included 22 patients with SqCC, and 22 age-, sex- and race-matched high-risk controls. As summarized in our previous study (20), 361 apparent lipid species in all 13 classes were detected in all of the training samples. Among the 361 apparent species, the highest median plasma concentration was that of the CE species (18:2; 12.71 nmol/µl in SqCC patients and 18.42 nmol/µl in controls). In the present study, 29 of the 390 lipid species were not detected in any of the training cohort samples; these primarily consisted of those from the lysoPE class (14 species) and PI class (9 species).
Identification of significantly different lipid species between patients with SqCC and high-risk individuals
To select individual apparent lipid biomarkers from the hundreds of species detected, a filtration strategy was employed to narrow down the 361 potential candidates of apparent lipid species. During this process, the following exclusion criteria were applied: i) Lipid species that could not be clinically used for the diagnosis of SqCC disease due to too low a concentration of detection; ii) there was an insignificant difference between the patient and control groups; and iii) the levels of plasma concentrations that were too close between the groups to interpret (although the difference may have been statistically significant). The inclusion criteria were as follows: i) A significant difference in the mean plasma lipid concentration (P≤0.05) was identified between the patient and control groups; ii) there was a change in the mean plasma lipid concentration of ≥10% (up or down); and iii) a mean plasma lipid concentration ≥10 nmol/µl. Using these criteria, 20 apparent lipid species were identified that fulfilled all 3 criteria and could be selected as potential plasma lipid biomarkers for SqCC (Table II). Among the 20 candidate lipid markers, decreasing levels were observed in 18 lipid species in SqCC patients; however, 2 lipid species, PC(38:6) and PI(38:4), presented an increased level in SqCC plasma when compared with the controls (Table II).
Table II.
Top 20 significantly differentiated lipid species for SqCC at the training stage.
| SqCC (n=22) | High risk (n=22) | |||||
|---|---|---|---|---|---|---|
| Lipids | Mean | SD | Mean | SD | Fold change | P-value |
| C16:0 CE | 0.9420 | 1.2608 | 1.0668 | 1.3492 | 1.1325 | 3.72×102 |
| C18:2 CE | 0.7879 | 1.2590 | 1.0644 | 1.3780 | 1.3509 | 1.00×104 |
| C20:4 CE | 0.8406 | 1.6088 | 1.3211 | 1.5260 | 1.5717 | 4.01×102 |
| C22:6 CE | 0.6812 | 1.7158 | 1.0093 | 1.6120 | 1.4815 | 1.01×102 |
| ePC (34:2) | 0.9868 | 1.2485 | 1.0113 | 1.3051 | 1.0248 | 2.84×102 |
| ePC (34:3) | 0.9634 | 1.3843 | 1.0373 | 1.3540 | 1.0768 | 1.31×102 |
| ePC (36:2) | 0.9727 | 1.3327 | 1.1001 | 1.2649 | 1.1310 | 1.80×103 |
| ePC (38:1) | 0.9520 | 1.3552 | 1.1049 | 1.2919 | 1.1605 | 1.07×102 |
| ePC (38:2) | 0.9768 | 1.3698 | 1.0456 | 1.2906 | 1.0705 | 1.60×102 |
| LPC (16:0) | 0.8984 | 1.6525 | 1.0497 | 1.4053 | 1.1685 | 1.88×102 |
| LPC (18:0) | 0.8089 | 1.7651 | 1.0159 | 1.3465 | 1.2559 | 3.80×103 |
| LPC (18:2) | 0.8585 | 1.8515 | 1.3798 | 1.5443 | 1.6072 | 2.00×104 |
| PC (34:2) | 1.0402 | 1.2913 | 1.1308 | 1.3081 | 1.0871 | 8.00×104 |
| PC (36:2) | 0.9491 | 1.3070 | 1.1489 | 1.3165 | 1.2105 | 2.00×104 |
| PC (36:4) | 0.9536 | 1.5849 | 1.2294 | 1.4101 | 1.2892 | 4.70×102 |
| PC(38:6) | 0.8903 | 1.4371 | 0.8598 | 1.5768 | 1.0355 | 1.54×102 |
| PC (40:7) | 0.8869 | 1.5719 | 0.9436 | 1.4884 | 1.0639 | 8.20×103 |
| PI (38:4) | 0.9769 | 1.4808 | 0.9588 | 1.3437 | 0.9814 | 4.59×102 |
| SM (16:1) | 0.9819 | 1.2954 | 1.1346 | 1.2007 | 1.1556 | 2.48×102 |
| SM (22:0) | 0.5988 | 1.3069 | 0.9607 | 4.3199 | 1.6045 | 1.30×103 |
Concentrations are presented as pmol/µl. The 2 lipids C18:2 CE and SM 22:0 shown in bold were selected for further study in the validation stage. SqCC, squamous cell lung cancer; CE, cholesterol esters; PC, phosphatidylcholine; ePC, PC with one ether-linked (alkyl or alkenyl) chain; LPC, lyso-PC; PI, phosphatidylinositol; SM, sphingomyelin;.
Identification of C18:2 CE and SM 22:0 as candidate biomarkers for early-stage SqCC
A second strategy that generated an additional differentiation of cancer and control samples was then utilized to ensure that the selected candidates were clinically useful and applicable, and also highly sensitive, specific and accurate in the differentiation of SqCC from high-risk controls. Following analysis using bioinformatics methods, any apparent lipid species of the selected potential candidates were selected as an individual plasma lipid biomarker for the diagnosis of SqCC cancer if the following criteria was fulfilled: i) Sensitivity, >80%; ii) specificity, >50%; and iii) area under the receiver operating characteristic curve, >80%.
Using these selection strategies, 2 lipid species, C18:2 CE and SM 22:0, were identified as candidate biomarkers for the early detection of SqCC disease during the training stage. The lipid C18:2 CE was detected at a mean concentration of 12.71 nmol/µl in 22 SqCC patients, with a 31.02% downregulation (P=0.0001) when compared with the demographically matched high-risk controls (18.42 nmol/µl; Fig. 1A). Similarly, SM 22:0 was downregulated in SqCC plasma samples (mean concentration, 95 pmol/µl), with a difference of 30.66% when compared with the high-risk cohort (mean concentration, 137 pmol/µl; P=0.0013; Fig. 1B). The lipid levels of C18:2 CE and SM 22:0 were then determined in one high-risk individual and one SqCC case, respectively. The predictive power of C18:2 CE in the diagnosis of early-stage SqCC exerted a sensitivity of 86.4%, a specificity of 54.5% and an area under the curve (AUC) value of 72.5% (Fig. 2A). In addition, SM 22:0 exerted a sensitivity of 90.9%, a specificity of 77.3% and an AUC of 84.1% (Table III). During the training stages, it was also observed that the combination of these 2 markers yielded the strongest predictive power with a higher degree of sensitivity (95.5%), specificity (90.9%) and accuracy (AUC, 95.2%), as shown in Table III and Fig. 2A.
Figure 1.
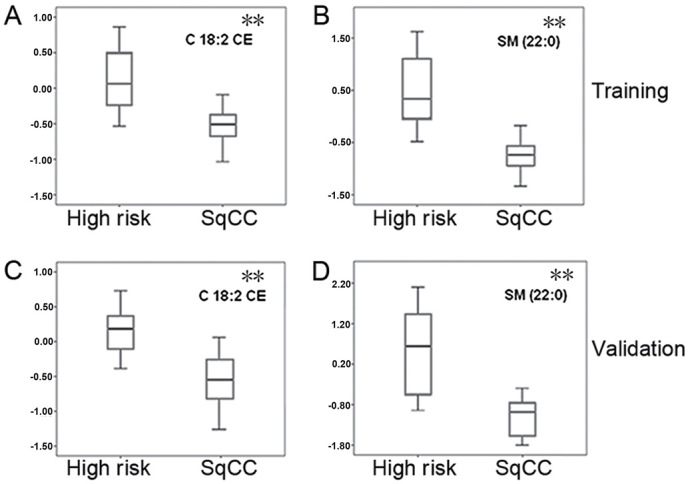
Box plots of C18:2 CE and SM 22:0 concentrations in the training and validation SqCC cohorts and high-risk controls. The mean concentrations of the 2 lipids C18:2 CE and SM 22:0 were measured in high-risk controls and patients with SqCC during the (A and B) training and (C and D) validation stages, respectively. The y-axis values were log2-transferred mean concentrations following normalization. CE, cholesterol esters; SM, sphingomyelin; SqCC, squamous cell lung cancer. A Student's unpaired t-test was performed to compare the two groups. **P<0.01.
Figure 2.
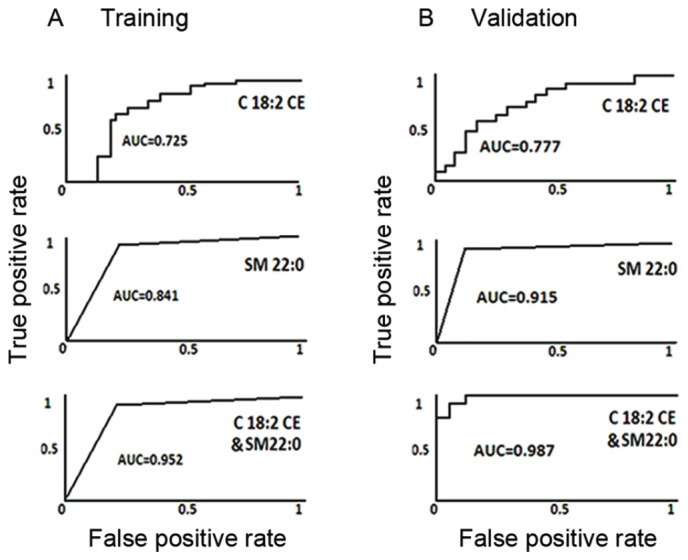
AUC values of C18:2 CE and SM 22:0 for SqCC prediction in the training and validation cohorts. The AUC values in SqCC prediction for C18:2 CE (top), SM 22:0 (middle) and the 2 combined lipid markers (bottom) in the (A) training and (B) validation samples. AUC, area under the curve; CE, cholesterol esters; SM, sphingomyelin; SqCC, squamous cell lung cancer.
Table III.
Predictive values of the 2 lipid markers at the training stage.
| Lipid markers | Sensitivity | Specificity | PPV | NPV | OR | AUC |
|---|---|---|---|---|---|---|
| C18:2 CE | 0.864 | 0.545 | 0.800 | 0.655 | 7.6 | 0.725 |
| SM (22:0) | 0.909 | 0.773 | 0.895 | 0.800 | 34.0 | 0.841 |
| C18:2 CE + SM (22:0) | 0.955 | 0.909 | 0.952 | 0.913 | 210.0 | 0.952 |
The predictive values of C18:2 CE and SM 22:0 were analyzed separately and combined. CE, cholesterol esters; SM, sphingomyelin; PPV, positive predictive value; NPV, negative predictive value; OR, odds ratio; AUC, area under the curve.
Validation of C18:2 CE and SM 22:0 in independent cohorts
In order to validate C18:2 CE and SM 22:0 as potential biomarkers for the early detection of SqCC, the 2 lipid molecules were further investigated using the ESI-MS lipid profiling technique in an independent cohort containing 22 SqCC cases and 22 high-risk controls. During this validation stage, as the biomarkers achieved in the training dataset are validated in any SqCC dataset, the controls were randomly selected without age-, sex- or ethnicity-matching with the SqCC patients (Table I). The box plots generated presented a similar pattern of the 2 lipid levels among the validation cohorts (Fig. 1C and D) and the training samples (Fig. 1A and B). A similar prediction power between C18:2 CE and SM 22:0 was observed in the validation samples. The sensitivity, specificity and AUC values for C18:2 CE were 62.5, 78.6 and 77.7%, respectively, and for SM 22:0 were 93.8, 89.3 and 91.5%, respectively. The strongest predictive power was produced by the combination of the 2 lipids, with markedly higher values of sensitivity (93.89%), specificity (92.9%) and AUC (98.7%) (Table IV; Fig. 2B).
Table IV.
Predictive values of the 2 lipid markers at the validation stage.
| Lipid markers | Sensitivity | Specificity | PPV | NPV | OR | AUC |
|---|---|---|---|---|---|---|
| C18:2 CE | 0.625 | 0.786 | 0.786 | 0.625 | 6.11 | 0.777 |
| SM (22:0) | 0.938 | 0.893 | 0.962 | 0.833 | 125.00 | 0.915 |
| C18:2 CE + SM (22:0) | 0.938 | 0.929 | 0.963 | 0.882 | 195.00 | 0.987 |
The predictive values of C18:2 CE and SM 22:0 were analyzed in the validation samples. CE, cholesterol esters; SM, sphingomyelin; PPV, positive predictive value; NPV, negative predictive value; OR, odds ratio; AUC, area under the curve.
Characteristics of the 2 identified plasma lipid biomarkers
To further reveal the characteristics of the 2 identified plasma lipid biomarkers for the diagnosis of SqCC, HCA was conducted in order to cluster the entity (the 2 lipid biomarkers) and condition (all 44 patients with SqCC and 44 high-risk individuals; training and validation cohorts) by combining dendrograms and a heatmap with panels of the characteristics at the top (Fig. 3). HCA analysis revealed that the 2 apparent lipid species C18:2 CE and SM 22:0 had a tendency to gradually decrease in mean plasma lipid concentration, from right (higher concentration, shown in red) to left (lower concentration, shown in blue; Fig. 3). This result further demonstrated that the concentrations of C18:2 CE and SM 22:0 lipid species in plasma samples were associated with the disease status of SqCC, with a decreasing tendency in patients with SqCC.
Figure 3.
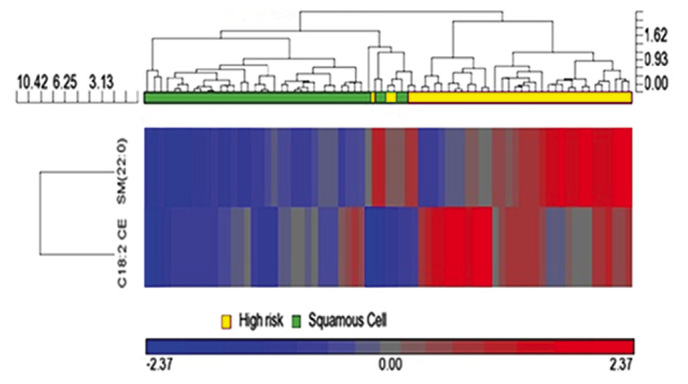
Hierarchical clustering analysis of C18:2 CE and SM 22:0 in all samples in the training and validation cohorts. The GeneSpring 12.6 program was used to perform hierarchical clustering analysis. The SqCC patients are represented in green on the left and the high-risk controls are in shown in yellow on the right. The C18:2 CE and SM 22:0 levels ranged from the lowest at −2.37 (deep blue) to the highest at 2.37 (deep red). CE, cholesterol esters; SM, sphingomyelin; SqCC, squamous cell lung cancer.
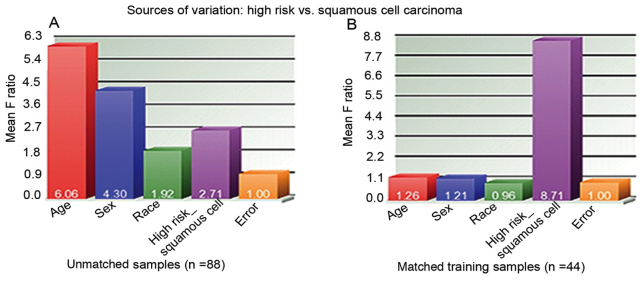
To assess the power of the sample size used during the training and validation stages, the present study conducted the source of variation analysis among all 88 unmatched subjects in the training and validation stages, and among age, sex, race and disease status (high risk vs. SqCC; Fig. 4). It was observed that the most significant difference was shown in age, with a mean F ratio of 6.06 in all 88 unmatched samples, which was much higher than the mean F ratio of 2.71 for disease status (high risk vs. SqCC; Fig. 4A). However, in the 44 matched training samples, the most significant factor was the disease status, with a mean F ratio of 8.71 (Fig. 4B). This observation indicated that the significant difference in disease status (high risk vs. SqCC) may contribute to the differentiated level of the 2 lipid species in plasma. As 70% of the cases (31 out of 44) who participated in the present study were patients with stage I or II SqCC, the panel of these 2 lipid biomarkers could also be applied to the prediction of early-stage disease.
Figure 4.
Sources of variation presented as the characteristic differences between the squamous cell lung cancer cohorts and high-risk cohorts used in the present study. (A) Unmatched samples (all 88 samples) and (B) matched training samples (n=44 training stage samples).
Discussion
In the present study, extensive plasma lipidomics profiling of patients with SqCC was performed and a panel of 2 lipid biomarkers that were able to detect SqCC during the early stages were identified. To the best of our knowledge, this is the first original report on plasma lipid biomarkers for the early detection of SqCC. The use of plasma is highly advantageous for the development of a quick, non-invasive blood test and is easy to obtain and use for the early diagnosis of SqCC. The results of the present study revealed the potential of using these 2 lipid markers in combination with LDCT-based screening methods in order to distinguish patients with SqCC from high-risk individuals.
In the present study, it was observed that patients with SqCC exhibited decreased plasma lipid concentrations of the 2 lipid molecules when compared with the high-risk controls. In a previous study, the cholesteryl linoleate C18:2 CE was revealed to be one of the three major cholesteryl esters that are present in human low-density lipoprotein, and C18:2 CE oxidization is thought to be associated with atherosclerosis (15). In addition, C18:2 CE can be oxidized to generate the compounds 9-oxononanoyl secosterol-A (9-ON-secoA) and 9-ON-secoB, which have been revealed to exhibit strong cytotoxic activities against human leukemia HL-60 cells (16). However, it remains unknown whether C18:2 CE oxidization is involved in lung cancer tumorigenesis. Although smoking history is thought to be a risk factor of SqCC, in a large-scale lipidomics analysis no associations were identified for the plasma concentration in cholesteryl esters between smokers and non-smokers (23). In agreement with these results, in the present study no associations between smoking history and the C18:2 lipid level were observed in any of plasma samples.
In humans, SM, also known as sphingophospholipid, is a type of sphingolipid observed in the cell membrane; it represents ~85% of all sphingolipids. SM is comprised of a phosphorylcholine head group, a sphingosine and a fatty acid tail; the sphingosine and fatty acid together are known as a ceramide. This composition enables SM to serve significant roles in signaling pathways (24) and SM degradation can produce ceramide, which is known to be involved in the apoptotic signaling pathway (25). In addition, ceramide easily converts into sphingosine 1-phosphate (S1P) or ceramide 1-phosphate (C1P). When compared with ceramide, S1P and C1P produce the opposite effect; in the regulation of cell growth and survival they serve as pro-survival or mitogenic signals in the majority of cell types, and they also induce different effects in controlling tumor progression and metastasis (26). The sphingolipid, ceramide, has been studied extensively under normal and pathological conditions, ranging from skin development to lung cancer. A previous nested case-control study demonstrated that higher concentrations of S1P and total ceramide in plasma were associated with an increased risk of lung cancer (24). A previous study has also revealed that high-risk smokers have increased ceramide levels; however, the underlying molecular mechanisms associated with how smoking and ceramide accumulation cause lung cancer remain unknown (27). Recently, 2 signaling pathways associated with neutral sphingomyelinase-2, an enzyme that hydrolyzes SM to ceramide, and the EGF receptor (EGFR), respectively, have been reported to be involved in the processes associated with cigarette smoke exposure in the lung airways; EGFR was also revealed to be co-localized in the plasma membrane in ceramide-enriched regions (28,29). In addition, it is thought that these 2 signaling pathways may converge and integrate with one another during these processes (27,28).
In conclusion, the present study identified a panel of 2 plasma lipid markers that may be able to distinguish patients with SqCC from high-risk individuals with a large prediction power. One limitation of the present study was that only relatively small cohorts were used in the training and validation studies; therefore, these 2 potential lipid biomarkers require further confirmation in a greater number of samples from different resources. Furthermore, the present study was only conducted in the SqCC subtype of lung cancer. As 80% of lung cancers are NSCLCs, of which ~30% are SqCC types, it is necessary to investigate plasma lipid profiling in patients with NSCLC. As such, a global lipidomics study is currently being performed at our laboratory in order to identify specific lipid biomarkers for the early stages of NSCLC.
Acknowledgements
Lipid analyses were conducted at the Kansas Lipidomics Research Center Analytical Laboratory.
Glossary
Abbreviations
- NSCLC
non-small cell lung cancer
- SqCC
squamous cell lung cancer
- LDCT
low-dose computed tomography
- MALDI-MS
matrix-assisted laser desorption/ionization-mass spectrometry
- ESI-MS
electrospray ionization-tandem mass spectrometry
- HCA
hierarchical clustering analysis
- CE
cholesteryl esters
- SM
sphingomyelin
Funding
The present study was supported, in part, by The National Institutes of Health (grant nos. R21CA164764, 5P30GM114737, P20GM103466, 2U54MD007601 and U54 MD007584) and Hawaii Community Foundation (grant no. 17ADVC-86288).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YD and WC designed and directed the research. ZY and HC performed the experiments and wrote the manuscript. JA performed the data analysis and prepared the tables. JB provided bio-specimens. YL and YZ prepared the figures and assisted with the data analysis. WG contributed to the analysis and interpretation of the data. BJ and JZ prepared the experimental study materials and revised the paper. All authors reviewed and approved the manuscript prior to submission.
Ethics approval and consent to participate
The Rush University Medical Center Institution Review Board approved the experimental protocol, including the human rights and protection aspects of the study, and all participants provided written informed consent.
Consent for publication
Written informed consent was obtained from all participants.
Competing interests
The authors declare they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Beadsmoore CJ, Screaton NJ. Classification, staging and prognosis of lung cancer. Eur J Radiol. 2003;45:8–17. doi: 10.1016/S0720-048X(02)00287-5. [DOI] [PubMed] [Google Scholar]
- 3.Papi A, Casoni G, Caramori G, Guzzinati I, Boschetto P, Ravenna F, Calia N, Petruzzelli S, Corbetta L, Cavallesco G, et al. COPD increases the risk of squamous histological subtype in smokers who develop non-small cell lung carcinoma. Thorax. 2004;59:679–681. doi: 10.1136/thx.2003.018291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janssen-Heijnen ML, Coebergh JW. Trends in incidence and prognosis of the histological subtypes of lung cancer in North America, Australia, New Zealand and Europe. Lung Cancer. 2001;31:123–137. doi: 10.1016/S0169-5002(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 5.Edwards BK, Brown ML, Wingo PA, Howe HL, Ward E, Ries LA, Schrag D, Jamison PM, Jemal A, Wu XC, et al. Annual report to the nation on the status of cancer, 1975–2002, featuring population-based trends in cancer treatment. J Natl Cancer Inst. 2005;97:1407–1427. doi: 10.1093/jnci/dji289. [DOI] [PubMed] [Google Scholar]
- 6.International Early Lung Cancer Action Program Investigators1, corp-author. Henschke CI, Yankelevitz DF, Libby DM, Pasmantier MW, Smith JP, Miettinen OS. Survival of patients with stage I lung cancer detected on CT screening. N Engl J Med. 2006;355:1763–1771. doi: 10.1056/NEJMoa060476. [DOI] [PubMed] [Google Scholar]
- 7.Roth JA, Ramsey SD. Computed tomography screening for lung cancer: A high-value proposition? JAMA. 2016;315:77–78. doi: 10.1001/jama.2015.17877. [DOI] [PubMed] [Google Scholar]
- 8.Sleeman MW, Wortley KE, Lai KM, Gowen LC, Kintner J, Kline WO, Garcia K, Stitt TN, Yancopoulos GD, Wiegand SJ, Glass DJ. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- 9.Cutler RG, Kelly J, Storie K, Pedersen WA, Tammara A, Hatanpaa K, Troncoso JC, Mattson MP. Involvement of oxidative stress-induced abnormalities in ceramide and cholesterol metabolism in brain aging and Alzheimer's disease. Proc Natl Acad Sci USA. 2004;101:2070–2075. doi: 10.1073/pnas.0305799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen NT, Magno CP, Lane KT, Hinojosa MW, Lane JS. Association of hypertension, diabetes, dyslipidemia, and metabolic syndrome with obesity: Findings from the National Health and Nutrition Examination Survey, 1999 to 2004. J Am Coll Surg. 2008;207:928–934. doi: 10.1016/j.jamcollsurg.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 11.Pendaries C, Tronchère H, Plantavid M, Payrastre B. Phosphoinositide signaling disorders in human diseases. FEBS Lett. 2003;546:25–31. doi: 10.1016/S0014-5793(03)00437-X. [DOI] [PubMed] [Google Scholar]
- 12.Ogretmen B, Hannun YA. Biologically active sphingolipids in cancer pathogenesis and treatment. Nat Rev Cancer. 2004;4:604–616. doi: 10.1038/nrc1411. [DOI] [PubMed] [Google Scholar]
- 13.Görke R1, Meyer-Bäse A, Wagner D, He H, Emmett MR, Conrad CA. Determining and interpreting correlations in lipidomic networks found in glioblastoma cells. BMC Syst Biol. 2010;4:126. doi: 10.1186/1752-0509-4-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee GK, Lee HS, Park YS, Lee JH, Lee SC, Lee JH, Lee SJ, Shanta SR, Park HM, Kim HR, et al. Lipid MALDI profile classifies non-small cell lung cancers according to the histologic type. Lung Cancer. 2012;76:197–203. doi: 10.1016/j.lungcan.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Suarna C, Dean RT, May J, Stocker R. Human atherosclerotic plaque contains both oxidized lipids and relatively large amounts of alpha-tocopherol and ascorbate. Arterioscler Thromb Vasc Biol. 1995;15:1616–1624. doi: 10.1161/01.ATV.15.10.1616. [DOI] [PubMed] [Google Scholar]
- 16.Miyoshi N, Iwasaki N, Tomono S, Higashi T, Ohshima H. Occurrence of cytotoxic 9-oxononanoyl secosterol aldehydes in human low-density lipoprotein. Free Radic Biol Med. 2013;60:73–79. doi: 10.1016/j.freeradbiomed.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Fhaner CJ, Liu S, Ji H, Simpson RJ, Reid GE. Comprehensive lipidome profiling of isogenic primary and metastatic colon adenocarcinoma cell lines. Anal Chem. 2012;84:8917–8926. doi: 10.1021/ac302154g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishikawa S, Tateya I, Hayasaka T, Masaki N, Takizawa Y, Ohno S, Kojima T, Kitani Y, Kitamura M, Hirano S, et al. Increased expression of phosphatidylcholine (16:0/18:1) and (16:0/18:2) in thyroid papillary cancer. PLoS One. 2012;7:e48873. doi: 10.1371/journal.pone.0048873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Mao J, Ai J, Deng Y, Roth MR, Pound C, Henegar J, Welti R, Bigler SA. Identification of plasma lipid biomarkers for prostate cancer by lipidomics and bioinformatics. PLoS One. 2012;7:e48889. doi: 10.1371/journal.pone.0048889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Z, Chen H, Ai J, Zhu Y, Li Y, Borgia JA, Yang JS, Zhang J, Jiang B, Gu W, Deng Y. Global lipidomics identified plasma lipids as novel biomarkers for early detection of lung cancer. Oncotarget. 2017;8:107899–107906. doi: 10.18632/oncotarget.22391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry. 2006;67:1907–1924. doi: 10.1016/j.phytochem.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Smith TC, Frank E. Introducing machine learning concepts with WEKA. Springer; New York, NY: 2016. pp. 353–378. [DOI] [PubMed] [Google Scholar]
- 23.Weir IM, Wong G, Barlow CK, Greeve MA, Kowalczyk A, Almasy L, Comuzzie AG, Mahaney MC, Jowett JB, Shaw J, et al. Plasma lipid profiling in a large population-based cohort. J Lipid Res. 2013;54:2898–2908. doi: 10.1194/jlr.P035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolesnick R. Signal transduction through the sphingomyelin pathway. Mol Chem Neuropathol. 1994;21:287–297. doi: 10.1007/BF02815356. [DOI] [PubMed] [Google Scholar]
- 25.Green DR. Apoptosis and sphingomyelin hydrolysis. The flip side. J Cell Biol. 2000;150:F5–F7. doi: 10.1083/jcb.150.1.F5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gangoiti P, Granado MH, Alonso A, Goñi FM, Gomez-Munoz A. Implication of ceramide, ceramide 1-phosphate and sphingosine 1-phosphate in tumorigenesis. Transl Oncogenomics. 2008;3:81–98. [PMC free article] [PubMed] [Google Scholar]
- 27.Alberg AJ, Armeson K, Pierce JS, Bielawski J, Bielawska A, Visvanathan K, Hill EG, Ogretmen B. Plasma sphingolipids and lung cancer: A population-based, nested case-control study. Cancer Epidemiol Biomarkers Prev. 2013;22:1374–1382. doi: 10.1158/1055-9965.EPI-12-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goldkorn T, Chung S, Filosto S. Lung cancer and lung injury: The dual role of ceramide. Handb Exp Pharmacol. 2013:93–113. doi: 10.1007/978-3-7091-1511-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goldkorn T, Filosto S. Lung injury and cancer: Mechanistic insights into ceramide and EGFR signaling under cigarette smoke. Am J Respir Cell Mol Biol. 2010;43:259–268. doi: 10.1165/rcmb.2010-0220RT. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.