Abstract
Lung cancer is one of the most frequently occurring and fatal cancer types worldwide. Cisplatin is widely used for chemotherapy of non-small cell lung cancer (NSCLC). However, the use of cisplatin has been met with the challenge of chemoresistance as a result of hypoxia, which is common in adult solid tumors and is a principal cause of a poor patient outcome. In the present study, the effects of hypoxia on the response of the NSCLC A549 cell line to the clinically relevant cytotoxic cisplatin were evaluated via regulating hypoxia inducible facor-1α (HIF-1α) and p53. Hypoxia exposure upregulated the expression levels of HIF-1α and p53, and promoted glycolysis in A549 cells, which was attenuated by HIF-1α knockdown by siRNA introduction, indicating the critical roles of HIF-1α in regulating glycolysis under hypoxic conditions. HIF-1α-knockdown also sensitized A549 cells to cisplatin in hypoxia-exposed, but not in normoxia-exposed A549 cells, suggesting that hypoxia-induced cisplatin resistance partially contributes toward the upregulation of HIF-1α by hypoxia exposure. The present study also determined that hypoxia-upregulated p53 activated its downstream target gene p21 transcriptionally and blocked the cell cycle at the G1-G0 phase, thereby leading to inhibition of cell proliferation. As a result, activated p53 desensitized A549 cells to cisplatin potentially through increasing the non-proliferation status of A549 cells and therefore minimizing the influence of cisplatin. Taken together, these results identified the exact effects of HIF-1α and p53 induced by hypoxia and potentially elucidated their protective effects on A549 cells against cisplatin.
Keywords: hypoxia, A549, hypoxia inducible facor-1α, p53, chemoresistance, cisplatin
Introduction
Non-small cell lung cancer (NSCLC) is one of the leading causes of cancer-associated mortality worldwide (1), and includes squamous cell carcinoma, adenocarcinoma and large cell carcinoma (2). Nearly 85% of all lung cancers are attributed to NSCLC, of which 65–80% of patients are diagnosed with an inoperable, locally advanced or metastatic disease (3,4), resulting in poorer patient outcomes. Platinum-based chemotherapy is considered efficient and is used as a standard treatment for completely resected advanced stage NSCLC (5). Cisplatin is one of the most widely used platinum-based chemoagents currently in use and different administration strategies have been developed as a single agent or in combination with other drugs for the treatment of different stages of NSCLC (6). However, the effectiveness of cisplatin-based NSCLC chemotherapy is facing a great clinical challenge from the intrinsic and acquired chemoresistance to cisplatin-related therapeutic strategies (3). Therefore, it is imperative to identify the potential regulatory factors that contribute toward cisplatin chemoresistance in NSCLC, which may be beneficial for the development of novel therapeutic strategies.
In normal tissue, the oxygen tension is in the region of 1–4%, while hypoxia is <1% (7); solid tumors, including lung cancer, frequently suffer from hypoxia. Hypoxia is caused by several mechanisms, including rapid proliferation of tumor cells. The fast-growing rate of tumors may result in the depletion of available oxygen, while erratically growing tumor cells can compress blood vessels (8). Tumors exposed to hypoxia switches on genetic pathways and results in the promotion of tumor malignancies, including chemoresistance, and patients with hypoxic tumors generally have a poorer prognosis (8). In solid tumors, numerous mechanisms were involved in chemoresistance induction via hypoxia exposure (8). In lung cancer, hypoxia can activate autophagy and mediate cisplatin resistance, which can be reversed by pre-treatment with an autophagy inhibitor, 3-methyladenine (9). Consistently, Lee et al (10) revealed that, in NSCLC, hypoxia-induced autophagy contributes toward chemoresistance.
The hypoxia-inducible factor-1 (HIF-1) protein, composed of a hypoxia-regulated α subunit and a non-hypoxia-regulated β subunit, is tightly activated by hypoxia in tumors suffering from hypoxia conditions (8,11,12). Under conditions of normoxia, HIF-1α degrades rapidly, and during hypoxia, HIF-1α protein accumulates and binds to hypoxia-regulated elements (HREs) contained within the promoter region of numerous genes, which regulate metabolism, cell survival, angiogenesis and invasion (13). Hypoxia rapidly stabilizes and activates p53 and is potentially one of the earliest driving forces to regulate p53 function during tumorigenesis (8,14). It has been reported that, under conditions of hypoxia, p53 is phosphorylated and stabilized by the ataxia telangiectasia mutated (ATM) and ATM and RAD3-related kinases (15,16). However, less is known about the exact p53 target genes responsible for the p53-regulated processes in hypoxia.
As an increasing amount of evidence has been revealed to demonstrate the importance of hypoxia in the development of chemoresistance, it is important to discover the potential mechanisms of inducing chemoresistance under exposure to hypoxia (17). For this purpose, an aim of the present study was to elucidate the regulation of hypoxia to the expression of HIF-1α and p53, and their contribution to cisplatin chemotreatment. An additional aim was to demonstrate the contribution of HIF-1α or p53 on the physiological processes of A549, including hypoxia-promoted glycolysis, apoptosis and cell proliferation.
Materials and methods
Cell culture
The NSCLC A549 cell line was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) in 95% air and 5% CO2 at 37°C.
For hypoxia exposure, cells were incubated and treated in an InVivo2 Hypoxia workstation 400 (Ruskinn Technology Ltd., Bridgend, UK) and flushed with 1% O2, 5% CO2 and 94% N2, which is referred to as hypoxia. Prior to specific treatments, cells were pre-incubated for 2, 4, 6 or 12 h under conditions of normoxia or hypoxia.
For PFTα treatment, 30 µM PFTα (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added to cells prior to normoxic or hypoxic exposure for 4 h at 37°C. Then cells were exposed to normoxic or hypoxic condition in RPMI-1640 medium supplemented with 30 µM PFTα.
Reactive oxygen species (ROS) measurement
In order to detect ROS accumulation, cells were co-incubated with 5-(and 6-)-chloromethy1-2-,7-dichlorofluorescin diacetate (DCHF-DA, Life Technologies; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Briefly, 5 mmol/l DCHF-DA was added into RPMI-1640 medium without serum for 15 min at 37°C in the dark. The cells were then washed in 1X PBS and resuspended in RPMI-1640 medium without serum followed by imaging under an X71 (U-RFL-T) fluorescence microscope (Olympus Corporation, Tokyo, Japan) under ×100 magnification. For quantitative measurement, stained cells were read by a microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek Instruments, Inc., Winooski, VT, USA). All experiments were performed with biological triplicates and the data are representative of at least three independent experiments.
CCK-8 assay
In order to evaluate cell viability, A549 cells were seeded in 96-well plates (2×103 cells/well). After 12 h normoxia or hypoxia exposure, 10 µl of CCK-8 solution (Beyotime Institute of Biotechnology, Beijing, China) was added to each well for an additional 4 h co-incubation at 37°C. The absorbance was measured at 620 nm.
EdU staining
To measure cell proliferation, the Click-iT EdU assay (Life Technologies; Thermo Fisher Scientific, Inc.) was performed according to the manufacturer's protocol. Briefly, 1×105 A549 cells were seeded in 12-well plate for 24 h. For labeling cells with 5-ethynyl-2′-deoxyuridine (EdU), an equal volume of 2X EdU solution was added to the wells and incubated for 2 h at 37°C. After three washes with ice-cold PBS, cells were fixed with 4% paraformaldehyde in PBS at room temperature for 10 min. Click-iT reaction cocktail was added to the fixed cells for 30 min incubation at room temperature in darkness. Then the cells were blocked with 1 ml 5% BSA (Sigma-Aldrich; Merck KGaA) in PBS at room temperature for 30 min and Vectashield mounting medium with 4′,6-diamidino-2-phenylindole (DAPI) was added to the cells for staining at room temperature for 10 min and protected from light. After three washes with ice-cold PBS, cells were imaged under a X71 (U-RFL-T) fluorescence microscope with magnification of ×100 (Olympus, Melville, NY, USA).
Western blotting
A549 cells were pelleted and washed three times with ice-cold PBS. Cells were re-suspended with lysis buffer containing 50 mM Tris, 150 mM NaCl, 1% Nonidet P40 (NP-40) and 0.25% sodium deoxycholate. Lysate was centrifuged for 5 min at 12,000 × g at 4°C to remove cell debris. The supernatant was removed into a fresh tube prior to the 200 µl of sample buffer (Beyotime Institute of Biotechnology) being added. Following boiling, sample concentration was determined by BCA assay kit (Sigma-Aldrich; Merck KGaA). A total of 20 µg of each sample was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Inc., Hercules, CA, USA), which were pre-treated with PBS containing 5% bovine serum albumin (BSA) and 0.3% Tween-20 at room temperature for 30 min. Membranes were probed with antibodies against β-actin (cat. no. ab8226), HIF-1α (cat. no. ab113642), p53 (cat. no. ab26) and p21 (cat. no. ab109520) which were purchased from Abcam (Cambridge, UK) at dilution of 1:1,000 at room temperature for 1 h. Then horseradish peroxidase conjugated goat anti-rabbit IgG H&L (cat. no. ab7090) or goat anti-mouse IgG H&L (cat. no. ab97040) secondary antibodies were incubated at a dilution of 1:5,000 at room temperature for 1 h. The signals were visualized using the enhanced chemiluminescence detection kit (GE Healthcare Life Sciences, Little Chalfont, UK) and blotted on X-ray films.
ATP production
A total of 2×105 A549 cells were plated in 12-well plates and allowed to adhere overnight. An ATP Lite assay kit (PerkinElmer, Inc., Waltham, MA, USA) was employed to measure ATP production according to the manufacturer's protocol. The luminescence produced was measured by a multimode microplate reader at a wavelength of 505 nm (Mithras, Mikrowin 2000 software; Berthold Technologies, GmbH, Wildbad, Germany).
Lactate measurements
A total of 2×105 A549 cells were plated in 12-well plates overnight and the medium was replaced with serum-free medium for 12 h. The supernatant was collected by centrifugation at 12,000 × g for 10 min, at 4°C and diluted with 500 µl PBS. For measuring lactate, an EnzyChrom L-lactate assay kit (BioAssay Systems, Hayward, CA, USA) was used according to the manufacturer's protocol and values were measured at 565 nm using a microplate reader Synergy 2 Multi-Mode Microplate Reader (BioTek, Instruments, Inc).
Glucose uptake assay
For measuring glucose uptake, 2×105 A549 cells were plated in 12-well plates overnight for attaching. Cells were incubated in glucose-free RPMI-1640 medium (Life Technologies; Thermo Fisher Scientific, Inc.) for 30 min at 37°C in a CO2 incubator to consume unlabeled intracellular glucose. Next, 2-(3H)deoxyglucose at 0.5 µCi was added into the medium for a 2-h incubation at 37°C. A total of 1×106 A549 cells were washed with ice-cold PBS three times to remove free glucose, and were transferred to scintillation vials for counting.
siRNA duplexes and transfection
siRNA-1 (5′-CAAAGTTCACCTGAGCCTA-3′), siRNA-2 (5′-GATTAACTCAGTTTGAACT-3′) and control siRNA (5′-TTAAATGCCAGCCACGTAC-3′) were synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). For siRNA transfection, Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) was employed according to the manufacturer's protocols. Briefly, 20 pmol siRNA was mixed with 5 µl Lipofectamine 2000 in 100 µl serum-free RPMI-1640 medium for 15 min, and the mixture was added into the refreshed culture medium without serum. A total of 4 h later, the medium was replaced with regular medium and the cells were cultured for an additional 48 h prior to the detection of HIF-1α expression.
Cell cycle analysis
The percentage of cells in each phase was analyzed using the CycleTEST™ PLUS DNA Reagent kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer's protocol. Briefly, cells were suspended using trypsin, prior to being pelleted and washed with ice-cold PBS three times. Cells were then incubated sequentially for 10 min each in solution A, solution B and solution C supplied in Cycletest™ Plus DNA Reagent kit (BD Biosciences). A flow cytometer was used to examine the percentage of cell phases in a BD FACSCanto II, (BD Biosciences). Data were analyzed using FlowJo V6.1.1. software (FlowJo LLC, Ashland, OR, USA).
Cell viability measurement
To measure cell viability, 20 µl 10 mg/ml MTT solution dissolved in PBS was added to each well. A total of 2–4 h later, cells were lysed with a lysis buffer (200 µl/well) containing 20% SDS in dimethyl formamide (DMSO)/H2O (1:1, v/v; pH 4.7) at 37°C for 6 h to dissolve the purple formazan. Next, the optical density (OD) was measured at an absorbance wavelength of 570 nm using a microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek, Instruments, Inc.).
Apoptosis assays
A total of 5×105 A549 cells were suspended and washed twice with PBS and the supernatant was discarded. The cell pellets were mixed with 185 µl binding buffer (BD Biosciences), 5 µl Annexin V-fluorescein isothiocyanate (FITC, BD Biosciences) and 10 µl propidium iodide (PI, Sigma-Aldrich; Merck KGaA) for 10 min in the dark. Subsequently, 300 µl binding buffer was added prior to measurement using a flow cytometer with the BD FACSCanto II (BD Biosciences). Data were analyzed using FlowJo V6.1.1. software (FlowJo LLC).
Statistical analysis
Data are presented as the mean ± standard deviation of three independent experiments. Data were analyzed using SPSS version 16 (SPSS, Inc., Chicago, IL, USA). The differences between two groups were compared using a paired Student's t-test and the differences between three or more groups using one-way analysis of variance followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Hypoxia exposure induced ROS generation, p53 activation and HIF-1α expression, and promoted glycolysis in A549 cells
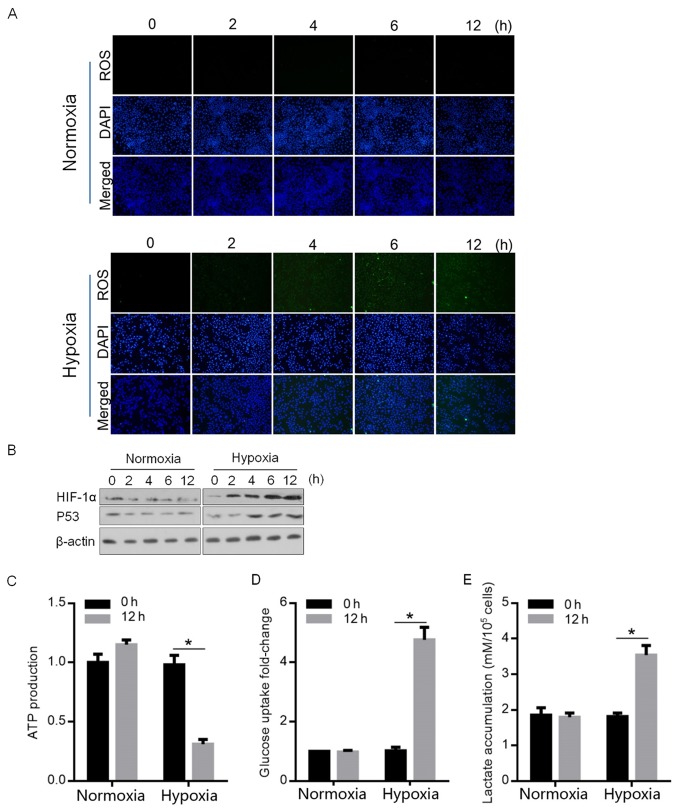
In order to determine the regulatory effects of hypoxia exposure on HIF-1α and p53, the accumulation of ROS was detected at 0, 2, 4, 6 and 12 h following hypoxia exposure. As demonstrated in Fig. 1A, the detectable accumulation of ROS was observed at 6 and 12 h (blue, stained nucleus; green, stained ROS), which is consistent with previous study (17). By performing semi-quantitative western blotting, the expression levels of HIF-1α and p53, which are tightly regulated by hypoxia exposure, were detected. As hypothesized, HIF-1α and p53 protein levels were detectably upregulated (Fig. 1B). A 12-h exposure to hypoxia was selected for further analysis due to the high levels of ROS accumulation, and HIF-1α and p53 expression. HIF-1α and p53 are two of the major transcription factors, which mainly regulate the enzymes that regulate the flow of glycolysis (18). The upregulation of HIF-1α and p53 by hypoxia prompted the detection of the modification of glycolysis by hypoxia exposure. The results demonstrated that hypoxia attenuated the ATP production, and upregulated glucose uptake and accumulated lactate, indicating the promoted glycolysis (Fig. 1C-E).
Figure 1.
Hypoxia exposure causes ROS accumulation, p53 activation and HIF-1α upregulation, and promoted glycolysis. (A) Normoxia- or hypoxia-exposed A549 cells were stained in order to detect ROS after 0, 2, 4, 6 and 12 h. ×40 magnification; blue, stained nucleus; green, stained ROS. (B) Western blotting was performed to detect HIF-1α and p53 following normoxia or hypoxia exposure at 0, 2, 4, 6 and 12 h. To evaluate glycolysis, (C) ATP production, (D) glucose uptake and (E) lactate accumulation were detected 12 h after hypoxia exposure. *P<0.05 vs. 0 h. ROS, reactive oxygen species; HIF-1α, hypoxia inducible facor-1α.
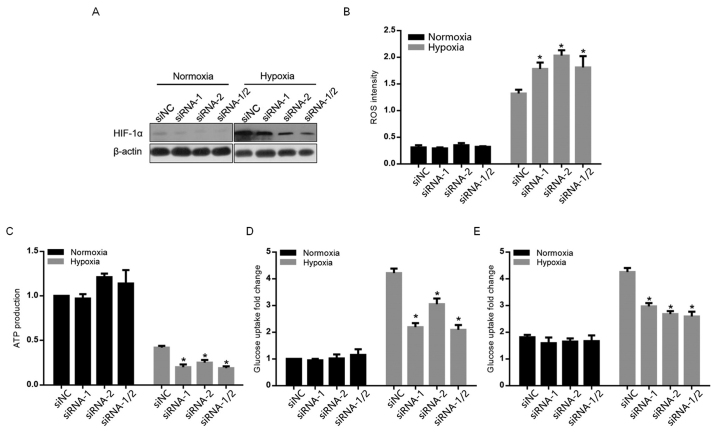
By knocking down HIF-1α, the present study attempted to elucidate the exact role of HIF-1α in glycolysis. As demonstrated in Fig. 2A, siRNA-1, siRNA-2 and siRNA-1/2 (mixture of equal amount of siRNA-1 and siRNA-2) targeting to HIF-1α efficiently knocked down HIF-1α in normoxia- and hypoxia-exposed A549 cells (Fig. 2A). Following HIF-1α knockdown, accumulated ROS and decreased ATP production, glucose uptake and lactate were observed (Fig. 2B-E), indicating that HIF-1α contributes critically for the promotion of glycolysis.
Figure 2.
Contribution of hypoxia-induced HIF-1α to ROS accumulation and glycolysis. (A) The knockdown efficiency of siRNA-1, siRNA-2 or siRNA-1/2 to HIF-1α was detected by western blot analysis. (B) ROS accumulation was detected following HIF-1α knockdown. Following HIF-1α knockdown, (C) ATP production, (D) glucose uptake and (E) lactate accumulation were detected following exposure to hypoxia after 12 h. *P<0.05 vs. normoxia. ROS, reactive oxygen species; siRNA, small interfering RNA; HIF-1α, hypoxia inducible facor-1α; NC, negative control.
Hypoxia-exposed A549 cells are resistant to cisplatin treatment via promoting glycolysis by upregulating HIF-1α
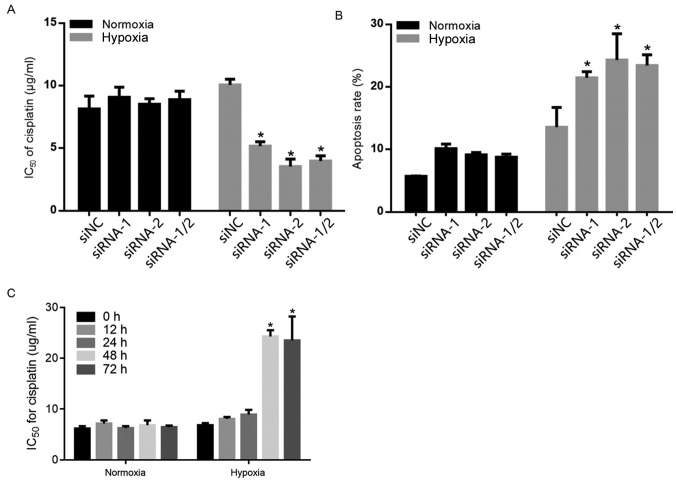
By considering the chemoresistant effects of glycolysis and accumulated ROS induced by hypoxia (17,19), the present study examined whether the presence of HIF-1α sensitized A549 cells to cisplatin. A total of 12 h after hypoxia exposure, compared with siNC-transfected cells, the IC50 value of cisplatin in siRNA-1, siRNA-2 and siRNA-1/2 transiently transfected A549 cells was significantly decreased (Fig. 3A). Additionally, the cells were double stained with Annexin V/PI followed by flow cytometric analysis. Consistently, HIF-1α knockdown significantly promoted apoptotic cell death under hypoxic conditions (Fig. 3B). Unexpectedly, 12-h exposure to hypoxia failed to affect sensitivity to cisplatin in A549 cells. In order to determine whether hypoxia exposure affects chemosensitivity in a time-dependent manner, A549 cells were exposed to hypoxia for 0, 12, 24, 48 or 72 h and were assessed for their chemosensitivity to cisplatin. As demonstrated in Fig. 3C, 48 and 72 h hypoxic-exposure significantly increased chemoresistance to cisplatin.
Figure 3.
The effects of HIF-1α on cisplatin-induced apoptotic cell death. (A) After HIF-1α knockdown, sensitization of A549 cells to cisplatin was measured following a 12-h exposure to normoxia or hypoxia. (B) After HIF-1α knockdown, the apoptotic cell death rate caused by 10 µg/ml cisplatin was measured by Annexin V/propidium iodide double staining followed by cytometric analysis following a 12-h exposure to normoxia or hypoxia. (C) Chemosensitivity was evaluated by measuring cell viability after 0, 12, 24, 48 and 72-h exposure to hypoxia or normoxia with the addition of cisplatin. *P<0.05 vs. normoxia. HIF-1α, hypoxia inducible facor-1α; NC, negative control.
Hypoxia exposure activated p53 and subsequently activated its downstream target gene p21 to block the cell cycle
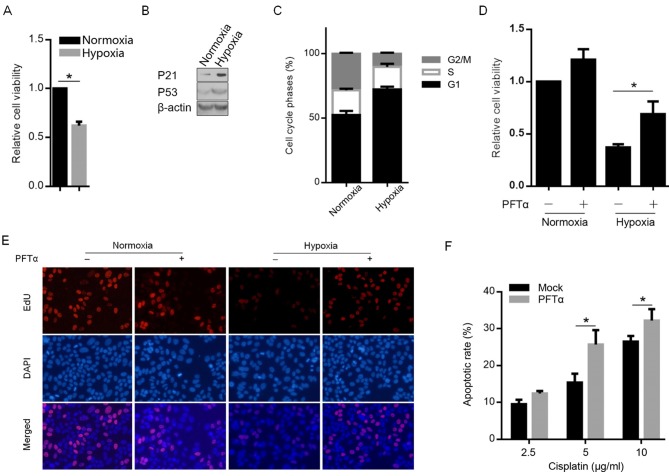
It has been demonstrated previously that hypoxia exposure upregulated and activated p53, which regulates numerous physiological processes in A549 cells (20). By performing MTT analysis, it was revealed that hypoxia exposure for 12 h inhibited cell viability 24 h later when compared with exposure to normoxia conditions (Fig. 4A). Therefore, the expression of p53's downstream target gene p21, which contributes toward cell cycle arrest at the G1 phase (21), was detected. As demonstrated in Fig. 4B, following 12-h hypoxia exposure, p53 and p21 were detectably upregulated. The cell cycle analysis results further indicated that the activated p21 by p53 block the cell cycle at the G2 phase. To further verify this, we pre-treated A549 cells with pifithrin-α (PFTα), a p53 inhibitor that downregulates p21 gene expression (22), followed by the hypoxia exposure for 12 h. The results demonstrated that pre-treatment of A549 cells with PFTα abolished the cell viability inhibition by hypoxia exposure (Fig. 4D). By performing EdU staining, as hypothesized, hypoxia exposure decreased EdU positively stained cells and pre-treatment of PFTα increased the number of EdU positively stained cells (Fig. 4E). The detection of the apoptotic cell death rate following cisplatin treatment (2.5, 5 and 10 µg/ml) also demonstrated that, following blockage of p53's transcriptional activity by PFTα treatment, the apoptotic rate increased significantly (Fig. 4F).
Figure 4.
Hypoxia-induced p53 upregulated p21 and blocked the cell cycle at G2 and thus inhibited proliferation following hypoxia exposure. (A) MTT assay was performed to detect the cell viability inhibition by hypoxia exposure. *P<0.05. (B) Semi-quantitative western blot analysis was performed to detect p53 and p21 protein levels. (C) Cell phase analysis by PI staining. (D) Cell viability was measured in A549 cells pre-treated with 30 µM PFTα followed by hypoxia exposure. *P<0.05. (E) EdU staining was performed to detect proliferating cells at magnification of ×100 (blue, stained nucleus; green, stained reactive oxygen species). (F) The apoptotic cell death rate was measured following PFTα treatment. *P<0.05. PFTα, pifithrin-α.
Discussion
The present study reported findings that revealed the potential effects of HIF-1α and p53 induced by hypoxia in NSCLC A549 cells on a number of physiological processes, including hypoxia-promoted glycolysis, apoptosis, cell proliferation and chemoresistance to cisplatin. Firstly, hypoxia exposure was demonstrated to induce HIF-1α and p53 expression and to promote ROS generation and glycolysis. By introducing siRNA targeting to HIF-1α for knockdown, it was demonstrated that hypoxia exposure promoted glycolysis and induced chemoresistance to cisplatin via activating HIF-1α. Next, the effects of hypoxia-induced p53 on cell proliferation were examined and it was identified that upregulated p53 transcriptionally activated its downstream target gene, p21, and blocked cell cycle progression at the G1-G0 phase, causing inhibition of cell proliferation. Notably, this also desensitized A549 cells to cisplatin, potentially through increasing the non-proliferation status of A549 cells and therefore minimizing the influence of cisplatin.
HIF-1α and HIF-2α are the most well studied members of the hypoxia-inducible factors (HIFs) family, which are master regulators of oxygen homeostasis (23,24). HIF-1α is the focus of the majority of studies due to its expression in all cells of all metazoan species and it transcriptionally regulates multiple genes that mediate glycolysis (24). HIF-1α activity is mainly regulated post-transcriptionally in response to hypoxia; consequently, HIF-1α serves a critical role in regulating glycolysis through its downstream target gene GAPDH under hypoxic conditions (25). In the present study, hypoxia was demonstrated to upregulate the expression of HIF-1α, and therefore partially promote glycolysis in A549 cells. Meanwhile, ROS accumulation induced by hypoxia exposure was partially inhibited by HIF-1α expression, potentially via promoting glycolysis. These results demonstrated that hypoxia-induced HIF-1α exerts a protective function in A549 cells by preventing ROS accumulation. Although no data directly indicated the protective role of HIF-1α against hypoxia stress, the direct regulation of glycolysis by HIF-1α has resulted in the hypothesis that HIF-1α protects NSCLC cells via regulating glycolysis.
Previous studies have demonstrated that hypoxia exposure induces and activates p53 and thus promotes apoptosis, indicating the primary role of p53 as a pro-apoptotic factor under hypoxic conditions (16,26,27). Leszczynska et al (28) reported that hypoxia-induced p53 targets several downstream target genes, including inositol polyphosphate-5-phosphatase (INPP5D), pleckstrin domain-containing A3 (PHLDA3), sulfatase 2 (SULF2), B cell translocation gene 2 (BTG2), cytoplasmic FMR1-interacting protein 2 (CYFIP2), and KN motif and Ankyrin repeat domains 3 (KANK3); consequently, p53 mediated tumor suppression (28). In the present study, similar activation of p53 following hypoxia exposure and the transcriptional activation of its downstream target gene, p21, was identified. Notably, inhibition of proliferation, by upregulated p21, exerted a protective effect against cisplatin treatment in A549 cells, which indicated that p53 exhibited a pro-survival role in this case. When considering that p53 exerts a protective effect at the comparative low dose of cisplatin, yet not the comparatively high dose, suggesting that p53's role potentially alters between pro-survival and pro-apoptotic, subject to the environmental conditions.
In conclusion, hypoxia-induced HIF-1α and p53 were demonstrated to significantly contribute toward cisplatin resistance, suggesting that hypoxia is a potential target for tumor therapy in NSCLC. p53 is widely believed to be required to prevent chemoresistance; however, the results of the present study indicated that the potential balance between the pro-survival and pro-apoptotic roles of p53 is dependent on the dose of cisplatin. This suggested that alternative approaches and appropriate dose of chemoagents are required in order to regulate chemoresistance. Further study is required in order to identify the precise roles and mechanisms of HIF-1α and p53 in hypoxia-induced chemoresistance.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Chengdu Medical College scientific grant (grant no. 17Z127).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author's contributions
QG and QZ designed part of the experiments. QG was involved in performing cell culture experiments. FL and XY performed the gene expression analysis, cell transfection and cell treatments. ZX and YW were involved in molecular experiments, data analysis and writing of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ma L, Huang Y, Zhu W, Zhou S, Zhou J, Zeng F, Liu X, Zhang Y, Yu J. An integrated analysis of miRNA and mRNA expressions in non-small cell lung cancers. PLoS One. 2011;6:e26502. doi: 10.1371/journal.pone.0026502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mountain CF, Lukeman JM, Hammar SP, Chamberlain DW, Coulson WF, Page DL, Victor TA, Weiland LH. Lung cancer classification: The relationship of disease extent and cell type to survival in a clinical trials population. J Surg Oncol. 1987;35:147–156. doi: 10.1002/jso.2930350302. [DOI] [PubMed] [Google Scholar]
- 3.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 4.Beadsmoore CJ, Screaton NJ. Classification, staging and prognosis of lung cancer. Eur J Radiol. 2003;45:8–17. doi: 10.1016/S0720-048X(02)00287-5. [DOI] [PubMed] [Google Scholar]
- 5.Merk J, Rolff J, Dorn C, Leschber G, Fichtner I. Chemoresistance in non-small-cell lung cancer: can multidrug resistance markers predict the response of xenograft lung cancer models to chemotherapy? Eur J Cardiothorac Surg. 2011;40:e29–e33. doi: 10.1016/j.ejcts.2011.02.010. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, Pu X, Wang Q, Cao J, Xu F, Xu LI, Li K. miR-96 induces cisplatin chemoresistance in non-small cell lung cancer cells by downregulating SAMD9. Oncol Lett. 2016;11:945–952. doi: 10.3892/ol.2015.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ebbesen P, Eckardt KU, Ciampor F, Pettersen EO. Linking measured intercellular oxygen concentration to human cell functions. Acta Oncol. 2004;43:598–600. doi: 10.1080/02841860410020220. [DOI] [PubMed] [Google Scholar]
- 8.Keith B, Simon MC. Hypoxia-inducible factors, stem cells, and cancer. Cell. 2007;129:465–472. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu HM, Jiang ZF, Ding PS, Shao LJ, Liu RY. Hypoxia-induced autophagy mediates cisplatin resistance in lung cancer cells. Sci Rep. 2015;5:12291. doi: 10.1038/srep12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee JG, Shin JH, Shim HS, Lee CY, Kim DJ, Kim YS, Chung KY. Autophagy contributes to the chemo-resistance of non-small cell lung cancer in hypoxic conditions. Respir Res. 2015;16:138. doi: 10.1186/s12931-015-0285-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galanis A, Pappa A, Giannakakis A, Lanitis E, Dangaj D, Sandaltzopoulos R. Reactive oxygen species and HIF-1 signalling in cancer. Cancer Lett. 2008;266:12–20. doi: 10.1016/j.canlet.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 12.Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brady CA, Attardi LD. p53 at a glance. J Cell Sci. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 15.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammond EM, Dorie MJ, Giaccia AJ. ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J Biol Chem. 2003;278:12207–12213. doi: 10.1074/jbc.M212360200. [DOI] [PubMed] [Google Scholar]
- 17.Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeung SJ, Pan J, Lee MH. Roles of p53, MYC and HIF-1 in regulating glycolysis the seventh hallmark of cancer. Cell Mol Life Sci. 2008;65:3981–3999. doi: 10.1007/s00018-008-8224-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai Z, Lu Y, Qiu S, Fan Z. Overcoming cisplatin resistance of ovarian cancer cells by targeting HIF-1-regulated cancer metabolism. Cancer Lett. 2016;373:36–44. doi: 10.1016/j.canlet.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan KD, Gallant-Behm CL, Henry RE, Fraikin JL, Espinosa JM. The p53 circuit board. Biochim Biophys Acta. 2012;1825:229–244. doi: 10.1016/j.bbcan.2012.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts JM, Koff A, Polyak K, Firpo E, Collins S, Ohtsubo M, Massagué J. Cyclins, cdks, and cyclin kinase inhibitors. Cold Spring Harb Symp Quant Biol. 1994;59:31–38. doi: 10.1101/SQB.1994.059.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 23.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92:967–1003. doi: 10.1152/physrev.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Del Rey MJ, Valín Á, Usategui A, García-Herrero CM, Sánchez-Aragó M, Cuezva JM, Galindo M, Bravo B, Cañete JD, Blanco FJ, et al. Hif-1α Knockdown reduces glycolytic metabolism and induces cell death of human synovial fibroblasts under normoxic conditions. Sci Rep. 2017;7:3644. doi: 10.1038/s41598-017-03921-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hammond EM, Mandell DJ, Salim A, Krieg AJ, Johnson TM, Shirazi HA, Attardi LD, Giaccia AJ. Genome-wide analysis of p53 under hypoxic conditions. Mol Cell Biol. 2006;26:3492–3504. doi: 10.1128/MCB.26.9.3492-3504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koumenis C, Alarcon R, Hammond E, Sutphin P, Hoffman W, Murphy M, Derr J, Taya Y, Lowe SW, Kastan M, Giaccia A. Regulation of p53 by hypoxia: Dissociation of transcriptional repression and apoptosis from p53-dependent transactivation. Mol Cell Biol. 2001;21:1297–1310. doi: 10.1128/MCB.21.4.1297-1310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leszczynska KB, Foskolou IP, Abraham AG, Anbalagan S, Tellier C, Haider S, Span PN, O'Neill EE, Buffa FM, Hammond EM. Hypoxia-induced p53 modulates both apoptosis and radiosensitivity via AKT. J Clin Invest. 2015;125:2385–2398. doi: 10.1172/JCI80402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.