Abstract
Keloids are a type of benign hyperplasia that cause dermatologic dysfunction and esthetic deformity by invading adjacent normal tissues. Little is known about their etiology, therefore, they are a challenge to treat using plastic surgery. In a previous study, it was demonstrated that the expression of the long non-coding RNA CACNA1G-AS1 (CAS1) is high in keloid tissue, suggesting that CAS1 is involved in keloid formation. In the present study, the aim was to identify potential keloid target proteins by exploring CAS1 biological function during cell proliferation and migration, cytokine secretion, collagen secretion and the control of calcium channel protein expression in human keloid fibroblasts. Three biopsy samples were collected from each patient with keloids at The Peking Union Medical College Hospital, which were then used to investigate the role of CAS1 in cell proliferation and migration. CAS1 silencing was also carried out using small interfering RNA; cell factors, collagen and calcium channel protein levels were compared with control cells. The interference of CAS1 expression reached 50% compared with the control group. CACNA1G and type I collagen expression was significantly downregulated by CAS1 knockdown, while the expression of transforming growth factor-β and type III collagen was not affected. Wound healing time was longer in the CAS1-knockdown group, but there was no visible change in cell proliferation. In conclusion, CAS1 appeared to promote calcium channel protein and type I collagen expression, and to have a positive effect on cell migration in human keloid fibroblasts. Therefore it has potential as a novel therapeutic target for keloids.
Keywords: keloid, fibroblast, long noncoding RNA, CACNA1G-AS1, calcium channel
Introduction
Keloids are a dermatologic condition that affects 1/10 people in Africa (1), and has an incidence of 0.15% in the Asian population (2). Although it is a benign hyperplasia, it causes dermatologic dysfunction and esthetic deformity by invading adjacent normal tissues (3). Frequent episodes of itching and pain also occur, resulting in physical and psychological distress (4). The majority of keloids are initiated by minor skin trauma, such as folliculitis and/or acne, and grow with chronic inflammation of the reticular layer of the dermis (5). A typical pathognomonic characteristic of keloids is the presence of thickened and hyalinized collagen (6). They are problematic in plastic surgery due to little being known about their etiology or optimal treatment.
Keloids are considered to result from prolonged, aberrant wound healing that involves excessive fibroblast participation and collagen deposition (7). Fibroblasts are the primary effector cells of keloid tissue and are characterized by excessive proliferation, disordered apoptosis, and increased secretion of extracellular matrix (8). Numerous previous studies have investigated the pathogenesis of keloids with respect to genetics, mechanics, endocrinology, immunology and nutrition (9–11). The skin injury-wound tension theory was a milestone in our understanding of keloid formation (9). Keloids are frequently observed on the anterior chest and scapular regions, but rarely on the scalp or anterior lower legs; this is closely associated with the frequency of local physical tension or movement in these regions (9). Reducing skin tension around wounds or scars can, therefore, be beneficial in the prevention and treatment of keloids (12). The characteristic shapes of keloid scars are largely determined by the direction of local mechanical forces on the skin (13). Other influences, besides these local factors, also promote keloid development, including genetic factors (single nucleotide polymorphisms) (14), systemic factors (hypertension (15) and estrogen levels) (16), endocrinal factors (physiological hyperactivity of the sebaceous gland) (17) and metabolic factors (higher adenosine triphosphate levels and insufficient oxygen levels) (18). Although keloid development is known to involve complex pathways, the exact mechanisms by which keloid formation is initiated and regulated remain to be elucidated.
In our previous study, an investigation of differential long non-coding RNA (lncRNA) expression between keloids and normal skin tissue was conducted, and it was revealed that the lncRNA CANCA1G-AS1 (CAS1) was significantly upregulated in keloid tissue (19). This suggested that CAS1 may be involved in the mechanism of keloid formation. In the present study, an investigation of the biological role of CAS1 in cell proliferation, cell migration, cytokine secretion and collagen secretion was performed, with controlled calcium channel protein expression in human keloid fibroblasts using loss-of-function studies.
Materials and methods
Patient samples
This study was approved by the institutional review board at Peking Union Medical College (Beijing, China), in accordance with the principles of the Declaration of Helsinki. Keloid and corresponding normal skin from 16 patients, who received surgery at The Peking Union Medical College Hospital (Beijing, China) (19), was previously obtained in 2014. Written informed consent was obtained from all patients prior to surgery. None of the patients in the study received any local treatment, such as corticosteroid injections, or radiation therapy prior to surgery.
Cell culture
Cultures were established from keloid specimens of three patients, processed within 4 h of post-surgical excision, using conventional methods (20). Briefly, these specimens were washed three times in 1X PBS and the epidermis and subdermal fat were removed. The remaining dermis was dissected and incubated in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) at 37°C in a 5% CO2 humidified atmosphere. The DMEM was replaced every 3 days and keloid fibroblasts were used during passages 3–5. Detailed information about primary keloid fibroblast isolation and culture was described in our previous report (19).
Transfection
Three small interfering RNAs (siRNAs) targeting different sites of lncRNA CAS1 (GenBank accession no. NR_038439.1) were designed and synthesized by Suzhou GenePharma Co., Ltd. (Jiangsu, China) to knock down CAS1 expression, as well as a control siRNA that did not target CAS1 as the negative control (NC). The sequences were as follows: Cas1 siRNA-1, 5-CCCUCAACCCAAGGAAGAUTT-3; Cas1 siRNA-2, 5-GCCUUCGCAACUCAUUCAUTT-3; Cas1 siRNA-3, 5-CCGUGUGAAGGGAGCAAUUTT-3; NC, 5-UUCUCCGAACGUGUCACGUTT-3. Keloid fibroblasts were seeded in 6-well plates (1×105 cells/well) and incubated overnight in an atmosphere of 5% CO2 at 37°C to allow their full extension and adherence prior to transfection. The cells were grown to 70–90% confluency, and the siRNAs were transfected using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. The cells were collected for further experiments 48 h after transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.) 48 h after transfection, according to the manufacturer's instructions. The expression of CAS1 and other target genes was quantified relative to endogenous GAPDH expression, using RT-qPCR with the following primer sets: CAS1 forward, 5′-TGTGCTTCACCATGCTCCAT-3′ and reverse, 5′-ATTAGTGCTCCGGCCAACAA-3′; GAPDH forward, 5′-GGTCACCAGGGCTGCTTTTA-3′ and reverse, 5′-GGATCTCGCTCCTGGAAGATG-3′; CACNA1G forward, 5′-CACGGTCATCTCGCCTATCT-3′ and reverse, 5′-TCCTTGTTGCTCTCCTCCAG-3′; transforming growth factor-β (TGF-β), forward, 5′-GCAACAATTCCTGGCGATAC-3′ and reverse, 5′-CTAAGGCGAAAGCCCTCAAT-3′; COL1A1, forward 5′-AAGACATCCCACCAATCACC-3′ and reverse, 5′-CGTCATCGCACAACACCTT-3′; and COL3A1 forward, 5′-CTGCCATCCTGAACTCAAGAGTGG-3′ and reverse, 5′-CCATCCTCCAGAACTGTGTAGG-3′. The PCR conditions were as follows: 95°C for 60 sec, followed by 40 cycles of 95°C for 10 sec, and finally 60°C for 45 sec. After amplification, real-time data acquisition and analysis were performed. The relative quantitative results were calculated using the 2−∆∆Cq method (21).
MTS assay
Cell proliferation was assessed using an MTS assay (Promega Corporation, Madison, WI, USA). Cells were plated at a density of 5×103 cells/well in 96-well plates and incubated overnight in an atmosphere of 5% CO2 at 37°C. After transfection, 20 µl MTS was added into each well containing 100 µl DMEM (Thermo Fisher Scientific, Inc.), and the cells were then incubated at 37°C for 1 h in a humidified 5% CO2 incubator. Absorbance was detected at 490 nm using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell cycle analysis by flow cytometry
Cells were collected 48 h after transfection, washed with PBS and trypsinized with 0.025% trypsin-EDTA to yield single cell suspensions. They were then fixed in ice-cold 70% ethanol and stained with 50 µg/ml propidium iodide solution (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) containing 10 µg/ml RNaseA (Tiangen Biotech Co., Ltd., Beijing, China). A BD Accuri™ C6 flow cytometer was used for flow cytometric analysis, and the cell cycle profiles were analyzed using ModFit LT software for Windows Version 3.2 (Verity Software House, Inc., Topsham, ME, USA).
Scratch wound migration assay
Keloid fibroblasts were seeded uniformly (1×104 cells/well) into 6-well plates and grown overnight. A total of 48 h after transfection, a scratch wound was made across the center of each confluent cell culture monolayer using a sterile 200 µl pipette tip, and any non-adherent cells were washed off with 1X PBS. Conditioned DMEM was then reapplied to the cells according to the previous treatment regimen, and the plates were incubated for a further 72 h in at atmosphere of 5% CO2 at 37°C). Each well was imaged every 24 h using objective inverted microscopy (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Statistical analysis was performed using the SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA). All results are presented as the mean ± standard deviation and were analyzed using a Student's t-test and one-way analysis of variance (ANOVA) to determine the levels of significance. Bonferroni correction was used to perform the post-hoc test following ANOVA. P<0.05 was considered to indicate a statistically significant difference.
Results
siRNA-3 downregulates CAS1 expression in keloid fibroblasts
A total of 48 h after transfection, RT-PCR was used to detect CAS1 mRNA expression in keloid fibroblasts. Of the three siRNA sequences, siRNA-3 had the greatest effect on reducing CAS1 expression (Fig. 1), and thus was selected as an effective means of interference for use in further experiments.
Figure 1.
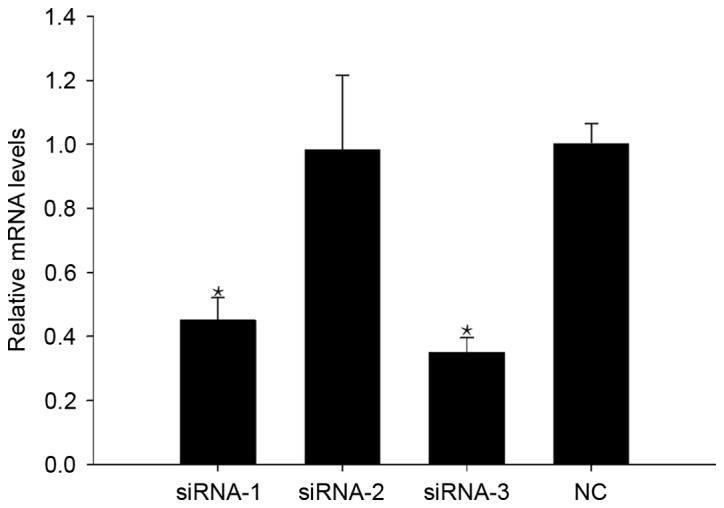
CAS1 expression in keloid fibroblasts 48 h after transfection with siRNAs. Results are presented as the mean ± standard deviation of three independent experiments. *P<0.05, compared with NC-treated cells. CAS1, CACNA1G-AS1; siRNA, small interfering RNA; NC, negative control.
CAS1 knockdown does not inhibit cell proliferation
To examine the effects of CAS1 on cell proliferation, an MTS assay, using keloid fibroblasts, was performed and cell viability was measured every 24 h (Fig. 2). The data revealed that CAS1 knockdown did not change the cell proliferative rate compared with NC-treated cells. This indicated that CAS1 may not be involved in cell proliferation.
Figure 2.
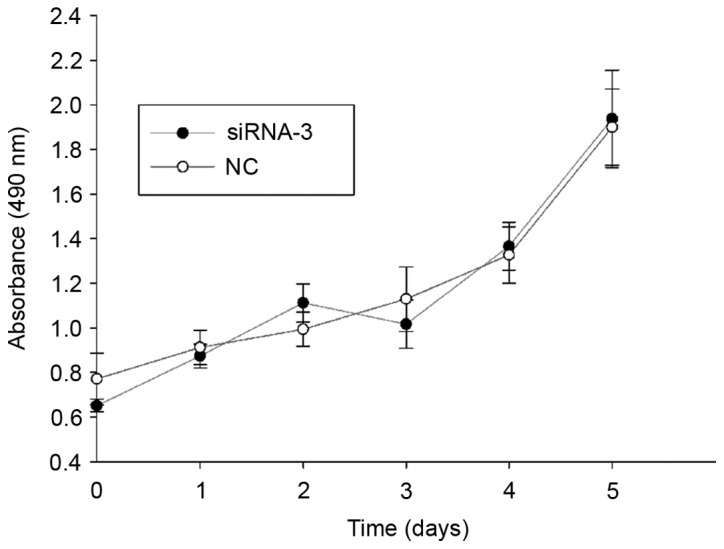
CAS1-knockdown effect on cell proliferation. No significant difference was detected in proliferation between cells transfected with siRNA-3 and NC-treated cells. CAS1, CACNA1G-AS1; siRNA, small interfering RNA; NC, negative control.
CAS1 knockdown does not change the cell mitotic index
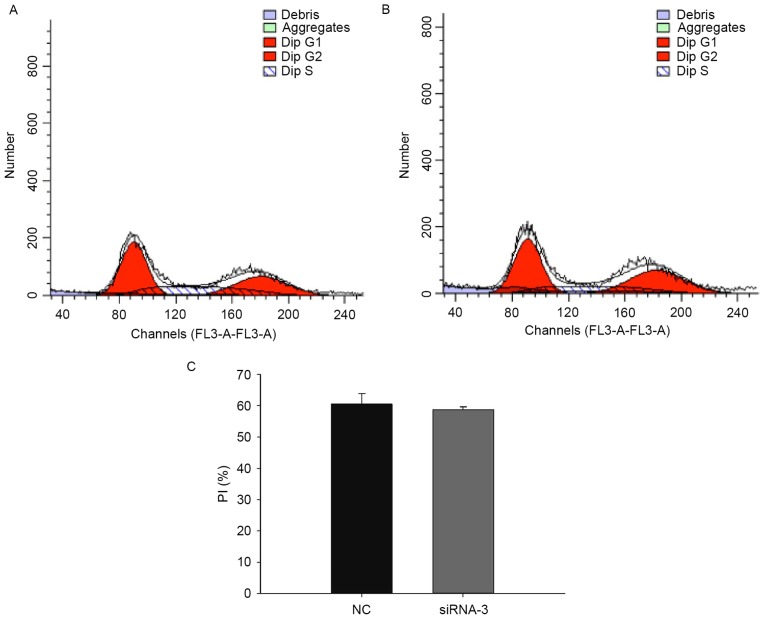
To assess the effects of CAS1 on the cell cycle, analysis of the cell cycle distribution 48 h after siRNA-3 transfection was conducted; this revealed no change in the cell cycle compared with NC-treated cells (Fig. 3). This suggested that downregulation of CAS1 expression does not alter the cell cycle in keloid fibroblasts, which implied that CAS1 does not participate in cell division.
Figure 3.
Effects of CAS1 knockdown on the cell cycle in keloid fibroblasts. (A) Cell cycle distribution in keloid fibroblasts 48 h after transfection with the control siRNA. G1, 39.49%; G2, 33.28%; S, 27.23%. (B) Cell cycle distribution in keloid fibroblasts 48 h after transfection with siRNA-3. G1, 41.31%; G2, 38.4%; S, 20.29%. (C) The PI (n=3) equals the sum of the G2 and S phases of the cell cycle, and is expressed as a percentage. Results are presented as the mean ± standard deviation of three independent experiments. P>0.05, compared with NC-treated cells. CAS1, CACNA1G-AS1; siRNA, small interfering RNA; PI, proliferation index; FL3, flavagline; NC, negative control.
CAS1 knockdown decreases the rate of wound closure
The scratch wound assay demonstrated that transfected cells had a slower closure rate than NC-treated cells (Fig. 4), suggesting that the downregulation of CAS1 expression reduced the fibroblast migration rate. This indicated that CAS1 may serve a role in keloid fibroblast migration.
Figure 4.
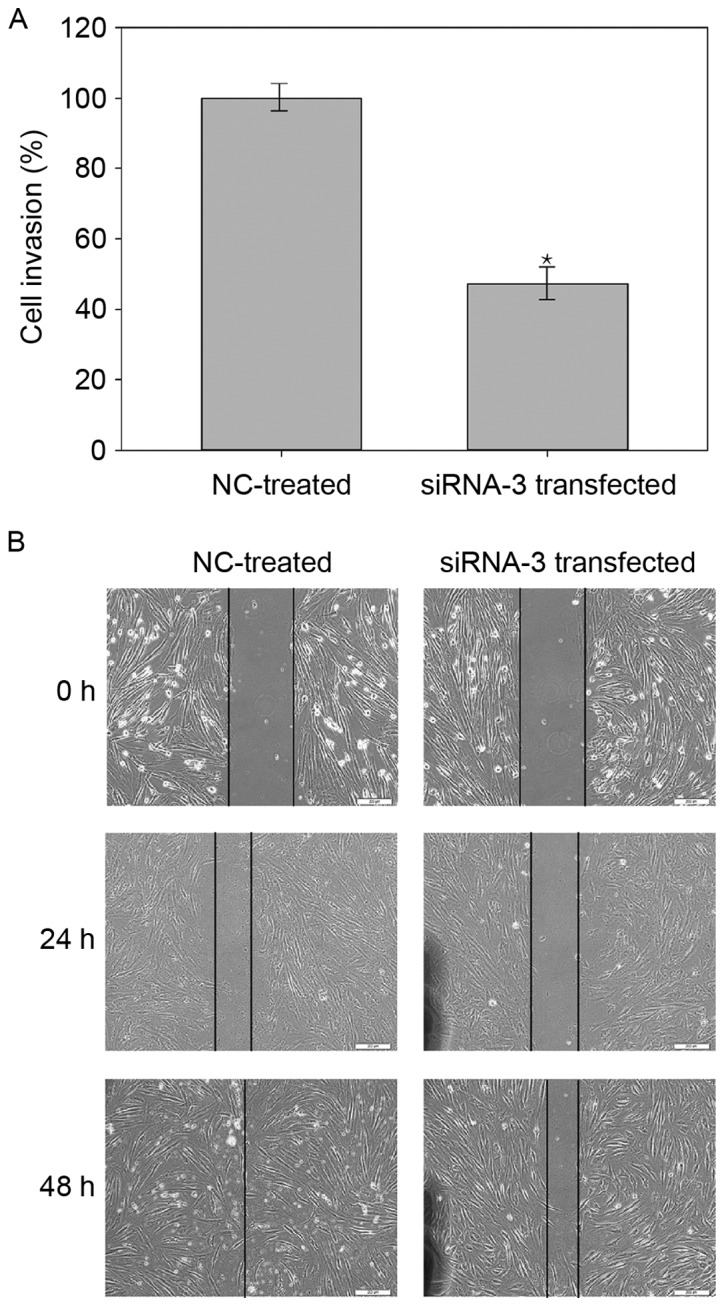
CAS1 knockdown decreases the rate of wound closure. (A) Effect of CAS1 knockdown on cell invasion. Cells migrated more slowly over the wound after CAS1 knockdown. Results are presented as the mean ± standard deviation of three independent experiments. *P<0.05, compared with NC-treated cells. (B) Representative light microscopy images of NC-treated and siRNA-3 transfected cells at 0, 24 and 48 h after scratching of the monolayer. Scale bar, 200 µm. CAS1, CACNA1G-AS1; siRNA, small interfering RNA; NC, negative control.
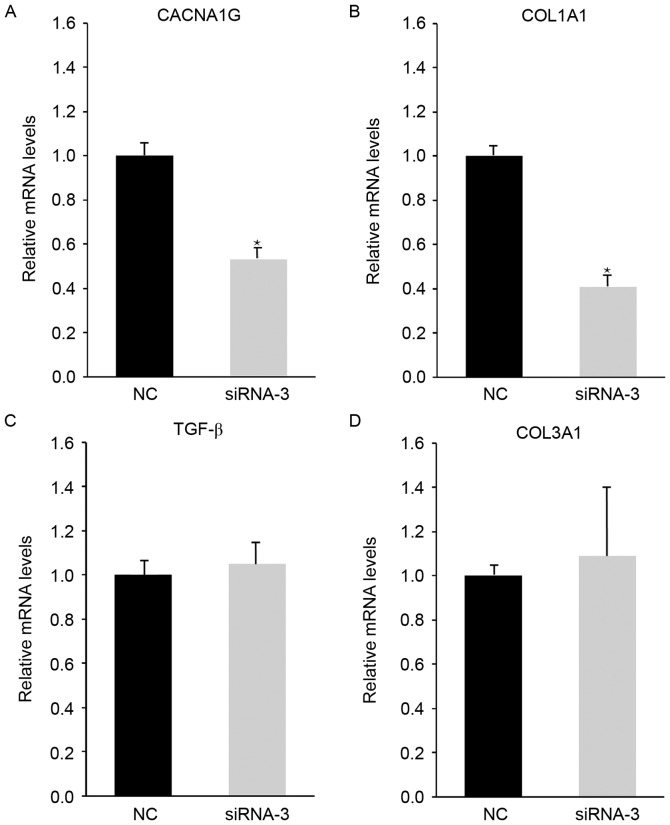
CAS1 knockdown alters CACNA1G and COL1A1 expression
To explore the interaction between CAS1 and the biological properties of keloid fibroblasts, analysis of CACNA1G, TGF-β, COL1A1 and COL3A1 expression using RT-PCR was conducted. Post-siRNA-3 transfection, CACNA1G and COL1A1 expression decreased, while TGF-β and COL3A1 expression was unchanged, compared with in the NC-treated cells (Fig. 5). These results suggested that CAS1 knockdown affects calcium channels and collagen secretion. The absence of a notable effect on TGF-β expression following CAS1 knockdown supports these findings, which suggests that CAS1 does not affect the cell cycle.
Figure 5.
CAS1 knockdown reduced (A) CACNA1G and (B) COL1A1 expression in keloid fibroblasts, whereas (C) TGF-β and (D) COL3A1 expression did not change. Results are presented as the mean ± standard deviation of three independent experiments. *P<0.05, compared with NC-treated cells. CAS1, CACNA1G-AS1; siRNA, small interfering RNA; NC, negative control.
Discussion
In the present study, it was demonstrated that CAS1 knockdown significantly downregulated CACNA1G and type I collagen expression, but that it had little effect on TGF-β and type III collagen expression. The wound healing assay also demonstrated that CAS1 knockdown inhibited keloid fibroblast invasion, but the MTS assay and cell cycle analysis revealed that it had a limited impact on cell proliferation.
lncRNAs are a group of RNA molecules that range in length from 200–100,000 nucleotides and do not encode proteins (22). Numerous studies have demonstrated that lncRNAs participate in various regulatory processes, including transcriptional activation, transcriptional interference and intranuclear transport (22–24). lncRNAs have also been identified to serve an important role in the development, growth and progression of human carcinomas, acting as oncogenic drivers through diverse mechanisms, including cell proliferation, invasion, apoptosis and the secretion of essential proteins (25). For example, several lncRNAs are differentially expressed in melanoma cell lines compared with their controls. One of these lncRNAs, SPRY4-IT1, is derived from an intron of the SPRY4 gene, and is predominantly localized in the cytoplasm of melanoma cells, as identified by RNA fluorescence in situ hybridization analysis (26). SPRY4-IT1 RNA interference impairs cell growth and differentiation, and leads to higher rates of apoptosis in melanoma cell lines. Thus, higher SPRY4-IT1 expression may be important to the molecular etiology of human melanoma, and could be used as an early biomarker for melanoma detection (26).
In an earlier study (19), microarray analysis was used to identify differential lncRNA expression between three pairs of keloid and normal skin tissues. A total of 1,731 lncRNAs were identified to be consistently upregulated and 782 were downregulated in keloids (fold-change ≥2.0). Validation of our findings using quantitative RT-PCR revealed consistency with these microarray results. It was also demonstrated that 11 signaling pathways were upregulated and 44 were downregulated in keloid tissues compared with the healthy controls. Within the co-expression network, one lncRNA was connected with numerous mRNAs, and vice versa. Bioinformatic analysis indicated that lncRNA CACNA1G-AS1 may be crucial for keloid formation (19).
Numerous studies (27–29) have demonstrated that lncRNAs may serve important roles in carcinomas and fibrotic diseases by regulating the process of cell proliferation, invasion, apoptosis, and the secretion of essential proteins.
One of the metastasis-associated lncRNAs, HOTAIR, was previously observed to be highly expressed in primary melanoma lymph node metastases (27). Knockdown of HOTAIR using siRNAs reduced motility and invasion of the human melanoma cell line A375. siHOTAIR also suppressed gelatin matrix degradation, suggesting that HOTAIR promotes gelatinase activity. These data indicate that lncRNAs may be involved in melanoma metastasis. In the present study, it was revealed that CAS1 may be involved in keloid fibroblast invasion, which provided new insights into the keloid pathology of invasion into adjacent normal tissues.
Mounting evidence exists for the deregulation of lncRNAs in fibrotic diseases, indicating that these molecules are differentially expressed during fibrotic remodeling (28,29). For example, it was recently demonstrated that Meg3 inhibits hepatic stellate cell activation and liver fibrogenesis. In vitro, Meg3 overexpression limits the TGF-β1-induced proliferation of hepatic stellate cells and activates p53-dependent apoptosis in fibrotic livers (28). Additionally, depletion of the lncRNA ZEB2NAT was demonstrated to decrease epithelial-mesenchymal transition (EMT)-associated gene expression and cancer cell invasion in urinary bladder cancer-associated fibroblasts. These results provide support for fibroblast induction of EMT and the invasion of urinary bladder cancer cells through the TGF-β1-ZEB2NAT axis (29). However, no significant association was identified between CAS1 and cell proliferation or TGF-β expression in the present study.
CAS1 is the antisense RNA of CACNA1G, which is the mRNA of the T-type channel protein Cav3.1 (30). Numerous antisense lncRNAs have been identified to be involved in the regulatory gene-net of diseases. For example, proliferating cell nuclear antigen (PCNA)-AS1, the antisense of PCNA, is significantly upregulated in hepatocellular carcinoma. It promotes tumor growth in vitro and in vivo through RNA hybridization, which increases PCNA mRNA stability (31).
Clinical trials (32–34) have demonstrated that verapamil, a calcium channel blocker that targets L-type and T-type channels, was effective at preventing and treating keloids with no major side effects. Calcium antagonists are reported to promote a change in cell shape from bipolar to spherical, which may reflect their calmodulin inhibitor-like behavior involving calcium-independent alteration or rearrangement of the actin cytoskeleton (35). Alternatively, the effects of calcium antagonists may be similar to that of cytochalasin B, which alters the cell shape by disrupting stress fibers, and inducing the expression of collagenase and protease (36). Further studies (37,38) revealed that verapamil induces procollagenase expression and increases collagenase; it also inhibits the synthesis of extracellular matrix molecules, including collagen, fibronectin and glycosaminoglycans. Fibroblasts in keloids exhibit elevated levels of interleukin 6 and vascular endothelial growth factor, which are decreased by verapamil, reducing cell proliferation and increasing apoptosis (39). Verapamil was also indicated to prevent keloid formation by inhibiting proliferation and TGF-β1 expression in fibroblasts (40). The finding that CACNA1G expression was downregulated by CAS1 knockdown suggests that CAS1 affects calcium channel expression, leading to a reduction in collagen levels and cell invasion.
In conclusion, CAS1 may promote the expression of the calcium channel protein CACNA1G and type I collagen, and also have a positive effect on cell migration in human keloid fibroblasts, rendering it a potential new therapeutic target for keloids.
Acknowledgements
The authors would like to thank Mr. Chongkai Li from Tongji University (Shanghai, China), the Center of Excellence in Tissue Engineering (Institute of Basic Medical Sciences) and the School of Basic Medicine (Chinese Academy of Medical Sciences) for technical support.
Glossary
Abbreviations
- CAS1
CACNA1G-AS1
- lncRNA
long non-coding RNA
- PCNA
proliferating cell nuclear antigen
- siRNA
small interfering RNA
- TGF-β
transforming growth factor-β
Funding
The present study was supported by Integrative Medicine Talents Program of Shanghai Municipal Commission of Health and Family Planning (grant no. ZY3-RCPY-4-2029).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
YL made substantial contributions to acquisition of data, and was involved in drafting the manuscript. XBL made substantial contributions to conception and design. PW and XJW made substatial contributions to interpretation of data and revised the manuscript critically for important intellectual content. XL made substantial contributions to analysis and interpretation of data. ZQM made substantial contributions to conception and design, gave final approval of the version to be published and agreed to be accountable for all aspects of the work. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the institutional review board at Peking Union Medical College (Beijing, China; approval no. ZS-1301), in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all patients for the use of their keloids and normal skin.
Consent for publication
Written informed consent was obtained from all patients for the publication of any associated data and accompanying images.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Robles DT, Berg D. Abnormal wound healing: Keloids. Clin Dermatol. 2007;25:26–32. doi: 10.1016/j.clindermatol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Sun LM, Wang KH, Lee YC. Keloid incidence in Asian people and its comorbidity with other fibrosis-related diseases: A nationwide population-based study. Arch Dermatol Res. 2014;306:803–808. doi: 10.1007/s00403-014-1491-5. [DOI] [PubMed] [Google Scholar]
- 3.Huang C, Murphy GF, Akaishi S, Ogawa R. Keloids and hypertrophic scars: Update and future directions. Plast Reconstr Surg Glob Open. 2013;1:e25. doi: 10.1097/GOX.0b013e31829c4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bock O, Schmid-Ott G, Malewski P, Mrowietz U. Quality of life of patients with keloid and hypertrophic scarring. Arch Dermatol Res. 2006;297:433–438. doi: 10.1007/s00403-006-0651-7. [DOI] [PubMed] [Google Scholar]
- 5.Ud-Din S, Volk SW, Bayat A. Regenerative healing, scar-free healing and scar formation across the species: Current concepts and future perspectives. Exp Dermatol. 2014;23:615–619. doi: 10.1111/exd.12457. [DOI] [PubMed] [Google Scholar]
- 6.Lee JY, Yang CC, Chao SC, Wong TW. Histopathological differential diagnosis of keloid and hypertrophic scar. Am J Dermatopathol. 2004;26:379–384. doi: 10.1097/00000372-200410000-00006. [DOI] [PubMed] [Google Scholar]
- 7.Huang C, Akaishi S, Hyakusoku H, Ogawa R. Are keloid and hypertrophic scar different forms of the same disorder? A fibroproliferative skin disorder hypothesis based on keloid findings. Int Wound J. 2014;11:517–522. doi: 10.1111/j.1742-481X.2012.01118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mofikoya BO, Adeyemo WL, Abdus-salam AA. Keloid and hypertrophic scars: A review of recent developments in pathogenesis and management. Nig Q J Hosp Med. 2007;17:134–139. doi: 10.4314/nqjhm.v17i4.12693. [DOI] [PubMed] [Google Scholar]
- 9.Ogawa R, Okai K, Tokumura F, Mori K, Ohmori Y, Huang C, Hyakusoku H, Akaishi S. The relationship between skin stretching/contraction and pathologic scarring: The important role of mechanical forces in keloid generation. Wound Repair Regen. 2012;20:149–157. doi: 10.1111/j.1524-475X.2012.00766.x. [DOI] [PubMed] [Google Scholar]
- 10.Naylor MC, Brissett AE. Current concepts in the etiology and treatment of keloids. Facial Plast Surg. 2012;28:504–512. doi: 10.1055/s-0032-1325644. [DOI] [PubMed] [Google Scholar]
- 11.Jin Z. Increased c-Met phosphorylation is related to keloid pathogenesis: Implications for the biological behaviour of keloid fibroblasts. Pathology. 2014;46:25–31. doi: 10.1097/PAT.0000000000000028. [DOI] [PubMed] [Google Scholar]
- 12.Ogawa R, Akaishi S, Huang C, Dohi T, Aoki M, Omori Y, Koike S, Kobe K, Akimoto M, Hyakusoku H. Clinical applications of basic research that shows reducing skin tension could prevent and treat abnormal scarring: The importance of fascial/subcutaneous tensile reduction sutures and flap surgery for keloid and hypertrophic scar reconstruction. J Nippon Med Sch. 2011;78:68–76. doi: 10.1272/jnms.78.68. [DOI] [PubMed] [Google Scholar]
- 13.Akaishi S, Akimoto M, Ogawa R, Hyakusoku H. The relationship between keloid growth pattern and stretching tension: Visual analysis using the finite element method. Ann Plast Surg. 2008;60:445–451. doi: 10.1097/SAP.0b013e3181238dd7. [DOI] [PubMed] [Google Scholar]
- 14.Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, Hosono N, Kubo M, Nakamura Y, Zembutsu H. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42:768–771. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- 15.Arima J, Huang C, Rosner B, Akaishi S, Ogawa R. Hypertension: A systemic key to understanding local keloid severity. Wound Repair Regen. 2015;23:213–221. doi: 10.1111/wrr.12277. [DOI] [PubMed] [Google Scholar]
- 16.Park TH, Chang CH. Keloid recurrence in pregnancy. Aesthetic Plast Surg. 2012;36:1271–1272. doi: 10.1007/s00266-012-9947-5. [DOI] [PubMed] [Google Scholar]
- 17.Fong EP, Bay BH. Keloids-the sebum hypothesis revisited. Med Hypotheses. 2002;58:264–269. doi: 10.1054/mehy.2001.1426. [DOI] [PubMed] [Google Scholar]
- 18.Ichioka S, Ando T, Shibata M, Sekiya N, Nakatsuka T. Oxygen consumption of keloids and hypertrophic scars. Ann Plast Surg. 2008;60:194–197. doi: 10.1097/SAP.0b013e318053ec1d. [DOI] [PubMed] [Google Scholar]
- 19.Liang X, Ma L, Long X, Wang X. LncRNA expression profiles and validation in keloid and normal skin tissue. Int J Oncol. 2015;47:1829–1838. doi: 10.3892/ijo.2015.3177. [DOI] [PubMed] [Google Scholar]
- 20.Russell SB, Russell JD, Trupin KM, Gayden AE, Opalenik SR, Nanney LB, Broquist AH, Raju L, WIlliams SM. Epigenetically altered wound healing in keloid fibroblasts. J Invest Dermatol. 2010;130:2489. doi: 10.1038/jid.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vance KW, Ponting CP. Transcriptional regulatory functions of nuclear long noncoding RNAs. Trends Genet. 2014;30:348–355. doi: 10.1016/j.tig.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: Past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khaitan D, Dinger ME, Mazar J, Crawford J, Smith MA, Mattick JS, Perera RJ. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res. 2011;71:3852–3862. doi: 10.1158/0008-5472.CAN-10-4460. [DOI] [PubMed] [Google Scholar]
- 27.Tang L, Zhang W, Su B, Yu B. Long noncoding RNA HOTAIR is associated with motility, invasion, and metastatic potential of metastatic melanoma. Biomed Res Int. 2013;2013:251098. doi: 10.1155/2013/251098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Wu YT, Huang C, Meng XM, Ma TT, Wu BM, Xu FY, Zhang L, Lv XW, Li J. Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis. Biochim Biophys Acta. 2014;1842:2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Zhuang J, Lu Q, Shen B, Huang X, Shen L, Zheng X, Huang R, Yan J, Guo H. TGFβ1 secreted by cancer-associated fibroblasts induces epithelial-mesenchymal transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep. 2015;5:11924. doi: 10.1038/srep11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukunaga K. Cognitive function and calcium. Cognitive improvement through T type calcium channel stimulation. Clin Calcium. 2015;25:247–254. (In Japanese) [PubMed] [Google Scholar]
- 31.Yuan SX, Tao QF, Wang J, Yang F, Liu L, Wang LL, Zhang J, Yang Y, Liu H, Wang F, et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349:87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 32.D'Andrea F, Brongo S, Ferraro G, Baroni A. Prevention and treatment of keloids with intralesional verapamil. Dermatology. 2002;204:60–62. doi: 10.1159/000051812. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Mao Y, Zhang Z, Li Z, Chen J, Cen Y. Role of verapamil in preventing and treating hypertrophic scars and keloids. Int Wound J. 2016;13:461–468. doi: 10.1111/iwj.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexandrescu D, Fabi S, Yeh LC, Fitzpatrick RE, Goldman MP. Comparative results in treatment of keloids with intralesional 5-FU/Kenalog, 5-FU/Verapamil, Enalapril Alone, verapamil alone, and laser: A case report and review of the literature. J Drugs Dermatol. 2016;15:1442–1447. [PubMed] [Google Scholar]
- 35.Grossman E, Messerli FH. Calcium antagonists. Prog Cardiovasc Dis. 2004;47:34–57. doi: 10.1016/j.pcad.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 36.Lee RC, Doong H, Jellema AF. The response of burn scars to intralesional verapamil. Report of five cases. Arch Surg. 1994;129:107–111. doi: 10.1001/archsurg.1994.01420250119015. [DOI] [PubMed] [Google Scholar]
- 37.Doong H, Dissanayake S, Gowrishankar TR, LaBarbera MC, Lee RC. The 1996 Lindberg Award. Calcium antagonists alter cell shape and induce procollagenase synthesis in keloid and normal human dermal fibroblasts. J Burn Care Rehabil. 1996;17:497–514. doi: 10.1097/00004630-199611000-00005. [DOI] [PubMed] [Google Scholar]
- 38.Lee RC, Ping JA. Calcium antagonists retard extracellular matrix production in connective tissue equivalent. J Surg Res. 1990;49:463–466. doi: 10.1016/0022-4804(90)90197-A. [DOI] [PubMed] [Google Scholar]
- 39.Giugliano G, Pasquali D, Notaro A, Brongo S, Nicoletti G, D'Andrea F, Bellastella A, Sinisi AA. Verapamil inhibits interleukin-6 and vascular endothelial growth factor production in primary cultures of keloid fibroblasts. Br J Plast Surg. 2003;56:804–809. doi: 10.1016/S0007-1226(03)00384-9. [DOI] [PubMed] [Google Scholar]
- 40.Xu SJ, Teng JY, Xie J, Shen MQ, Chen DM. Comparison of the mechanisms of intralesional steroid, interferon or verapamil injection in the treatment of proliferative scars. Zhonghua Zheng Xing Wai Ke Za Zhi. 2009;25:37–40. (In Chinese) [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.