Abstract
Rationale: Contacts of patients with tuberculosis (TB) constitute an important target population for preventive measures because they are at high risk of infection with Mycobacterium tuberculosis and progression to disease.
Objectives: We investigated biosignatures with predictive ability for incident TB.
Methods: In a case–control study nested within the Grand Challenges 6-74 longitudinal HIV-negative African cohort of exposed household contacts, we employed RNA sequencing, PCR, and the pair ratio algorithm in a training/test set approach. Overall, 79 progressors who developed TB between 3 and 24 months after diagnosis of index case and 328 matched nonprogressors who remained healthy during 24 months of follow-up were investigated.
Measurements and Main Results: A four-transcript signature derived from samples in a South African and Gambian training set predicted progression up to two years before onset of disease in blinded test set samples from South Africa, the Gambia, and Ethiopia with little population-associated variability, and it was also validated in an external cohort of South African adolescents with latent M. tuberculosis infection. By contrast, published diagnostic or prognostic TB signatures were predicted in samples from some but not all three countries, indicating site-specific variability. Post hoc meta-analysis identified a single gene pair, C1QC/TRAV27 (complement C1q C-chain / T-cell receptor-α variable gene 27) that would consistently predict TB progression in household contacts from multiple African sites but not in infected adolescents without known recent exposure events.
Conclusions: Collectively, we developed a simple whole blood–based PCR test to predict TB in recently exposed household contacts from diverse African populations. This test has potential for implementation in national TB contact investigation programs.
Keywords: tuberculosis, gene expression, biomarkers
At a Glance Commentary
Scientific Knowledge on the Subject
Reliable predictors of incident tuberculosis that can guide preventive treatment are urgently needed.
What This Study Adds to the Field
This study describes a small gene expression signature that predicts incident tuberculosis in household contacts across different African sites.
Tuberculosis (TB), caused by infection with Mycobacterium tuberculosis (Mtb) (1, 2), is the leading cause of death caused by a single pathogen globally (3). Before development of symptomatic disease, latent Mtb infection can be detected by measuring immunological sensitization using the tuberculin skin test (TST) and/or IFN-γ release assays (IGRAs) (4). Most infected individuals have effective defense mechanisms to control Mtb (5); only 5 to 10% will progress to TB during their lifetime. Despite this, each year over 10 million new cases of TB caused by either Mtb reactivation or direct transmission are diagnosed, and almost 2 million people die of the disease (3). Although recent Mtb exposure and TST- or IGRA-confirmed conversion are associated with higher risk of TB progression (6), the positive predictive values of these tests are low (1.5% and 2.7%, respectively) (7), falling short of current World Health Organization–supported guidelines for incipient TB (8). Thus, the number of TST+ or IGRA+ individuals requiring treatment to prevent progression to a single incident case of TB is prohibitively high (9).
Factors associated with elevated risk of progression to TB include age, sex, HIV (10, 11), and especially being in recent contact with a patient with active pulmonary TB (12, 13). A biomarker that identifies household contacts (HHCs) who will progress to TB would provide an opportunity to arrest disease progression through targeted prophylactic intervention (14, 15). Such prognostic biomarkers would have the most impact in point-of-care tests for resource-limited settings, such as those in Sub-Saharan Africa. Test performance should not be adversely affected by diversity of ethnic background (16) and circulating Mtb lineage (17), as seen in Africa. A TB risk test must be practical for field application and therefore based on accessible biological samples routinely used in clinical settings, such as peripheral blood (18).
Transcriptional profiling of blood cells has emerged as a powerful platform for discovery of potential TB biomarkers discriminating patients with TB from healthy uninfected and/or individuals with latent Mtb infection (19–24). We previously defined a 16-gene blood transcriptional correlate of risk (COR) signature that predicts risk of progression to TB in HIV-negative South African adolescents with Mtb infection and HHCs from South Africa and the Gambia (25). However, given that this COR signature was developed using a single cohort of South African adolescents with latent Mtb infection, the predictive accuracy for HHCs in diverse African populations may be suboptimal (25). It would also be desirable to reduce the number of transcripts in the signature to facilitate implementation of a point-of-care test. These tests pave the way for simple identification of individuals at highest risk for progression. Some of the results of this study were previously reported in the form of abstracts (26, 27).
Methods
Study Design and Participants
All clinical sites adhered to the Declaration of Helsinki principles and good clinical practice guidelines. Ethical approval was obtained from institutional review boards (see Table E1 in the online supplement). The HHC study included participants from four African sites (South Africa, the Gambia, Ethiopia, and Uganda) as part of the Bill and Melinda Gates Foundation Grand Challenges 6-74 (GC6-74) program (Figure 1 and Table E2). Samples were collected at enrollment/baseline and at 6 and 18 months, with the exception of South Africa, where samples at 6 months were not available. The ACS (Adolescent Cohort Study) was described previously (25, 28) and included IGRA+ and/or TST+ South African adolescents aged 12–18 years with Mtb infection occurring at unspecified times. Adult participants, or legal guardians of participants aged 10–17 years old, provided written or thumbprinted informed consent to participate after careful explanation of the study and potential risks.
Figure 1.
Consolidated Standards of Reporting Trials diagram describing the inclusion and exclusion of participants from the different African cohorts in the Grand Challenges 6-74 (GC6-74, GC6) household contact (HHC) study for training predictive transcriptomic biomarker for tuberculosis (TB) progression: Stellenbosch University in South Africa (SUN), Armauer Hansen Research Institute in Ethiopia (AHRI), Makerere University in Uganda (MAK), Medical Research Council in the Gambia (MRC), and the external validation natural history study of South African adolescents (Adolescent Cohort Study [ACS]). “QC Excluded” refers to samples excluded because they did not meet the minimum quality control requirement for RNA sequencing of an RNA yield greater than or equal to 200 ng and an RNA integrity number greater than or equal to 7. RISK4 = four-gene correlate-of-risk signature.
Sample Processing and RNA Sequencing
PAXgene (PreAnalytiX) blood RNA samples were collected from all participants. Progressors were defined as individuals who developed TB 3–24 months after household contact. Nonprogressor samples were matched to the prediagnosis time points of each progressor by site, sex, age, and recruitment year (see the online supplement). RNA sequencing (RNA-seq) was performed by the Beijing Genomics Institute (Shenzhen, China); additional details regarding processing and quality control are provided in the online supplement. FASTQ files have been deposited in the Gene Expression Omnibus database under accession number GSE94438.
Identification of Predictive Signatures
The hypothesis of the study was that gene expression signatures exist that can predict incident TB in recent HHCs of patients with active TB. Candidate site-specific signatures of risk for TB disease progression and final, simplified qRT-PCR–based candidate signatures were developed using the pair ratios algorithm (see the online supplement), which was previously described (29) and is a variation of the pairwise approach used to discover the ACS COR signature (25). To summarize, the step-by-step procedure for computing the four-gene COR signature (RISK4) scores using sample qRT-PCR measurements was as follows:
Measure the cycle thresholds for the four primer-probe sets (TaqMan assays; Applied Biosystems) listed in Table E3.
For each of the four pairs of primer-probe sets, compute the difference in raw cycle thresholds, which produces the log-transformed ratio of expression.
Compare the measured ratio to ratios in the lookup table for the given pair of transcripts listed in Tables E4–E7. Find the minimal ratio in column 1 of the table that is greater than or equal to the measured ratio.
Assign the corresponding score in the second column of the lookup table to the ratio. If the measured ratio is larger than all ratios in column 1 of the lookup table, then assign a score of 1 to the ratio.
Compute the average of the scores generated from the set of pairs. If any assay failed on the sample, compute the average score of all ratios, not including the failed assays. The resulting average is the final score for that sample.
Adaptation of Published Diagnostic Signatures to qRT-PCR
The previously published signatures reported by Maertzdorf and colleagues (30) and Sweeney and colleagues (31) were adapted to the qRT-PCR platform, where we refer to them as DIAG4 and DIAG3, respectively. Primer-probe sets were selected for each gene in the respective signatures, and overall scores were computed for each sample as the difference in the mean of the upregulated and downregulated transcripts (Tables E8 and E9). All statistical analyses were performed in R (version 3.1.0) using the R package pROC (32). Details are provided in the online supplement.
Results
We enrolled 4,466 HIV-negative healthy HHCs of 1,098 index TB cases between 2006 and 2010 into the GC6-74 cohorts across four African sites (Figure 1 and Table E2). Samples from Uganda were not available in sufficient quantities for this analysis (Figure 1). TB incidence in HIV-negative healthy HHCs was highest in South Africa and lowest in Ethiopia (Table 1), as defined by TB case classifications A–K in Table E10. Incident cases (progressors) were defined as those who developed TB between 3 and 24 months after index case diagnosis. “Co-incident” cases (i.e., diagnosed with TB within 3 mo of contact with the index case; see Methods above) were not included in the analysis. Prior TB was an exclusion criterion (see the online supplement); thus, progressors likely had their first TB episode during follow-up. The median age of progressors was comparable across the four African sites (Kruskal-Wallis P = 0.92) (Table 1). Median times to progression were 7 months in both South Africa and Uganda, 10.5 months in the Gambia, and 10 months in Ethiopia (Tables 1 and E11A). Progressors, as defined by clinical symptoms, chest and other radiographs consistent with TB, and response to chemotherapy, without microbiological confirmation, comprised 25% (4 of 12) of progressors in Ethiopia, 2% (1 of 43) in South Africa, and 6% (3 of 34) in the Gambia (TB classification K in Tables E10 and E11A).
Table 1.
Baseline Demographic Characteristics of Progressors and Matched Nonprogressor Control Subjects in the Four African Household Contact Cohorts
| South Africa | The Gambia | Ethiopia | Uganda | |
|---|---|---|---|---|
| HIV-negative HHC, n | 1,197 | 1,948 | 818 | 499 |
| Progressors, n | 43 | 34 | 12 | 11 |
| Incidence, % | 3.6 | 1.7 | 1.5 | 2.2 |
| Age, yr, median (IQR) | ||||
| Progressors | 25 (18–41) | 22.5 (20–30.75) | 23 (19.75–27) | 23 (18–36) |
| Nonprogressors | 24 (18–38) | 24 (18–30.25) | 25 (20–35) | 27 (19–38.75) |
| Male sex, % | ||||
| Progressors | 41.9 | 44.1 | 33.3 | 54.5 |
| Nonprogressors | 40.7 | 44.1 | 35.4 | 54.5 |
| Time to TB, mo, median (IQR) | ||||
| Progressors | 7 (5–17) | 10.5 (7–18.75) | 10 (6.5–15) | 7 (5–11) |
Definition of abbreviations: HHC = household contacts; IQR = interquartile range; TB = tuberculosis.
RISK4, a Four-Gene COR Signature Predicts TB Progression in HHCs
We divided South African and Gambian HHC cohorts into training and test sets, whereas the entire Ethiopian cohort was assigned to the test set owing to its small sample size (Figure 1 and Tables E11A and E11B). We used the South African and Gambian training sets to construct site-specific signatures of TB risk using RNA-seq transcriptomes and the pair ratio approach, which uses ratios of transcripts regulated in opposite directions during TB progression (see online supplement text and Tables E12 and E13). Leave-one-out cross-validation analysis (applied to all samples from specific individuals) indicated strong potential for predicting TB progression in both cohorts (South Africa [Figure 2A], area under the receiver operating characteristic curve [AUC], 0.86 [95% confidence interval {CI}, 0.79–0.94], P = 8.4 × 10−10; the Gambia [Figure 2B], AUC, 0.77 [95% CI, 0.66–0.88], P = 2.5 × 10−10). By applying the algorithm to the South African and Gambian cohorts, we generated two distinct, site-specific risk signatures (Figures 2C and 2D). When measured by qRT-PCR using primer-probe sets that corresponded to the exons, predictive accuracy was maintained (Figure E1). The Gambia signature failed to validate in samples from South Africa (Figure 2A) (AUC, 0.59 [95% CI, 0.46–0.73]; P = 0.061), whereas the South Africa signature weakly validated in Gambian samples (Figure 2B) (AUC, 0.66 [95% CI, 0.54–0.76]; P = 8.8 × 10−3), suggesting site-specific progression signatures in South Africa and the Gambia.
Figure 2.
Site-specific feature selection and translation to RT-PCR. (A) Receiver operating characteristic curve for leave-one-out cross-validation (LOOCV) of South Africa (blue; area under the receiver operating characteristic curve [AUC], 0.86 [95% confidence interval (CI), 0.79–0.94]; P = 8.4 × 10−10) versus the Gambia–trained prospective signature (red; AUC, 0.59 [95% CI, 0.46–0.73]; P = 0.06) in the South African training set (samples listed in Tables E11A and E11B). (B) Receiver operating characteristic curves for LOOCV of the Gambia (blue; AUC, 0.77 [95% CI, 0.66–0.88]; P = 2.5 × 10−5) versus South Africa prospective signature (red; AUC, 0.66 [95% CI, 0.54–0.77]; P = 8.8 × 10−3) in the Gambia training set containing 26 progressor and 76 nonprogressor samples. (C and D) Heat maps showing the expression of each splice junction in (C) the South Africa and (D) the Gambia signatures in nonprogressors (left columns), progressors 1–2 years before diagnosis (middle columns), and progressors 0–1 year before diagnosis (right columns). For each group of samples, the central column is the mean fold expression change versus nonprogressors, whereas left/right columns in each group correspond to mean ± SEM. Each row corresponds to a splice junction, and genes with multiple rows are represented by multiple splice junctions in the signature.
The poor cross-prediction of South African and Gambian signatures motivated explicit development of a multicohort signature using a training set that combined samples from both sites. We pooled PCR-based transcript pairs that comprised all of the South Africa (38 transcripts), and the Gambia (35 transcripts) signatures (Figures 2C and 2D and Tables E12 and E13) to identify top transcript pairs that were significantly predictive of TB progression in both cohorts, and we successively added the next best pair to the ensemble and reassessed the predictive power at each stage until the gain in predictive performance reached a plateau (see online supplement text and Table E14). This resulted in the RISK4 signature comprising four unique genes: GAS6 (growth arrest–specific 6) and SEPT4 (septin 4), which were upregulated, and CD1C (cluster of differentiation 1C) and BLK (B lymphocyte kinase), which were downregulated, in progressors versus matched control subjects (Figure 3A).
Figure 3.
Validation of a multicohort four-gene signature (RISK4) derived from the South African and Gambian training sets. (A) Expression ratio of gene pairs in the RISK4 signature in the South African (top) and Gambian (bottom) training sets: nonprogressors (left columns), progressors 1–2 years before diagnosis (middle columns), and progressors 0–1 (right columns) year before diagnosis. In each group, the central column is the mean fold expression over nonprogressors, whereas left/right columns in each group correspond to mean ± SEM. (B) Receiver operating characteristic (ROC) curves for blind predictions of RISK4 on test set samples of all sites (black; area under the receiver operating characteristic curve [AUC], 0.67 [95% confidence interval (CI), 0.57–0.77]; P = 2.6 × 10−4), South Africa (red; AUC, 0.72 [95% CI, 0.53–0.92]; P = 6.3 × 10−3), the Gambia (blue; AUC, 0.72 [95% CI, 0.55–0.88]; P = 5.4 × 10−3), and Ethiopia (green; AUC, 0.67 [95% CI, 0.5–0.83]; P = 0.02). (C) Performance of RISK4 signature in test set samples obtained within 1 year of diagnosis (red; AUC, 0.66 [95% CI, 0.55–0.78], P = 2 × 10−3; 30 progressor samples, 201 nonprogressor samples) or 1–2 years before diagnosis (blue; AUC, 0.69 [95% CI, 0.51–0.86]; P = 0.02; 12 progressor samples, 201 nonprogressor samples). (D) ROC curve of RISK4 on all baseline test set samples (AUC, 0.69 [95% CI, 0.52–0.86]; P = 5 × 10−3). (E) ROC curve blind prediction of RISK4 in South African adolescents with latent Mycobacterium tuberculosis (M.tb) infection (AUC, 0.69 [95% CI, 0.62–0.76]; P = 3.4 × 10−7). BLK = B lymphocyte kinase; CD1C = cluster of differentiation 1C; GAS6 = growth arrest–specific 6; SEPT4 = septin 4; TB = tuberculosis.
Having developed a multisite PCR-based signature of risk, we validated it by blind prediction of TB progression on the multicohort test sets from South Africa, the Gambia, and Ethiopia (Figure 1). The RISK4 signature significantly predicted progression in the entire combined test set (AUC, 0.67 [95% CI, 0.57–0.77]; P = 2.6 × 10−4) (Figure 3B) and on each individual site (South Africa, the Gambia, and Ethiopia with AUCs of 0.66–0.72; P < 0.03) (Figure 3B). Surprisingly, performance of the signature on combined test set samples within one year of TB diagnosis (AUC, 0.66 [95% CI, 0.55–0.78]; P = 1.9 × 10−3) (Figure 3C) was comparable to samples collected more than one year before diagnosis (AUC, 0.69 [95% CI, 0.51–0.86]; P = 0.015). Because deploying a risk signature in a screen-and-treat strategy in TB HHCs would most likely entail testing early after exposure, we assessed the predictive performance of RISK4 on samples from HHCs collected within two months of diagnosis of the index case. RISK4 also was validated in this setting (Figure 3D) (AUC, 0.69 [95% CI, 0.52–0.86]; P = 4.8 × 10−3). Finally, to test the robustness of RISK4, we performed blinded predictions on samples from an external cohort of IGRA+/TST+ South African adolescents (the ACS cohort), in whom the time of TB exposure was unknown (25). RISK4 also significantly predicted risk of TB progression in this cohort (Figure 3E) (AUC, 0.69 [95% CI, 0.62–0.76]; P = 3.4 × 10−7).
Comparison of RISK4 with Published Diagnostic TB Signatures
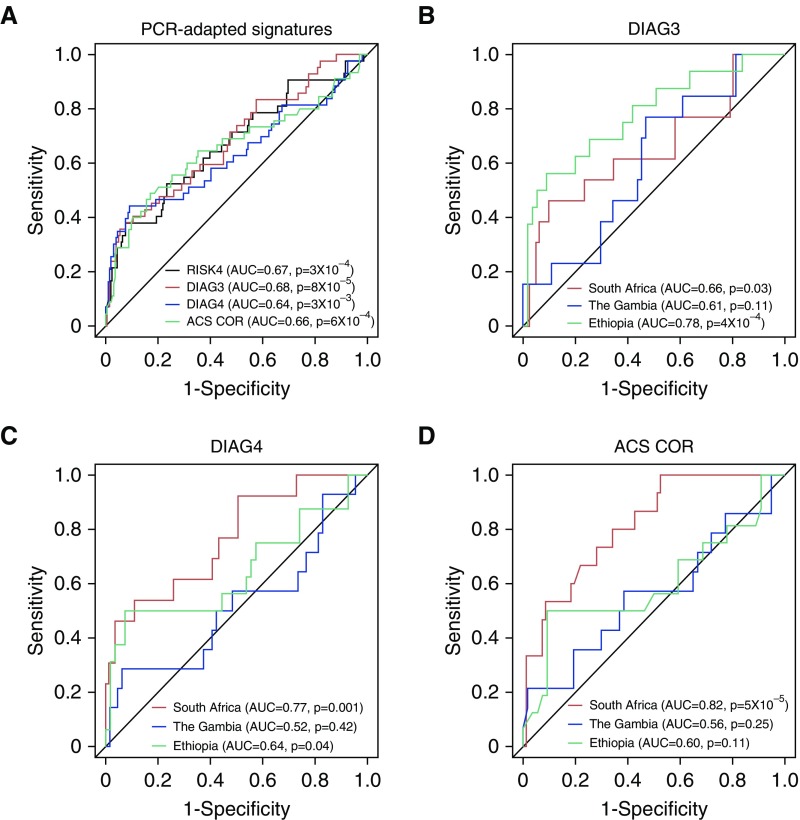
To benchmark the predictive performance of the RISK4 signature, we compared it with qRT-PCR–based versions of three published transcriptional signatures for TB diagnosis: “DIAG3,” the three-gene diagnostic signature reported by Sweeney and colleagues (31); “DIAG4,” the four-gene diagnostic signature reported by Maertzdorf and colleagues (30); and our own previously reported 16-gene COR signature for TB progression (“ACS COR”; Zak and colleagues [25]). Because HHC training set samples were used to discover the RISK4 signature, we compared the performance of PCR-adapted published signatures with that of RISK4 in the HHC test set only. The three signatures predicted TB progression in the combined test set with accuracy comparable to that of RISK4 (Figure 4A) (AUCs, 0.64–0.68; P < 3 × 10−3). When we compared the predictive accuracy of RISK4 with each of the three PCR-adapted signatures, the AUCs were not statistically different (Table E15). However, unlike RISK4 (Figure 3B), the three other signatures did not validate on all sites when evaluated individually (Figures 4B–4D), suggesting that RISK4 represents a more generalizable prognostic signature.
Figure 4.
Comparison of PCR-adapted signatures: RISK4, ACS 16-gene correlate of risk (ACS COR), and tuberculosis diagnostic signatures (DIAG3 and DIAG4). (A) Receiver operating characteristic curves for blind predictions of RISK4 (black; area under the receiver operating characteristic curve [AUC], 0.67 [95% confidence interval (CI), 0.57–0.77]; P = 3 × 10−4), DIAG3 (red; AUC, 0.68 [95% CI, 0.59–0.78]; P = 8 × 10−5), DIAG4 (blue; AUC, 0.64 [95% CI, 0.53–0.74]; P = 3 × 10−3), and ACS COR (green; AUC, 0.66 [95% CI, 0.55–0.76]; P = 6 × 10−4) in all test set samples. (B–D) Blind prediction of PCR-adapted signatures: (B) DIAG3 (South Africa AUC, 0.66 [95% CI, 0.47–0.84]; the Gambia AUC, 0.61 [95% CI, 0.45–0.77]; and Ethiopia AUC, 0.78 [95% CI, 0.64–0.92]), (C) DIAG4 (South Africa AUC, 0.77 [95% CI, 0.62–0.91]; the Gambia AUC, 0.52 [95% CI, 0.33–0.71]; and Ethiopia AUC, 0.64 [95% CI, 0.46–0.83]), and (D) ACS COR (South Africa AUC, 0.82 [95% CI, 0.71–0.92]; the Gambia AUC, 0.56 [95% CI, 0.37–0.75]; and Ethiopia AUC, 0.6 [95% CI, 0.41–0.79]). South Africa, the Gambia, and Ethiopia AUCs are depicted in red, blue, and green, respectively. ACS = Adolescent Study Cohort; RISK4 = four-gene correlate-of-risk signature.
After unblinding the South African, Gambian, and Ethiopian test sets, we interrogated whether the RISK4 signature could be reduced to a single pair of transcripts without a loss of predictive accuracy. We applied each of the four ratios in the RISK4 signature to each of the test set cohorts individually, and we compared the performance with that of the entire RISK4 signature (Table E16). The ratio between the SEPT4 and BLK primers reproduced the performance of the RISK4 signature on all three test set cohorts, demonstrating feasibility of a highly simplified two-gene host RNA-based signature for identifying HHCs at greatest risk of progressing to active TB.
Meta-Analysis Identifies Gene Pairs That Predict TB Progression across Africa
Overall, predictions for TB progression were the least accurate for the Ethiopian cohort, which was not used to develop the initial RISK4 signature (Figures 1, 3, and 4). To determine whether accuracy could be further improved for a signature performing well at all sites, we performed a meta-analysis of RNA-seq profiles for the combined training and test datasets from all of our three cohorts. This post hoc analysis was performed after unblinding of the test set and was focused on identifying better predictive gene pairs, given that the single transcript pair SEPT4/BLK performed equivalently to the RISK4 signature (Table E16).
We combined RNA-seq data from all training and test cohorts, thus merging the three independent cohorts from South Africa, the Gambia, and Ethiopia. Pairs of upregulated and downregulated transcripts were formed from all transcripts that individually discriminated progressors from control subjects in at least one cohort (Tables E17 and E18; Wilcoxon false discovery rate <0.05 in at least one of the three cohorts). Each pair was then analyzed on each of the three sites. We identified nine transcript pairs that discriminated progressors from control subjects with AUC greater than 0.75 on all three sites (Table E19). The optimal pair consisted of C1QC (complement C1q C-chain) (upregulated) and TRAV27 (T-cell receptor-α variable gene 27) (downregulated) and achieved AUC greater than 0.76 on all three sites. We performed logistic regression analysis to determine whether the remaining eight pairs (Table E20 and Supplementary Methods in the online supplement) captured information about TB progression that was redundant or complementary to the signals detected by C1QC/TRAV27. The ratio between ANKRD22 (ankyrin repeat domain 22; upregulated with TB progression) and OSBPL10 (oxysterol binding protein like 10; downregulated with progression) led to significantly increased discrimination between progressors and control subjects when it was combined with the C1QC/TRAV27 ratio in HHC cohorts (Figures 5A–5C), increasing the AUC on all three HHC cohorts individually to greater than 0.79 (Table E21). Thus, the ratios C1QC/TRAV27 and ANRKD22/OSBPL10 capture distinct aspects of TB progression signals in HHC that are shared across three distinct African sites.
Figure 5.
Gene pairs to predict tuberculosis progression in African cohorts. Ratios of C1QC/TRAV27 and ANKRD22/OBSPL10 plotted on samples from (A) South Africa, (B) the Gambia, and (C) Ethiopia, together with an optimal discriminant (dashed line; optimizes sum of sensitivity and specificity) separating progressors (orange) from nonprogressors (blue). In each cohort, the two pairs provide complementary information. P values correspond to chi-square complementation analysis in Table E16. (D) Receiver operating characteristic curves showing the ability of the Grand Challenges 6-74 (GC6)-trained C1QC/TRAV27 (solid line; area under the receiver operating characteristic curve [AUC], 0.57 [95% confidence interval (CI), 0.49–0.64]; P = 0.042), ANKRD22/OBSPL10 (dashed line; AUC, 0.75 [95% CI, 0.68–0.81]; P = 2.86 × 10−11), and a linear combination of C1QC/TRAV27 and ANKRD22/OBSPL10 (dotted line; AUC, 0.69 [95% CI, 0.61–0.76]; P = 4.3 × 10−7) models to predict tuberculosis disease progression in the South African Adolescent Cohort Study (ACS) cohort. (E) Receiver operating characteristic curves showing the ability of the GC6-trained C1QC/TRAV27 (solid line; AUC, 0.77 [95% CI, 0.71–0.83]; P = 4.5 × 10−16), ANKRD22/OBSPL10 (dashed line; AUC, 0.76 [95% CI, 0.7–0.82]; P = 3.7 × 10−15), and a linear combination of C1QC/TRAV27 and ANKRD22/OBSPL10 (dotted line; AUC, 0.79 [95% CI, 0.74–0.85]; P = 6.1 × 10−19) models to predict TB disease progression in the GC6 household contact (HHC) cohort. (F and G) Logarithmic ratios of expression (mean ± 95% CI) for (F) ANKRD22/OBSPL10 and (G) C1QC/TRAV27 are plotted as a function of time to diagnosis for both GC6-74 (blue) and ACS (red) progressor samples. Comparison of C1QC/TRAV27 expression at 19–24 months before diagnosis between the GC6-74 HHC and ACS cohorts was statistically significantly different (P = 3 × 10−3) using the Mann-Whitney U test. ANKRD22 = ankyrin repeat domain 22; C1QC = complement C1q C-chain; OBSPL10 = oxysterol binding protein like 10; TB = tuberculosis; TRAV27 = T-cell receptor-α variable gene 27.
To determine whether the C1QC/TRAV27 and ANKRD22/OSBPL10 signatures captured universal aspects of TB progression rather than HHC-associated biology, we evaluated them using data from the cohort of IGRA+ TST+ South African adolescents (25). The ANKRD22/OBSPL10 ratio strongly predicted TB progression among the adolescents with Mtb infection (Figure 5D) (AUC, 0.75 [95% CI, 0.68–0.81]; P = 2.86 × 10−11), but the C1QC/TRAV27 ratio was poorly predictive in the adolescent cohort (Figure 5D) (AUC, 0.57 [95% CI, 0.49–0.64]; P = 0.042). In contrast to the HHCs, combining the two ratios did not lead to improved discrimination of progressors and control subjects in the adolescent cohort (AUC, 0.69 [95% CI, 0.61–0.76]) (Figures 5D and E2A). To further understand the disparity in the predictive performance for the HHC cohorts and the adolescents with Mtb infection, we evaluated the longitudinal behavior of the transcript ratios for progressor samples in the HHC and adolescent cohorts (Figures 5F and 5G). The ANKRD22/OSBPL10 pair exhibited similar behavior in the HHC and ACS cohorts, with a steady upregulation during progression and no significant difference between GC6-74 and adolescent participants in any 6-month time window preceding TB diagnosis (Figure 5F). In contrast, the C1QC/TRAV27 ratio was significantly higher in HHC progressors than in adolescents with Mtb infection 19–24 months before TB diagnosis (P = 3 × 10−3) (Figure 5G). Importantly, samples from HHC progressors were collected mostly at enrollment, immediately after exposure to the respective TB index cases, thus possibly representing a signature of recent Mtb exposure.
Discussion
We identified and validated a simple PCR-based transcriptomic signature, “RISK4,” to predict the risk of progression to active TB disease in diverse African cohorts of recently exposed HHCs of index TB cases. This four-gene signature predicted risk of progression with similar accuracy in four cohorts from three sub-Saharan African populations with heterogeneous genetic backgrounds, TB epidemiology, and circulating Mtb strains (33). Importantly, RISK4 exhibited consistent predictive performance in all test set cohorts, whereas previously reported signatures (25, 30, 31) exhibited cohort-specific variability in performance. We previously reported that the ACS COR signature was validated in the combined South African and Gambian HHC cohorts (25). In the present analysis, the stochastic partitioning of HHC samples into training and test sets resulted in ACS COR performance measurements different from the previously reported results.
The signatures we report represent significant and translational improvements over currently used biomarkers for predicting risk of TB, such as IGRAs or the TST (14, 15). Recent estimates suggest the TB incidence rates in South Africa and the Gambia to be 0.8% (3) and 0.3% (34), respectively. However, IGRA+ and TST+ prevalence can reach up to 50% in the Gambia and 80% in South Africa (3), and although IGRAs and the TST have a high (approximately 80%) sensitivity for Mtb infection, they have poor positive predictive values of 2.7% and 1.5%, respectively, for TB progression. Therefore, dozens of individuals would require prophylactic treatment to prevent progression to TB in a single individual (35, 36). The World Health Organization recently published guidelines for an incipient TB target product profile to predict TB progression (8) in order to ensure that individuals at high risk of TB progression are not falsely excluded (7, 18) but instead are referred for additional investigation for TB or offered prophylactic treatment (37). At sensitivities of 81%, 71%, 62%, and 50%, the RISK4 signature achieved specificities of 34%, 52%, 63%, and 77%, respectively, in healthy asymptomatic individuals by selection of different thresholds (Table E22). Although RISK4 has a poor positive predictive value of 3%, similar to IGRA or TST, owing to its lower sensitivity at higher specificity thresholds (Table E23), it importantly has lower positivity rates in the target population. To achieve test performance similar to IGRAs (between 70 and 80% sensitivity and the number to harm to prevent one case of approximately 85), the RISK4 threshold would identify between 38 and 54% of HHCs for preventive measures, compared with 78% for IGRA (Table E22).
There are several limitations of this study, including the small sample size (only 100 of the 4,460 HHCs progressed to active TB). Furthermore, although the intended application is for a trans-Africa test, we could sample from only three regions. The poor RISK4 performance in Ethiopia calls for larger multicenter studies, particularly of underrepresented populations throughout Africa and other TB endemic areas. Although we defined two-transcript signatures that have broader application, the sample size was not adequate to have unblinded validation sets or to ensure that transcript pairs were not selected by chance during our post hoc analyses. The test is based on blood samples, which are easily and routinely obtained in laboratory diagnostics. However, the test still requires translation into field-friendly instrumentation to bypass the multistep processing involved in RNA extraction and RT-PCR. Encouragingly, recent advances in point-of-care PCR technologies offer promise for developing rapid diagnostics. We envision platforms where blood from a finger prick can be translated using field-friendly handheld PCR instruments into interpretable scores. Field staff would then triage near-patient contacts into low-risk and higher-risk groups for further assessment and potential treatment for subclinical or active TB disease (8). One advantage of the calculation of RISK4 or the two-transcript scores is the pairs ratio structure, which eliminates the need for housekeepers or other standardization methods. As a proof of principle, a clinical trial (registered with www.clinicaltrials.gov [NCT02735590]) stratifying participants by ACS COR positivity (25) will provide real-world data on the efficacy of a strategy that screens South African adults with the COR signature and provides preventive therapy to those who are COR positive. Evaluation of the costs and benefits of such strategies should be carefully evaluated in future implementation studies of RISK4 (36). One benefit might be a strong motivation for both healthcare personnel and patients alike to initiate and complete preventive treatment in the face of a positive COR test. The potential need for repeat test performance also needs to be evaluated.
We identified several transcript pairs that recapitulated the predictive performance of the RISK4 signature and reflected complementary signals in predicting risk of TB progression. RISK4 comprises GAS6 and SEPT4 (upregulated) and BLK and CD1C (downregulated). Interestingly, CD1C and GAS6 (activating ligand of AXL) are expressed in two distinct dendritic cell (DC) subsets where GAS6 expression defines a newly characterized AXL (AXL receptor tyrosine kinase)+SIGLEC6 (sialic acid binding Ig like lectin 6)+ DC population (38), suggesting that TB pathogenesis may involve redistribution of circulating DC subsets. The SEPT4 protein has antiapoptotic functions, and its deletion improved wound healing in mice (39), suggesting a possible association with lung healing during TB progression. BLK is a B-cell receptor kinase, and its downregulation is consistent with reduced B-cell proportions in blood during TB (19, 40). The most generalizable pair defined in our meta-analysis showed upregulation of C1QC and downregulation of TRVAV27. Interestingly, complement pathway genes are markedly upregulated after Mtb infection of nonhuman primates (41), consistent with the upregulation of C1QC/TRAV27 at baseline in the HHCs. Complement activation is also observed early during human progression to TB (40), whereas C1q is downregulated early after starting TB treatment (22), suggesting that C1q may be a proxy of early TB pathology. Conversely, downregulation of TRAV27 and several other T-cell genes (Table E18) is likely associated with the overall decrease in peripheral T-cell frequencies and their associated gene expression modules during TB progression, potentially due to migration of T cells to the disease site (19, 21, 40). The simple C1QC/TRAV27 signal may thus be a readout of TB risk after initial exposure to an individual with pulmonary TB, which is more synchronized in an HHC study design, even though prior exposure to Mtb cannot be ruled out in our GC6-74 study and individuals with progression to TB disease within the first three months of the observation period were excluded from the analysis. This may explain why C1QC/TRAV27 signal was less predictive in the natural history cohort of adolescents with Mtb infection, where the time of Mtb exposure was unspecified. Early clinical studies suggest that recent exposure to Mtb, indicated by TST conversion, can correlate with symptoms consistent with febrile disease, such as fever and erythema nodosum (42, 43), which are markers of systemic inflammation. C1QC/TRAV27 may reflect this inflammatory response induced by failed containment of Mtb after recent exposure.
Overall, our study identifies and validates a simple PCR-based test with accessible blood samples that predicts TB in heterogeneous African populations with intermediate to high TB burdens (14, 15). Such a test can potentially be developed into a screening test for risk of progression during TB contact investigation implemented by national public health structures (13, 35, 36). The next steps include assessment of the performance of RISK4 and the two-transcript C1QC/TRAV27 signature in other settings, including non-African populations, and determination of the feasibility of developing a near-patient test for targeted intervention.
Acknowledgments
GC6-74 cohort study team
DST/NRF Centre of Excellence for Biomedical TB Research and MRC Centre for TB Research, Division of Molecular Biology and Human Genetics, Faculty of Medicine and Health Sciences, Stellenbosch University, Tygerberg, South Africa: Gerhard Walzl, Gillian F. Black, Gian van der Spuy, Kim Stanley, Magdalena Kriel, Nelita Du Plessis, Nonhlanhla Nene, Teri Roberts, Leanie Kleynhans, Andrea Gutschmidt, Bronwyn Smith, Nonhlanhla Nene, Andre G. Loxton, Novel N. Chegou, Gerhardus Tromp, and David Tabb
Department of Infectious Diseases, Leiden University Medical Centre, Leiden, the Netherlands: Tom H. M. Ottenhoff, Michel R. Klein, Marielle C. Haks, Kees L. M. C. Franken, Annemieke Geluk, Krista E. van Meijgaarden, and Simone A. Joosten
Tuberculosis Research Unit, Department of Medicine, Case Western Reserve University School of Medicine and University Hospitals Case Medical Center, Cleveland, Ohio: W. Henry Boom, Bonnie Thiel
Department of Medicine and Department of Microbiology, College of Health Sciences, Faculty of Medicine, Makerere University, Kampala, Uganda: Harriet Mayanja-Kizza, Moses Joloba, Sarah Zalwango, Mary Nsereko, Brenda Okwera, and Hussein Kisingo
Department of Immunology, Max Planck Institute for Infection Biology, Berlin, Germany: Stefan H. E. Kaufmann (GC6-74 principal investigator), Shreemanta K. Parida, Robert Golinski, Jeroen Maertzdorf, January Weiner 3rd, and Marc Jacobson
Department of Immunology and Infection, Faculty of Infectious and Tropical Diseases, London School of Hygiene and Tropical Medicine, London, United Kingdom: Hazel M. Dockrell, Maeve Lalor, Steven Smith, Patricia Gorak-Stolinska, Yun-Gyoung Hur, and Ji-Sook Lee
Karonga Prevention Study, Chilumba, Malawi: Amelia C. Crampin, Neil French, Bagrey Ngwira, Anne Ben-Smith, Kate Watkins, Lyn Ambrose, Felanji Simukonda, Hazzie Mvula, Femia Chilongo, Jacky Saul, and Keith Branson
South African Tuberculosis Vaccine Initiative, Institute of Infectious Disease and Molecular Medicine and Division of Immunology, Department of Pathology, University of Cape Town, Cape Town, South Africa: Sara Suliman, Thomas J. Scriba, Hassan Mahomed, E. Jane Hughes, Nicole Bilek, Mzwandile Erasmus, Onke Xasa, Ashley Veldsman, Katrina Downing, Michelle Fisher, Adam Penn-Nicholson, Humphrey Mulenga, Brian Abel, Mark Bowmaker, Benjamin Kagina, William Kwong Chung, and Willem A. Hanekom
Aeras, Rockville, Maryland: Jerry Sadoff, Donata Sizemore, S. Ramachandran, Lew Barker, Michael Brennan, Frank Weichold, Stefanie Muller, and Larry Geiter
Ethiopian Health and Nutrition Research Institute, Addis Ababa, Ethiopia: Desta Kassa, Almaz Abebe, Tsehayenesh Mesele, and Belete Tegbaru
University Medical Centre, Utrecht, the Netherlands: Debbie van Baarle and Frank Miedema
Armauer Hansen Research Institute, Addis Ababa, Ethiopia: Rawleigh Howe, Adane Mihret, Abraham Aseffa, Yonas Bekele, Rachel Iwnetu, Mesfin Tafesse, and Lawrence Yamuah
Vaccines and Immunity Theme, Medical Research Council Unit, Fajara, the Gambia: Martin Ota, Jayne Sutherland, Philip Hill, Richard Adegbola, Tumani Corrah, Martin Antonio, Toyin Togun, Ifedayo Adetifa, and Simon Donkor
Department of Infectious Disease Immunology, Statens Serum Institute, Copenhagen, Denmark: Peter Andersen, Ida Rosenkrands, Mark Doherty, and Karin Weldingh
Department of Microbiology and Immunology, Stanford University, Stanford, California: Gary Schoolnik, Gregory Dolganov, and Tran Van
The ACS cohort study team
South African Tuberculosis Vaccine Initiative, Institute of Infectious Disease and Molecular Medicine and Division of Immunology, Department of Pathology, University of Cape Town, Cape Town, South Africa: Fazlin Kafaar, Leslie Workman, Humphrey Mulenga, Thomas J. Scriba, E. Jane Hughes, Nicole Bilek, Mzwandile Erasmus, Onke Xasa, Ashley Veldsman, Yolundi Cloete, Deborah Abrahams, Sizulu Moyo, Sebastian Gelderbloem, Michele Tameris, Hennie Geldenhuys, Willem Hanekom, and Gregory Hussey
School of Public Health and Family Medicine, University of Cape Town, Cape Town, South Africa: Rodney Ehrlich
KNCV Tuberculosis Foundation, The Hague, and Amsterdam Institute of Global Health and Development, Academic Medical Centre, Amsterdam, the Netherlands: Suzanne Verver
Aeras, Rockville, Maryland: Larry Geiter
Footnotes
Supported by Bill and Melinda Gates Foundation grants OPP1065330, OPP1023483, and OPP1055806; GC6-74 grant 37772; and NIH grants R01AI087915 (W.A.H.), U01AI115619 (G.W.), and N01AI095383/AI070022 (W.H.B.). The study was also supported by the Strategic Health Innovation Partnerships (SHIP) Unit of the South African Medical Research Council with funds received from the South African Department of Science and Technology. Support was also provided by Postdoctoral Research Awards from The Carnegie Corporation of New York (A.P.-N. and S. Suliman); the South African National Research Foundation (S. Suliman); the Claude Leon Foundation and the Columbia University–Southern African Fogarty AIDS International Training and Research Program (AITRP) through the Fogarty International Center, NIH grant D43 TW000231 (A.P.-N.); and TBVAC Horizon2020 (Tuberculosis Vaccine Initiative under the European Commission’s Horizon 2020 research and innovation program) grant agreement 643381 (T.H.M.O. and S.H.E.K.).
Complete lists of members of the GC6-74 and ACS groups may be found before the beginning of the References.
Author Contributions: S. Suliman, E.G.T., S.H.E.K., P.C.H., W.A.H., G.W., T.J.S., and D.E.Z.: designed the study; S. Suliman, E.G.T., S.H.E.K., G.W., T.J.S., and D.E.Z.: drafted the manuscript; S. Suliman, E.G.T., J.S., J.W., S. Shankar, B.T., A.P.-N., M.E., J.M., F.J.D., E.J.H., K.S., K.D., M.L.F., J.V., S.K.P., G.v.d.S., G.T., I.M.O.A., S.D., R.H., H.M.-K., and W.H.B.: contributed to sample and data management as well as data acquisition; and S. Suliman, E.G.T., J.S., J.W., M.O.C.O., S. Shankar, B.T., A.P.-N., M.E., J.M., F.J.D., H.M.D., T.H.M.O., M.H., A.A., W.A.H., T.J.S., S.H.E.K., D.E.Z., G.W., and various members of the Grand Challenges 6-74 study and the Adolescent Cohort Study groups: contributed to data analysis and interpretation. All authors reviewed, provided feedback on, and approved the manuscript and are accountable for the accuracy and integrity of the work.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201711-2340OC on April 6, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
Collaborators: the Grand Challenges 6-74 (GC6-74) and Adolescent Cohort Study (ACS) groups, Gerhard Walzl, Gillian F. Black, Gian van der Spuy, Kim Stanley, Magdalena Kriel, Nelita Du Plessis, Nonhlanhla Nene, Teri Roberts, Leanie Kleynhans, Andrea Gutschmidt, Bronwyn Smith, Nonhlanhla Nene, Andre G. Loxton, Novel N. Chegou, Gerhardus Tromp, David Tabb, Tom H. M. Ottenhoff, Michel R. Klein, Marielle C. Haks, Kees L. M. C. Franken, Annemieke Geluk, Krista E. van Meijgaarden, Simone A. Joosten, W. Henry Boom, Bonnie Thiel, Harriet Mayanja-Kizza, Moses Joloba, Sarah Zalwango, Mary Nsereko, Brenda Okwera, Hussein Kisingo, (GC6-74 principal investigator), Shreemanta K. Parida, Robert Golinski, Jeroen Maertzdorf, January Weiner, 3rd, Marc Jacobson, Hazel Dockrell, Steven Smith, Patricia Gorak-Stolinska, Yun-Gyoung Hur, Maeve Lalor, Ji-Sook Lee, Amelia C. Crampin, Neil French, Bagrey Ngwira, Anne Ben-Smith, Kate Watkins, Lyn Ambrose, Felanji Simukonda, Hazzie Mvula, Femia Chilongo, Jacky Saul, Keith Branson, Sara Suliman, Thomas J. Scriba, Hassan Mahomed, E. Jane Hughes, Nicole Bilek, Mzwandile Erasmus, Onke Xasa, Ashley Veldsman, Katrina Downing, Michelle Fisher, Adam Penn-Nicholson, Humphrey Mulenga, Brian Abel, Mark Bowmaker, Benjamin Kagina, William Kwong Chung, Willem A. Hanekom, Jerry Sadoff, Donata Sizemore, S. Ramachandran, Lew Barker, Michael Brennan, Frank Weichold, Stefanie Muller, Larry Geiter, Desta Kassa, Almaz Abebe, Tsehayenesh Mesele, Belete Tegbaru, Debbie van Baarle, Frank Miedema, Rawleigh Howe, Adane Mihret, Abraham Aseffa, Yonas Bekele, Rachel Iwnetu, Mesfin Tafesse, Lawrence Yamuah, Martin Ota, Jayne Sutherland, Philip Hill, Richard Adegbola, Tumani Corrah, Martin Antonio, Toyin Togun, Ifedayo Adetifa, Simon Donkor, Peter Andersen, Ida Rosenkrands, Mark Doherty, Karin Weldingh, Gary Schoolnik, Gregory Dolganov, Tran Van, Fazlin Kafaar, Leslie Workman, Humphrey Mulenga, Thomas J. Scriba, E. Jane Hughes, Nicole Bilek, Mzwandile Erasmus, Onke Xasa, Ashley Veldsman, Yolundi Cloete, Deborah Abrahams, Sizulu Moyo, Sebastian Gelderbloem, Michele Tameris, Hennie Geldenhuys, Willem Hanekom, Gregory Hussey, Rodney Ehrlich, Suzanne Verver, and Larry Geiter
References
- 1.Riley F. Tuberculosis in hospital nurses; five years figures. Mon Bull Minist Health Public Health Lab Serv. 1959;18:38–45. [PubMed] [Google Scholar]
- 2.Yates TA, Khan PY, Knight GM, Taylor JG, McHugh TD, Lipman M, et al. The transmission of Mycobacterium tuberculosis in high burden settings. Lancet Infect Dis. 2016;16:227–238. doi: 10.1016/S1473-3099(15)00499-5. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) Global tuberculosis report 2017. [accessed 2018 Jan 26]. Available from: http://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf?u a=1.
- 4.Rieder H. Annual risk of infection with Mycobacterium tuberculosis. Eur Respir J. 2005;25:181–185. doi: 10.1183/09031936.04.00103804. [DOI] [PubMed] [Google Scholar]
- 5.O’Garra A, Redford PS, McNab FW, Bloom CI, Wilkinson RJ, Berry MP. The immune response in tuberculosis. Annu Rev Immunol. 2013;31:475–527. doi: 10.1146/annurev-immunol-032712-095939. [DOI] [PubMed] [Google Scholar]
- 6.Andrews JR, Noubary F, Walensky RP, Cerda R, Losina E, Horsburgh CR. Risk of progression to active tuberculosis following reinfection with Mycobacterium tuberculosis. Clin Infect Dis. 2012;54:784–791. doi: 10.1093/cid/cir951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cobelens F, Kik S, Esmail H, Cirillo DM, Lienhardt C, Matteelli A. From latent to patent: rethinking prediction of tuberculosis. Lancet Respir Med. 2017;5:243–244. doi: 10.1016/S2213-2600(16)30419-2. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization (WHO) Development of a Target Product Profile (TPP) and a framework for evaluation for a test for predicting progression from tuberculosis infection to active disease. [accessed 2018 Jan 26]. Available from: http://apps.who.int/iris/bitstream/10665/259176/1/WHO-HTM-TB-2017.18-eng.pdf.
- 9.Sester M, van Crevel R, van Leth F, Lange C. Numbers needed to treat to prevent tuberculosis. Eur Respir J. 2015;46:1836–1838. doi: 10.1183/13993003.01047-2015. [DOI] [PubMed] [Google Scholar]
- 10.Blaser N, Zahnd C, Hermans S, Salazar-Vizcaya L, Estill J, Morrow C, et al. Tuberculosis in Cape Town: an age-structured transmission model. Epidemics. 2016;14:54–61. doi: 10.1016/j.epidem.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wood R, Lawn SD, Caldwell J, Kaplan R, Middelkoop K, Bekker LG. Burden of new and recurrent tuberculosis in a major South African city stratified by age and HIV-status. PLoS One. 2011;6:e25098. doi: 10.1371/journal.pone.0025098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasaie P, Andrews JR, Kelton WD, Dowdy DW. Timing of tuberculosis transmission and the impact of household contact tracing: an agent-based simulation model. Am J Respir Crit Care Med. 2014;189:845–852. doi: 10.1164/rccm.201310-1846OC. [DOI] [PubMed] [Google Scholar]
- 13.Fox GJ, Barry SE, Britton WJ, Marks GB. Contact investigation for tuberculosis: a systematic review and meta-analysis. Eur Respir J. 2013;41:140–156. doi: 10.1183/09031936.00070812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abu-Raddad LJ, Sabatelli L, Achterberg JT, Sugimoto JD, Longini IM, Jr, Dye C, et al. Epidemiological benefits of more-effective tuberculosis vaccines, drugs, and diagnostics. Proc Natl Acad Sci USA. 2009;106:13980–13985. doi: 10.1073/pnas.0901720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dye C, Glaziou P, Floyd K, Raviglione M. Prospects for tuberculosis elimination. Annu Rev Public Health. 2013;34:271–286. doi: 10.1146/annurev-publhealth-031912-114431. [DOI] [PubMed] [Google Scholar]
- 16.Coussens AK, Wilkinson RJ, Nikolayevskyy V, Elkington PT, Hanifa Y, Islam K, et al. Ethnic variation in inflammatory profile in tuberculosis. PLoS Pathog. 2013;9:e1003468. doi: 10.1371/journal.ppat.1003468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comas I, Coscolla M, Luo T, Borrell S, Holt KE, Kato-Maeda M, et al. Out-of-Africa migration and Neolithic coexpansion of Mycobacterium tuberculosis with modern humans. Nat Genet. 2013;45:1176–1182. doi: 10.1038/ng.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denkinger CM, Kik SV, Cirillo DM, Casenghi M, Shinnick T, Weyer K, et al. Defining the needs for next generation assays for tuberculosis. J Infect Dis. 2015;211(Suppl 2):S29–S38. doi: 10.1093/infdis/jiu821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry MP, Graham CM, McNab FW, Xu Z, Bloch SA, Oni T, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maertzdorf J, Repsilber D, Parida SK, Stanley K, Roberts T, Black G, et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 2011;12:15–22. doi: 10.1038/gene.2010.51. [DOI] [PubMed] [Google Scholar]
- 21.Joosten SA, Fletcher HA, Ottenhoff TH. A helicopter perspective on TB biomarkers: pathway and process based analysis of gene expression data provides new insight into TB pathogenesis. PLoS One. 2013;8:e73230. doi: 10.1371/journal.pone.0073230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cliff JM, Lee JS, Constantinou N, Cho JE, Clark TG, Ronacher K, et al. Distinct phases of blood gene expression pattern through tuberculosis treatment reflect modulation of the humoral immune response. J Infect Dis. 2013;207:18–29. doi: 10.1093/infdis/jis499. [DOI] [PubMed] [Google Scholar]
- 23.Cliff JM, Kaufmann SH, McShane H, van Helden P, O’Garra A. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev. 2015;264:88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ottenhoff TH, Dass RH, Yang N, Zhang MM, Wong HE, Sahiratmadja E, et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS One. 2012;7:e45839. doi: 10.1371/journal.pone.0045839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, et al. ACS and GC6-74 cohort study groups. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet. 2016;387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Churchyard G, Gupta A, Swindells S, Hesseling A, Opollo V, Suryavanshi N, et al. A5300/I2003 Study Team. Accessing MDR-TB exposed households: the PHOENiX MDR-TB feasibility study. Presented at the 48th Union Conference on Lung Health. October 14, 2017, Guadalajara, Mexico. [Google Scholar]
- 27.Kaufmann SHE. Predictive biosignatures to improve tuberculosis vaccine development. Presented at the Fifth Global Forum on TB Vaccines; February 22, 2018, New Delhi, India. [Google Scholar]
- 28.Mahomed H, Hawkridge T, Verver S, Geiter L, Hatherill M, Abrahams DA, et al. SATVI Adolescent Study Team. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. Int J Tuberc Lung Dis. 2011;15:331–336. [PubMed] [Google Scholar]
- 29.Thompson EG, Du Y, Malherbe ST, Shankar S, Braun J, Valvo J, et al. Catalysis TB–Biomarker Consortium. Host blood RNA signatures predict the outcome of tuberculosis treatment. Tuberculosis (Edinb) 2017;107:48–58. doi: 10.1016/j.tube.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maertzdorf J, McEwen G, Weiner J, III, Tian S, Lader E, Schriek U, et al. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol Med. 2016;8:86–95. doi: 10.15252/emmm.201505790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweeney TE, Braviak L, Tato CM, Khatri P. Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir Med. 2016;4:213–224. doi: 10.1016/S2213-2600(16)00048-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, et al. Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis. 2008;198:1037–1043. doi: 10.1086/591504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adetifa IM, Kendall L, Bashorun A, Linda C, Omoleke S, Jeffries D, et al. A tuberculosis nationwide prevalence survey in Gambia, 2012. Bull World Health Organ. 2016;94:433–441. doi: 10.2471/BLT.14.151670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petruccioli E, Scriba TJ, Petrone L, Hatherill M, Cirillo DM, Joosten SA, et al. Correlates of tuberculosis risk: predictive biomarkers for progression to active tuberculosis. Eur Respir J. 2016;48:1751–1763. doi: 10.1183/13993003.01012-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiore-Gartland A, Carpp LN, Kogieleum N, Thompson E, Zak DE, et al. Considerations for biomarker-targeted intervention strategies for tuberculosis disease prevention. Tuberculosis (Edinb) 2018;109:61–68. doi: 10.1016/j.tube.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penn-Nicholson A, Scriba TJ, Hatherill M, White RG, Sumner T. A novel blood test for tuberculosis prevention and treatment. S Afr Med J. 2016;107:4–5. doi: 10.7196/SAMJ.2016.v107.i1.12230. [DOI] [PubMed] [Google Scholar]
- 38.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. doi: 10.1126/science.aah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fuchs Y, Brown S, Gorenc T, Rodriguez J, Fuchs E, Steller H. Sept4/ARTS regulates stem cell apoptosis and skin regeneration. Science. 2013;341:286–289. doi: 10.1126/science.1233029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scriba TJ, Penn-Nicholson A, Shankar S, Hraha T, Thompson EG, Sterling D, et al. other members of the ACS cohort study team. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 2017;13:e1006687. doi: 10.1371/journal.ppat.1006687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gideon HP, Skinner JA, Baldwin N, Flynn JL, Lin PL. Early whole blood transcriptional signatures are associated with severity of lung inflammation in cynomolgus macaques with Mycobacterium tuberculosis infection. J Immunol. 2016;197:4817–4828. doi: 10.4049/jimmunol.1601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wallgren A. The time-table of tuberculosis. Tubercle. 1948;29:245–251. doi: 10.1016/s0041-3879(48)80033-4. [DOI] [PubMed] [Google Scholar]
- 43.Poulsen A. Some clinical features of tuberculosis. 1. Incubation period. Acta Tuberc Pneumol Scand. 1950;24:311–346. [PubMed] [Google Scholar]