Abstract
A number of studies have indicated that thyroid hormone receptor β1 (TRβ1) functions as a tumor suppressor. TRs mediate transcriptional responses through a highly conserved DNA-binding domain (DBD). A novel rat TRβ isoform (rTRβΔ) was previously identified, in which a novel exon, N (108 bp), is located between exons 3 and 4 within the DBD; this exon represents the only difference between rTRβΔ and rTRβ1. In vitro, rTRβΔ exhibits a stronger tumor-suppressive capacity than rTRβ1, and further analysis revealed a high level of conservation between the rat and human DBD sequences. In the present study, an artificially modified human TRβ1 (m-hTRβ1) was constructed via the introduction of the 108-bp sequence into the corresponding position of the wild-type human TRβ1 (wt-hTRβ1) DBD. An electrophoretic mobility shift assay and transfection experiments confirmed that m-hTRβ1 is functional. Overexpression of m-hTRβ1 inhibits the proliferation of MDA-MB-468 cells in the presence of triiodothyronine by promoting apoptosis, which may be associated with the upregulation of Caspase-3 and Bak gene expression and the activation of the Caspase-3 protein. In addition, the pro-apoptotic effect of m-hTRβ1 was stronger, compared with wt-hTRβ1. These results indicated that m-hTRβ1 may act as a tumor suppressor in MDA-MB-468 cells. These data provided a novel insight into gene therapy for breast cancer.
Keywords: thyroid hormone receptor β1, caspase-3, tumor suppressor, MDA-MB-468 cells
Introduction
Thyroid hormone receptors (TRs) are members of a superfamily of nuclear receptors and ligand-dependent transcription factors. TRs are encoded by two genes, TRα (THRA) and TRβ (THRB) (1), and four triiodothyronine (T3) and DNA-binding receptor isoforms, TRα1, TRβ1, TRβ2 and TRβ3, that are only present in rats, are generated by alternative mRNA splicing (2). These isoforms are considered functional receptors and share a common structure with several regions from the N- to the C-terminus, including: A N-terminal activation domain (A and B regions); a conserved DNA-binding domain (DBD) (C region); a hinge domain (D region); and a ligand-binding domain (E and F regions) (3). TRs can bind to thyroid hormone response elements (TREs) as homodimers or heterodimers via the retinoid X receptor (4). In addition to the regulation of metabolism, growth and development, increasing evidence has indicated that TRs have an important role in cell proliferation and malignant transformation (5). Low or no expression of TRs and alterations in TR genes, particularly TRβ, have been identified in a number of cancer types (6,7). Studies have also demonstrated that mutations in TRβ are closely associated with a number of cancer types (8,9).
Cell-based studies and xenograft models have confirmed that TRβ, particularly TRβ1, functions as a tumor suppressor to suppress tumor cell proliferation, migration and tumorigenesis (5,10). A novel rat TRβ isoform have confirmed TRβΔ (GenBank number: DQ191165) was previously identified. rTRβΔ is homologous to rTRβ1 and only contains an additional 108 nucleotides or 36 amino acids (11). Structural analysis has indicated that these additional 36 amino acids are located in the DBD, which alters this highly conserved domain (11). Nonetheless, previous studies (12,13) have demonstrated that rTRβΔ is a functional TR with transcription factor characteristics that exhibit greater tumor-suppressive ability in vitro, compared with rTRβ1. In the present study, the additional 36 amino acids were introduced into the corresponding position of wild-type human TRβ1 (wt-hTRβ1), which was termed modified human TRβ1 (m-hTRβ1); then, the transcription factor characteristics and in vitro anti-tumor effects of m-hTRβ1 were examined in human breast cancer MDA-MB-468 cells without endogenous TRβ.
Materials and methods
Cells and reagents
MDA-MB-468 and MCF-7 human breast cancer cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Chitosan (CS), with a molecular mass of 60,000–100,000 Da (degree of deacetylation ~85%) was purchased from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). L-15 cell culture medium, an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit, NE-PER™ Nuclear and Cytoplasmic Extraction Reagents and an anti-TRβ-1 antibody (cat. no. MA1-216; dilution, 1:1,000) were purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA). A Caspase-3 spectrophotometric assay kit (cat. no. G007) was purchased from Nanjing KeyGen Biotech Co., Ltd. (Nanjing, Jiangsu, China). Antibodies against Histone H3 (cat. no. bs-0349R; dilution, 1:1,000), active Caspase-3 (cat. no. bsm-33199M; dilution, 1:200) and Bak (cat. no. bs-1284R; dilution, 1:200) were purchased from BIOSS (Beijing, China). M-MLV reverse transcriptase, TRIzol® total RNA extraction reagent and quantitative real-time polymerase chain reaction (PCR) detection kits (SYBR® Premix Ex Taq™ II; cat. no. RR820B were obtained from Takara Bio, Inc. (Otsu, Japan). KOD-Plus-Ver polymerase and the KOD-Plus-Mutagenesis kit were obtained from Toyobo Life Science (Osaka, Japan).
Construction of pcDNA3.1-wt-hTRβ1 and pcDNA3.1-m-hTRβ1 vectors
MCF-7 cells were cultured and collected. Total RNA was extracted using TRIzol reagent and was used for reverse transcription to generate cDNA. This ‘total cDNA’ was used as a template for PCR to obtain the full-length wt-hTRβ1 cDNA. The forward primer 5′-CCCAAGCTTATGACTCCCAACAGTATGAC-3′ contains a restriction endonuclease site for HindIII (underlined), whilst the reverse primer 5′-CCGGAATTCCTAATCCTCGAACACTTCC-3′ contains an EcoRI site (underlined). The HindIII-EcoRI PCR product was subcloned into pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) at the corresponding sites. The constructed expression plasmid, pcDNA3.1-wt-hTRβ1, was confirmed by sequence analysis.
Mutant constructs were generated by site-directed mutagenesis. The following primers were used to generate mutations: Primer 1 5′-TCTACCTCTCTGCATTCGGTCTGGATGTTGTCCTCAAGGCAGTGGGACCCTAGGTTTCTTTAGAAGAACCATTCAGAAAAAT-3′; Primer 2 5′-ACTGGTGTCTGTATGGAACCAAATCCCTGTCTTCTCGTCTCTGGTGTGAGAAGGCTTGCAGCCTTCACACGTGATACAGCGGT-3′. The underlined portion of the primers indicates the novel introduced 108-bp exon, and the non-underlined portion indicates the partial wt-hTRβ1 DBD sequence. Site-directed mutagenesis was performed using a KOD-Plus-Mutagenesis kit. This procedure involved PCR amplification using the pcDNA3.1-wt-hTRβ1 plasmid as the template and mutation primers were performed according to the manufacturer's instructions. The desired mutations were confirmed by DNA sequencing of the entire gene. The recombinant plasmid carrying the 108-bp exon was termed pcDNA3.1-m-hTRβ1.
Preparation of pDNA/nucleic kinase substrate short peptide (NNS)CS complexes
The nuclear localization signal-linked NNS was fused to CS to form the gene carrier NNSCS as previously described (14). The empty plasmid pcDNA3.1 and the plasmids pcDNA3.1-wt-hTRβ1, pcDNA3.1-m-hTRβ1 and pGL3-TRE (the palindromic TRE is located upstream of the SV40 promoter in the pGL3-promoter vector, which contains a Luciferase reporter gene) (11) were extracted, and the pDNA/NNSCS complex (pcDNA3.1/NNSCS, pcDNA3.1-wt-hTRβ1/NNSCS, pcDNA3.1-m-hTRβ1/NNSCS and pGL3-TRE/NNSCS) was prepared as previously described (14).
Western blotting
MDA-MB-468 cells were seeded at a density of 1×106 cells/ml in the wells of a 6-well plate in L-15 medium supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.). When the cells had grown to 75% confluence, transfection were performed as below, and pcDNA3.1/NNSCS, pcDNA3.1-wt-hTRβ1/NNSCS and pcDNA3.1-m-hTRβ1/NNSCS were added separately to the 6 wells to achieve DNA concentrations of 4 µg/well. Cells were collected following 48 h of transfection, and nuclear proteins were extracted using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (cat. no. 78833; Thermo Fisher Scientific, Inc.). The nuclear protein levels were measured by BCA assay (cat. no. CW0014; CWBIO, Beijing, China) and each sample containing 30 µg nuclear protein were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with TBS-T [25 mM Tris-Cl (pH 7.4), 150 mM NaCl and 0.05% Tween-20] containing 5% skimmed milk for 2 h at room temperature, and then incubated with primary antibodies against TRβ1 (cat. no. MA1-216; dilution, 1:1,000) and Histone H3 (cat. no. bs-0349R; dilution, 1:1,000). TRβ1 recognizes an epitope in the A/B domain of hTRβ1, namely, residues 1–101, which are common sequences of wt-hTβ1 and m-hTRβ1. After being thoroughly washed in TBS-T, the membranes were incubated with the secondary antibodies, HRP-conjugated mouse anti-rabbit IgG (cat. no. bs-0295M-HRP; dilution, 1:5,000; BIOSS) or Goat Anti-Mouse IgG, HRP Conjugated (cat. no. bs-0368G-HRP, dilution 1:5,000, BIOSS) for 1.5 h at room temperature. After thorough washing with TBS-T, the membranes were visualized using an enhanced chemiluminescence kit (Merck KGaA) and Kodak films (Kodak, Rochester, NY, USA).
Electrophoretic mobility shift assay (EMSA)
The DNA-binding abilities of wt-hTRβ1 and m-hTRβ1 were first determined using a synthetic DR4 TRE; the oligonucleotide sequences and labeling of the DR4 TRE probe were reported previously (11). Following the aforementioned transfection of MDA-MB-468 cells with pcDNA3.1-wt-hTRβ1/NNSCS and pcDNA3.1-m-hTRβ1/NNSCS for 72 h, nuclear proteins were extracted and subjected to standard EMSA analysis, as previously described (11).
Transcriptional activity analysis
pcDNA3.1/NNSCS, pcDNA3.1-wt-hTRβ1/NNSCS and pcDNA3.1-m-hTRβ1/NNSCS were co-transfected with pGL3-TRE/NNSCS to determine the transcriptional activity of wt-hTRβ1 and m-hTRβ1. A Renilla luciferase plasmid, pRL-TK (Promega Corporation, Madison, WI, USA), was included to correct for transfection efficiency. Following MDA-MB-468 cell transfection for 6 h, the transfection solution was discarded and replaced with serum-free medium, and 10 nM T3 (Sigma-Aldrich; Merck KGaA) was added to each T3 intervention group. Following 48 h of transfection (T3 exposure for 42 h), luciferase and Renilla activities were assayed using a Dual-Glo® Luciferase Assay System (Promega Corporation) according to the manufacturer's instructions. Firefly luciferase activity was normalized to the Renilla controls. At the same time, each group of nuclear proteins was extracted using NE-PER® Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions, and the overexpression of wt-hTRβ1 and m-hTRβ1 protein was measured using a standard western blotting assay as aforementioned. The results demonstrated that the expression levels of wt-hTRβ1 and m-hTRβ1 for each condition are identical.
Cell proliferation and apoptosis assays
MDA-MB-468 cells were seeded at a density of 1×104 cells/ml in the wells of a 96-well plate. When the cells had grown to 75% confluence, pcDNA3.1/NNSCS, pcDNA3.1-wt-hTRβ1/NNSCS and pcDNA3.1-m-hTRβ1/NNSCS were added to the wells. Following 6 h of transfection, the transfection solution was discarded and replaced with serum-free medium IL-15, and 10 nM T3 was added to the intervention groups. At 48 h following transfection (T3 exposure for 42 h), MTT (Promega Corporation) solution (20 µl, 5 mg/ml) was added to each well, and the plate was incubated at 37°C for 4 h, following which the formazan crystals were solubilized in dimethyl sulfoxide (200 µl/well). The absorbance at 570 nm was recorded using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA), and background absorbance at 630 nm was subtracted.
MDA-MB-468 cells were seeded in wells of a 12-well plate and transfected as aforementioned. At 48 h following transfection (T3 exposure for 42 h), the cells were collected and stained with FITC-conjugated Annexin V and propidium iodide for 10 min at room temperature. The stained cells were then detected by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Bak, Bax and Caspase-3 expression assays
For the expression assay, MDA-MB-468 cells were transfected as aforementioned. At 48 h following transfection (T3 exposure for 42 h), the cells were collected and total RNA was extracted using TRIzol reagent. mRNA (2 µg) was reverse transcribed into total cDNA in a 20 µl reaction mixture using the PrimeScript™ RT reagent kit with gDNA Eraser (cat. no. RR047A; Takara Bio, Inc.). Bak, Bax and Caspase-3 mRNA levels were analyzed by reverse transcription-quantitative PCR (RT-qPCR) and were detected with SYBR® Green using an iQ5TM system (Bio-Rad Laboratories, Inc.) according to the manufacturer's protocol. The thermocycling conditions were as follows: Initial denaturation at 95°C for 30 sec, followed by 35 cycles at 95°C for 5 sec, 56°C for 20 sec and 72°C for 20 sec. Primers for each gene were as follows: Bak, forward 5′-TACCGCCATCAGCAGGAAC-3′ and reverse 5′-TCTGAGTCATAGCGTCGGTTG-3′; Bax, forward 5′-GGAGCTGCAGAGGATGATTG-3′ and reverse 5′-GGCCTTGAGCACCAGTTTG-3′; Caspase-3, forward 5′-GGAAGCGAATCAATGGACTC-3′ and reverse 5′-TTCCCTGAGGTTTGCTGC-3′; and Gapdh, forward 5′-GCATCCTGGGCTACACTGAG-3′ and reverse 5-CCACCACCCTGTTGCTGTAG-3′. The Cq values for all genes from different samples were collected. Raw data were normalized to GAPDH expression, and the relative expression level of each gene is represented as 2−ΔΔCq (15).
The expression levels of active Caspase-3 and Bak proteins were analyzed by western blot analysis as aforementioned. Antibodies against active Caspase-3 (cat. no. bs-0081R; dilution, 1:300), Bak (cat. no. bs-1284R; dilution, 1:300) and GAPDH (cat. no. sc-32233; dilution 1:1,000) (all Santa Cruz Biotechnology, Inc., Dallas, TX, USA) were used.
Caspase-3 activity assay
MDA-MB-468 cells were seeded in a 6-well plate and transfected as aforementioned. At 48 h after transfection (T3 exposure for 42 h), the cells were lysed using the Lysis Buffer in the Caspase-3 Spectrophotometric assay kits and the Caspase-3 activity was assessed using this kit according to the manufacturer's instructions.
Statistical analysis
For all measurements, the data are expressed as the mean ± standard deviation based on three independent experiments. The results were analyzed using SPSS 17.0 (SPSS, Inc., Chicago, IL, USA). One-way analysis of variance was used for multi-group comparisons with the Student-Newman-Keuls as the post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
m-hTRβ1 construction, expression and functional analysis
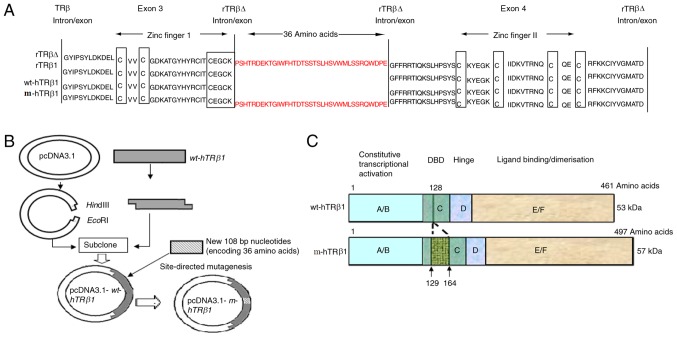
The amino acid sequences of the human and rTRβ1 DBDs are highly conserved (16). rTRβ∆ has an additional 108 bp sequence between exons 3 and 4, and the segment containing the additional 36 amino acids was located on the C-terminal side of the first two amino acids of the P-box (Fig. 1A) of the first zinc finger motif. This increased the length of the linker between the two zinc fingers by 3.1-fold (17 to 53 amino acids). The full-length coding sequence of wt-hTRβ1 (1368 bp) was successfully cloned into pcDNA3.1 to obtain the expression plasmid pcDNA3.1-wt-hTRβ1 (Fig. 1B). The coding sequence of the additional 36 residues (108 bp) was successfully introduced into a similar position in wt-hTRβ1 by site-directed mutagenesis, and this mutant was termed pcDNA3.1-m-hTRβ1 (Fig. 1B). The DBD of m-hTRβ1 is only 36 amino acids longer than wt-hTRβ1 (Fig. 1C).
Figure 1.
Schematic of the wt-hTRβ1 and m-hTRβ1 protein structure and expression vector construction. (A) Amino acid sequences around the P box in the DBD of TRβs. The amino acid sequences are presented from exon 3–4 of TRβ. Homologous sequences are relative to the zinc finger motifs. A long gap is introduced for best alignment. The P box and the cysteines of the zinc fingers are boxed. The coding sequence of the 36 amino acids was introduced between exon 3 and 4 of wt-hTRβ. The mutant was termed m-hTRβ1 (B) Map of the pcDNA3.1-wt-hTRβ1 and pcDNA3.1-m-hTRβ1 plasmids, vector construction and the mutation are depicted. (C) Schematic diagram representing the protein structure of wt-hTRβ1 and m-hTRβ1. TRβ, thyroid hormone receptor β; wt-hTRβ1, wild-type human TRβ1; m-hTRβ1, modified human TRβ1; rTRβΔ, rat TRβ isoform; DBD, DNA-binding domain.
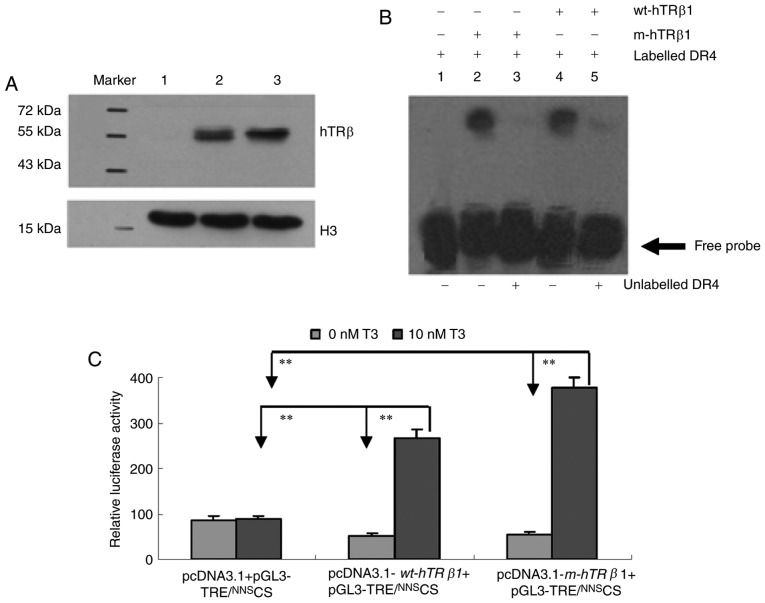
To examine wt-hTRβ1 and m-hTRβ1 expression and activity, pcDNA3.1 (control), pcDNA3.1-wt-hTRβ1 and pcDNA3.1-m-hTRβ1 were transiently transfected into MDA-MB-468 cells. Protein expression was assessed by western blot. The two proteins (m-hTRβ1 and wt-hTRβ1) were ~57 and ~53 kDa, respectively, (Fig. 2A, lanes 2 and 3), and no TRβ was detected in the control transfection group (Fig. 2A, lane 1). The lower band depicted in Fig. 2 is Histone H3, which was used as a control. DNA binding was assessed by EMSA using a DR4 TRE. Nuclear proteins containing TRβ (wt-hTRβ1 and m-hTRβ1) can bind to DR4, as demonstrated by a shift in DNA electrophoretic mobility. These DNA binding assays were sequence-specific, as the receptors competed with a 100-fold excess of unlabeled DR4 (Fig. 2B). The EMSA results indicated that TRβ (wt-hTRβ1 and m-hTRβ1) receptors may bind to DR4.
Figure 2.
Expression and properties of the wt-hTRβ1 and m-hTRβ1 proteins. (A) Western blot detection of wt-hTRβ1 and m-hTRβ1 expression in MDA-MB-468 cells. Lane 1, pcDNA3.1/NNSCS; Lane 2, pcDNA3.1-m-hTRβ1/NNSCS; and Lane 3, pcDNA3.1-wt-hTRβ1/NNSCS. (B) DNA binding activity was measured by EMSA. Lane 1, digoxin-labeled DR4 (17 fmol/µl); lanes 2 and 4: digoxin-labeled DR4 (17 fmol/µl) incubated with m-hTRβ1 and wt-hTRβ1 in the absence of competitor (100-fold excess of unlabeled DR4); lanes 3 and 5: digoxin-labeled DR4 (17 fmol/µl) incubated with m-hTRβ1 and wt-hTRβ1 in the presence of competitor (100-fold excess of unlabeled DR4). (C) Luciferase activity in MDA-MB-468 cell lysates. Firefly luciferase activity was normalized to Renilla controls and data are presented as the mean ± standard deviation. **P<0.01. wt-hTRβ1, wild-type human TRβ1; m-hTRβ1, modified human TRβ1; TRE, thyroid hormone response elements; NNS, nucleic kinase substrate short peptide; CS, chitosan; T3, triiodothyronine.
The transcriptional activities of wt-hTRβ1 and m-hTRβ1 as T3 receptors were compared using a transient transfection system. Relative luciferase activity levels were assessed in each group. Transfection with pcDNA3.1+pGL3-TRE/NNSCS resulted in basic luciferase activity levels in the presence or absence of T3 (P>0.05). The pcDNA3.1-wt-hTRβ1 + pGL3-TRE/NNSCS and pcDNA3.1-m-hTRβ1 + pGL3-TRE/NNSCS transfection group demonstrated a similar increase in luciferase activity levels in the absence of T3 (P>0.05); these levels were increased 5.4- and 6.9-fold, respectively, following treatment with 10 nM T3 (Fig. 2C).
hTRβ1 inhibits MDA-MB-468 cell proliferation and promotes MDA-MB-468 cell apoptosis
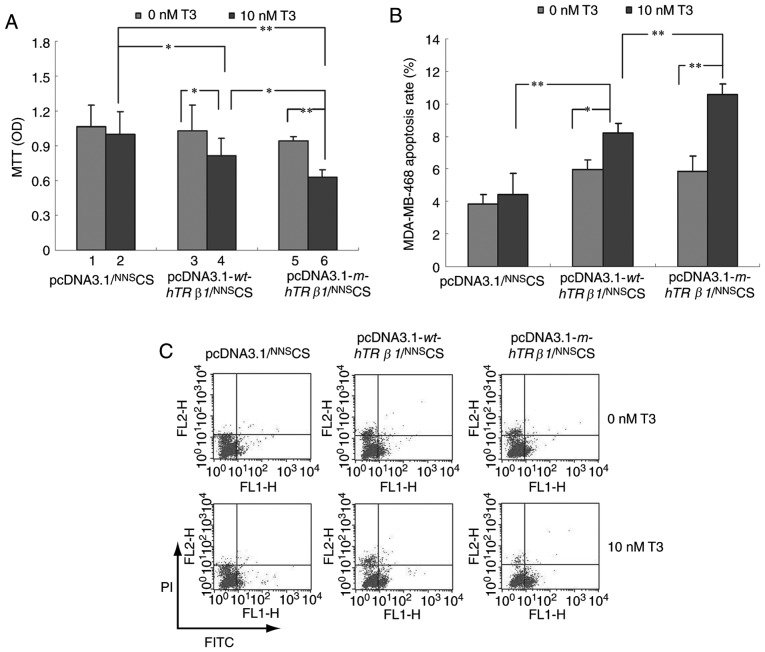
To examine the effect of wt-hTRβ1 and m-hTRβ1 on cell proliferation, MDA-MB-468 cells were transiently transfected with pcDNA3.1/NNSCS, pcDNA3.1-wt-hTRβ1/NNSCS or pcDNA3.1-m-hTRβ1/NNSCS, and subjected to MTT assays. Fig. 3A depicted that T3 had no significant effect on MDA-MB-468 cell growth. However, the expression of wt-hTRβ1 and m-hTRβ1 inhibited cell proliferation in the presence of T3. Furthermore, the effect of m-hTRβ1 on cell proliferation inhibition was stronger, compared with wt-hTRβ1; whereas, wt-hTRβ1 and m-hTRβ1 expression had no significant effect on proliferation in the absence of T3.
Figure 3.
Effect of wt-hTRβ1 and m-hTRβ1 on the proliferation and apoptosis of MDA-MB-468 cells. (A) The proliferation of MDA-MB-468 cells following wt-hTRβ1 or m-hTRβ1-gene transfection was detected by MTT assay. Data are presented as the mean ± SD. (B and C) The apoptosis of MDA-MB-468 cells following wt-hTRβ1 or m-hTRβ1-gene transfection was detected by Annexin V-FITC assay. The total number of apoptotic cells was determined by calculating the sum of early apoptotic cells (Annexin V-FITC+/PI-) and late apoptotic cells (Annexin V-FITC+/PI+) detected by flow cytometry (C); Analysis of the apoptosis rate of MDA-MB-468 cell (B); the data are presented as the mean ± SD. *P<0.05; **P<0.01. OD, optical density; wt-hTRβ1, wild-type human TRβ1; m-hTRβ1, modified human TRβ1; NNS, nucleic kinase substrate short peptide; CS, chitosan; T3, triiodothyronine; PI, propidium iodide; FITC, fluorescein isothiocyanate; SD, standard deviation.
To understand how wt-hTRβ1 and m-hTRβ1 overexpression inhibits cancer cell proliferation in vitro, flow cytometry was conducted to evaluate apoptosis in MDA-MB-468 cells transfected with pcDNA3.1/NNSCS, pcDNA3.1-wt-hTRβ1/NNSCS and pcDNA3.1-m-hTRβ1/NNSCS. wt-hTRβ1 and m-hTRβ1 enhanced apoptosis in the presence of 10 nM T3, and the effects of m-hTRβ1 were stronger, compared with wt-hTRβ1. By contrast, wt-hTRβ1 and m-hTRβ1 expression had no significant effect on apoptosis in the absence of T3 (Fig. 3B and C).
Effects of hTRβ1 on Bak, Bax and Caspase-3 expression in MDA-MB-468 cells
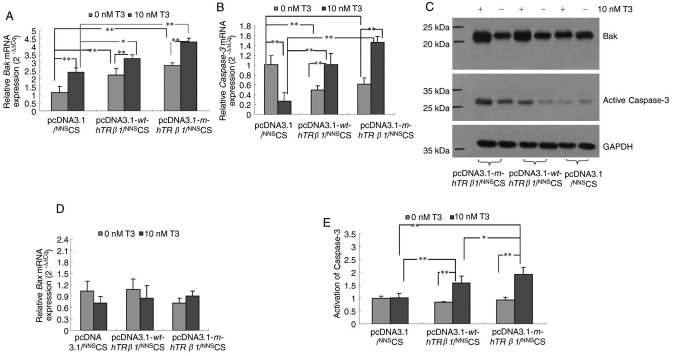
wt-hTRβ1 and m-hTRβ1 reduced cell proliferation mainly through the promotion of apoptosis (Fig. 3). To determine the possible pathways that wt-hTRβ1 and m-hTRβ1 act to promote apoptosis, the relative expression levels of key apoptotic regulators, including Bak, Bax and Caspase-3, were detected in the different transfection groups. Expression of wt-hTRβ1 and m-hTRβ1 resulted in a gradual but significant upregulation of Bak and Caspase-3 mRNA and protein expression in the presence of T3. However, wt-hTRβ1 and m-hTRβ1 expression downregulated Caspase-3 gene expression in the absence of T3 (Fig. 4A-C). Expression of wt-hTRβ1 and m-hTRβ1 had no effect on Bax mRNA expression regardless of T3 presence or absence (Fig. 4D).
Figure 4.
Expression of Bak, Bax and Caspase-3 and activity of Caspase-3 in MDA-MB-468 cells. Reverse transcription-quantitative polymerase chain reaction analysis of the relative gene expression of (A) Bak and (B) Caspase-3 in MDA-MB-468 cells following wt-hTRβ1 or m-hTRβ1-gene transfection. The expression levels were normalized to those of GAPDH; the relative expression level of each gene was calculated using 2−ΔΔCq. The data are presented as the mean ± SD. (C) Western blot analysis of the effect of wt-hTRβ1 and m-hTRβ1 on the expression of Bak and Caspase-3. The results are from a single experiment that is representative of three independent experiments. (D) Reverse transcription-quantitative polymerase chain reaction analysis of the relative gene expression of Bax in MDA-MB-468 cells following wt-hTRβ1 or m-hTRβ1-gene transfection. (E) Colorimetric assay of Caspase-3 activation. Data are presented as the mean ± SD. *P<0.05; **P<0.01. wt-hTRβ1, wild-type human TRβ1; m-hTRβ1, modified human TRβ1; NNS, nucleic kinase substrate short peptide; CS, chitosan; T3, triiodothyronine; SD, standard deviation.
hTRβ1 increases the activity of Caspase-3 in MDA-MB-468 cells
As the expression of wt-hTRβ1 and m-hTRβ1 upregulated Caspase-3 expression in MDA-MB-468 cells in the presence of T3 (Fig. 4B and D), Caspase-3 activity was evaluated in these cells. Fig. 4E depicted that the expression of wt-hTRβ1 and m-hTRβ1 also increased the activity of Caspase-3 in the presence of T3. Increased Caspase-3 activity is indicative of elevated apoptotic activity (17), and elevated Bak promotes apoptosis via the activation of caspases (18). These data indicated that wt-hTRβ1 and m-hTRβ1 can increase the expression of Bak to promote apoptosis through the activation of Caspase-3. In addition, the effect of m-hTRβ1 is stronger, compared with wt-hTRβ1.
Discussion
TRs can function as a tumor suppressor (5,10). Evidence from in vivo and in vitro studies supports the tumor-suppressive function of TRβ1 in breast cancer (1,19,20). Nonetheless, controversial conclusions are abundant regarding the role of thyroid hormones and their receptor TRβ1 in the occurrence and development of breast cancer (21). Thyroid hormones and breast cancer and normal mammary glands (1). Decreased expression, promoter hypermethylation or expression of truncated TRβ1 has been observed in human breast cancer (22), and truncated TRβ1 has been associated with cancer occurrence (23). rTRβΔ is an extended receptor (11), and to explore the association between the extended receptor rTRβΔ and cancer, TRβΔ was overexpressed in rat breast cancer SHZ-88 cells in vitro. The results demonstrated that rTRβΔ clearly inhibited proliferation of SHZ-88 cells. Although rTRβΔ is highly homologous to rTRβ1, the former contains a larger DBD (Fig. 1A); thus, the strong inhibitory effect of rTRβΔ on breast cancer cells may be closely associated with this additional 108 bp. Sequence analysis has revealed high conservation between the DBD region of rats and humans (Fig. 1A). Accordingly, this extra 108-bp exon from the DBD of rTRβΔ was introduced into the corresponding location of wt-hTRβ1 by site-directed mutagenesis, but did not change the original reading frame of wt-hTRβ1 (Fig. 1B and C).
wt-hTRβ1 contains an ‘EG…G’ P-box sequence in the DBD (24), and studies have indicated that the first two amino acids (EG) of the P-box are the most crucial for TRE binding specificity (25,26). If the first two amino acids are mutated, TRβ binding ability to TRE is significantly reduced or completely lost, though the requirements of the third amino acid are not strict (24,27). The additional 36 amino acids were introduced to the C-terminal EG of the P-box, and the third amino acid was changed from G to P (Fig. 1A). In the present study, the aim was to clarify whether artificial m-hTRβ1 is a functional receptor and to evaluate the effect of m-hTRβ1 on MDA-MB-468 cells.
Overall, the results of the EMSA and transcriptional activity analysis demonstrated that m-hTRβ1 can bind TREs (DR4) with transactivation by T3. An in vitro analysis demonstrated that the overexpression of m-hTRβ1 in MDA-MB-468 cells could inhibit their proliferation via the promotion of apoptosis in the presence of T3; the effect of m-hTRβ1 was stronger, compared with wt-hTRβ1. One study has demonstrated that the levels of the thyroid hormone T3 are elevated in the serum of patients with breast cancer (28), where this hormone promotes the growth and motility of breast cancer cells (18,29,30). However, since Beatson (31) described the treatment of metastatic breast tumors with thyroid extracts, a number of in vitro and in vivo studies have demonstrated that T3 can inhibit breast tumor cell proliferation and metastasis (1,32). These data indicated that thyroid hormones may have a bidirectional effect on breast cancer cells, particularly among different breast cancer cell lines. The results indicated that T3 inhibits the proliferation of MDA-MB-468 cells that overexpress m-hTRβ1.
Previous studies have demonstrated that TRβ expression reduces MCF-7 tumor growth through the activation of apoptosis in vivo (20) and the induction of apoptosis in human breast cancer MCF-7 cells in the presence of T3 in vitro (1). To explore the underlying mechanism that m-hTRβ1 promotes MDA-MB-468 cell apoptosis, the expression of pro-apoptotic Caspase-3, Bax and Bak, was detected and the Caspase-3 activity was examined. TRβ expression induces apoptosis in breast cancer cells via an increase in cleaved PARP and Caspase-3 (20,33). In the present study, it was determined that in the presence of T3, the overexpression m-hTRβ1 promoted MDA-MB-468 cell apoptosis via the upregulation of Caspase-3 and Bak mRNA and protein expression and via an increase in Caspase-3 activity. In addition, the pro-apoptotic effect of m-hTRβ1 was stronger, compared with wt-hTRβ1. These results indicated that modification of the DBD region may alter the TRβ activation intensity of target genes.
To conclude, an artificial m-hTRβ1 was constructed through the introduction of a novel 108-bp exon into the DBD of wt-hTRβ1. m-hTRβ1 was functional in MDA-MB-468 cells and inhibited MDA-MB-468 cell proliferation, as it promoted apoptosis in the presence of T3. In the future, the role of m-hTRβ1 in other cancer cell lines in vitro and its role in tumorigenesis in vivo, using xenograft models, will be ascertained. This information may provide a novel possibility of gene therapy for TR-deficient breast cancer. Therefore, inhibition of the growth of cancer cells (TR-deficient) and improvements in patient survival may be possible via TR transgenes.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science Foundation of China (grant nos. 81770915 and 81301737) and the Shandong Provincial Natural Science Foundation, China (grant nos. ZR2012HQ034 and ZR2015HL025).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
RLZ designed the project, and contributed to all experiments, analysis of data and to writing the manuscript. XXP contributed to all experiments and to writing the manuscript. YYZ, YLS, LJW, WS and QL conducted the experiments. All authors read and provided their approval for the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Sar P, Peter R, Rath B, Das Mohapatra A, Mishra SK. 3,3′5 Triiodo L thyronine induces apoptosis in human breast cancer MCF-7 cells, repressing SMP30 expression through negative thyroid response elements. PLoS One. 2011;6:e20861. doi: 10.1371/journal.pone.0020861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng SY, Leonard JL, Davis PJ. Molecular aspects of thyroid hormone actions. Endocr Rev. 2010;31:139–170. doi: 10.1210/er.2009-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassett JH, Harvey CB, Williams GR. Mechanisms of thyroid hormone receptor-specific nuclear and extra nuclearactions. Mol Cell Endocrinol. 2003;213:1–11. doi: 10.1016/j.mce.2003.10.033. [DOI] [PubMed] [Google Scholar]
- 4.Paquette MA, Atlas E, Wade MG, Yauk CL. Thyroid hormone response element half-site organization and its effect on thyroid hormone mediated transcription. PLoS One. 2014;9:e101155. doi: 10.1371/journal.pone.0101155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu L, Tian G, Yang Q, De G, Zhang Z, Wang Y, Nie H, Zhang Y, Yang X, Li J. Thyroid hormone receptor β1 suppresses proliferation and migration by inhibiting PI3K/Akt signaling in human colorectal cancer cells. Oncol Rep. 2016;36:1419–1426. doi: 10.3892/or.2016.4931. [DOI] [PubMed] [Google Scholar]
- 6.Ling Y, Shi X, Wang Y, Ling X, Li Q. Down-regulation of thyroid hormone receptor β1 gene expression in gastric cancer involves promoter methylation. Biochem Biophys Res Commun. 2014;444:147–152. doi: 10.1016/j.bbrc.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Rosignolo F, Maggisano V, Sponziello M, Celano M, Di Gioia CR, D'Agostino M, Giacomelli L, Verrienti A, Dima M, Pecce V, Durante C. Reduced expression of THRβ in papillary thyroid carcinomas: Relationship with BRAF mutation, aggressiveness and miR expression. J Endocrinol Invest. 2015;38:1283–1289. doi: 10.1007/s40618-015-0309-4. [DOI] [PubMed] [Google Scholar]
- 8.Guigon CJ, Kim DW, Willingham MC, Cheng SY. Mutation of thyroid hormone receptor-β in mice predisposes to the development of mammary tumors. Oncogene. 2011;30:3381–3390. doi: 10.1038/onc.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park JW, Zhao L, Willingham M, Cheng SY. Oncogenic mutations of thyroid hormone receptor β. Oncotarget. 2015;6:8115–8131. doi: 10.18632/oncotarget.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim WG, Zhao L, Kim DW, Willingham MC, Cheng SY. Inhibition of tumorigenesis by the thyroid hormone receptor β in xenograft models. Thyroid. 2014;24:260–269. doi: 10.1089/thy.2013.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao RL, Sun B, Liu Y, Li JH, Xiong WL, Liang DC, Guo G, Zuo AJ, Zhang JY. Cloning and identification of a novel thyroid hormone receptor β isoform expressed in the pituitary gland. Mol Cell Biochem. 2014;389:141–150. doi: 10.1007/s11010-013-1935-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhao RL, Song W, Sun YL, Li Q, Li M, Chu HR, Peng XX. Effects of thyroid hormone receptor βΔ on apoptosis and proliferation of hepatoma RH-35 cells. Chin J Endocrinol Metab. 2016;32:691–695. [Google Scholar]
- 13.Zhang YY, Peng XX, Sun YL, Song W, Zhao RL. Effect of thyroid hormone receptor βΔ on proliferation and apoptosis of rat breast cancer cells SHZ-88. Cancer Res Prev Treat. 2016;43:1018–1022. [Google Scholar]
- 14.Zhao R, Sun B, Liu T, Liu Y, Zhou S, Zuo A, Liang D. Optimize nuclear localization and intra-nucleus disassociation of the exogenefor facilitating transfection efficacy of the chitosan. Int J Pharm. 2011;413:254–259. doi: 10.1016/j.ijpharm.2011.04.039. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Jones I, Srinivas M, Ng L, Forrest D. The thyroid hormone receptor beta gene: Structure and functions in the brain and sensory systems. Thyroid. 2003;13:1057–1068. doi: 10.1089/105072503770867228. [DOI] [PubMed] [Google Scholar]
- 17.Cryns V, Yuan J. Proteases to die for. Genes Dev. 1988;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 18.Guo W, Zhang Y, Ling Z, Liu X, Zhao X, Yuan Z, Nie C, Wei Y. Caspase-3 feedback loop enhances Bid-induced AIF/endoG and Bak activationin Bax and p53-independent manner. Cell Death Dis. 2015;6:e1919. doi: 10.1038/cddis.2015.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruiz-Llorente L, Ardila-González S, Fanjul LF, Martínez-Iglesias O, Aranda A. microRNAs 424 and 503 are mediators of the anti-proliferative and anti-invasive action of the thyroid hormone receptor beta. Oncotarget. 2014;5:2918–2933. doi: 10.18632/oncotarget.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JW, Zhao L, Cheng SY. Inhibition of estrogen-dependent tumorigenesis by the thyroid hormonereceptor β in xenograft models. Am J Cancer Res. 2013;3:302–311. [PMC free article] [PubMed] [Google Scholar]
- 21.de Sibio MT, de Oliveira M, Moretto FC, Olimpio RM, Conde SJ, Luvizon AC, Nogueira CR. Triiodothyronine and breast cancer. World J Clin Oncol. 2014;5:503–508. doi: 10.5306/wjco.v5.i3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Meng ZH, Chandrasekaran R, Kuo WL, Collins CC, Gray JW, Dairkee SH. Biallelic inactivation of the thyroid hormone receptor beta1 gene in earlystage breast cancer. Cancer Res. 2002;62:1939–1943. [PubMed] [Google Scholar]
- 23.Jazdzewski K, Boguslawska J, Jendrzejewski J, Liyanarachchi S, Pachucki J, Wardyn KA, Nauman A, de la Chapelle A. Thyroid hormone receptor beta (THRB) is a major target gene for microRNAs deregulated in papillary thyroid carcinoma (PTC) J Clin Endocrinol Metab. 2011;96:E546–E553. doi: 10.1210/jc.2010-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson CC, Hendy SC, Faris JS, Romaniuk PJ. Retinoid X receptor alters the Determination of DNA binding specificity by the P-box amino acids of the thyroid hormone receptor. J Biol Chem. 1996;271:19464–19474. doi: 10.1074/jbc.271.32.19464. [DOI] [PubMed] [Google Scholar]
- 25.Figueira AC, Lima LM, Lima LH, Ranzani AT, Mule Gdos S, Polikarpov I. Recognition by the thyroid hormone receptor of canonical DNA response elements. Biochemistry. 2010;49:893–904. doi: 10.1021/bi901282s. [DOI] [PubMed] [Google Scholar]
- 26.Berglund H, Wolf-Watz M, Lundbäck T, van den Berg S, Härd T. Structure and dynamics of the glucocorticoid receptor DNA-binding domain: Comparison of wild type and a mutant with altered specificity. Biochemistry. 1997;36:11188–11197. doi: 10.1021/bi970343i. [DOI] [PubMed] [Google Scholar]
- 27.Shibusawa N, Hashimoto K, Nikrodhanond AA, Liberman MC, Applebury ML, Liao XH, Robbins JT, Refetoff S, Cohen RN, Wondisford FE. Thyroid hormone action in the absence of thyroid hormone receptor DNA-binding in vivo. J Clin Invest. 2003;112:88–97. doi: 10.1172/JCI18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi XZ, Jin X, Xu P, Shen HM. Relationship between breast cancer and levels of serum thyroid hormones and antibodies: A meta-analysis. Asian Pac J Cancer Prev. 2014;15:6643–6647. doi: 10.7314/APJCP.2014.15.16.6643. [DOI] [PubMed] [Google Scholar]
- 29.Rasool M, Naseer MI, Zaigham K, Malik A, Riaz N, Alam R, Manan A, Sheikh IA, Asif M. Comparative study of alterations in Tri-iodothyronine (T3) and Thyroxine (T4) hormone levels in breast and ovarian cancer. Pak J Med Sci. 2014;30:1356–1360. doi: 10.12669/pjms.306.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flamini MI, Uzair ID, Pennacchio GE, Neira FJ, Mondaca JM, Cuello-Carrión FD, Jahn GA, Simoncini T, Sanchez AM. Thyroid hormone controls breast cancer cell movement via integrin αV/β3/SRC/FAK/PI3-Kinases. Horm Cancer. 2017;8:16–27. doi: 10.1007/s12672-016-0280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beatson GT. On the etiology of cancer, with a note of some experiments. Br Med J. 1899;1:399–400. doi: 10.1136/bmj.1.1990.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Llorente L, Martínez-Iglesias O, García-Silva S, Tenbaum S, Regadera J, Aranda A. The thyroid hormone receptors as tumor suppressors. Horm Mol Biol Clin Investig. 2011;5:79–89. doi: 10.1515/HMBCI.2010.045. [DOI] [PubMed] [Google Scholar]
- 33.Gu G, Gelsomino L, Covington KR, Beyer AR, Wang J, Rechoum Y, Huffman K, Carstens R, Andò S, Fuqua SA. Targeting thyroid hormone receptor beta in triple-negative breast cancer. Breast Cancer Res Treat. 2015;150:535–545. doi: 10.1007/s10549-015-3354-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.