Abstract
This study aimed to investigate the expression of microRNA (miRNA) 299 and miRNA-7706 in patients with hepatocellular carcinoma (HCC), and to explore their effects on proliferation of SK-HEP-1 HCC cells. Expression of miRNA-299 and miRNA-7706 in tumor tissue (HCC group) and adjacent healthy tissue (>30 mm away from the tumor tissue) of 179 patients with HCC was determined by real-time polymerase chain reaction (qRT-PCR). miR-299 mimics and miR-7706 mimics were transfected into SK-HEP-1 HCC cells by RNA transfection. The proliferation and invasion of SK-HEP-1 cells were detected by CCK-8 kit and Transwell kit, respectively. Compared with adjacent tissues, expression levels of miRNA-299 and miRNA-7706 in HCC group were significantly downregulated. Analyses on the correlation between the expression of miRNA-299 and miRNA-7706 and clinical factors showed that expression levels of miRNA-299 and miRNA-7706 were significantly correlated with pathological stages and lymph node metastasis. ROC curve analysis showed that the areas under the curve were 0.837 and 0.845 for miRNA-299 and miRNA-7706 in the prediction of HCC, respectively. Survival analysis showed that the 5-year overall survival rate of patients with high expression levels of miRNA-299 and miRNA-7706 was significantly different from that of patients with low expression levels (P=0.016). Compared with cells transfected with scramble mimics, proliferation and invasion abilities of SK-HEP-1 cells transfected with miR-299 mimics and miRNA-7706 were significantly weakened. Results suggested that downregulation of miRNA-299 and miRNA-7706 can inhibit the proliferation of HCC cells and can be used as a new target for the treatment of HCC.
Keywords: miRNA-299, miRNA-7706, HCC, PCR, ROC
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignant cancer in the world (1,2). HCC is also the third most common cause of cancer-related death (3,4). The early diagnosis of HCC is usually difficult and there is a lack of effective treatments, so the prognosis of patients with HCC is usually poor (5). The development of HCC is a multi-step and multi-stage pathology process with multiple genes involved (6,7). To date, no specific and sensitive tumor markers have been clinically used for early diagnosis of HCC. Therefore, this study was carried out to identify markers for the diagnosis of HCC.
MicroRNA (miRNA) is a group of endogenous single-stranded non-coding RNA with a length of about 17–25 nucleotides. miRNAs mainly target the 3′ untranslated region (3′UTR) of target genes to degrade mRNA or inhibit the translation of mRNA (8–10). miRNAs also participate in the formation of organs, cell proliferation and apoptosis, and tumorigenesis (11,12). It has been found that miRNAs are closely related to the development of different types of cancer. Differential expression of some miRNAs may affect the growth and metastasis of tumors (13).
We screened the differential expression genes of patients with HCC through the TCGA database and found that miRNA-299 and miRNA-7706 were differentially expressed in HCC patients (data not shown). This finding has not been reported before. Therefore, this study was carried out to investigate the expression of miRNA-299 and miRNA-7706 in HCC and their effects on SK-HEP-1 cells, so as to promote the early diagnosis and prognosis of HCC.
Materials and methods
Sample collection
Tumor tissues and adjacent healthy tissues were collected from 179 HCC patients who were treated in The Third Affiliated Hospital of Sun Yet-sen University (Guangzhou, China) from June 2010 to February 2015. Those patients included 127 males (70.9%) and 52 females (29.1%) with a mean age of 47.2±8 years. None of the patients received radiotherapy and chemotherapy before surgery. TNM staging was performed in accordance with the 2009 seventh edition of the UICC (Union for International Cancer Control). HCC TNM staging: 36 cases of stage I, 69 cases of stage II, and 74 cases of stage III–IV. Pathological types of all specimens were confirmed as HCC. This study was approved by the Ethics Committee of The Third Affiliated Hospital of Sun Yet-sen University, and all patients signed informed consent.
Reagents and instruments
SK-HEP-1 cells were purchased from Shanghai Institutes for Biological Sciences of the Chinese Academy of Sciences (Shanghai, China). Total RNA extraction reagent TRIzol, miScript Reverse Transcription kit, PCR kit, fetal bovine serum, DMEM, PCR amplification primers and transfection reagent were purchased from Invitrogen (Thermo Fisher Scientific, Inc., Waltham, MA, USA). miRNA-299, miRNA-7706 and negative control scramble mimics were purchased from GenePharma Co., Ltd. (Shanghai, China). 7900HT fluorescence quantitative PCR instrument was purchased from ABI (Thermo Fisher Scientific, Inc.).
Primer design
miRNA-199 and miRNA-7706 reverse transcription primers and PCR specific primers were designed and synthesized by GenePharma Co., Ltd. (Table I).
Table I.
Primers of miRNA-199, miRNA-7706 and endogenous control.
| Items | Primer sequences |
|---|---|
| miRNA-299 | |
| Reverse transcription primers | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAGCGGTT-3′ |
| Sense | 5′-TCAAGCTTTGAAGCGCCTGTGC-3′ |
| Antisense | 5′-GCCTAAGGAGGGTCCGAGGTATTC-3′ |
| miRNA-7709 | |
| Reverse transcription primers | 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCTCGGCA-3′ |
| Sense | 5′-TCAAGCTTTGAAGCGCCTGTGC-3′ |
| Antisense | 5′-GCCTAAGGAGGGTCCGAGGTATTC-3′ |
| U6 | |
| Sense | 5′-GGAACGCTTCACGAATTTG −3′ |
| Antisense | 5′-ATTGGAACGATACAGAGAAGATT-3′ |
Experimental methods
Cell culture
SK-HEP-1 cells were cultured with DMEM containing 10% fetal bovine serum and 100 g/l penicillin-streptomycin double antibiotics in an incubator (37°C, 5% CO2). SK-HEP-1 and SK-HEP-1 cells were collected at logarithmic growth phase. After digestion with 0.25% trypsin, 3×105 cells were transferred to each well of 6-well plates. Cells were cultured in medium containing no antibiotics before transfection. Transfection mixture was prepared according to the instructions of Lipofectamine™ 2000 kit. Transfection mixture was added to each well of 6-well plate and was shaken. After incubation at 37°C for 6 h, culture medium and transfection reagent were removed. Finally, 2 ml medium containing 10% fetal bovine serum was added into each well for further culture.
Total RNA extraction
Total RNA was extracted from the cells using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the instructions. After RNA extraction, the concentration of each sample RNA was determined by ultraviolet spectrophotometer. All RNA samples were stored at −80°C before use.
Reverse transcription
Reverse transcription was performed using miScript Reverse Transcription kit and 1 µg of total RNA was used to synthesize cDNA. Reverse transcription conditions: 42°C for 3 min, 42°C for 60 min and 95°C for 3 min. All cDNA samples were stored at −80°C before use.
RT-PCR
Real-time fluorescence quantitative PCR was performed on 7900HT fluorescence quantitative PCR instrument using cDNA and SYBR premix Ex Taq™ II PCR kit. Reaction system was: 10 µl of SYBR premix Ex Taq™ II PCR mix, 1 µl of cDNA, 0.5 µl of each primer and 8 µl of RNase-free water. Reaction conditions were: 95°C for 5 min, followed by 45 cycles of 95°C for 15 sec, 60°C for 45 sec and 72°C for 25 sec. U6 was used as an endogenous control (Table II). Results were analyzed using the 2−ΔΔCt method.
Table II.
Correlation between the expression of miRNA-299 and miRNA-7706 and clinical factors.
| Clinical factors | N (%) | Expression level of miRNA-299 | P-value | Expression level of miRNA-7706 | P-value |
|---|---|---|---|---|---|
| Age (years) | 0.684 | 0.841 | |||
| <60 | 72 (40.2) | 0.94±0.24 | 1.12±0.41 | ||
| ≥60 | 107 (59.8) | 0.87±0.31 | 1.24±0.47 | ||
| Sex | 0.714 | 0.614 | |||
| Male | 127 (70.9) | 0.67±0.51 | 0.91±0.47 | ||
| Female | 52 (29.1) | 0.88±0.45 | 0.98±0.51 | ||
| Pathological stage | 0.039 | 0.042 | |||
| I | 58 (32.4) | 0.94±0.22 | 1.22±0.44 | ||
| II | 79 (44.1) | 0.81±0.39 | 0.98±0.39 | ||
| III | 42 (23.5) | 0.73±0.16 | 0.81±0.33 | ||
| TNM stage | 0.057 | 0.061 | |||
| I | 36 (20.1) | 0.94±0.29 | 1.14±0.41 | ||
| II | 69 (38.5) | 0.92±0.32 | 1.07±0.37 | ||
| III/IV | 74 (41.4) | 0.85±0.34 | 0.92±0.46 | ||
| Lymph node metastasis | 0.031 | 0.044 | |||
| Positive | 89 (49.7) | 0.73±0.23 | 0.84±0.32 | ||
| Negative | 90 (50.3) | 0.91±0.18 | 1.27±0.39 | ||
| History of smoking | 0.062 | 0.784 | |||
| Yes | 100 (55.8) | 0.82±0.33 | 1.09±0.36 | ||
| No | 79 (44.2) | 0.87±0.36 | 1.01±0.44 |
Cell proliferation assay
miRNA-299, miRNA-7706 and scramble mimics were transfected into 1×106 SK-HEP-1 cells, followed by incubation for 24 h. After that, SK-HEP-1 cells were inoculated into 96-well plates with 6×103 cells per well, followed by incubation for 3–5 h. DMEM (100 µl) was added after cell adhesion, followed by incubation in an incubator (37°C, 5% CO2) for 6 days. CCK-8 (10 µl) was added daily, and OD values at 450 nm were measured using a microplate reader.
Transwell invasion assay
Matrigel was kept at 4°C overnight (12 h). Matrigel (50 µl; 1:10 dilution) was added to the upper chamber, followed by incubation at 37°C for 6 h. After intervention with serum-free medium for 48 h, cells were digested with trypsin and resuspended in DMEM containing 1% fetal bovine serum to adjust the cell density to 1×106/ml. Cell suspension (100 µl) was transferred into the upper chamber, while the lower chamber was filed with 500 µl of culture medium containing 10% FBS. After incubation for 24 h in an incubator, Matrigel and the cells that failed to invade were removed. The membrane was fixed for 30 min, and staining with 0.1% crystal violet was performed for 15 min. The results were observed under an inverted microscope. Each experiment was repeated three times.
Statistical analysis
Statistical analysis was performed using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). Measurement data were expressed as mean ± SD. ANOVA was used for comparisons between multiple groups and SNK test was used for post hoc test. Student's t-test was used for comparison of clinical factors. Correlation analyses were performed by Pearson's correlation analysis. Survival analysis was performed using Kaplan-Meier curve method. P<0.05 was considered to indicate a statistically significant difference.
Results
miRNA-299 and miRNA-7706 expression in HCC tissues and adjacent tissues
The relative expression levels of miRNA-299 and miRNA-7706 in tumor tissue were 0.71±0.41 and 1.2±0.57, respectively, which were significantly lower than those in adjacent healthy tissue (P<0.05) (Fig. 1).
Figure 1.

Expression of miRNA-299 and miRNA-7706 in HCC tissues and adjacent tissues. Expression of miRNA-299 and miRNA-7706 in different tissues was determined by RT-PCR. Expression levels of of miRNA-299 and miRNA-7706 in HCC tissues were significantly lower than those in adjacent tissues (*P<0.05). HCC, hepatocellular carcinoma.
Correlation between miRNA-299 and miRNA-7706 expression and clinical factors
Expression levels of miRNA-299 and miRNA-7706 were correlated with pathological stage and lymph node metastasis (P<0.05). With the increase in pathological stage, expression levels of miRNA-299 and miRNA-7706 significantly decreased (P=0.039; P=0.042). Patients with lymph node metastasis showed significantly reduced expression levels of miRNA-299 and miRNA-7706 (P=0.031; P=0.044). No significant correlation were found between expression levels of miRNA-299 and miRNA-7706 and other clinical and pathological factors including age, sex, TNM stages and history of smoking (P>0.05) (Table II).
Diagnostic value of miRNA-299 and miRNA-7706 for HCC
Diagnostic value of miRNA-299 and miRNA-7706 for HCC was analyzed by ROC curve. As shown in Fig. 2, AUC=0.804, AUC=0.781; confidence interval 95% CI: 0.724–0.842, 95% CI: 0.754–0.876.
Figure 2.
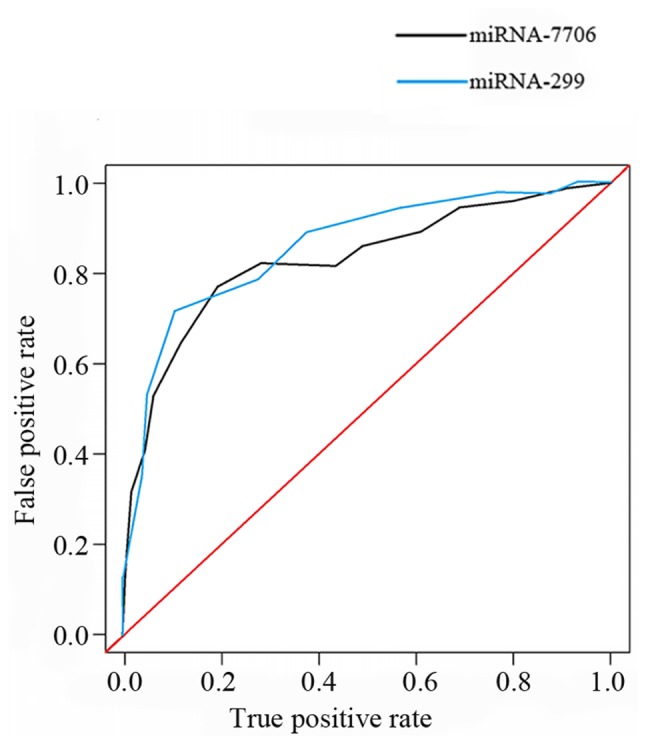
ROC curve analysis of the diagnostic values of miRNA-299 and miRNA-7706 for HCC. The areas under the curve of miRNA-299 and miRNA-7706 were 0.804 and 0.781, respectively. HCC, hepatocellular carcinoma.
Correlation between expression of miRNA-299 and miRNA-7706 and prognosis of HCC patients
Patients were divided in to two groups according to the median value of the expression levels of miRNA-299 and miRNA-7706. Follow-up study showed that survival rate of miRNA-299 and miRNA-7706 high expression group was significantly higher than that of low expression group (P<0.05, P=0.027). As shown in Figs. 3 and 4, Kaplan-Meier survival curve analysis showed that expression levels of miRNA-299 and miRNA-7706 were correlated with the prognosis of patients with HCC.
Figure 3.

Correlation between miRNA-299 and prognosis of HCC patients. Kaplan-Meier survival curve analysis showed that the survival rate of miRNA-299 high expression group was significantly higher than that of low expression group (P=0.027). HCC, hepatocellular carcinoma.
Figure 4.
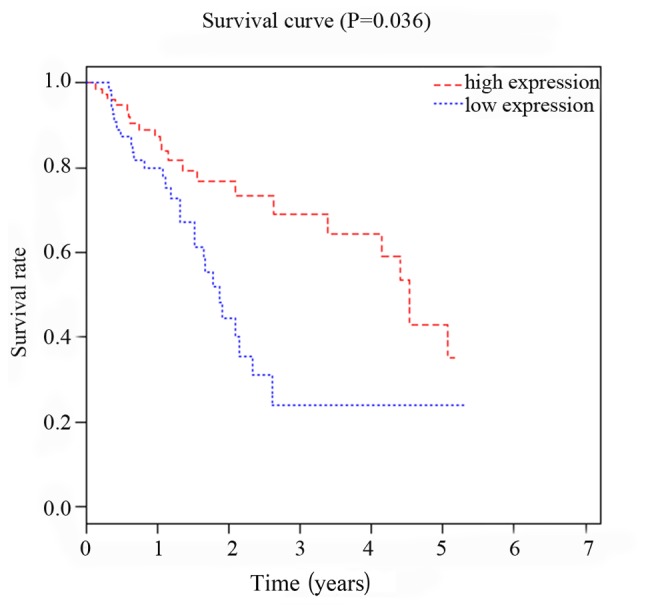
Correlation between miRNA-7706 and prognosis of HCC patients. Kaplan-Meier survival curve analysis showed that the survival rate of miRNA-7706 high expression group was significantly higher than that of low expression group (P=0.027). HCC, hepatocellular carcinoma.
Effects of miRNA-299 mimics and miRNA-7706 mimics transfection on expression of miRNA-299 and miRNA-7706 in HCC cells
After transfection of miRNA-299-mimics-SK-HEP-1 and miRNA-299-scramble-mimics-SK-HEP-1 into SK-HEP-1 cells, expression levels of miRNA-299 were 7.89±2.87 and 1.17±0.45, respectively. After transfection of miRNA-7706-mimics-SK-HEP-1 and miRNA-7706-scramble-mimics-SK-HEP-1 into SK-HEP-1 cells, expression levels of miRNA-7706 were 11.58±2.25 and 1.53±0.57. After transfection of miRNA-299-mimics-SK-HEP-1 and miRNA-7706-mimics-SK-HEP-1, expression levels of miRNA-299 and miRNA-7706 in SK-HEP-1 cells were significantly increased (t=4.01, P<0.05; t=7.50, P<0.05).
Effects of miRNA-299 and miRNA-7706 on proliferation of SK-HEP-1 cells
miRNA-299-mimics-SK-HEP-1, miRNA-7706-mimics-SK-HEP-1, miRNA-299-scramble-mimics -SK-HEP-1 and miRNA-7706-scramble-mimics-SK-HEP-1 were transfected into 1×106 SK-HEP-1 cells, followed by incubation for 24 h. After that, SK-HEP-1 cells were inoculated into 96-well plates with 6×103 cells per well, followed by incubation in an incubator (37°C, 5% CO2) for 6 days. CCK-8 (10 µl) was added daily, and OD values at 450 nm were measured using a microplate reader. GraphPad Prism 5 software (GraphPad Software, Inc., La Jolla, CA, USA) was used to plot the proliferation curve of SK-HEP-1 HCC cells after transfection (Fig. 5).
Figure 5.
Effects of miRNA-299 and miRNA-7706 on the proliferation of SK-HEP-1 cells. CCK-8 method was used to detect cell proliferation after miR-299 mimics and miRNA-7706 mimics transfection. After 6 days of culture, proliferation of SK-HEP-1was significantly inhibited after transfection with miRNA-299-mimics-SK-HEP-1 and miRNA-7706-mimics-SK-HEP-1 (*P<0.05).
Effects of miRNA-299 and miRNA-7706 on invasion of SK-HEP-1 cells
Compared with SK-HEP-1 cells transfected with scramble mimics, invasion ability of SK-HEP-1 cells was significantly weakened after transfection with miRNA-299 and miRNA-7706 mimics (P<0.05) (Fig. 6).
Figure 6.
Effects of miRNA-299 and miRNA-7706 on the invasion of SK-HEP-1 cells. Results of Transwell assay showed that invasion of SK-HEP-1 cells was significantly reduced after transfection with miRNA-299 mimics and miRNA-7706 mimics (*P<0.05).
Discussion
In this study, expression of miRNA-299 and miRNA-7706 in tumor tissue and adjacent healthy tissue of HCC patients was determined by qRT-PCR. Studies have found that the expression level of miRNA-299 is low in a variety of tumor tissues, such as endometrial rectal, colon and ovarian cancers (14–16). However, studies on miRNA-7706 are relatively insufficient. Expression pattern and function of two miRNAs in HCC still haven't been reported. Analysis on the correlation between expression levels of miRNA-299 and miRNA-7706 and patients' clinical data showed that expression levels of miRNA-299 and miRNA-7706 were significantly reduced in patients with lymph node metastasis. Other clinical and pathological factors such as age, sex, TNM grade and history of smoking showed no significant correlation with miRNA-299 and miRNA-7706 expression. Expression levels of miRNA-299 and miRNA-7706 in tumor tissues were significantly lower than that in adjacent tissues. At the same time, expression levels of those two miRNAs in HCC cells SK-HEP-1 were also lower than that in control cells. These results indicate that miRNA-299 and miRNA-7706 can inhibit tumor cell proliferation. This is consistent with the finding of Göhring et al (17) that miRNA-299 can inhibit the proliferation and invasion of tumor cells. Based on those studies, we speculate that miRNA-299 and miRNA-7706 can also regulate cell proliferation and invasion in HCC. In this study, results of CCK-8 cell proliferation assay and Transwell cell invasion assay showed that miRNA-299 and miRNA-7706 could significantly inhibit the proliferation and invasion of SK-HEP-1 cells. Many studies (18–20) found that inhibition of tumor cell proliferation and invasion can significantly inhibit tumor growth, which is consistent with the results in our study. ROC curve analysis showed that AUCs of miRNA-299 and miRNA-7706 AUC were 0.804 and 0.781, respectively, which is consistent with the finding reported by Ren et al (21), indicating that two miRNAs can be used as markers for clinical diagnoses. Besides that, analyses on the correlation between the expression of miRNA-299 and miRNA-7706 and clinical factors showed that expression levels of miRNA-299 and miRNA-7706 have no significant correlations with age, sex, TNM stage and history of smoking of patients (P<0.05), indicating that miRNA-299 and miRNA-7706 can potentially be new markers for HCC. Kaplan-Meier survival curve analysis showed that the survival rate of patients in miRNA-299 and miRNA-7706 high expression group was significantly higher than that of low expression group, indicating that patients with low expression levels of those two miRNAs tend to have short-term survival. In this study, we detected the expression of miRNA-299 and miRNA-7706 in HCC. Correlation analysis was performed to explore the correlations between the expression of those two miRNAs and clinical factors, which were indicated by the P-value. Chi-square test showed that pathological stage and lymph node metastasis were significantly correlated with the expression of those two miRNAs in different stages. Those data may provide new indexes for clinical staging and prediction of lymph node metastasis. In order to provide references for clinical practice, effects of those two miRNAs on proliferation and invasion of HCC cells were explored. We found that those two miRNAs could effectively inhibit the proliferation and invasion of cancer cells. ROC analysis showed that those two miRNAs has promising clinical diagnostic value for HCC, which is expected to be applied clinically in future.
Results of our study showed that miR-299 and miR-7706 have similar functionality in hepatoma carcinoma cell line SK-HEP-1. However, our study also has some shortcomings. Most patients in this study were from local region. Therefore regional difference may affect the results of our study. In addition, this study is also limited by the small sample size. The interaction between miRNA-299 and miRNA-7706 is still unknown. Therefore, our future studies will focus on the interactions between those two miRNAs and their roles in other diseases.
In conclusion, expression levels of miRNA-299 and miRNA-7706 were significantly reduced in tumor tissue of HCC patients, suggesting that they may play important roles in the development of HCC. Detection of expression levels of miRNA-299 and miRNA-7706 may provide references for the diagnosis of HCC. Our study provided new insights for the diagnosis and treatment of HCC.
Acknowledgements
Not applicable.
Funding
This study was funded by Special Scientific Expenditure for Business Construction of State TCM Clinical Research Base, State Administration of TCM of the People's Republic of China. State TCM Scientific Task [2016] 20, no. JDZX2015173.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
MD and FW wrote the manuscript and was responsible for cell culture. HC collected the sample. YL performed reverse transcription. JZ contributed to RT-PCR. ZZ and HY helped cell proliferation assay and Transwell assay. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of The Third Affiliated Hospital of Sun Yet-sen University (Guangzhou, China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499:97–101. doi: 10.1038/nature12347. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol. 2013;47:S2–6. doi: 10.1097/MCG.0b013e3182872f29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YC, Xu Z, Zhang TF, Wang YL. Circulating microRNAs as diagnostic and prognostic tools for hepatocellular carcinoma. World J Gastroenterol. 2015;21:9853–9862. doi: 10.3748/wjg.v21.i34.9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kokudo N, Hasegawa K, Akahane M, Igaki H, Izumi N, Ichida T, Uemoto S, Kaneko S, Kawasaki S, Ku Y, et al. Evidence-based clinical practice guidelines for hepatocellular carcinoma: The Japan society of hepatology 2013 update (3rd JSH-HCC guidelines) Hepatol Res. 2015;45:45. doi: 10.1111/hepr.12464. [DOI] [PubMed] [Google Scholar]
- 7.Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du Y, Luo X, Zheng F, Liu R, Zhang H, et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer. 2012;12:111. doi: 10.1186/1471-2407-12-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. miRTarBase update 2014: An information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(D1):D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee YJ, Kim V, Muth DC, Witwer KW. Validated microRNA target databases:an evaluation. Drug Dev Res. 2015;76:389–396. doi: 10.1002/ddr.21278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudo M, Matsui O, Izumi N, Iijima H, Kadoya M, Imai Y, Okusaka T, Miyayama S, Tsuchiya K, Ueshima K, et al. Liver Cancer Study Group of Japan: JSH consensus-based clinical practice guidelines for the management of hepatocellular carcinoma: 2014 update by the Liver Cancer Study Group of Japan. Liver Cancer. 2014;3:458–468. doi: 10.1159/000343875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi: 10.1186/s12935-015-0185-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang H, Li RP, Liang P, Zhou YL, Wang GW. miR-125a inhibits the migration and invasion of liver cancer cells via suppression of the PI3K/AKT/mTOR signaling pathway. Oncol Lett. 2015;10:681–686. doi: 10.3892/ol.2015.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Chen X, Zhang Y, Hu Y, Shen X, Zhu W. Long non-coding RNA TUG1 promotes endometrial cancer development via inhibiting miR-299 and miR-34a-5p. Oncotarget. 2017;8:31386–31394. doi: 10.18632/oncotarget.15607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fateh A, Feizi MAH, Safaralizadeh R, Azarbarzin S. Importance of miR-299-5p in colorectal cancer. Ann Gastroenterol. 2017;30:322–326. doi: 10.20524/aog.2017.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xing Y, Cui D, Wang S, Wang P, Xing X, Li H. Oleuropein represses the radiation resistance of ovarian cancer by inhibiting hypoxia and microRNA-299-targetted heparanase expression. Food Funct. 2017;8:2857–2864. doi: 10.1039/C7FO00552K. [DOI] [PubMed] [Google Scholar]
- 17.Göhring AR, Reuter S, Clement JH, Cheng X, Theobald J, Wölfl S, Mrowka R. Human microRNA-299-3p decreases invasive behavior of cancer cells by downregulation of Oct4 expression and causes apoptosis. PLoS One. 2017;12:e0174912. doi: 10.1371/journal.pone.0174912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HS, Lee KS, Bae HJ, Eun JW, Shen Q, Park SJ, Shin WC, Yang HD, Park M, Park WS, et al. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6:8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li WF, Dai H, Ou Q, Zuo GQ, Liu CA. Overexpression of microRNA-30a-5p inhibits liver cancer cell proliferation and induces apoptosis by targeting MTDH/PTEN/AKT pathway. Tumour Biol. 2016;37:5885–5895. doi: 10.1007/s13277-015-4456-1. [DOI] [PubMed] [Google Scholar]
- 20.Yuan Q, Loya K, Rani B, Möbus S, Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology. 2013;57:299–310. doi: 10.1002/hep.25984. [DOI] [PubMed] [Google Scholar]
- 21.Ren S, Xin Z, Xu Y, Xu J, Wang G. Construction and analysis of circular RNA molecular regulatory networks in liver cancer. Cell Cycle. 2017;6:2204–2211. doi: 10.1080/15384101.2017.1346754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.




