Abstract
Long non-coding RNA Sox2 overlapping transcript (lncRNA Sox2ot) expression has been demonstrated to be upregulated in a number of types of tumor, and may act as an oncogene. The present study aimed to evaluate the clinical role of lncRNA Sox2ot and its association with the epithelial-mesenchymal transition in hepatocellular carcinoma (HCC). Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was performed to determine the expression of lncRNA Sox2ot in 86 cases of HCC tissues and adjacent non-tumor tissues. Kaplan-Meier and log-rank test was used to analyze the association between lncRNA Sox2ot expression and disease-free (DFS) or overall (OS) survival time. In addition, the capacity of HCC cells with lncRNA Sox2ot knockdown for invasion was evaluated via transwell cell invasion assays. RT-qPCR and western blot analyses were also performed to determine the mRNA and protein expression of Twist1, E-cadherin and N-cadherin in the HCC cells. It was indicated that lncRNA Sox2ot expression levels were significantly higher in HCC tissues compared with adjacent non-tumor tissues. Increased expression levels of lncRNA Sox2ot were associated with the tumor size, the tumor number and vein invasion in patients with HCC. An association was observed between lncRNA Sox2ot and the DFS and OS of patients with HCC. Furthermore, it was determined that cell invasion was inhibited following the siRNA knockdown of lncRNA Sox2ot in MHCC97H and SMCC-7721 cells via transwell cell invasion assays. Furthermore, it was demonstrated that knockdown of lncRNA Sox2ot downregulated the mRNA and protein expression of Twist1 and N-cadherin, but upregulated the E-cadherin expression levels in MHCC97H and SMCC-7721 cells. Thus, it was indicated that lncRNA Sox2ot may be a novel predictive biomarker and a potential therapeutic target for HCC.
Keywords: hepatocellular carcinoma, Sox2 overlapping transcript, epithelial-mesenchymal transition, tumor invasion
Introduction
Hepatocellular carcinoma (HCC) ranks as the third leading cause of cancer-associated mortality, and accounts for ~80% of primary liver cancers (1,2). Despite the development of surgical techniques and advances in molecularly targeted drugs, therapeutic approaches for the treatment of HCC provide an unsatisfactory 5-year overall survival (OS) rate due to late detection as a result of the lack of specific symptoms in the early stages of the disease (3). Thus, the investigation of potential predictive biomarkers and therapeutic targets is required to improve the survival rate for patients with HCC.
Long non-coding RNAs (lncRNAs) are ~200 nt to 100 kb long and have emerged as key regulators in the majority of biological processes (4,5). Previous studies have demonstrated that lncRNAs participate in a number of tumor processes and may be suitable as therapeutic targets and biomarkers for predicting the prognosis of patients (6,7). For example, the expression of PCAT-1 was observed to be significantly increased in HCC tissues, and was significantly associated with the OS time of patients with HCC (8). ZFAS1 may function as an oncogene in HCC progression, as it has been demonstrated to bind miR-150 and abrogate its tumor suppressive function, thus promoting the expression of ZEB1 and the matrix metalloproteinases MMP14 and MMP16 (9). CARLo-5 has been identified as an independent risk factor for OS and disease-free survival (DFS) in HCC; it may promote the proliferation and metastasis of HCC, and is a potential novel therapeutic target (10). The decreased expression of the lncRNA GAS5 indicates a relatively poor prognosis, and promotes cell proliferation and invasion in HCC, via the regulation of vimentin (11).
The association between lncRNA Sox2 overlapping transcript (lncRNA Sox2ot; on human chromosome 3q26.33) expression and the epithelial-mesenchymal transition (EMT) in HCC was not previously examined. In the present study, it was determined that the expression level of lncRNA Sox2ot was significantly higher in HCC tissues compared with adjacent non-tumor tissues. Furthermore, it was demonstrated that cell invasion was inhibited following the knockdown of lncRNA Sox2ot in HCC cells. The knockdown of lncRNA Sox2ot in the cells downregulated the expression levels of Twist1 and N-cadherin, but upregulated the E-cadherin expression level. Thus, the results of the study indicated that Sox2ot could be a novel biomarker and a potential therapeutic target for patients with HCC.
Materials and methods
Human tissue samples
A total of 86 HCC tissue samples paired with adjacent normal tissue samples were obtained from patients who underwent surgical resection between November 2009 and March 2014 at the Shandong Provincial Hospital Affiliated to Shandong University (Jinan, China). The fresh tissue samples were immediately frozen in liquid nitrogen and stored at −80°C. The clinical stage was assessed using the 2010 6th edition Tumor Node Metastasis (TNM) system (International Union of Cancer Control/American Joint Committee of Cancer (12). The present study was approved by the Ethical Committee of Shandong Provincial Hospital Affiliated to Shandong University, and written informed consent was obtained from all patients.
Cell lines and culture
The MHCC97H and SMMC-7721 human HCC cell lines and the LO2 normal liver cell line were purchased from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) and supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin and streptomycin in a humidified atmosphere containing 5% CO2 at 37°C.
Cell transfection and RNAi
The cells were transfected with siRNA against lncRNA Sox2ot purchased from Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The siRNA sequences were as follows: siR-Sox2ot, 5′-CAAAAUAGGUCAUAGCAAATT-3′; si-negative control, 5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were seeded into 6-well plate and cultured for 48 h, then transfected with Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) as per the manufacturer's protocol. At 48 h post-transfection, the efficiency of transfection was assessed using reverse transcription-quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR
Total RNA was extracted from tissues and cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RNA were reverse transcribed into cDNA using a reverse transcription reagent kit (Takara Biotechnology Co., Ltd., Dalian, China). qPCR was performed using the SYBR® Green PCR kit (Takara Biotechnology Co., Ltd.). The PCR reaction conditions were as follows: Preliminary denaturation at 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 20 sec. Primer sequences were as follows: lncRNA Sox2ot forward, 5′-GCTCGTGGCTTAGGAGATTG-3′ and reverse 5′-CTGGCAAAGCATGAGGAACT-3′; GAPDH forward, 5′-GTCAACGGATTTGGTCTGTATT-3′ and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′. The relative quantification (2−ΔΔCt) method was used for calculating fold change (13).
Transwell invasion assay
Cell invasion was detected using Transwell chambers (8-µm pore size; Corning, Inc., Corning, NY, USA) with Matrigel (BD Biosciences, San Jose, CA, USA). At 48 h post-cell transfection, 1×105 cells MHCC97H or SMMC-7721 cells were added into the upper chamber in 400 µl FBS-free culture medium. A total of 500 µl culture medium containing 10% FBS was added to the lower chamber. Cells were cultured for 48 h in a humidified atmosphere containing 5% CO2 at 37°C. The invasive cells on the lower chamber were fixed with 100% ethanol for 15 min at room temperature, then stained with 0.1% crystal violet for 15 min at room temperature. Cells were counted under a light microscope in 5 random fields (magnification, ×200).
Western blot analysis
The total protein was extracted from cells using radioimmunoprecipitation assay lysis buffer (Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). Protein concentration was measured using a bicinchoninic acid protein assay kit (EMD Millipore, Billerica, MA, USA). Equal quantities of protein (40 µg/lane) were separated via 10% SDS-PAGE and then transferred onto a polyvinylidene fluoride membrane (EMD Millipore). The membranes were blocked using 5% non-fat milk at room temperature for 1 h. The blotted membranes were incubated with antibodies against Twist1 (cat. no. sc-6269; dilution, 1:1,000), E-cadherin (cat. no. sc-21791; dilution, 1:1,000), N-cadherin (cat. no. sc-31031; dilution, 1:500) and GAPDH (cat. no. sc-69778; dilution, 1:2,000; all from Santa Cruz Biotechnology, Inc., Dallas, TX, USA) antibodies and were incubated at 4°C overnight. The membrane was incubated with a secondary goat anti-rabbit horseradish peroxidase-conjugated antibody (cat no. CW0102S; dilution, 1:2,000; Jiangsu Kangwei Century Biotechnology Co., Ltd., Beijing, China) for 1 h at room temperature. The protein bands were detected using an enhanced chemiluminescence system (Pierce; Thermo Fisher Scientific, Inc.).
Statistical analysis
All data were analyzed using SPSS 18.0 (SPSS, Inc., Chicago, IL, USA). All continuous variable values were represented as the mean ± standard deviation from ≥3 independent experiments. Differences between two groups were analyzed using Student's t-test, and data from multiple groups were analyzed with a one-way analysis of variance (ANOVA). The Student-Newman-Keuls test was used as a post-hoc test following ANOVA. The associations between lncRNA Sox2ot expression and clinicopathological factors were analyzed by the χ2 test. The Kaplan-Meier method and a log-rank test were used to analyze the association between lncRNA Sox2ot expression and DFS or OS time. P<0.05 was considered to indicate a statistically significant difference.
Results
lncRNA Sox2ot is upregulated in HCC tissues and associated with a poor prognosis for patients with HCC
To investigate whether lncRNA Sox2ot affected hepatocarcinogenesis, the expression level of lncRNA Sox2ot was measured in HCC tissues and adjacent normal tissues using RT-qPCR. The results demonstrated that lncRNA Sox2ot was upregulated in HCC tissues compared with the adjacent normal tissues (Fig. 1A; P<0.05). HCC tissue samples were divided into higher- and lower-expression groups based on the mean expression level for all the samples. The patients with the higher expression of lncRNA Sox2ot had a significantly increased tumor size (P=0.007), tumor number (P=0.036) and vein invasion rate (P=0.011; Table I; Fig. 1B-D). There was no significant association between the expression of lncRNA Sox2ot and other clinicopathological features, including age, sex, hepatitis B infection status, histological grade and serum α-fetoprotein level (Table I). Furthermore, it was demonstrated that higher lncRNA Sox2ot expression was predictive for a relatively poor DFS and OS time (Fig. 1E and F; log-rank, 8.567, P<0.05 and log-rank, 8.339, P<0.05, respectively) compared with lower lncRNA Sox2ot expression.
Figure 1.
lncRNA Sox2ot was upregulated in HCC tissues and associated with the poor prognosis of patients with HCC. (A) lncRNA Sox2ot expression was measured via reverse transcription-quantitative polymerase chain reaction in HCC tissues and adjacent normal tissues (n=86). (B) Association between lncRNA Sox2ot expression and tumor size (≥5 cm) or tumor size (<5 cm). (C) Association between lncRNA Sox2ot expression and tumor number. (D) Association between lncRNA Sox2ot expression and vein invasion. Kaplan-Meier survival analysis and the log-rank test were performed to analyze the (E) DFS and (F) OS time of patients with HCC in higher and lower lncRNA Sox2ot expression groups. *P<0.05. lncRNA Sox2ot, long non-coding RNA Sox2 overlapping transcript; HCC, hepatocellular carcinoma; DFS, disease-free survival; OS, overall survival.
Table I.
The association between lncRNA Sox2ot expression and clinicopathological features.
| lncRNA Sox2ot expression, n | |||||
|---|---|---|---|---|---|
| Clinicopathological features | All | Low | High | χ2 test | P-value |
| Total | 86 | 41 | 45 | ||
| Sex | 0.255a | 0.059 | |||
| Female | 23 | 12 | 11 | ||
| Male | 63 | 29 | 34 | ||
| Age, years | 0.100a | 0.752 | |||
| ≤60 | 56 | 26 | 30 | ||
| >60 | 30 | 15 | 15 | ||
| Tumor size, cm | 7.274a | 0.007b | |||
| <5 | 50 | 30 | 20 | ||
| ≥5 | 36 | 11 | 25 | ||
| Hepatitis B infection | 0.081a | 0.776 | |||
| Positive | 60 | 28 | 32 | ||
| Negative | 26 | 13 | 13 | ||
| Histological grade | 1.325a | 0.250 | |||
| Well | 32 | 20 | 12 | ||
| Moderate | 24 | 12 | 12 | ||
| Low | 30 | 9 | 21 | ||
| Tumor number | 2.628a | 0.036b | |||
| Single | 49 | 26 | 23 | ||
| Multiple | 37 | 15 | 22 | ||
| α-fetoprotein, ng/ml | 2.098a | 0.148 | |||
| <400 | 31 | 18 | 13 | ||
| ≥400 | 55 | 23 | 32 | ||
| Vein invasion | 6.539a | 0.011b | |||
| Negative | 34 | 22 | 12 | ||
| Positive | 52 | 19 | 33 | ||
| AJCC stage | 0.078a | 0.780 | |||
| I–II | 49 | 24 | 25 | ||
| III–IV | 37 | 17 | 20 | ||
χ2 test.
P<0.05. JACC, American Joint Committee on Cancer.
lncRNA Sox2ot promotes cell invasion in MHCC97H and SMCC-7721 cells
It was determined that lncRNA Sox2ot expression was higher in MHCC97H and SMCC-7721 HCC cells compared with LO2 non-cancer cells (Fig. 2A). lncRNA Sox2ot was knocked down with siRNA in MHCC97H and SMCC-7721 cells. It was verified that the expression of lncRNA Sox2ot was reduced by the transfection with the lncRNA Sox2ot siRNA compared with a control oligonucleotide (Fig. 2B and C). Transwell cell invasion assays demonstrated that the cell invasion ability was inhibited following the knockdown of lncRNA Sox2ot in MHCC97H and SMCC-7721 cells (Fig. 2D-F). Thus, the results indicated that lncRNA Sox2ot promoted invasion by HCC cells.
Figure 2.
Knockdown of lncRNA Sox2ot inhibited cell invasion ability in hepatocellular carcinoma. (A) lncRNA Sox2ot expression level, as determined with RT-qPCR, for LO2, SMCC-7721 and MHCC97H cells. lncRNA Sox2ot expression was measured via RT-qPCR following lncRNA Sox2ot silencing in (B) MHCC97H or (C) SMCC-7721 cells, compared with the si-NC group. (D) Cell invasion ability was analyzed via transwell cell invasion assays following Sox2ot silencing by si-Sox2ot in MHCC97H or SMCC-7721 cells, compared with the si-NC group. (E) Quantification of MHCC97H cell invasion. (F) Quantification of SMCC-7721 cell invasion. The mean values and standard deviation were calculated from triplicates of the representative experiment. *P<0.05. lncRNA Sox2ot, long non-coding RNA Sox2 overlapping transcript; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; si-NC, small interfering RNA negative control.
lncRNA Sox2ot promotes cell EMT in MHCC97H and SMCC-7721 cells
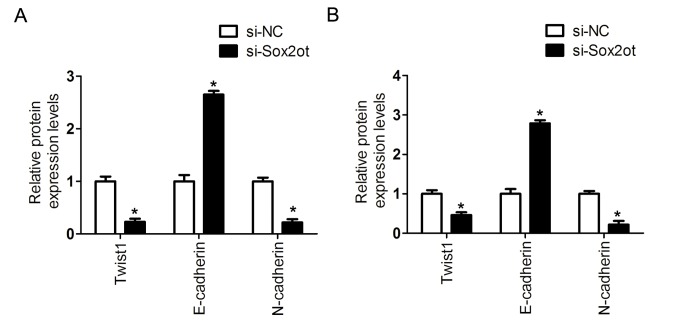
Furthermore, to evaluate the association between lncRNA Sox2ot expression and the EMT process, the expression of EMT-associated factors was assessed. It was identified that the mRNA level of E-cadherin was upregulated, and those of Twist1 and N-cadherin downregulated following the knockdown of lncRNA Sox2ot in MHCC97H or SMCC-7721 cells (Fig. 3A and B). In addition, the protein expression was detected following the knockdown of lncRNA Sox2ot in MHCC97H or SMCC-7721 cells. It was demonstrated that the protein level of E-cadherin was upregulated, whereas Twist1 and N-cadherin were downregulated (Figs. 3C, D and 4). Thus, these results indicate that lncRNA Sox2ot may promote EMT in HCC cells.
Figure 3.
Knockdown of lncRNA Sox2ot suppressed the epithelial-mesenchymal transition in HCC cells. The mRNA expression levels of Twist1, E-cadherin and N-cadherin were analyzed via reverse transcription-quantitative polymerase chain reaction following lncRNA Sox2ot silencing in (A) MHCC97H or (B) SMCC-7721 cells, compared with the si-NC group. Data are presented as the means ± standard deviation from three independent experiments. The protein expression levels of Twist1, E-cadherin and N-cadherin were analyzed via western blot analysis following lncRNA Sox2ot silencing in (C) MHCC97H and (D) SMCC-7721 cells, compared with the si-NC group. GAPDH was used as an internal control. *P<0.05. lncRNA Sox2ot, long non-coding RNA Sox2 overlapping transcript; HCC, hepatocellular carcinoma; si-NC, small interfering RNA negative control.
Figure 4.
The protein expression of Twist1, E-cadherin and N-cadherin were shown. (A) The protein expression levels of Twist1, E-cadherin and N-cadherin were analyzed via western blot analysis following lncRNA Sox2ot silencing in=MHCC97H and SMCC-7721 cells, compared with the si-NC group. GAPDH was used as an internal control. Presenting the mean ± SD of three quantified experiments, *P<0.05. NC, negative control.
Discussion
lncRNAs participate in the progression of a range of tumor types and may be suitable as biomarkers for predicting prognosis (14). For example, HOTAIRM1 may function as a tumor suppressor, and is a potential biomarker for the diagnosis of colorectal cancer (15). The high expression of lncRNA HULC is a predictor of a relatively poor prognosis and promotes cell proliferation in glioma (16). The high expression of lncRNA Sox2ot is associated with the aggressive progression of gastric cancer, and poor OS and DFS times (17). The increased expression of lncRNA Sox2ot has been demonstrated to promote cell proliferation and motility in colorectal cancer via promoting Cyclin B1 and CDC25C expression (18).
In the present study, it was determined that lncRNA Sox2ot was upregulated in HCC tissues, and that higher lncRNA Sox2ot expression was associated with the tumor size, tumor number and vein invasion. The patients with a higher level of lncRNA Sox2ot expression had a reduced DFS and OS time. A previous meta-analysis of the prognostic value of various abnormally expressed lncRNAs in HCC demonstrated that the transcription level of various lncRNAs was significantly associated with the tumor size, microvascular invasion and portal vein tumor thrombus, and may serve in the prognostic evaluation of patients with HCC (6). In another previous study, lncRNA Sox2ot expression was associated with T stage, distant metastasis, differentiation and a poorer OS and DFS time in breast cancer (17). Tang et al (19) reported that lncRNA Sox2ot was overexpressed in breast cancer tissues, and that a higher expression of lncRNA Sox2ot increased the risk of breast cancer for Chinese women. Shi et al (20) determined that the high expression of lncRNA Sox2ot was associated with the histological grade, the Tumor-Node-Metastasis stage, vein invasion and a relatively poor 5-year OS time in HCC, which was consistent with the results from the present study. Thus, lncRNA Sox2ot may be a prognostic biomarker for patients with HCC.
Various lncRNAs were previously determined to serve crucial roles in cancer invasion and metastasis via regulating critical biological events, particularly the EMT. Increased expression of lncRNA ZFAS1 is associated with EMT in gastric cancer progression (21). lncRNA HULC may promote the EMT, tumorigenesis and metastasis of HCC via the mediation of the miR-200a-3p/ZEB1 signaling pathway (22). lncRNA CPS1-IT1 may inhibit HCC invasion and metastasis via regulating HIF-1α activity and suppressing EMT (23). The upregulation of H19 may indicate a poorer prognosis in gallbladder carcinoma and promote EMT via upregulating Twist1 expression (24). Overexpression of prostate lncRNA-1 significantly increases cell proliferation, migration and invasion via enhancing EMT signaling (25). In the present study, it was determined that the knockdown of lncRNA Sox2ot inhibited the cell invasion ability and the EMT process via upregulating E-cadherin expression and downregulating Twist1 and N-cadherin expression. In the EMT process, activating Twist upregulates N-cadherin expression and downregulates E-cadherin expression, which are considered hallmarks of EMT (26). Thus, lncRNA Sox2ot may have promoted cell invasion and the EMT process via promoting Twist1 and N-cadherin expression. As the underlying molecular regulatory mechanisms between lncRNA Sox2ot and Twist or N-cadherin are unknown, further investigation is required.
In conclusion, the results of the present study demonstrated that lncRNA Sox2ot expression was increased in HCC tissue, and higher lncRNA Sox2ot expression was predictive of a poorer prognosis in patients with HCC. Furthermore, it was determined that the knockdown of lncRNA Sox2ot inhibited cell invasion and the EMT; thus, lncRNA Sox2ot may be a novel biomarker of HCC prognosis and a potential target for HCC treatment.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used in the present study are available from the corresponding author on reasonable request.
Author's contributions
JS and XW conceived and designed the study. JS, XW and LX performed the experiments. JS analyzed and interpreted the data. LX wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethical Committee of Shandong Provincial Hospital Affiliated to Shandong University.
Consent for publication
Written informed consent was obtained from all patients in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Gramantieri L, Fornari F, Callegari E, Sabbioni S, Lanza G, Croce CM, Bolondi L, Negrini M. MicroRNA involvement in hepatocellular carcinoma. J Cell Mol Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rich N, Singal AG. Hepatocellular carcinoma tumour markers: Current role and expectations. Best Pract Res Clin Gastroenterol. 2014;28:843–853. doi: 10.1016/j.bpg.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 4.Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma. Cancer Lett. 2015;360:119–124. doi: 10.1016/j.canlet.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Chen J, Zhang K, Feng B, Wang R, Chen L. Progress and prospects of long noncoding RNAs (lncRNAs) in hepatocellular carcinoma. Cell Physiol Biochem. 2015;36:423–434. doi: 10.1159/000430109. [DOI] [PubMed] [Google Scholar]
- 6.Qu Z, Yuan CH, Yin CQ, Guan Q, Chen H, Wang FB. Meta-analysis of the prognostic value of abnormally expressed lncRNAs in hepatocellular carcinoma. Onco Targets Ther. 2016;9:5143–5152. doi: 10.2147/OTT.S108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L, Peng F, Tao Y, Fan X, Li N. Roles of long noncoding RNAs in hepatocellular carcinoma. Virus Res. 2016;223:131–139. doi: 10.1016/j.virusres.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Yan TH, Yang H, Jiang JH, Lu SW, Peng CX, Que HX, Lu WL, Mao JF. Prognostic significance of long non-coding RNA PCAT-1 expression in human hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:4126–4131. [PMC free article] [PubMed] [Google Scholar]
- 9.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, et al. Amplification of long noncoding RNA ZFAS1 promotes metastasis in hepatocellular carcinoma. Cancer Res. 2015;75:3181–3191. doi: 10.1158/0008-5472.CAN-14-3721. [DOI] [PubMed] [Google Scholar]
- 10.Wang F, Xie C, Zhao W, Deng Z, Yang H, Fang Q. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med. 2017;17:33–43. doi: 10.1007/s10238-015-0395-9. [DOI] [PubMed] [Google Scholar]
- 11.Chang L, Li C, Lan T, Wu L, Yuan Y, Liu Q, Liu Z. Decreased expression of long non-coding RNA GAS5 indicates a poor prognosis and promotes cell proliferation and invasion in hepatocellular carcinoma by regulating vimentin. Mol Med Rep. 2016;13:1541–1550. doi: 10.3892/mmr.2015.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bosman FT, Carneiro F, Hruban RH, Theise ND. Fourth edition. Lyon: World Health Organization; 2010. WHO classification of tumours of the digestive system; pp. 205–227. [Google Scholar]
- 13.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 14.He Y, Meng XM, Huang C, Wu BM, Zhang L, Lv XW, Li J. Long noncoding RNAs: Novel insights into hepatocelluar carcinoma. Cancer Lett. 2014;344:20–27. doi: 10.1016/j.canlet.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 15.Wan L, Kong J, Tang J, Wu Y, Xu E, Lai M, Zhang H. HOTAIRM1 as a potential biomarker for diagnosis of colorectal cancer functions the role in the tumour suppressor. J Cell Mol Med. 2016;20:2036–2044. doi: 10.1111/jcmm.12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan H, Tian R, Zhang M, Wu J, Ding M, He J. High expression of long noncoding RNA HULC is a poor predictor of prognosis and regulates cell proliferation in glioma. Onco Targets Ther. 2016;10:113–120. doi: 10.2147/OTT.S124614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou JH, Li CY, Bao J, Zheng GQ. High expression of long noncoding RNA Sox2ot is associated with the aggressive progression and poor outcome of gastric cancer. Eur Rev Med Pharmacol Sci. 2016;20:4482–4486. [PubMed] [Google Scholar]
- 18.Liu S, Xu B, Yan D. Enhanced expression of long non-coding RNA Sox2ot promoted cell proliferation and motility in colorectal cancer. Minerva Med. 2016;107:279–286. [PubMed] [Google Scholar]
- 19.Tang X, Gao Y, Yu L, Lu Y, Zhou G, Cheng L, Sun K, Zhu B, Xu M, Liu J. Correlations between lncRNA-SOX2OT polymorphism and susceptibility to breast cancer in a Chinese population. Biomark Med. 2017;11:277–284. doi: 10.2217/bmm-2016-0238. [DOI] [PubMed] [Google Scholar]
- 20.Shi XM, Teng F. Up-regulation of long non-coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int J Clin Exp Pathol. 2015;8:4008–4014. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou H, Wang F, Chen H, Tan Q, Qiu S, Chen S, Jing W, Yu M, Liang C, Ye S, Tu J. Increased expression of long-noncoding RNA ZFAS1 is associated with epithelial-mesenchymal transition of gastric cancer. Aging (Albany NY) 2016;8:2023–2038. doi: 10.18632/aging.101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, Wang CY, Zhang HM, Zhang RX, Zhang JJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget. 2016;7:42431–42446. doi: 10.18632/oncotarget.9883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang TH, Yu CC, Lin YS, Chen TC, Yeh CT, Liang KH, Shieh TM, Chen CY, Hsueh C. Long noncoding RNA CPS1-IT1 suppresses the metastasis of hepatocellular carcinoma by regulating HIF-1α activity and inhibiting epithelial-mesenchymal transition. Oncotarget. 2016;7:43588–43603. doi: 10.18632/oncotarget.9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SH, Wu XC, Zhang MD, Weng MZ, Zhou D, Quan ZW. Upregulation of H19 indicates a poor prognosis in gallbladder carcinoma and promotes epithelial-mesenchymal transition. Am J Cancer Res. 2015;6:15–26. [PMC free article] [PubMed] [Google Scholar]
- 25.Dong L, Ni J, Hu W, Yu C, Li H. Upregulation of long non-coding RNA PlncRNA-1 promotes metastasis and induces epithelial-mesenchymal transition in hepatocellular carcinoma. Cell Physiol Biochem. 2016;38:836–846. doi: 10.1159/000443038. [DOI] [PubMed] [Google Scholar]
- 26.Khan MA, Chen HC, Zhang D, Fu J. Twist: A molecular target in cancer therapeutics. Tumour Biol. 2013;34:2497–2506. doi: 10.1007/s13277-013-1002-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the present study are available from the corresponding author on reasonable request.