Abstract
Cervical cancer remains one of the most frequent gynecological malignancies among females around the world. Therefore, fully understanding the molecular mechanisms underlying the progression of cervical cancer may be critical for the development of effective therapeutic strategies against cervical cancer. The object was to evaluate the potential effect of miR-211 and verify its influence on the function of secreted protein acidic and rich in cysteine (SPARC) in cervical cancer. It was demonstrated that miR-211 was downregulated in cervical cancer cell lines (HeLa and C33A) and cervical cancer specimens, while SPARC expression level was higher in tumor tissues. We also revealed miR-211 upregulated expression could inhibit cells proliferation, migration and invasion in vivo. SPARC was confirmed as a direct and functional target of miR-211 and the inverse relationship between them was also observed. The results of the present study suggest that miR-211 reduced cancer growth, migration and invasion, and suppresses the SPARC expression in cervical cancer. This newly identified miR-211 may provide further insight into the progression and offers a promising target for cervical cancer therapy.
Keywords: miR-211, SPARC, proliferation, migration, invasion, cervical cancer
Introduction
Cervical cancer remains one of the most frequent gynecological malignancies in females around the world, which leads to the highest morbidity and mortality in young women, particularly in developing countries (1). Approximately 98,900 new cases and 30,500 cancer deaths due to cervical cancer are estimated to occur in China each year (2). Cervical cancers frequently infiltrate into the neighboring tissue and metastasize to other organs resulting in a poor prognosis. Although great advanced protocols including operation, chemotherapy and postoperative radiotherapy have been established in the therapy of cervical cancer, the long-term outcomes of clinical therapy remain unsatisfactory and most patients succumb to metastasis (3). Accumulating research on the molecular mechanism of progression and carcinogenesis has produced a number of new potential molecular biomarkers which are likely to be the targets of cervical cancer (4), but there are few biomarkers used in predicting of the disease. Therefore, it is critical to identify specific types of molecular biomarkers which may accurately predict disease outcome before the standard treatment. The molecular biomarkers for the early identification of cervical cancer patients, may contribute to understanding cervical carcinogenesis and ascertaining diagnostic and therapeutic strategies, assessing prognosis, and monitoring response to therapy.
The group of highly conserved, small non-coding RNAs (18–24 nucleotides) (5) the microRNAs (miRNAs) have critical roles in modulating in length the physiological process or pathogenesis by regulating the translation or degradation of target messenger RNA (6). Accumulating evidence has indicated that the dysfunction of miRNAs occur in a wide range of human tumors and can regulate cancer progressions that are involved in cell differentiation, proliferation, metastasis, and apoptosis (7–9). In addition, the dysfunctions of miRNAs act as either tumor suppressors or promoters (10), and studies have suggested miR-211 could act as boh a suppressor and a promoter, and it can be considered quite contradictory. miR-211 was detected at a significantly low level in various types of cancers, such as hepatocellular carcinoma, melanoma, glioma, epithelial ovarian cancer (11–14), and on the contrary, miR-211 was upregulated in other tumors, for example human non-small cell lung cancer, and colorectal cancer (15,16). Previous studies have indicated that miR-211 inhibited cervical cancer cell invasion and epithelial-to-mesenchymal transition (EMT) through targeting MUC4 (17). However, the miR-211 expression remains unclear in cervical cancer tissues. So the regulatory mechanism of miR-211 on cervical cancer needs further investigation.
Secreted protein acidic and rich in cysteine (SPARC), as a member of the matricellular family of secreted proteins, can modulate cell matrix interactions and affect cell progression, such as cell adhesion, tissue repair and remodeling (18,19). SPARC is taken to be an oncogene on account of its high expression in a great range of tumors including gastric carcinomas, prostate cancer, glioma, and cervical cancer (20–23). A previous study has demonstrated that overexpression of SPARC has significant connection with negative prognostic clinicopathological characteristics and might function as an important point in EMT of cervical cancer (23).
miR-211 was frequently downregulated and the miR-211 overexpression remarkably suppressed cell proliferation, invasion and migrationx in vivo as shown in our study. Furthermore, we confirmed SPARC is a direct and functional target of miR-211. The newly identified miR-211/SPARC partially elucidates the cervical cancer molecular mechanism of proliferation, invasion and migration, and represents a therapeutic point for cervical cancer treatment.
Materials and methods
Tissue samples
Cervical cancer and the corresponding normal tissues from 52 patients who underwent surgery were obtained at Yuhuangding Hospital (Yantai, China) from 2014 to 2016. Histopathological diagnoses were based on the WHO classification and clinical stages classification followed the International Federation of Gynecology and Obstetrics criteria (FIGO). The characteristics of patients with cervical cancer are described in Table I. The patients signed informed consent and the study was approved by the Ethics Committee of Yuhuangding Hospital (Yantai, China).
Table I.
Clinicopathological variables and miR-211 expression in 52 cervical cancer patients.
| miR-211 expression | ||||
|---|---|---|---|---|
| Characteristics | Cases (n=52) | Low (%) | High (%) | P-value |
| Age (years) | 0.7232 | |||
| ≤40 | 16 | 12 (75.0) | 4 (25.0) | |
| >40 | 36 | 28 (77.8) | 8 (22.2) | |
| Tumor size (cm) | 0.4989 | |||
| <3 | 26 | 18 (69.2) | 8 (30.8) | |
| ≥3 | 26 | 16 (61.5) | 10 (38.5) | |
| Local relapse | 0.0264a | |||
| Yes | 10 | 5 (50.0) | 5 (50.0) | |
| No | 42 | 39 (92.9) | 3 (7.1) | |
| Distant metastasis | 0.0253a | |||
| Yes | 22 | 12 (54.5) | 10 (45.5) | |
| No | 30 | 25 (83.3) | 5 (16.7) | |
| FIGO stage | 0.0395a | |||
| IB | 28 | 16 (57.1) | 12 (42.9) | |
| IIA | 24 | 16 (66.7) | 8 (33.3) | |
| Differentiation | 0.3158 | |||
| Well | 14 | 8 (57.1) | 6 (42.9) | |
| Moderate/poor | 38 | 26 (68.4) | 12 (31.6) | |
P<0.05.
Cell culture
Primary normal cervical squamous cells (NCSC) and two human cervical cancer cell lines (HeLa and C33A) were obtained from ATCC (Manassas, VA, USA). The cells were cultured in RPMI-1640 medium supplemented with 10%, fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Cell transfection
The cells were transfected with miR-211 mimics/inhibitor as well as the corresponding control using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.), respectively. Cells were used for cell proliferation, migration and invasion assays after transfection. All transfection was conducted in three times. miR-211 mimic 5′-UUCCCUUUGUCAUCCUUCGCCU-3′, miR-211 inhibitor 5′-AGGCGAAGGAUGACAAAGGGAA-3′, and a negative control oligonucleotide were designed and synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). In order to construct SPARC overexpressing plasmid, the gene without 3′-UTR was amplified from cDNA of human normal cervical tissues by polymerase chain reaction (PCR).
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA of cervical cells and tissues were extracted using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers instructions. Then, 5 ng of total RNA was reverse transcribed using a TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA), and the expression levels of mature forms of miR-211 was determined using an miRNA-specific TaqMan MiRNA Assay Kit. To determine the mRNA levels of SPARC, total RNA (500 ng) was reverse transcribed using PrimeScript® RT Master Mix (Takara Biotechnology Co., Ltd., Dalian, China). reverse transcriptase according to the manufacturers instructions, and RT-qPCR analyses were performed using SYBR-Premix Ex Taq™ (Takara Biotechnology Co., Ltd., Dalian, China). U6 and GAPDH acted as the internal control for the expression of miR-211 and SPARC. The primers of SPARC and GAPDH were synthesized by Invitrogen; Thermo Fisher Scientific, Inc. The experiments were performed in triplicate. The transcription primer and PCR primer of miR-211 and U6 were purchased from Guangzhou RiboBio Co., Ltd. The sequences of the primers were as follows: miR-211 F, 5′-TCGGCAGGTCCCTTTGTCATCC-3′ and R, 5′-TGCAGGTCAACTGGTGTCGT-3′; U6 F, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and R, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; SPARC F, 5′-AGCACCCCATTGACGGGTA-3′ and R, 5′-GGTCACAGGTCTCGAAAAAGC-3′, GAPDH F, 5′-CTGGGCTACACTGAGCACC-3′ and R, 5′-AAGGGTCGTTGAGGGCAATG-3′.
Western blot analysis
Proteins were isolated from cervical cells with different transfection using RIPA lysis buffer (Thermo Fisher Scientific, Inc.) phenylmethylsulfonyl fluoride. The measurement of protein concentration used bicinchoninic acid assay kit (Beyotime Institute of Biotechnology, Haimen, China). The protein was transferred onto polyvinylidene fluoride membrane (Bio-Rad Laboratories, Inc., Hercules, CA, USA). Then, the membranes were blocked by 5% bovine serum albumin and incubated with specific primary antibody rabbit polyclonal anti-SPARC antibody (dilution 1:1,000; cat. no. ab209556; Abcam, Cambridge, MA, USA). After that, the membrane was incubated in the secondary antibody goat polyclonal anti-rabbit IgG H&L secondary antibody (dilution 1:2,000; cat. no. ab150077; Abcam). The bands were subjected to quantification using the ImageJ imaging processing program (National Institutes of Health, Bethesda, MD, USA).
CCK-8 assay
Cell Counting kit-8 (CCK-8) (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) was performed to detect cell proliferation. Cervical cells transfected with miR-211 mimics/inhibitor were seeded into 96-well plates. Then each plate was added with 10 µl of CCK-8 reagent. The absorbance of each well at 0, 24, 48, 72 and 96 h was detected at 450 nm. All experiments were performed three times.
Transwell assay
The Transwell assay with or without Matrigel (Clontech Laboratories, Inc., Mountainview, CA, USA) was chosen to measure the migration and invasion. Cervical cells (5×105) were seeded on the top 24-well Transwell chamber. RPMI-1640 medium with 20% FBS was added to the lowerchamber. The cells on the low chambers were removed using cotton swab. Then the cells on the top chamber were fixed with methanol, stained with 0.05% crystal violet. Finally, the migrated or invaded cells were counted under an inverted microscope (IX31; Olympus Corporation, Tokyo, Japan) and images were captured at ×200 magnification.
Luciferase assay
The bioinformatics analysis software TargerScan (http://www.targetscan.org/) and miRanda (http://www.microrna.org/microrna/home.do) were chosen for predicting the targets of miR-211. The 3′-UTRs of SPARC were amplified with PCR from genomic DNA. The wild-type (WT) and mutant type (MT) 3′-UTR of SPARC were cloned into the pMIR-REPORT luciferase vector (Ambion; Thermo Fisher Scientific, Inc.) and verified by sequencing. For the luciferase assay, the cells were co-transfected with miR-211 mimics and WT or MT 3′-UTR of SPARC luciferase reporter plasmid. Then we used dual-luciferase reporter assay system (Promega Corporation, Madison, WI, USA) for measuring the reporter activities. Renilla luciferase activity was used as normalization.
Statistical analysis
The results are presented as the mean ± standard deviation (SD). Statistical analysis was conducted using the SPSS 19.0 statistical software (IBM Corp., Armonk, NY, USA). Each experiment was repeated at least three times. Student's t-test or Tukey-Kramer post hoc test after one-way analysis of variance (ANOVA) in SPSS were used to analyze the differences between the groups. Correlation between mRNA and miRNA were estimated using the Spearman's correlation method. P<0.05 was considered to be a significant difference.
Results
miR-211 expression is downregulated and inversely correlates with SPARC expression
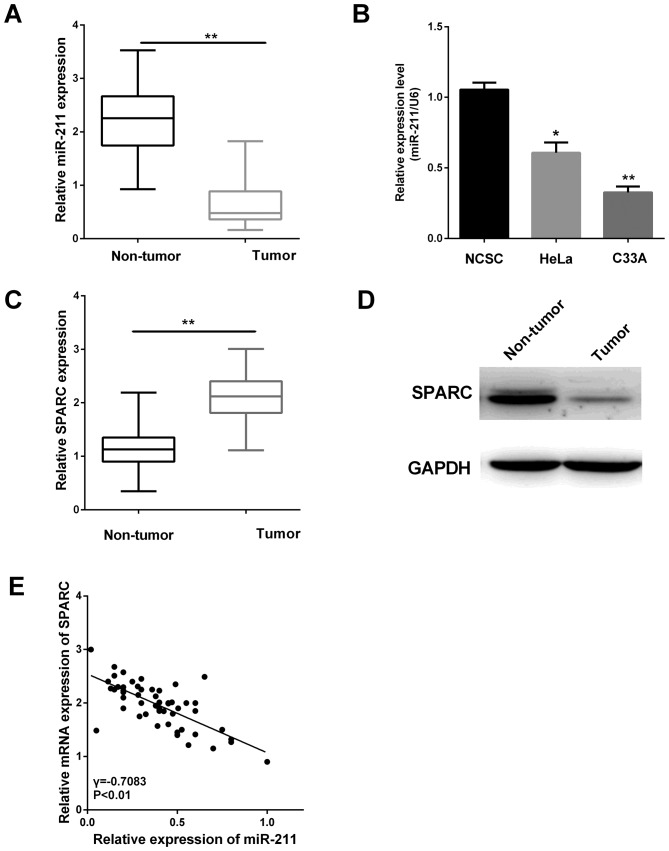
To investigate whether the miR-211 expression was altered in cervical tissues, RT-qPCR was performed in 52 pairs of cervical tissues. The result suggested the miR-211 level was lower in cervical tissues (Fig. 1A, P<0.01). We further measured miR-211 expression in cervical cancer cell lines (HeLa and C33A) compared with the normal NCSC cells. The results showed the miR-211 expression in HeLa and C33A were significantly reduced relative to that in the normal NCSC cells (Fig. 1B, P<0.05, P<0.01).
Figure 1.
miR-211 expression is downregulated and inversely connected with SPARC. (A) miR-211 expression in 52 pairs of samples and their corresponding adjacent normal tissues. The miR-211 expression was detected by RT-qPCR. (B) miR-211 levels in two cancer cell lines and NCSC. (C) Relative mRNA expression of SPARC in cervical cancer tissues. (D) Relative protein expression of SPARC in cervical cancer tissues. (E) Spearman's correlation analysis of miR-211 and SPARC expression in cervical cancer tissues. **P<0.01; *P<0.05; SPARC, secreted protein acidic and rich in cysteine; NCSC, primary normal cervical squamous cells; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Furthermore, the SPARC expression levels in 52 pairs of cervical tissues were also assessed by RT-qPCR and western blot analysis. The relative expression of SPARC was higher in cancer tissues as shown in Fig. 1C and D (P<0.01). Then, we found there was an inverse correlation between miR-211 expression and the SPARC level in these clinical specimens (Fig. 1E). This correlation may play an essential part in the cervical cancer progression.
miR-211 inhibits cervical cell proliferation, migration and invasion
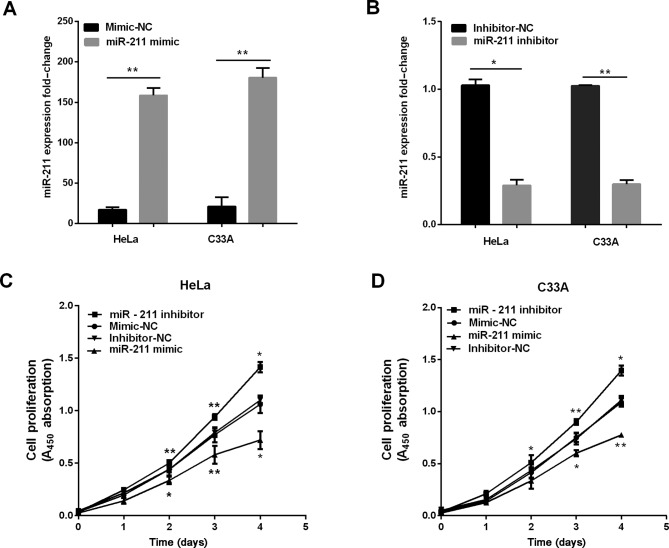
We established stable cells to investigate the effect of miR-211 in the progression of cervical cancer in both HeLa and C33A cells, and the overexpression of miR-211 was detected using RT-qPCR. As shown in Fig. 2A and B, the miR-211 expression was higher when cells were transfected with miR-211 mimics, and the expression of miR-211 was lower when transfected with miR-211 inhibitor. Subsequently, CCK-8 was used to measure the potential of miR-211 in cervical cell proliferation. The results demonstrated overexpression of miR-211 inhibited cervical cell proliferation, and knockdown of miR-211 dramatically promoted proliferative abilities as determined by CCK-8 assay (Fig. 2D).
Figure 2.
Overexpression of miR-211 induces growth inhibition in cervical cancer. (A and B) miR-211 was re-expressed in HeLa and C33A cell lines and miR-211 levels were determined by RT-qPCR after transfection with miR-211 mimic/inhibitor and NC. (C and D) CCK-8 assay results showed that miR-211 suppressed the viability of two cells lines after transfection of miR-211 mimic/inhibitor/NC. **P<0.01; *P<0.05; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCK-8, Cell Counting kit-8.
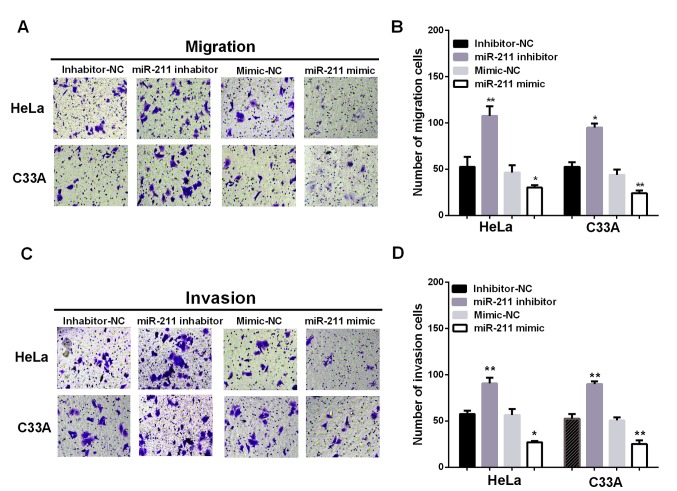
Additionally, to investigate the effect of miR-211 in metastasis, Transwell chambers were conducted to test the migration and invasion ability of cervical cells with different transfection. Ectopic expression of miR-211 significantly suppressed HeLa and C33A cell migration, whereas inhibiting miR-211 expression promoted HeLa and C33A cell migration (Fig. 3A and B; P<0.05). In the cell invasion assay, overexpression of miR-211 dramatically suppressed HeLa and C33A cell invasion, whereas inhibition of miR-211 expression significantly promoted HeLa and C33A cell invasion (Fig. 3C and D; P<0.05). These results suggested miR-211 can inhibit the migratory and invasive ability of cervical cancer cells.
Figure 3.
miR-211 inhibits cervical cell migration and invasion. (A and B) Ectopic expression of miR-211 significantly suppressed HeLa and C33A cell migration, whereas inhibiting miR-211 expression promoted HeLa and C33A cell migration. (C and D) Overexpression of miR-211 dramatically inhibited HeLa and C33A cell invasion, whereas inhibition of miR-211 expression significantly promoted HeLa and C33A cell invasion. **P<0.01; *P<0.05.
SPARC is the direct target of miR-211 in cervical cells
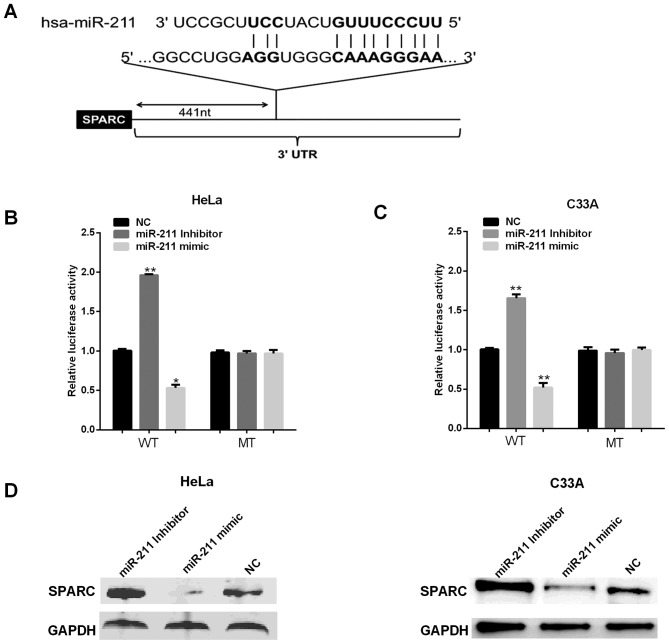
Bioinformatics analysis software TargetScan and miRanda were used for predicting the targets of miR-211. The putative binding sites for miR-211 were found at the 3′-UTR of SPARC (Fig. 4A). To further confirm whether the 3′-UTR of SPARC can be directly targeted by miR-211, WT pmirGLO-UTR of SPARC 3′-UTR or MT pmirGLO-UTR of SPARC 3′-UTR vector was co-transfected into two cell lines with either miR-211 mimic/inhibitor or negative controls (NC), followed by the measurement with luciferase reporter assays. Fig. 4B and C shows that reductions were approximately 40% in HeLa cell and 45% in C33A cells of the luciferase activity in all cells transfected with the miR-211 mimic and SPARC 3′-UTR WT vector, whereas the repressive effect of miR-211-inhibitor increased luciferase activity in WT SPARC by approximately 46 and 35%, but the luciferase activity had no significant change in cells transfected with SPARC 3′UTR-containing mutant miR-211-binding sites. We next assessed the miR-211 effect on its potential target SPARC expression at protein level. To this end, miR-211 mimics/inhibitor were transfected into HeLa and C33A cells, respectively, SPARC expression was measured using western blot analysis. The results show miR-211 mimics decreased the protein level of SPAR (Fig. 4C and D), while miR-211 inhibitor enhanced SPARC expression. Together, these results demonstrated miR-211 negatively regulated endogenous SPARC expression in HeLa and C33A cells.
Figure 4.
miR-211 suppresses SPARC expression by targeting its 3′-UTR. (A) The binding sites of miR-211 on SPARC 3′-UTR. (B) Luciferase reporter assay with the pGL3-SPARC-3′-UTR-WT or pGL3-SPARC-3′-UTR-MT were co-transfected with miR-211 mimics/inhibitor/NC. (D) SPARC expression in HeLa cells transfected with miR-211 mimic/inhibitor/NC. **P<0.01; *P<0.05; SPARC, secreted protein acidic and rich in cysteine; WT, wild-type; MT, mutant type.
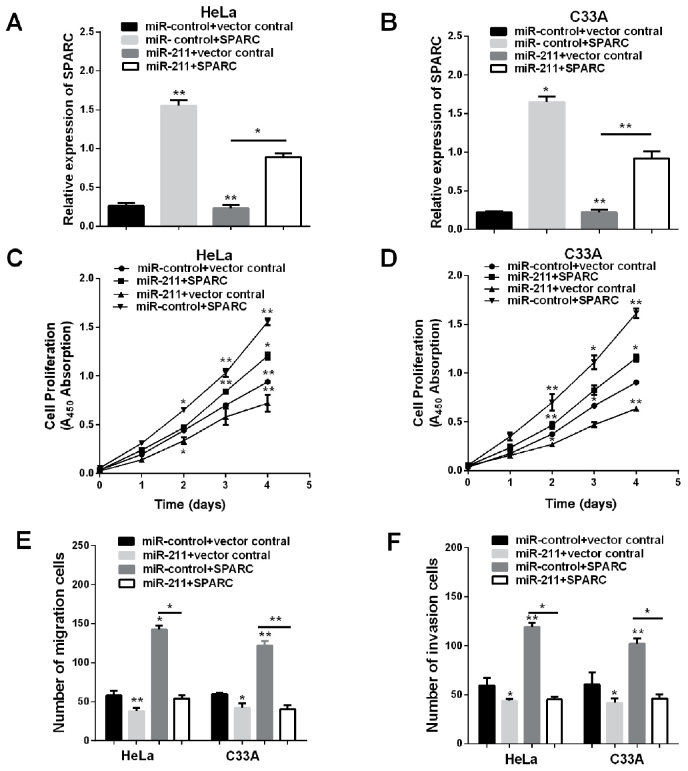
Overexpression of SPARC restores the inhibited proliferation, migration and invasion function of miR-211
The evidence given indicates that miR-211 may have various types of potential targets in HeLa and C33A cells, the anti-proliferative effect of miR-211 may not be limited to repression of SPARC. To detect whether overexpression of SPARC would simulate miR-211-mediated effects, transfection of pmirGLO-SPARC and co-transfection of pmirGLO-SPARC and miR-211 increased the mRNA expression of SPARC in HeLa and C33A cells, co-transfection of pmirGLO-SPARC and miR-211 abrogated the effects of miR-211 on SPARC expression (Fig. 5A and B), and then we calculated the capabilities of cell proliferation, migration and invasion. Restoration of SPARC eliminated cell viability, invasion and migration that was reduced by miR-211 mimic (Fig. 5C-F). These results demonstrated that SPARC is a direct functional target gene of miR-211 and that miR-211 functions as a tumor suppressor through SPARC.
Figure 5.
Upregulation of SPARC reverses the inhibitory effects of miR-211. (A and B) HeLa and C33A cells were co-transfected with control miR (miR-control) or miR-211 either with vector control or SPARC and the expression of SPARC was checked by RT-qPCR. (C and D) CCK-8 assay was conducted in HeLa and C33A cell lines. (E and F) Transwell assay was performed in HeLa and C33A cells lines in different conditions. **P<0.01; *P<0.05; SPARC, secreted protein acidic and rich in cysteine; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCK-8, Cell Counting kit-8.
Discussion
Mounting evidence has demonstrated that miR-211 acts as an inhibitor in the progression of cancer cell growth, invasion and migration in many kinds of cancers (12,14,24,25). However, the molecular mechanism by which miR-211 regulates the progression and development of cervical cancer has not been fully investigated. This investigation demonstrated the synthesized analysis of miR-211 effects on cervical cancer. The miR-211 expression is downregulated in cervical cancer in vivo, and reduced miR-211 levels have inverse correlation with SPARC expression. The results indicated miR-211 might act as a suppressor and play a significantly important role of cervical cancer growth and metastasis.
There is increasing evidence demonstrating that miRNAs can function as an crucial point in gene expressions, and then influence the tumor development and procession (26). The miR-211 expression was downregulated in various tumors, for example glioma (13), and ovarian cancer (14). However, the biological effect of miR-211 remains poorly understood in cervical cancer. In the study, the findings showed the miR-211 can inhibit growth, migration and invasion, and indicated the crucial point of miR-211 in cervical cancer progression.
This research revealed the relative expression level of SPARC was higher in cervical cancer tissues. SPARC can be found widely expressed in tumors and could function as an important member in regulating cell invasiveness, survival and tumor-stroma interactions to expedite cancer progression. Two different bioinformatics analysis software were used for predicting the targets of miR-211. SPARC was looked for as the potential gene effector which may participate in the biological function of miR-211. We confirmed that SPARC was a direct target, and the higher SPARC expression was confirmed taking significant part in tumor invasion and metastasis in certain cancers, such as gastric cancer (20), melanoma (27), and hepatocellular carcinoma (28). In our study, overexpression of SPARC restored the inhibited proliferation, migration and invasion function of miR-211, these results demonstrated that SPARC is a direct functional target gene of miR-211 and that miR-211 functions as a tumor suppressor through SPARC.
In conclusion, miR-211 acts as a tumor suppressor by reducing cell growth, migration and invasion in cervical cancer. Furthermore, we noted that miR-211 has an inverse correlation with SPARC and targets it by binding to its 3′-UTR. This newly identified miR-211 may provide further insight into progression and offers a promising therapeutic target for the treatment of cervical cancer. Further study to investigate the function of miR-211/SPARC in tumorigenesis and progression of cervical cancer is needed.
Acknowledgements
Not applicable.
Funding
This study did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
XQ and DG contributed to the conception of the study. QR contributed significantly to the data analysis and study preparation. XJ and JB performed the data analyses and wrote the study. LS helped perform the analysis with constructive discussions. All authors have read and approved the final study.
Ethics approval and consent to participate
The study was approved by the Ethics Committee of Yuhuangding Hospital (Yantai, China) and the patients signed the informed consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pimple S, Mishra G, Shastri S. Global strategies for cervical cancer prevention. Curr Opin Obstet Gynecol. 2016;28:4–10. doi: 10.1097/GCO.0000000000000241. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Sun Y, Yang X, Liu M, Tang H. B4GALT3 up-regulation by miR-27a contributes to the oncogenic activity in human cervical cancer cells. Cancer Lett. 2016;375:284–292. doi: 10.1016/j.canlet.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 4.Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V, Lee RC. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol Biol. 2004;265:131–158. doi: 10.1385/1-59259-775-0:131. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 7.Saumet A, Lecellier CH. microRNAs and personalized medicine: Evaluating their potential as cancer biomarkers. Adv Exp Med Biol. 2015;888:5–15. doi: 10.1007/978-3-319-22671-2_2. [DOI] [PubMed] [Google Scholar]
- 8.Srivastava SK, Arora S, Averett C, Singh S, Singh AP. Modulation of microRNAs by phytochemicals in cancer: Underlying mechanisms and translational significance. BioMed Res Int. 2015;2015:848710. doi: 10.1155/2015/848710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virant-Klun I, Ståhlberg A, Kubista M, Skutella T. MicroRNAs: From female fertility, germ cells, and stem cells to cancer in humans. Stem Cells Int. 2016;2016:3984937. doi: 10.1155/2016/3984937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ventura A, Jacks T. MicroRNAs and cancer: Short RNAs go a long way. Cell. 2009;136:586–591. doi: 10.1016/j.cell.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazar J, Qi F, Lee B, Marchica J, Govindarajan S, Shelley J, Li JL, Ray A, Perera RJ. MicroRNA 211 functions as a metabolic switch in human melanoma cells. Mol Cell Biol. 2016;36:1090–1108. doi: 10.1128/MCB.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng B, Qu L, Li J, Fang J, Yang S, Cao Z, Mei Z, Sun X. MiRNA-211 suppresses cell proliferation, migration and invasion by targeting SPARC in human hepatocellular carcinoma. Sci Rep. 2016;6:26679. doi: 10.1038/srep26679. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Zhang J, Lv J, Zhang F, Che H, Liao Q, Huang W, Li S, Li Y. MicroRNA-211 expression is down-regulated and associated with poor prognosis in human glioma. J Neurooncol. 2017;133:553–559. doi: 10.1007/s11060-017-2464-2. [DOI] [PubMed] [Google Scholar]
- 14.Xia B, Yang S, Liu T, Lou G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting Cyclin D1 and CDK6. Mol Cancer. 2015;14:57. doi: 10.1186/s12943-015-0322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye L, Wang H, Liu B. miR-211 promotes non-small cell lung cancer proliferation by targeting SRCIN1. Tumour Biol. 2016;37:1151–1157. doi: 10.1007/s13277-015-3835-y. [DOI] [PubMed] [Google Scholar]
- 16.Sümbül AT, Göğebakan B, Bayram S, Batmacı CY, Öztuzcu S. MicroRNA 211 expression is upregulated and associated with poor prognosis in colorectal cancer: A case-control study. Tumour Biol. 2015;36:9703–9709. doi: 10.1007/s13277-015-3708-4. [DOI] [PubMed] [Google Scholar]
- 17.Xu D, Liu S, Zhang L, Song L. MiR-211 inhibits invasion and epithelial-to-mesenchymal transition (EMT) of cervical cancer cells via targeting MUC4. Biochem Biophys Res Commun. 2017;485:556–562. doi: 10.1016/j.bbrc.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 18.Bornstein P, Sage EH. Matricellular proteins: Extracellular modulators of cell function. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/S0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Shi D, Liu X, Fang S, Zhang J, Zhao Y. Targeting SPARC by lentivirus-mediated RNA interference inhibits cervical cancer cell growth and metastasis. BMC Cancer. 2012;12:464. doi: 10.1186/1471-2407-12-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao ZS, Wang YY, Chu YQ, Ye ZY, Tao HQ. SPARC is associated with gastric cancer progression and poor survival of patients. https://doi.org/10.1158/1078-0432.CCR-09-1247. Clin Cancer Res. 2010;16:260–268. doi: 10.1158/1078-0432.CCR-09-1247. [DOI] [PubMed] [Google Scholar]
- 21.Derosa CA, Furusato B, Shaheduzzaman S, Srikantan V, Wang Z, Chen Y, Seifert M, Ravindranath L, Young D, Nau M, et al. Elevated osteonectin/SPARC expression in primary prostate cancer predicts metastatic progression. Prostate Cancer Prostatic Dis. 2012;15:150–156. doi: 10.1038/pcan.2011.68. [DOI] [PubMed] [Google Scholar]
- 22.Shi Q, Bao S, Maxwell JA, Reese ED, Friedman HS, Bigner DD, Wang XF, Rich JN. Secreted protein acidic, rich in cysteine (SPARC), mediates cellular survival of gliomas through AKT activation. J Biol Chem. 2004;279:52200–52209. doi: 10.1074/jbc.M409630200. [DOI] [PubMed] [Google Scholar]
- 23.Shi D, Jiang K, Fu Y, Fang R, Liu XI, Chen J. Overexpression of SPARC correlates with poor prognosis in patients with cervical carcinoma and regulates cancer cell epithelial-mesenchymal transition. Oncol Lett. 2016;11:3251–3258. doi: 10.3892/ol.2016.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang CY, Hua L, Sun J, Yao KH, Chen JT, Zhang JJ, Hu JH. MiR-211 inhibits cell proliferation and invasion of gastric cancer by down-regulating SOX4. Int J Clin Exp Pathol. 2015;8:14013–14020. [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Shen YF, Shi ZM, Shang XJ, Jin DL, Xi F. Overexpression miR-211-5p hinders the proliferation, migration, and invasion of thyroid tumor cells by downregulating SOX11. J Clin Lab Anal e22293. 2017 doi: 10.1002/jcla.22293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manikandan J, Aarthi JJ, Kumar SD, Pushparaj PN. Oncomirs: The potential role of non-coding microRNAs in understanding cancer. Bioinformation. 2008;2:330–334. doi: 10.6026/97320630002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An XJ, Li YQ, Qu XY, Zhang J, Zhang LY, Wang M, Zhu L, Chen SY, Chen HX, Tu YT, et al. Silencing endothelin-3 expression attenuates the malignant behaviors of human melanoma cells by regulating SPARC levels. https://doi.org/10.1007/s11596-013-1162-3. J Huazhong Univ Sci Technolog Med Sci. 2013;33:581–586. doi: 10.1007/s11596-013-1162-3. [DOI] [PubMed] [Google Scholar]
- 28.Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66:454–458. doi: 10.1016/j.biopha.2012.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.