Abstract
Hybrids of RNA and arabinonucleic acid (ANA) as well as the 2′-fluoro-ANA analog (2′F-ANA) were recently shown to be substrates of the enzyme RNase H. Although RNase H binds to double-stranded RNA, no cleavage occurs with such duplexes. Therefore, knowledge of the structure of ANA/RNA hybrids may prove helpful in the design of future antisense oligonucleotide analogs. In this study, we have determined the NMR solution structures of ANA/RNA and DNA/RNA hairpin duplexes and compared them to the recently published structure of a 2′F-ANA/RNA hairpin duplex. We demonstrate here that the sugars of RNA nucleotides of the ANA/RNA hairpin stem adopt the C3′-endo (north, A-form) conformation, whereas those of the ANA strand adopt a ‘rigid’ O4′-endo (east) sugar pucker. The DNA strand of the DNA/RNA hairpin stem is flexible, but the average DNA/RNA hairpin structural parameters are close to the ANA/RNA and 2′F-ANA/RNA hairpin parameters. The minor groove width of ANA/RNA, 2′F-ANA/RNA and DNA/RNA helices is 9.0 ± 0.5 Å, a value that is intermediate between that of A- and B-form duplexes. These results rationalize the ability of ANA/RNA and 2′F-ANA/RNA hybrids to elicit RNase H activity.
INTRODUCTION
Over the past few years, synthetically modified oligodeoxynucleotides (ODN) have been widely used for artificially regulating gene expression through the targeting of RNA (1,2). Substitutions at the 2′-position of ribose yield nuclease-resistant RNA analogs with increased binding affinity towards target RNAs (3). This is because alkylation of the ribose 2′-position generally leads to structures that are conformationally biased or conformationally locked in the north or C3′-endo sugar pucker (3,4). However, preorganization of the ODN into a pure north conformation (A-DNA-like) produces an ODN/RNA duplex not generally recognized by RNase H, a critical step in the mechanism of action of antisense compounds (5). Such a lack of recognition has been generally solved by the use of the so-called ‘gapmer’ technology, where 2′-deoxynucleotide ‘gaps’ are flanked at either end of the ODN with north-biased nucleotide units (e.g. RNA-DNA-RNA chimeras) (6–8). With this modification, the ‘gap’ of the resulting chimeric duplex has a hybrid-like conformation which is then efficiently recognized by the enzyme RNase H.
Recently, we showed that the stereochemistry at the 2′-position of the sugar of antisense oligonucleotides is a key determinant in the target RNA binding affinity and the activation of RNase H (9,10). We demonstrated that both arabinonucleic acid (ANA) and the corresponding 2′-fluoro-arabinonucleic acid analog (2′F-ANA; Fig. 1) form hybrids with RNA, and that they are able to induce RNase H degradation of the target RNA. Since RNA/RNA duplexes are not substrates of RNase H (11), these findings provided an important advancement towards understanding the catalytic mechanism and substrate selectivity of RNase H.
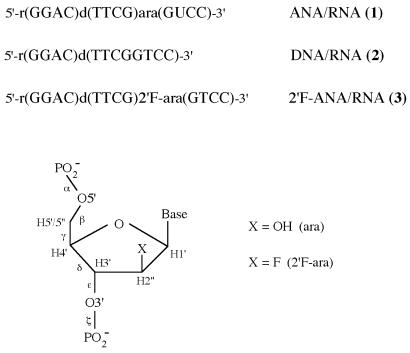
Figure 1.
Chemical structures of the hairpins. The first four RNA nucleotides, r(GGAC), are base paired to the last four ANA, DNA or 2′F-ANA nucleotides. The loop, d(TTCG), is of constant base and sugar composition.
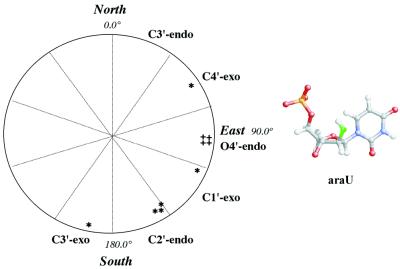
Relatively little is known about the structure of duplexes with ANA modifications. Previous studies have been mostly restricted to DNA duplexes containing one or two neighboring ANA or 2′F-ANA residues (12–20). X-ray crystallographic data revealed the adoption of an O4′-endo (east) sugar pucker by 2′F-ANA residues (20), a conformation that lies halfway between the C2′-endo (south, B-DNA) and C3′-endo (north, A-RNA) puckers (Fig. 2). This is consistent with the results of more recent molecular dynamics (MD) calculations of the conformation of 2′F-ANA-containing duplexes (21). Modeling studies of two other ANA analogs, namely [3.3.0]-bicyclo-ANA (bc-ANA) and [3.2.0]-bicyclo-ANA were also suggestive of the adoption of east-type (O4′-endo) sugar puckers (22). Recently, Egli and co-workers (23) crystallized a DNA dodecamer duplex containing bc-ANA modifications and found that the bc-ANA residues display sugar puckers that fall uniformly within the O4′-endo range. They also concluded that steric constraints within the bicyclic ANA framework prohibited adoption of either the C2′-endo or C3′-endo conformations. Our recently published NMR structure of a 2′F-ANA/RNA hairpin duplex also revealed an O4′-endo pucker for the 2′F-ANA residues (24). This is in contrast to NMR and MD studies of monomeric nucleoside units (e.g. araF-A,T), which identified the C2′-endo/C1′-exo puckers as the most stable conformations (25,26).
Figure 2.
Schematic of the pseudorotation phase angle (P) cycle with the positions of selected pucker types indicated. The P angles of arabino-residues in average structure of ANA/RNA hairpin (1) (+) are compared with the P angles of arabinonucleotides in X-ray and NMR structures of B- and Z-DNA duplexes (*) (12–18). In the O4′-endo conformation, the 2′-OH of arabinose is pseudoaxial and gauche to the ring oxygen O4′.
While 2′-fluoroarabino and bc-arabinonucleotides exhibit a preference for the east pucker, arabinonucleotides (β-2′-OH) adopt a variety of conformations. The actual conformation appears to depend on the environment (crystal, solution) and other factors. For example, computational simulations have shown that the arabinose (2′OH) sugars of ANA/RNA hybrids prefer the south C2′-endo pucker due to an intranucleotide O2′-H···O5′ hydrogen bond (21). This feature is also observed in the crystal structure of arabinocytidine (ara-C) (27). Crystal structures of Z-DNA duplexes that contain ANA residues also show a preference for the south conformational range C2′-endo/C3′-exo (17,18). Other studies on duplex structures containing ANA residues have detected the south C2′-endo (12,15,16), C1′-exo (13) and north C4′-exo (14) conformations (Fig. 2). Conformational variations have also been noted at the monomeric nucleoside level (27–30).
To date, no NMR structures of ANA (2′OH)/RNA hybrids have been reported, although the conformation of such hybrids has been predicted on the basis of molecular modeling studies (21). This fact, combined with the knowledge that ANA/RNA hybrids are thermodynamically less stable than DNA/RNA (10,31) and 2′F-ANA/RNA hybrids (9), prompted us to carry out an NMR-based structure study of a chimeric hairpin containing an ANA/RNA stem (Fig. 1). The results suggest striking similarities in the global and local structure of ANA/RNA, 2′F-ANA/RNA and DNA/RNA hybrids.
MATERIALS AND METHODS
Sample preparation
Chimeric hairpin oligonucleotides were synthesized as described previously (9,10,31). The samples at 1 mM concentration were dissolved in 0.4 ml of 100% D2O or 9:1 H2O/D2O (v/v) (for imino-proton spectra). The solution contains 0.1 mM EDTA sodium salt, and the pH was adjusted to 7.0 by adding NaOH.
The hairpin melting temperatures (Tm) determined at concentrations of ∼1 OD/ml were confirmed qualitatively by recording proton NMR spectra at different temperatures. As observed for unimolecular transitions in similar all-RNA or all-DNA hairpin studies (32,33), the Tm values were independent of oligonucleotide concentration. The Tm values for the ANA/RNA hairpin (1) and for the DNA/RNA hairpin (2) are 44 and 60°C, respectively. These are less than the Tm of 67°C for the previously studied 2′F-ANA/RNA hairpin duplex (3) (24).
NMR spectroscopy
The NMR spectra were recorded on a Bruker DRX-500 spectrometer equipped with a 1H/13C/31P triple resonance (x, y, z) gradient probe operating at 500.13 MHz of proton resonance. The proton chemical shifts were measured relative to internal DSS, and the phosphorus resonances were indirectly referenced to 85% H3PO4 by multiplying the DSS proton resonance absolute frequency by 0.404807356 (24). All 2D spectra were collected in the phase-sensitive mode via the TPPI method and presaturation. The 3-9-19 pulse sequence (34) was used to suppress the water signal for samples in 9:1 H2O/D2O.
NOESY experiments in D2O were performed at 7 and 15°C [hairpin (1)] and at 20°C [hairpin (2)] using mixing times tm of 50, 70, 200 and 400 ms. The final spectral size was 2K × 2K points after Fourier transformation. The volumes of crosspeaks of NOESY spectra were calculated using XWINNMR (Bruker). NOESY experiments in 9:1 H2O/D2O (v/v) were performed at 15°C with a mixing time of 180 ms. DQF-COSY spectra were collected with and without phosphorus decoupling (final data size of 8K × 2K points). Proton MLEV-17 TOCSY experiments were performed with a mixing time of 84 ms. H,C-correlation HMQC spectra were recorded using GARP heteronuclear decoupling (1JCH = 180 Hz). Inverse (H,P)-HetCOSY and HetTOCSY spectra were collected with final spectral sizes of 2K × 1K data points. In 2D NMR experiments, relaxation delays were 1.5–2.5 s, the number of t1 experiments was 256 or 512, and the number of scans was 32–128 for each FID. Correlation times (τc = 2.0 ns) were determined using the H5-H6 NOE of deoxyribo, ribo and arabinocytidine residues of the various hairpins as reference.
Structural modeling
The starting coordinates of the hairpins were generated using Sybyl 6.5 software (Tripos Inc.) from both A- and B-type DNA structures. The X-PLOR 3.843 package (35) with standard nucleic acid force field (parallhdg.dna) was used for hairpin molecular modeling. Partial charges for arabinonucleotides were set to be the same as for ribonucleotides.
At the first stage of refinement, 10 ‘randomized’ structures (five with canonical A-type and five with B-type of hairpin stem) were generated by 1–2 ps MD at 300 K without experimental constraints. Subsequent stages of simulated annealing with NOE-distance and torsion angle constraints were similar to those described previously for hairpins (32,33,36,37). The X-PLOR refinement involved an initial energy minimization and MD simulation of 14 ps (2 ps of heating from 300 to 1000 K, 5 ps at 1000 K, 4 ps of cooling back to 300 K and then 3 ps at 300 K). A distance-dependent dielectric constant was used to mimic the solvent. During the last 2 ps, the atomic coordinates were saved at 200 fs interval, and 10 conformers collected. These conformers were then averaged and energy minimized. Atomic coordinates of the hairpin structures have been deposited in the PDB under accession numbers 1HO6 for the ANA/RNA hairpin (1) and 1HOQ for the DNA/RNA hairpin (2).
Distance restraints were derived from NOESY spectra at different mixing times by crosspeak volume integration, using the r–6 distance relationship (38) and average crosspeak volume values for calibration for H5-H6 in cytidines (r = 2.46 Å) and for Me-H6 in thymidines (r = 2.70 Å). The distance constraints in the hairpin stems were given 10% of lower and 15% of upper bounds. Due to loop mobility, the NOEs for loops were classified as strong, medium, weak and absent with corresponding distance ranges of 1.8–3.2, 2.0–5.0, 3.0–7.0 and >4.0 Å (33). No constraints were applied for loop NOEs involving exchangeable protons. Sugar puckering was determined by the PSEUROT 3B program (39) from vicinal JHH coupling constants. The five ν0–ν4 torsion angles for hairpin sugars were constrained (with ±10° bounds) according to sugar conformations determined from J-couplings; in the case of stem DNA nucleotides [hairpin (2)] torsion angles were constrained according to intrasugar NOE distances. Backbone torsion angle constraints were set as described in the text. During refinement, the NOE distance and hydrogen bond force constants were gradually built up to final values of 30–40 kcal/(mol Å2) and the backbone torsion angle constants to 60 kcal/(mol rad2). Calculation of helical parameters was carried out with CURVES 5.2 program (40).
RESULTS
Resonance assignments
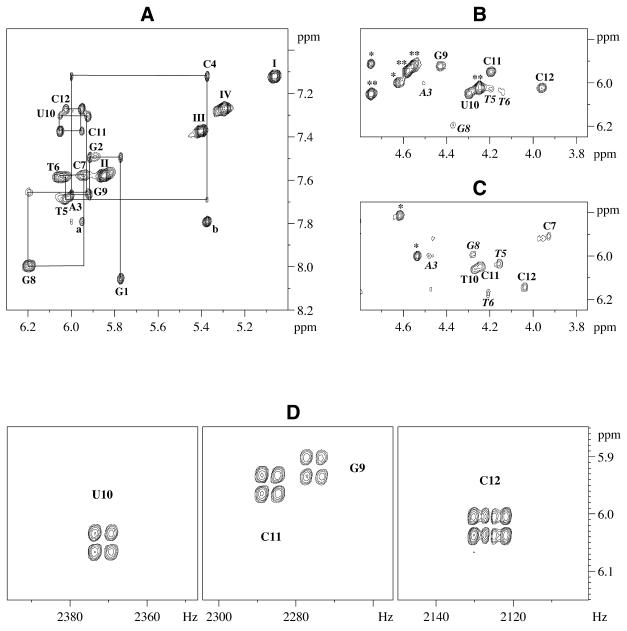
The assignment of oligonucleotide non-exchangable proton resonances in NOESY spectra of hairpins (1) and (2) was carried out using the H1′(i)-H6/H8(i)-H1′(i-1) pathway in the standard manner employed for right-handed DNA duplexes (Fig. 3A) (38). The DQF-COSY, TOCSY and (H,C)-HMQC spectra based on different 13C chemical shifts for C2′/C3′, C4′ and C5′ were also used to assign the sugar protons. All assignments were further corroborated by the H6/H8(i)-H2′, H2″, H3′, H4′(i,i-1) pathways. The adenine H2 resonances were identified by inversion–recovery experiments and confirmed by NOESY intranucleotide H2-H1′ and interstrand H2(A3)-H1′(C11) crosspeaks. It should be mentioned that some sugar crosspeaks for the dG9 nucleotide in hairpin (2) were broadened at room temperature. The assignment of stem imino-protons was made from NOESY spectra in H2O/D2O using the crosspeaks of NH(G) with NH2(C) or NH(T,U10) with H2(A3) of complementary base pairs. Signals of NH2(C) were easily identified from their strong crosspeaks with H5(C) of the same nucleotide. Phosphorus signals were determined from strong H3′-P crosspeaks in (H,P)-HetCOSY spectra.
Figure 3.
Expanded plots of NOESY spectra at 500 MHz. (A) ANA/RNA hairpin (1), 7°C, mixing time tm = 200 ms, the assignment of oligonucleotide protons are shown by solid lines and nucleotide name with number; I-IV-H5-H6(C4,7,11,12) crosspeaks, respectively. (B and C) ANA/RNA hairpin (1), 7°C, and DNA/RNA hairpin (2), 20°C, tm = 50 ms in both cases, H1′-H4′ crosspeaks are labeled by nucleotide name with number, H1′-H2′ (RNA) crosspeaks (*) and H1′-H2″ (ANA) crosspeaks (**). (D) Expanded plots of DQF-COSY spectrum of ANA/RNA hairpin (1) at 500 MHz, 15°C, H1′-H2″ (ANA) crosspeaks are marked by nucleotide name with number.
Proton and phosphorus chemical shifts of harpins (1) and (2) are provided in the Supplementary Material. The proton chemical shifts for loop nucleotides are close to the all-DNA (33) or 2′F-ANA/RNA (24) hairpins, both having flexible loop conformations. The set of sequential NOE contacts between loop residues C4, T5 and T6 was complicated by medium–weak H2′(T5)-H5(C7) crosspeaks in both hairpins (1) and (2). No strong or medium sequential NOEs were found between T6 and C7, or between C7 and G8. As a result, we were unable to build a single loop conformation.
Sugar ring conformation
Sugar conformations were determined from 3JHH data obtained from DQF-COSY experiments (Fig. 3D and Table 1). Coupling constants J1′2′ and J1′2′′ were determined from H1′-H2″ or H1′-H2′ crosspeak splitting, whereas J2′3′, J2′′3′ and J3′4′ were extracted from sums of coupling constants as previously described (38,41,42). The phase angles of pseudorotation (P) and mole fractions of conformers (f) were determined using the PSEUROT program (39) with fixed puckering amplitude of 37°. The ranges for pseudorotation parameters were calculated from the uncertainty of measured values of J-couplings and are given in Table 1.
Table 1. Coupling constants (Hz) in hairpins (1) and (2) and calculated sugar puckersa.
| Residue |
Hairpin |
J1′2′ |
J1′2′′ |
J2′3′ |
J2′′3′ |
J3′4′ |
J4′5′ + J4′5′′ |
JH3′-P |
f1 (%) |
P1 |
P2 |
| G1 | (1) | 2 | – | 4.5 | – | 8 | 7 | 9.5 | 0 | – | 26–58 |
| (2) | <2 | – | 5.5 | – | 7.5 | 6.5 | 9 | 0 | – | 21–49 | |
| G2 | (1) | <2 | – | 4 | – | 10 | <4 | 7.5 | 0 | – | 24–45 |
| (2) | <2 | – | 5 | – | 9 | <4 | 9 | 0 | – | 28–49 | |
| A3 | (1) | <2 | – | 4 | – | 10 | <4 | 8.5 | 0 | – | 24–45 |
| (2) | <2 | – | 5 | – | 8.5 | 4 | 9.5 | 0 | – | 22–48 | |
| C4 | (1) | 2 | – | 4.5 | – | 9.5 | <4 | 9 | 0 | – | 30–58 |
| (2) | 2 | – | 4.5 | – | 9.5 | <4 | 8 | 0 | – | 30–58 | |
| T5 | (1) | 4.2 | 7.4 | 7 | 5 | 5 | <4 | 5 | 30–43 | 156–170 | 2–37 |
| (2) | 4.5 | 7.8 | 7 | 5 | 5 | <4 | 5.5 | 28–42 | 150–165 | 2–42 | |
| T6 | (1) | 10.2 | 5.2 | 7 | <2 | <2 | n.d. | 5.5 | 99–100 | 132–154 | – |
| (2) | 9.8 | 5.5 | 7.5 | <2 | <2 | n.d. | 6 | 98–100 | 130–149 | – | |
| C7 | (1) | 9.8 | 6.0 | 6 | <2 | 2 | n.d. | 6 | 96–100 | 151–188 | – |
| (2) | 9.8 | 5.7 | 6.5 | <2 | <2 | n.d. | 6.5 | 96–100 | 141–171 | – | |
| G8 | (1) | 9.5 | 5.3 | 7.5 | <2 | 3 | n.d. | 4 | 95–100 | 122–143 | – |
| (2) | 6.3 | 7.5 | 6 | 4.5 | 4.5 | 8 | 3.5 | 44–61 | 158–198 | 0–48 | |
| G9 | (1) | – | 4.3 | – | <2 | 5 | n.d. | 4 | 82–100 | 91–115 | – |
| (2) | n.d.b | n.d.b | n.d. | n.d. | n.d. | n.d. | 4 | – | – | – | |
| U10 | (1) | – | 4.5 | – | <2 | 5 | 5 | 4 | 80–100 | 90–115 | – |
| T10 | (2) | 4.5 | 8.0 | 8 | 5 | 5 | <4 | 4 | 21–40 | 147–161 | 10–49 |
| C11 | (1) | – | 4.5 | – | <2 | 5.5 | n.d. | 4 | 87–100 | 83–106 | – |
| (2) | 4.5 | 7.8 | 7.5 | 5 | 5 | 4 | 3 | 26–45 | 150–165 | 4–45 | |
| C12 | (1) | – | 5.1 | – | 3.1 | 6 | 4 | – | 46–64 | 93–122 | – |
| (2) | 5.5 | 7.0 | 7 | 5.5 | 4.5 | <4 | – | 33–50 | 142–190 | 0–40 |
The data show that, in both hybrids, the ribose sugars adopt the north (mostly C3′-endo) pucker, whereas the arabinonucleotide (2′OH) residues prefer the east (mostly O4′-endo) sugar pucker (Fig. 2). We conclude that ANA and 2′F-ANA adopt the same pucker when hybridized to complementary RNA (24). Consistent with the O4′-endo conformation is the very strong H1′-H4′ NOE crosspeaks of the arabinose residues (d = 2.5 Å) (Fig. 3B). Only the sugar moiety of terminal arabinonucleotide residue (C12) exists as a 1:1 mixture of east and probably north conformers. The coupling constant PSEUROT and H1′-H4′ NOE crosspeak analysis for sugar conformations in the DNA loops revealed a mostly north pucker for T5 loop residue probably stacked with neighboring C4 ribonucleotide (33) and south puckers for the remaining loop nucleotides. Nevertheless, the PSEUROT analysis of sugar puckering for G9-C12 DNA nucleotides in DNA/RNA hairpin is at variance with measured values of H1′-H4′ NOE distances (d ≈ 2.8 Å) due to the presence of multiple conformational states for stem DNA sugars (see Discussion).
Backbone β, γ , ɛ and glycosidic χ torsion angles
The β torsion angles were constrained using the information about H5′/H5″-P and H4′-P crosspeaks in H,P-HetCOSY spectra. Except for G9 nucleotides, the β angles were found to be in the trans conformation (180 ± 45°) as determined by the symmetry and low intensity of the H5′/H5″-P crosspeaks and detectable 4JH4′-P W-pathway coupling constants (29,38,43). In the case of G9 [hairpins (1) and (2)] very strong coupling was detected between the 5′-phosphate and one of the 5′-protons, and therefore we did not constrain the value of β(G9).
The γ angles were constrained using the sums of JH4′H5′ and JH4′H5′′, which were available from phosphorus decoupled DQF-COSY spectra (Table 1), and the NOE H1′/H6/H8-H4′ crosspeak linewidths (43). The sums of these coupling constants in most nucleotides were <5 Hz, which is consistent with a γ angle range of 60 ± 20°. For T6, C7, G9 and araC11 nucleotides the linewidths of H4′ lie in the range 10–14 Hz and, hence, in these cases, the range of γ angles can be constrained to 60 ± 40°. For G8 nucleotide, the sum of JH4′H5′ and JH4′H5′′ coupling constants (8 Hz) and large H4′ linewidths (>15 Hz) did not allow constraint of the γ(G8) angle in the flexible loop of both hairpins (1) and (2).
The ɛ angles have been estimated from the vicinal 3JH3′-P coupling constants (Table 1). These values of coupling constants were determined from comparison of DQF-COSY crosspeak splittings in spectra with and without phosphorus decoupling. The 3JH3′-P coupling constants lie in the range of 8–10 Hz for nucleotides in RNA strands and ɛ angles were constrained to 240 ± 50° from the Karplus equation (43) in accordance with data for DNA/RNA duplexes (44–48). In contrast, the small 3JH3′-P values observed for the stem arabino and deoxyribonucleotides (3–4 Hz) suggest an ɛ value of 170 ± 50°. The ɛ angles for the flexible loop nucleotides were constrained in the range of both BI- and BII-type phosphate conformations, i.e. 240 ± 120° (49).
Glycosidic χ angles were not constrained in both hairpins (1) and (2); rather, they were fixed indirectly by intranucleotide aromatic–sugar distance constraints. Most nucleotides are in anti-conformation except for G8 nucleotides where NOESY crosspeak volumes correspond to syn/anti conformational exchange with an H8-H1′ NOE distance of 2.7–2.8 Å. Aromatic–sugar intranucleotide distance constraints for G8 were omitted during structure refinement calculations.
Structure refinement and analysis
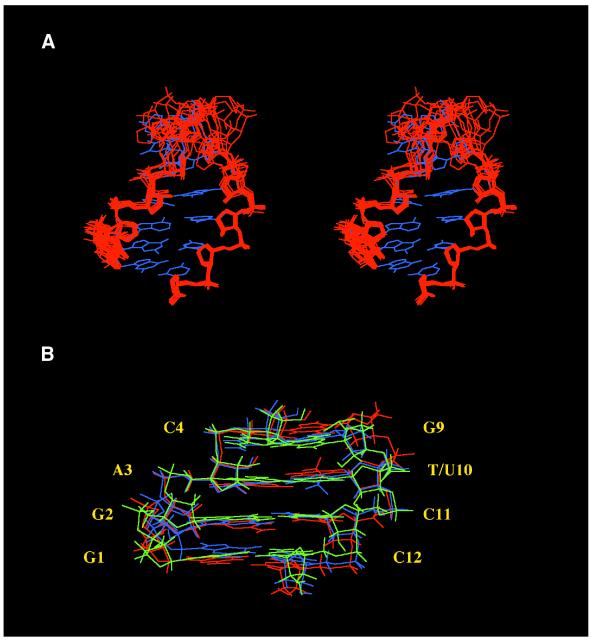
In order to determine hairpin structures that are consistent with the experimental data, NMR-restrained MD calculations were performed. The structural statistics of X-PLOR refinement are presented in Table 2, and superimposition of final individual structures for ANA/RNA hairpin (1) is shown in Figure 4A. Ten A- and B-type starting structures (an overall r.m.s.d. of 1.8 Å for stem heavy atoms) with randomly organized loop structures for both hairpins (1) and (2) were refined by X-PLOR to r.m.s.d. of 0.3–0.8 Å for stem nucleotides. As expected from 2′F-ANA/RNA (24) and all-DNA hairpin (33) studies, the loops in both hairpins are flexible and do not significantly influence stem structures. The stacking between stem C4 and loop T5 bases was confirmed in both hairpin structures leading to a smaller r.m.s.d. for T5 (1.1–1.7 Å) in comparison with other loop nucleotides (up to 4.4 Å).
Table 2. Structural statistics for 10 final individual structures of hairpins (1) and (2).
| Parameter |
ANA/RNA (1) |
DNA/RNA (2) |
| Number of NOE distance restraints | 146 | 175 |
| Intranucleotide | 96 | 118 |
| Internucleotide | 50 | 57 |
| Stem residues | 72 | 105 |
| Torsion angle restraints | 91 | 71 |
| Hydrogen bond restraints | 11 | 11 |
| NOE violation (>0.2 Å) | 1–3 | 1–3 |
| Torsion angle violation (>5) | 0 | 0 |
| Average R1/6-factor for stem residuesa | 0.078 ± 0.004 | 0.088 ± 0.004 |
| r.m.s.d. for all heavy atoms (Å)b | ||
| For stem residues | 0.3–0.8 | 0.3–0.6 |
| For loop residues | 1.1–4.4 | 1.7–4.4 |
| Average r.m.s.d. from covalent geometry | ||
| Bond lengths (Å) | 0.0092 ± 0.0003 | 0.0098 ± 0.0003 |
| Angles (˚) | 1.58 ± 0.03 | 1.58 ± 0.02 |
| Impropers (˚) | 0.64 ± 0.02 | 0.72 ± 0.02 |
aComparison of NOESY experimental and back-calculated from final structure (by X-PLOR) crosspeak volumes (tm = 200 ms and τc = 2.0 ns).
bRelative to the average structure with stem superposition.
Figure 4.
Spatial structures of the hairpins. (A) Stereoview of the superimposition of 10 final individual structures (sugar-phosphate heavy atoms only; red) and average minimized structure (blue) for ANA/RNA hairpin (1). (B) View of the stem superimposition of average minimized structures of ANA/DNA (1) (green), DNA/RNA (2) (blue) and 2′F-ANA/RNA (3) (red) hairpins.
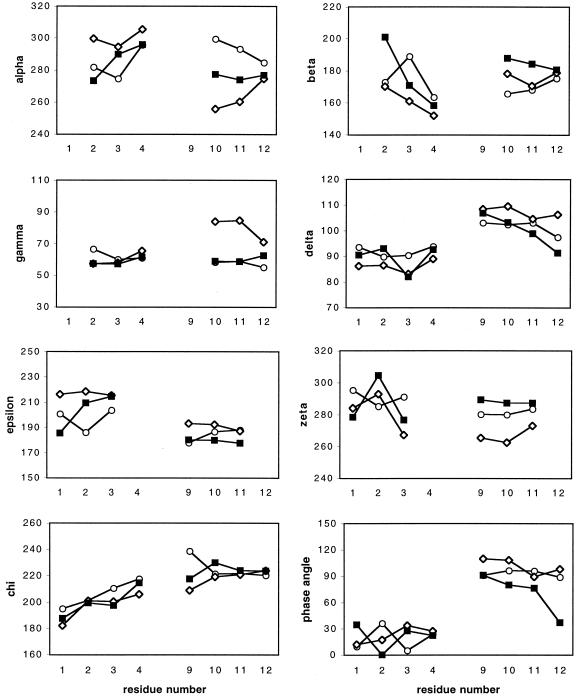
The final individual structures of hairpins (1), (2) and previously published (24) hairpin (3) were averaged and energy minimized for the purpose of comparison of average structural parameters for ANA/RNA, DNA/RNA and 2′F-ANA/RNA hairpin stems. The view of stem superimposition for these hairpin structures is shown in Figure 4B. The r.m.s.d. are 0.92 Å between stem heavy atoms of ANA/RNA and DNA/RNA hairpins, and 1.05 Å between ANA/RNA and 2′F-ANA/RNA hairpins. Backbone torsion angles in hairpin stems (Fig. 5) are close and reflect the strong similarity of the hairpin structures. The phase angles of sugar puckers for four G9-C12 arabinonucleotides in average structure of hairpin (1) are in the range 89–96° (Fig. 2).
Figure 5.
Backbone torsion angles (α, β, γ, δ, ɛ, ζ), glycosidic torsion angles (χ) and pseudorotation phase angles (P) (in degrees) for stem residues in average minimized structures. Open circles, ANA/RNA; filled squares, DNA/RNA; open diamonds, 2′F-ANA/RNA.
DISCUSSION
There is considerable interest in the contribution made by the 2′-hydroxyl group of ribose residues to the overall stability of nucleic acids (50). In addition to stabilizing the C3′-endo conformation of the ribose, the 2′-hydroxyl seems to play a significant role in stabilizing A-helices through interactions with water molecules in the minor groove (50,51). This feature may be responsible for the very different thermodynamic stability of DNA and RNA duplexes.
Unlike RNA, ANA and its 2′-fluoro derivative (2′F-ANA) form hybrids with RNA that are capable of activating RNase H, resulting in cleavage of the RNA strand (9,10). In this work, we determined the solution NMR structure of a hybrid duplex formed between ANA and a complementary RNA strand. We hoped these studies would provide a basis for the recognition and cleavage of ANA/RNA hybrids by RNase H, and explain the lower thermal stability of ANA/RNA hybrids relative to RNA/RNA duplexes (10,31). The loss of stability on inverting the configuration at C2′ is significant (10). This destabilization might well arise from the fact that, in arabinonucleosides, the 2′-hydroxyl group is cis with respect to the heterocyclic base, leading to changes about the N-glycosidic bond conformation which, in turn, would result in local deformation (unstacking) of the base pairs (17,52). In fact, slight steric contacts have been noted between arabino C2′-OMe groups and C6/C8 carbons of the flanking aromatic bases in a hybrid duplex (21). Steric effects can be probed by replacing the 2′-OH group with a smaller substituent (fluorine). We showed that 2′-F substitution results in significantly more stabilization than observed with the ANA (2′-OH) analog (9,25). The relative order of stability of hybrid duplexes is: 2′F-ANA/RNA > RNA/RNA > DNA/RNA > thioate-DNA/RNA > ANA/RNA. This is an unusual finding in that the 2′F-ANA/RNA hybrid is significantly more stable than the DNA/RNA hybrid and therefore steric effects cannot fully account for the observed trends. Consistent with this notion is the fact that our solved ANA/RNA structure showed no evidence of severe steric interactions between the flanking bases and the arabinose 2′-OH groups (Fig. 2).
Other possible explanations may be considered. The 2′-OH groups in RNA duplexes are prominent in the minor groove of the helix, where they propagate stable and conserved networks of water molecules (51). In addition, the 2′-OH groups lock the ribose-phosphate backbone in a conformation (C3′-endo) that allows water molecules in the major groove to bridge adjacent phosphates, while mediating hydrogen bonding to adjacent sugars (O4′). In this work we show that inversion of stereochemistry at C2′ is accompanied by a conformational change in the ribose sugar from the north (C3′-endo) to an east (O4′-endo) pucker. This pucker, combined with the β orientation of the C2′-substituents, orients the arabinose 2′-OH groups into the major groove. We therefore propose that the low stability observed for duplexes containing ANA strands, e.g. Tm of ANA/ANA < ANA/RNA < RNA/RNA, may be attributed at least in part to the differential hydration pattern of these helices. The differential stability observed for ANA (O4′-endo)/RNA versus 2′F-ANA (O4′-endo)/RNA hybrids may be due to different extents of hydration of the 2′-hydroxy versus 2′-fluorine groups. In fact, Egli and co-workers (20) have suggested that the increased stability of 2′F-ANA/RNA hybrids relative to DNA/RNA hybrids originates not only from the conformational preorganization of the sugar moieties, but also from clathrate-like ordered water molecules around the fluorine atoms. A potential hydrogen bonding interaction between the ‘up’ 2′-fluorine atom and the base protons (H6 or H8) may provide additional preorganization to 2′F-ANA relative to ANA strands (24,25).
The ANA/RNA hybrid mimics quite closely the structure 2′F-ANA/RNA hybrids and average structure of the DNA/RNA hybrids. The minor groove widths and principal helical parameters (such as X-displacement and inclination presented in Table 3) for all three hairpin stems are close to one another and between A- and B-type DNA parameters. Similar structural parameters were observed in DNA/RNA duplexes earlier (44–48). The precise conformation adopted by deoxyriboses in DNA/RNA duplexes is somewhat controversial. Both possibilities—a mixture of south (C2′-endo) and north (C3′-endo) puckers (44,47,53) as well as a more east (O4′-endo) sugar pucker—have been proposed (45,46). NMR studies suggest that the deoxyribose sugars are very flexible and that the complete set of NMR experimental parameters do not fit a single east pucker mode, or an equilibrium between south and north conformers (44,45,47,48). These findings are consistent with the results obtained in this study. PSEUROT analysis of our DNA/RNA hybrid (2) is consistent with of a north–south equilibrium for the deoxy G9-C12 residues with some predominance of north form typical for DNA (Py-rich)/RNA (Pu-rich) hybrids (47,54); however, the NOE data clearly indicate that the O4′-endo conformer is populated as well. Based on our work on the 2′F-ANA/RNA duplex, we recently suggested that the DNA strands in DNA/RNA hybrids should be modelled as a mixture of three conformers; C2′-endo, C3′-endo and O4′-endo (24). In the case of ANA/RNA and 2′F-ANA/RNA hybrids, both H1′-H4′ NOE distances (d = 2.35–2.5 Å) and J-coupling data imply the existence of a single O4′-endo sugar pucker for the antisense strand. We note that a 20% O4′-endo is necessary to yield 2.8 Å average H1′-H4′ NOE distance as observed in the DNA strand of our DNA/RNA hybrids (Fig. 3C) (24). Assuming a three-conformer equilibrium model, the remaining 80% would be a mixture of C2′-endo and C3′-endo, in order to account for the observed scalar couplings. For comparison, a pure south–north equilibrium (as seen for the loop DNA residues) would yield H1′-H4′ NOE distances of 3.0–3.3 Å.
Table 3. Rangesa of minor groove widths and global helical parameters for stem base pairs in average minimized structures of the hairpins and in canonical A- and B-types of DNA.
| Structure |
Minor groove width (Å) |
X-displacement (Å) |
Inclination (˚) |
Rise (Å) |
Helical twist (˚) |
| ANA/RNA (1) | 9.0 ± 0.2 | –2.5 ± 0.4 | 5.5 ± 2.5 | 2.8 ± 0.2 | 33.7 ± 4.5 |
| DNA/RNA (2) | 9.2 ± 0.2 | –3.3 ± 0.5 | 10.5 ± 1.5 | 2.7 ± 0.2 | 35.0 ± 4.2 |
| 2′F-ANA/RNA (3) | 8.8 ± 0.3 | –3.0 ± 0.2 | 2.9 ± 1.3 | 3.1 ± 0.2 | 34.0 ± 2.3 |
| A-type DNA | 11.1 | –5.4 | 19.3 | 2.6 | 32.7 |
| B-type DNA | 5.9 | –0.7 | –6.0 | 3.4 | 36.1 |
aAverage values for these ranges with corresponding deviations for the set of stem base pairs are shown. Helical parameters were calculated by CURVES program (40).
Reid and co-workers (45,46) have shown that DNA/RNA hybrid duplexes have a minor groove width that is intermediate between A- and B-form duplexes. The calculated values for the minor groove widths of ANA/RNA, 2′F-ANA/RNA and the DNA/RNA studied here are ∼9.0 ± 0.5 Å. Since ANA/RNA, 2′F-ANA/RNA and the native DNA/RNA hybrids are substrates of RNase H, this supports the hypothesis made earlier by Reid and co-workers (46) that the uniqueness of the minor groove width in hybrids is highly important for RNase H recognition and cleavage of the RNA strand.
While DNA/RNA and ANA/RNA (and 2′F-ANA/RNA) hybrids have many conformational and biochemical properties in common, there are some differences when examined in sufficient detail. DNA/RNA hybrids are generally (but not always) better substrates for RNase H than ANA/RNA (10) and 2′F-ANA/RNA hybrids (M.J.Damha, M.A.Parniak, K.Viazovkina, C.J.Wilds, C.-N.Lok and K.-C.Min, unpublished results). Combined analysis of NOE and J-coupling data have indicated that the arabinose sugars (2′OH or 2′F) are conformationally ‘rigid’ and adopt almost exclusively the O4′-endo pucker. This contrasts with the ‘dynamic’ sugar conformation observed in the DNA strands of DNA/RNA hybrid duplexes, where a mixture of conformers likely co-exist (O4′, C2′ and C3′-endo) (24). The energy barrier for interconversion between ANA, and particularly 2′F-ANA, sugar conformers, is higher due to the large gauche effect from the 2′-substituent (OH or F) and the furanose ring oxygen (O4′) (25). These observations hint at a correlation between dynamics of the antisense strand and the efficiency with which RNase H can cleave the RNA complement. We therefore propose that the greater conformational freedom (‘flexibility’) of 2′-deoxyribose relative to the arabinose sugars account, at least in part, for the more rapid rate of cleavage of DNA/RNA hybrids relative to ANA/RNA and 2′F-ANA/RNA hybrids. Although we have no formal proof of this hypothesis, we note that the conformationally locked bc-ANA analog (O4′-endo) forms a hybrid with RNA that does not activate RNase H (23). An induced-fit model based on the flexibility of deoxyribose and on the hydration patterns of the duplexes was also proposed (55).
We conclude that for an antisense oligonucleotide compound to elicit RNase H activity, it must fulfill the following criteria: (i) adopt an east DNA-like conformation (O4′-endo); (ii) possess phosphodiester linkages (PNA and methylphosphonate-DNA do not elicit RNase H activity); (iii) display sugar mobility (flexibility), although some preorganization is desirable (e.g. 2′F-ANA) to increase binding affinity towards the target RNA; (iv) form hybrid duplexes with a proper characteristic pattern of hydration (55,56), and with a minor groove width that is intermediate between those of pure A- and B-form duplexes (46). This knowledge may prove helpful in the design of future antisense modifications.
CONCLUSIONS
Structural parameters of the ANA/RNA hybrid, especially minor groove widths, were found to be similar to structural parameters in 2′F-ANA/RNA and average structural parameters in the native DNA/RNA hybrids. The structural similarities between DNA/RNA hybrids and the present ANA/RNA hybrid suggest that the overall structure assumed by these duplexes may in fact be an important factor responsible for RNase H recognition and cleavage of hybrid duplexes (46,57).
Contrary to what was recently proposed through computational studies (21), we find that the ANA residues of the hybrid assume conformations in the O4′-endo (east) range. The same is observed for the arabinose sugars of 2′F-ANA/RNA hybrid duplexes. This contrasts strongly with the ‘dynamic’ sugar conformation in the DNA strands of DNA/RNA hybrids, where a mixture of conformers is likely to co-exist (O4′ + C2′ + C3′-endo) (24). Since all these hybrids are substrates of RNase H, our findings support the hypothesis that the geometry of the antisense strand that is processed by RNase H falls within the O4′-endo range (23,24). The rather low stability observed for duplexes containing ANA strands, e.g. Tm of ANA/ANA < ANA/RNA < RNA/RNA, may be attributed to the different hydration pattern of these helices. The differential stability observed for ANA/RNA versus 2′F-ANA/RNA hybrids may be due to different extents of hydration of the 2′-hydroxy versus 2′-fluorine groups. Differential rates of RNase H cleavage as observed for ANA/RNA versus DNA/RNA hybrids (10) are likely related to differences in the binding affinity and sugar flexibility associated with ANA versus DNA strands.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by grants from the Natural Sciences and Engineering Research Council of Canada (M.J.D.) and the Canadian Institutes of Health Research (K.G. and M.J.D.).
References
- 1.Uhlmann E. and Peyman,A. (1990) Chem. Rev., 90, 543–584. [Google Scholar]
- 2.Knorre D.G., Vlassov,V.V., Zarytova,V.F., Lebedev,A.V. and Fedorova,O.S. (1994) Design and Targeted Reactions of Oligonucleotide Derivatives. CRC Press, Boca Raton, FL.
- 3.Manoharan M. (1999) 2′-Carbohydrate modifications in antisense oligonucleotide therapy: importance of conformation, configuration and conjugation. Biochim. Biophys. Acta, 1489, 117–130. [DOI] [PubMed] [Google Scholar]
- 4.Wengel J. (1999) Synthesis of 3′-C- and 4′-C-branched oligodeoxynucleotides and the development of locked nucleic acid (LNA). Acc. Chem. Res., 32, 301–310. [Google Scholar]
- 5.Walder R.Y. and Walder,J.A. (1988) Role of RNase H in hybrid-arrested translation by antisense oligonucleotides. Proc. Natl Acad. Sci. USA, 85, 5011–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue H., Hayase,Y., Iwai,S. and Ohtsuka,E. (1987) Sequence-dependent hydrolysis of RNA using modified oligonucleotide splints and RNase H. FEBS Lett., 215, 327–330. [DOI] [PubMed] [Google Scholar]
- 7.Monia B.P., Lesnik,E.A., Gonzalez,C., Lima,W.F., McGee,D., Guinosso,C.J., Kawasaki,A.M., Cook,P.D. and Freier,S.M. (1993) Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 8.Zhou W. and Agrawal,S. (1998) Mixed-backbone oligonucleotides as second-generation antisense agents with reduced phosphorothioate-related side effects. Bioorg. Med. Chem. Lett., 8, 3269–3274. [DOI] [PubMed] [Google Scholar]
- 9.Damha M.J., Wilds,C.J., Noronha,A., Brukner,I., Borkow,G., Arion,D. and Parniak,M.A. (1998) Hybrid of RNA and arabinonucleic acids (ANA and 2′F-ANA) are substrates of ribonuclease H. J. Am. Chem. Soc., 120, 12976–12977. [Google Scholar]
- 10.Noronha A.M., Wilds,C.J., Lok,C.N., Viazovkina,K., Arion,D., Parniak,M.A. and Damha,M.J. (2000) Synthesis and biophysical properties of arabinonucleic acids (ANA): circular dichroic spectra, melting temperatures, and ribonuclease H susceptibility of ANA–RNA hybrid duplexes. Biochemistry, 39, 7050–7062. [DOI] [PubMed] [Google Scholar]
- 11.Lima W.F. and Crooke,S.T. (1997) Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide–RNA hybrids. Biochemistry, 36, 390–398. [DOI] [PubMed] [Google Scholar]
- 12.Pierters J.M., de Vroom,E., van der Marel,G.A., van Boom,J.H., Koning,T.M.G., Kaptein,R. and Altona,C. (1990) Hairpin structures in DNA containing arabinofuranosylcytosine. A combination of nuclear magnetic resonance and molecular dynamics. Biochemistry, 29, 788–799. [DOI] [PubMed] [Google Scholar]
- 13.Gao Y.G., van der Marel,G.A., van Boom,J.H. and Wang,A.H. (1991) Molecular structure of a DNA decamer containing anticancer nucleoside arabinosylcytosine: conformational perturbation by arabinosylcytosine in B-DNA. Biochemistry, 30, 9922–9931. [DOI] [PubMed] [Google Scholar]
- 14.Schweitzer B.I., Mikita,T., Kellogg,G.W., Gardner,K.H. and Beardsley,G.P. (1994) Solution structure of a DNA dodecamer containing the anti-neoplastic agent arabinocytosine: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement. Biochemistry, 33, 11460–11475. [PubMed] [Google Scholar]
- 15.Gotfredsen C.H., Spielmann,H.P., Wengel,J. and Jacobsen,J.P. (1996) Structure of a DNA duplex containing a single 2′-O-methyl-β-d-araT: combined use of NMR, restrained molecular dynamics, and full relaxation matrix refinement. Bioconjug. Chem., 7, 680–688. [DOI] [PubMed] [Google Scholar]
- 16.Gmeiner W.H., Konerding,D. and James,T.L. (1999) Effect of Cytarabine on the NMR structure of a model Okazaki fragment from the SV40 genome. Biochemistry, 38, 1166–1175. [DOI] [PubMed] [Google Scholar]
- 17.Teng M.K., Liaw,Y.C., van der Marel,G.A., van Boom,J.H. and Wang,A.H. (1989) Effects of the O2′ hydroxyl group on Z-DNA conformation: structure of Z-RNA and (araC)-[Z-DNA]. Biochemistry, 28, 4923–4928. [DOI] [PubMed] [Google Scholar]
- 18.Zhang H., van der Marel,G.A., van Boom,J.H. and Wang,A.H. (1992) Conformational perturbation of the anticancer nucleotide arabinosylcytosine on Z-DNA: molecular structure of (araC-dG)3 at 1.3 Å resolution. Biopolymers, 32, 1559–1569. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda H., Fernandez,R., Wilk,A., Barchi,J.J., Huang,X. and Marquez,V.E. (1998) The effect of two antipodal fluorine-induced sugar puckers on the conformation and stability of the Dickerson–Drew dodecamer duplex [d(CGCGAATTCGCG)]2. Nucleic Acids Res., 26, 2237–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berger I., Tereshko,V., Ikeda,H., Marques,V.E. and Egli,M. (1998) Crystal structures of B-DNA with incorporated 2′-deoxy-2′-fluoro-arabino-furanosyl thymines: implications of conformational preorganization for duplex stability. Nucleic Acids Res., 26, 2473–2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venkateswarlu D. and Ferguson,D.M. (1999) Effect of C2′-substitution on arabinonucleic acid structure and conformation. J. Am. Chem. Soc., 121, 5609–5610. [Google Scholar]
- 22.Christensen N.K., Petersen,M., Nielsen,P., Jacobsen,J.P., Olsen,C.E. and Wengel,J. (1998) A novel class of oligonucleotide analogues containing 2′-O,3′-C-linked [3.2.0]bicycloarabinonucleoside monomers: synthesis, thermal affinity studies, and molecular modeling. J. Am. Chem. Soc., 120, 5458–5463. [Google Scholar]
- 23.Minasov G., Teplova,M., Nielsen,P., Wengel,J. and Egli,M. (2000) Structure basis of cleavage by RNase H of hybrids of arabinonucleic acids and RNA. Biochemistry, 39, 3525–3532. [DOI] [PubMed] [Google Scholar]
- 24.Trempe J.-F., Wilds,C.J., Denisov,A.Y., Pon,R.T., Damha,M.J. and Gehring,K. (2001) NMR solution structure of an oligonucleotide hairpin with a 2′F-ANA/RNA stem: implications for RNase H specificity toward DNA/RNA hybrid duplexes. J. Am. Chem. Soc., 123, 4896–4903. [DOI] [PubMed] [Google Scholar]
- 25.Wilds C.J. and Damha,M.J. (2000) 2′-Deoxy-2′-fluoro-β-d-arabinonucleotides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res., 28, 3625–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barchi J.J., Jeong,L.S., Siddiqui,M.A. and Marquez,V.E. (1997) Conformational analysis of the complete series of 2′ and 3′ monofluorinated dideoxyuridines. J. Biochem. Biophys. Methods, 34, 11–29. [DOI] [PubMed] [Google Scholar]
- 27.Chwang A.K. and Sundaralingam,M. (1973) Intramolecular hydrogen bonding in 1-β-d-arabinofuranosylcytosine (ara-C). Nature New Biol., 243, 78–79. [PubMed] [Google Scholar]
- 28.Ekiel I., Remin,M., Darzynkiewich,E. and Shugar,D. (1979) Correlations of conformational parameters and equilibrium conformational states in a variety of β-d-arabinonucleosides and their analogues. Biochim. Biophys. Acta, 562, 177–191. [DOI] [PubMed] [Google Scholar]
- 29.Saenger W. (1984) Principles of Nucleic Acids Structure. Springer-Verlag, New York, NY.
- 30.Ford H., Dai,F., Mu,L., Siddiqui,M.A., Nickaus,M.C., Anderson,L., Marquez,V.E. and Barchi,J.J. (2000) Adenosine deaminase prefers a distinct sugar ring conformation for binding and catalysis: kinetic and structural studies. Biochemistry, 39, 2581–2592. [DOI] [PubMed] [Google Scholar]
- 31.Guinnares P.A. and Damha,M.J. (1994) Hybridization properties of oligoarabinonucleotides. Can. J. Chem., 72, 909–918. [Google Scholar]
- 32.Varani G., Cheong,C. and Tinoco,I. (1991) Structure of an unusually stable RNA hairpin. Biochemistry, 30, 3280–3289. [DOI] [PubMed] [Google Scholar]
- 33.James J.K. and Tinoco,I. (1993) The solution structure of a d[C(TTCG)G] DNA hairpin and comparison to the unusually stable RNA analogue. Nucleic Acids Res., 21, 3287–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piotto M., Saudek,V. and Sklenar,V. (1992) Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR, 2, 661–666. [DOI] [PubMed] [Google Scholar]
- 35.Brünger A.T. (1992) X-PLOR 3.1, A System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT.
- 36.Klinck R., Sprules,T. and Gehring,K. (1997) Structural characterization of three RNA hexanucleotide loops from the internal ribosome entry site of polioviruses. Nucleic Acids Res., 25, 2129–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim C.H., Kao,C.C. and Tinoco,I. (2000) RNA motifs that determine specificity between a viral replicase and its promoter. Nature Struct. Biol., 7, 415–422. [DOI] [PubMed] [Google Scholar]
- 38.Wüthrich K. (1986) NMR of Protein and Nucleic Acids. John Wiley & Sons, New York, NY.
- 39.de Leeuw F.A.A.M. and Altona,C. (1983) Quantum Chemistry Program Exchange, no. 463: PSEUROT 3B. Indiana University, Bloomington, IN.
- 40.Lavery R. and Sklenar,H. (1997) Curves 5.2, Helical Analysis of Irregular Nucleic Acids. Laboratory de Biochimie Theorique, CNRS URA 77, Paris.
- 41.van Wijk J., Huckriede,B.D., Ippel,J.H. and Altona,C. (1992) Furanose sugar conformations in DNA from NMR coupling constants. Methods Enzymol., 211, 286–306. [DOI] [PubMed] [Google Scholar]
- 42.Rinkel L.J. and Altona,C. (1987) Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton–proton coupling constants: a graphical method. J. Biomol. Struct. Dyn., 4, 621–649. [DOI] [PubMed] [Google Scholar]
- 43.Kim S.G., Lin,L.J. and Reid,B.R. (1992) Determination of nucleic acid backbone conformation by 1H NMR. Biochemistry, 31, 3564–3574. [DOI] [PubMed] [Google Scholar]
- 44.Gonzalez C., Stec,W., Reynolds,M.A. and James,T.L. (1995) Structure and dynamics of a DNA–RNA hybrid duplex with a chiral phosphorothionate moiety: NMR and molecular dynamics with conventional and time-averaged restraints. Biochemistry, 34, 4969–4982. [DOI] [PubMed] [Google Scholar]
- 45.Fedoroff O.Y., Ge,Y. and Reid,B.R. (1997) Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis. J. Mol. Biol., 269, 225–239. [DOI] [PubMed] [Google Scholar]
- 46.Fedoroff O.Y., Salazar,M. and Reid,B.R. (1993) Structure of a DNA–RNA hybrid duplex. Why RNase H does not cleave pure RNA. J. Mol. Biol., 233, 509–523. [DOI] [PubMed] [Google Scholar]
- 47.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) Solution structures of DNA–RNA hybrids with purine-rich and pyrimidine-rich strands: comparison with the homologous DNA and RNA duplexes. Biochemistry, 37, 73–80. [DOI] [PubMed] [Google Scholar]
- 48.Bachelin M., Hessler,G., Kurtz,G., Hacia,J.G., Dervan,P.B. and Kessler,H. (1998) Structure of a stereoregular phosphorothionate DNA/RNA duplex. Nature Struct. Biol., 5, 271–275. [DOI] [PubMed] [Google Scholar]
- 49.Gorenstein D.G., Meadows,R.P., Metz,J.T., Nikonowicz,E. and Post,C.B. (1990) 31P and 1H 2-dimensional NMR and NOESY-distance restrained molecular dynamics methodologies for defining sequence-specific variations in duplex oligonucleotides. A comparison of NOESY two-spin and relaxation rate matrix analyses. Adv. Biophys. Chem., 1, 47–124. [Google Scholar]
- 50.Wang A.H., Fujii,S., van Boom,J.H., van der Marel,G.A., van Boeckel,S.A. and Rich,A. (1982) Molecular structure of r(GCG)d(TATACGC): a DNA–RNA hybrid helix joined to double helical DNA. Nature, 299, 601–604. [DOI] [PubMed] [Google Scholar]
- 51.Egli M., Portmann,S. and Usman,N. (1996) RNA hydration: a detailed look. Biochemistry, 35, 8489–8494. [DOI] [PubMed] [Google Scholar]
- 52.Mikita T. and Beardsley,G.P. (1988) Functional consequences of the arabinosylcytosine structural lesion in DNA. Biochemistry, 27, 4698–4705. [DOI] [PubMed] [Google Scholar]
- 53.Lane A.N., Ebel,S. and Brown,T. (1993) NMR assignments and solution conformation of the DNA.RNA hybrid duplex d(GTGAACTT).r(AAGUUCAC). Eur. J. Biochem., 215, 297–306. [DOI] [PubMed] [Google Scholar]
- 54.Xiong Y. and Sundaralingam,M. (2000) Crystal structure oa a DNA–RNA hybrid duplex with a polypurine RNA r(gaagaagag) with a complementary polypyrimidine DNA d(CTCTTCTTC). Nucleic Acids Res., 28, 2171–2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szyperski T., Gotte,M., Billeter,M., Perola,E., Cellai,L., Heumann,H. and Wüthrich,K. (1999) NMR structure of the chimeric hybrid duplex r(gcaguggc).r(gcca)d(CTGC) comprising the tRNA–DNA junction formed during initiation of HIV-1 reverse transcription. J. Biomol. NMR, 13, 343–355. [DOI] [PubMed] [Google Scholar]
- 56.Hsu S.T., Chou,M.T. and Cheng,J.W. (2000) The solution structure of [d(CGC)r(aaa)d(TTTGCG)]2: hybrid junctions flanked by DNA duplexes. Nucleic Acids Res., 28, 1322–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nakamura H., Oda,Y., Iwai,S., Inore,H., Ohtsuka,E., Kanaya,S., Kimura,S., Katsuda,C., Katayannagi,K., Morikawa,K., Miyashiro,H. and Ikehara,M. (1991) How does RNase recognize a DNA–RNA hybrid? Proc. Natl Acad. Sci. USA, 88, 11535–11539. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.