Abstract
Public availability of biological sequences is essential for their widespread access and use by the research community. The Universal Protein Resource (UniProt) is a comprehensive resource for protein sequence and functional data. While most protein sequences entering UniProt are imported from other source databases containing nucleotide or 3D structure data, protein sequences determined at the protein level can be submitted directly to UniProt. To this end, UniProt provides a web interface called SPIN. This service enables researchers to make their de novo-sequenced proteins available to the scientific community and acquire UniProt accession numbers for use in publications. This unit explains the process of submitting a protein sequence to UniProt using SPIN. The basic protocol describes all the necessary steps for a single sequence. A support protocol gives guidance on how best to deal with exceptionally large datasets.
Keywords: UniProt, submission, direct protein sequencing
INTRODUCTION
UniProt provides a web interface called SPIN for researchers to submit protein sequences for which there is evidence at the protein level. SPIN covers an important gap in a data submission landscape dominated by nucleotide data. Any protein sequence determined via Edman degradation or the manual interpretation of mass spectrometry data is acceptable. However, sequence data derived from peptide mass fingerprinting or any other mass spectrometry technique reliant on database searches or any sequence resulting from the translation of nucleotide sequences will not be accepted and should be submitted to the appropriate dedicated databases.
SPIN is hosted at the European Bioinformatics Institute and can be accessed at https://www.ebi.ac.uk/swissprot/Submissions/spin/. The following basic protocol describes all the necessary steps to submit a protein sequence whose primary structure has been determined at the protein level to UniProt via the web tool SPIN and receive an accession number in return. Researchers can send any UniProt-related questions they might have prior to submitting data to help@uniprot.org. While SPIN was originally designed for small-scale submissions, large-scale submissions are possible and the mechanism for that is described in the support protocol.
BASIC PROTOCOL 1: SUBMIT A SINGLE PROTEIN SEQUENCE
UniProt SPIN is a dedicated interface for protein sequence submissions and can be accessed at https://www.ebi.ac.uk/swissprot/Submissions/spin/. As some communication between submitter and UniProt curators is always necessary, registration is required prior to submission. Once registered and signed in, SPIN presents a web form for entering data. The minimum amount of information required for a valid submission is small (see Table 1) although submitters can provide a wealth of data pertaining to a given protein should they choose to. Once a sequence is submitted, it is processed by UniProt curators, who will contact the submitter with any questions they might have and/or send a UniProt accession.
Table 1.
Minimum required information for a submission
| Type of information | Example |
|---|---|
| Protein name | Pyrokinin |
| Sequencing method | Edman degradation |
| Amino acid sequence | DPKFSPRL |
| Species of origin | Bacteria ferula |
| Citation | Warner K., Pichler K., New pyrokinin from a stick insect (In preparation) |
| Confidentiality preference | Publish without further notice |
Necessary Resources
An up-to-date Web browser such as Firefox, Chrome or Safari
A valid email address
Protocol Steps
Create an account to use SPIN
Step 1: Visit the SPIN web page at https://www.ebi.ac.uk/swissprot/Submissions/spin/ (Fig. 1).
Figure 1.

SPIN login page. Sign in as an existing user or register as a new user. Note the explained scope of SPIN submissions. Use one of the other options for submitting sequences that do not qualify for SPIN.
Step 2: Click on ‘Create an account’ to register as a SPIN user (Fig. 2). Take care to provide a valid email address.
Figure 2.

Web form for registering as a SPIN user. Mandatory fields are marked with an asterisk.
In the form, mandatory fields are marked with an asterisk. Please provide a valid email address as the system will send a confirmation email as part of the registration process. The email address is also vital for communication during the submission process as UniProt curators send questions and accession numbers via email. Any registration details provided will not be used outside SPIN submissions.
Step 3: Activate your account by clicking on the link provided in the registration email.
The email sender will be uniprot-submissions@ebi.ac.uk. Following the link will take you to the SPIN start page with a green banner near the top stating that you have successfully activated your account.
Step 4: Use the registered email address and password to sign in to SPIN (Fig. 1).
This will take you to a new page where you can start a new submission. The start page will also list any saved unfinished submissions (Fig. 3).
Figure 3.

SPIN start page after signing in. This page lists saved unfinished submissions if there are any. Start a new submission by clicking on the button ‘Create a new submission’. Edit saved submissions by clicking on their name.
Start a new submission
Step 5: Click on the button ‘Create a new submission’ (Fig. 3). This will take you to a web form.
Some fields of this form are mandatory but most are optional (see table 1 for the minimal information needed for a submission). Once started, submissions can be suspended and finished later at any time; to do this just click the button ‘Finish later’ at the bottom of the web form. You will then be taken back to the top-level page where your submission(s) in progress are now listed and waiting to be finished or discarded.
Step 6: Provide a name for the amino acid sequence you are submitting. Paste or type the name into the field ‘Protein name’ (Fig. 4).
Figure 4.
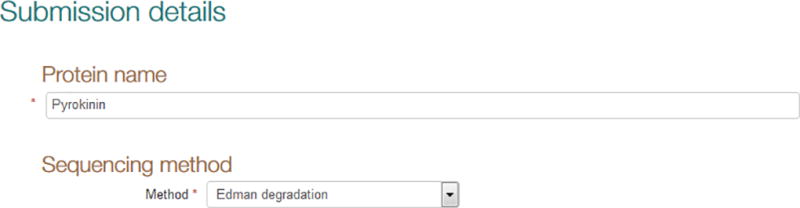
Providing protein name and sequencing method.
Step 7: Select the method used to determine the amino acid sequence you are submitting from the drop-down menu in the field ‘Sequencing method’ (Fig. 4). If the menu does not list the method you employed, choose ‘Other’ and provide a short description in the text box.
Depending on which value you choose from the drop-down menu, a warning may be displayed reminding you that only amino acid sequences determined at the protein level are valid for submission to UniProt via SPIN; options for other resources to submit to are displayed as part of such a warning as appropriate.
Step 8: Specify whether you are submitting a complete sequence or (a) fragment(s) (Fig. 5).
Figure 5.
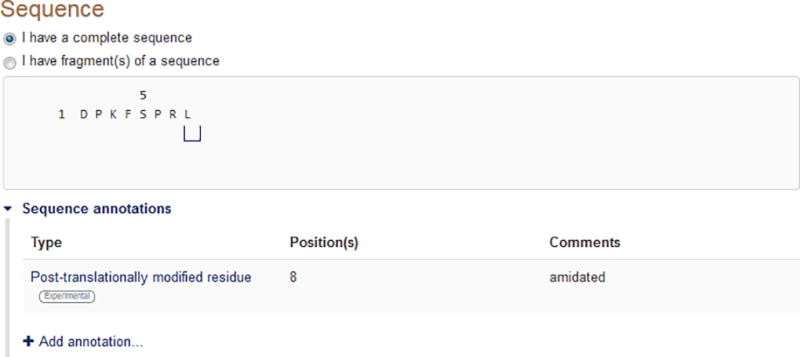
Providing the protein sequence and residue-specific annotations, if any.
A complete sequence can mean either a complete precursor sequence or, more often, a mature and functional protein or peptide. For example, the amino acid sequence of a secreted protein minus its signal sequence would be considered complete as it constitutes a mature functional protein/entity. However, if you have determined only the N-terminal ten amino acids of a protein that is much longer, this is considered a fragment.
Step 9: Provide the amino acid sequence. You can either type the sequence into the text box, copy-paste it in or upload it from a file. Click ‘Save’ when you are done.
When copy-pasting the sequence, only paste the amino acid string. When uploading from a file, the sequence can be in FASTA format; if the file contains several FASTA-formatted sequences, SPIN will take only the first one. SPIN accepts the standard amino acid one-letter codes as well as the non-standard U (selenocysteine) and O (pyrrolysine) codes.
Step 10: (optional) Add residue-specific sequence annotations (Fig. 6). Annotations will be marked on the sequence as well as in a table (Fig. 5). They can be edited or deleted.
Figure 6.

Specifying residue-specific annotations.
Step 11: If you specified ‘fragment’ in step 8 and you would like to add more fragments then click ‘Add fragment’ and repeat steps 9 and 10 as often as required.
Step 12: State the scientific name of the species from which the amino acid was isolated. Once you start typing the species name, SPIN will offer suggestions for auto-completion (Fig. 7).
Figure 7.

Providing the species of origin of the submitted protein sequence.
Based on the species name, SPIN will try to populate the field ‘taxonomy ID’ with an identifier from the NCBI taxonomy database. If the field remains empty, the curator processing your submission will add it for you. You can also provide additional information on a species common name, a strain and/or tissue from which you have isolated the protein.
Step 13: Click on ‘Add citation’ to provide a citation for the submitted sequence (Fig. 8).
Figure 8.

Providing a citation.
In many cases, the submission of protein sequences to UniProt accompanies or even precedes publication in a peer-reviewed journal so the citation provided can consist of just a single author name together with the default citation type ‘Unpublished’ and a generic title. For convenience, you can copy citation details from a previous submission if one exists.
Step 14: (optional) Add protein properties and data (Fig. 9). Some common data types such as the mass spectrometry derived molecular weight or the function of the protein are listed at the top of this section. The subsection ‘Additional properties’ provides further options. Enzyme-specific data can be entered in the subsection ‘Is this protein an enzyme?’. A UniProt curator will review all data provided to ensure correctness and proper formatting.
Figure 9.

Adding additional data in the ‘Protein properties’ section.
Finish or save your submission
Step 15: Specify your preferences regarding the confidentiality of the submitted sequence (Fig. 10). Following checks by a curator, your data can be released immediately or be kept confidential. If you would like your sequence(s) to be kept confidential, you can either inform us once an associated paper describing the sequence(s) has been published or define a date after which your data can be made public.
Figure 10.

Stating preferences with regard to confidentiality and adding final remarks, if any. Click ‘Submit’ to finish a submission, click ‘Finish later’ to save without finishing.
Publishing of non-confidential sequences and release of previously confidential sequences is not instantaneous. It usually takes several weeks for any sequence submitted to us to become publicly available on the UniProt website due to the length of the UniProt release cycle.
Step 16: (optional) Add a comment to highlight any issues you think important for a UniProt curator to know about (Fig. 10).
Step 17: Send the submission to a UniProt curator by clicking ‘Submit’ at the bottom of the web form. If you would rather save the submission and come back to it later, click ‘Finish later’ (Fig. 10).
Clicking ‘Finish later’ will take you back to the screen shown in Fig. 3, which lists saved unfinished submissions. To continue working on them, click on a protein name or click on the pencil icon, which appears when hovering over a submission entry.
Wait for feedback
Step 18: Check email for an automated notification of a successful submission.
The email sender will be uniprot-submissions@ebi.ac.uk and the subject line will read ‘Your SPIN submission SPIN<number> has been completed’ where <number> will be an 8-digit number. The email body will also list details of the submission you have provided.
Step 19: Wait for a UniProt curator to contact you with any questions they might have. If there are no questions and the submission is valid we will send you a UniProt accession number once your submission has been processed by a curator.
SUPPORT PROTOCOL 1: SUBMIT A LARGE SET OF PROTEIN SEQUENCES
The basic protocol for SPIN submissions is geared towards submitting single or, when followed repeatedly, several sequences. Using it for large numbers of sequences may become tedious. If you have a large set of sequences, please consider the following support protocol.
Necessary Resources
Same as for basic protocol 1
Protocol Steps
Step 1: Submit one representative sample sequence as described in the basic protocol, taking care to explain in a comment (see basic protocol step 16) that you would like to submit more sequences of the same kind and indicating how many there are (Fig. 10).
This submission should contain data for all the fields you would like to provide data for in the remainder of the sequences.
Step 2: The UniProt curator dealing with your submission will contact you with any questions regarding your test sequence. Once we have established the validity of your submission, we will ask you for a FASTA-formatted file containing the rest of the sequences with individual data points included in the header of each sequence. If, for example, you have many neuropeptides from different insects together with mass spectrometry data and tissue specificity for each sequence the header specification might look like this: >protein_name,species,mass,tissue#1,…,tissue#n
COMMENTARY
Background Information
UniProt aids scientific discovery by collecting, interpreting and organizing information so that it is easy to access and use. It saves researchers countless hours of work in monitoring and collecting this information themselves. UniProt provides access to most of the protein sequences in the public domain and sequences can be referenced unambiguously using accession numbers. The UniProt Knowledgebase (UniProtKB) is the central hub for the collection of functional information and contains the expertly curated UniProtKB/Swiss-Prot section and the un-reviewed automatically annotated UniProtKB/TrEMBL section. The vast majority of protein sequences in UniProtKB are derived from translations from nucleotide sequences with only a fraction having evidence at protein level (ca. 17.5% of sequences in UniProtKB/Swiss-Prot and ca. 0.1% of sequences in UniProtKB/TrEMBL). For this reason, sequence data generated by direct protein sequencing are valuable as they can prove the existence of a protein. Depending on methods used, such data can also describe mature peptides generated from a precursor or report post-translational modifications. Commonly employed direct protein sequencing methods are Edman degradation and the manual interpretation of tandem mass spectrometry data where, in both cases, the analyzed proteins are isolated directly from tissue samples.
Using SPIN, researchers can submit direct protein sequencing data, together with any associated functional data, for inclusion in UniProtKB/Swiss-Prot. Accession numbers are assigned as part of the submission process and these can be used in publications. Thus, SPIN is an important tool for researchers to ensure public availability of their valuable direct protein sequencing data and to contribute to enhancing existing protein sequences.
Significance Statement.
Public availability of biological sequences is essential for their usefulness to the research community. The SPIN web interface enables researchers to submit protein sequences determined at the protein level directly to UniProt. Assigned accession numbers can be used in publications and sequences will be made available on the UniProt website along with any associated submitted functional data, thus directly expanding the publicly available sequence space. Submitted protein sequences provide valuable evidence for the existence of proteins, their isoforms, mature products and post-translational modifications. In short, SPIN is an important service for sharing protein sequence data and ensuring that it is made freely available to the research community.
Acknowledgments
UniProt has been prepared by Alex Bateman, Maria Jesus Martin, Sandra Orchard, Michele Magrane, Emanuele Alpi, Benoit Bely, Mark Bingley, Ramona Britto, Borisas Bursteinas, Hema Bye-A-Jee, Alan Da Silva, Tunca Dogan, Leyla Garcia Castro, Luis Figueira, Penelope Garmiri, George Georghiou, Leonardo Gonzales, Emma Hatton-Ellis, Alexandr Ignatchenko, Vishal Joshi, Dushyanth Jyothi, Jie Luo, Yvonne Lussi, Alistair MacDougall, Mahdi Mahmoudy, Andrew Nightingale, Joseph Onwubiko, Klemens Pichler, Sangya Pundir, Guoying Qi, Alexandre Renaux, Rabie Saidi, Tony Sawford, Aleksandra Shypitsyna, Elena Speretta, Edward Turner, Nidhi Tyagi, Preethi Vasudev, Vladimir Volynkin, Tony Wardell, Kate Warner, Xavier Watkins, Ying Yan, Rossana Zaru and Hermann Zellner at the European Bioinformatics Institute; Ioannis Xenarios, Alan Bridge, Sylvain Poux, Nicole Redaschi, Lucila Aimo, Ghislaine Argoud-Puy, Andrea Auchincloss, Kristian Axelsen, Parit Bansal, Delphine Baratin, Marie-Claude Blatter, Brigitte Boeckmann, Jerven Bolleman, Emmanuel Boutet, Lionel Breuza, Cristina Casal-Casas, Edouard de Castro, Elisabeth Coudert, Beatrice Cuche, Mikael Doche, Dolnide Dornevil, Severine Duvaud, Anne Estreicher, Livia Famiglietti, Marc Feuermann, Elisabeth Gasteiger, Sebastien Gehant, Vivienne Gerritsen, Arnaud Gos, Nadine Gruaz-Gumowski, Ursula Hinz, Chantal Hulo, Nevila Hyka-Nouspikel, Florence Jungo, Guillaume Keller, Arnaud Kerhornou, Vicente Lara, Philippe Lemercier, Damien Lieberherr, Thierry Lombardot, Xavier Martin, Patrick Masson, Anne Morgat, Teresa Neto, Nevila Nouspikel, Ivo Pedruzzi, Sandrine Pilbout, Monica Pozzato, Manuela Pruess, Catherine Rivoire, Christian Sigrist, Karin Sonesson, Sylvie Staehli, Andre Stutz, Shyamala Sundaram, Michael Tognolli, Laure Verbregue, Anne-Lise Veuthey and Daniel Walther at the SIB Swiss Institute of Bioinformatics; Cathy H. Wu, Cecilia N. Arighi, Leslie Arminski, Chuming Chen, Yongxing Chen, John S. Garavelli, Hongzhan Huang, Kati Laiho, Peter McGarvey, Darren A. Natale, Karen Ross, C.R. Vinayaka, Qinghua Wang, Yuqi Wang, Lai-Su Yeh and Jian Zhang at the Protein Information Resource.
FUNDING
National Institutes of Health [U41HG007822, U41HG002273, R01GM080646, P20GM103446, U01GM120953]; British Heart Foundation [RG/13/5/30112]; Parkinson's Disease United Kingdom [G-1307]; Swiss Federal Government through the State Secretariat for Education, Research and Innovation and European Molecular Biology Laboratory core funds.
Footnotes
INTERNET RESOURCES
https://www.ebi.ac.uk/swissprot/Submissions/spin/help
Documentation for all fields in the SPIN web form including examples.
This is the URL where data submitted via SPIN will ultimately become available.


