Abstract
Leishmaniasis are infectious diseases caused by parasites of genus Leishmania that affects 12 million people in 98 countries mainly in Africa, Asia, and Latin America. Effective treatments for this disease are urgently needed. In this study, we present a computer-aided approach to investigate a set of 32 recently synthesized chalcones and chalcone-like compounds to act as anti-leishmanial agents. As a result, nine most promising compounds and three potentially inactive compounds were experimentally evaluated against Leishmania infantum amastigotes and mammalian cells. Four compounds exhibited EC50 in the range of 6.2–10.98 μM. In addition, two compounds, LabMol-65 and LabMol-73, exhibited cytotoxicity in macrophages >50 μM that resulted in better selectivity than Amphotericin B. These two compounds also demonstrated low cytotoxicity and high selectivity towards Vero cells. The results of target fishing followed by homology modeling and docking suggest that these chalcone compounds could act in Leishmania because of their interaction with cysteine proteases, such as procathepsin L. Finally, we have provided structural recommendations for designing of new anti-leishmanial chalcones.
Keywords: Antileishmanial agents, Nitroheterocycle chalcones, Selectivity, Molecular modeling, Target fishing
TOC image
Endemic in 88 countries, leishmaniasis are infectious diseases caused by parasites of genus Leishmania.1,2 According to World Health Organization (WHO), around 1.3 million new cases occur per year.3 Visceral leishmaniasis (VL) is the most severe form, in which vital organs are affected causing chronic fever, liver issues, spleen enlargement, anemia, and other blood problems.4,5
The first-line drugs for treatment of leishmaniasis are the pentavalent antimonials, meglumine antimoniate (Glucantime®) and sodium stibogluconate (Pentosan®). If they fail, second-line drugs such as pentamidine, amphotericin B and miltefosine are used.2 However, the treatment is lengthy, toxic and painful for patients. Moreover, the resistance against the available drugs has increased over the years. Additionally, the high cost of some therapies has limited their use. Thus, there is an urgent need for the discovery of new drugs and targets for this neglected disease.4
Recent advances in genomics have triggered a shift in drug discovery from the paradigm of focusing on strong single-target interaction to more global and comparative analysis of multi-targets network.6,7 In silico methods, including target- and ligand-based strategies, are widely used in industry and academia complementary to experimental techniques.8 For instance, in silico target fishing can enable the discovery of a number of putative targets for a given set of small molecules with known biological effects.9
Chalcones are biologically classified as secondary metabolites of low molecular weight. In medicinal chemistry, they are considered privileged structures for research and development of new drugs, due to the diversity of substituents that can be linked to conjugated system scaffold.10 Chemically, chalcones are classified as 1,3-diaryl-2-propen-1-ones and possess a broad spectrum11–24 of properties including antileishmanial activity.25
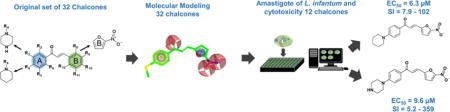
The goal of this study was to identify novel antileishmanial compounds among 32 previously synthesized chalcones and chalcone-like compounds26. The general workflow is shown in Figure 1. Initially, 32 compounds have been submitted to a target fishing approach using pharmacophore modelling, then the 3D structures of selected targets were obtained by homology modeling and we performed molecular docking between the 32 chalcones and the selected targets. Finally, the in vitro biological activity on Leishmania infantum, cytotoxicity on macrophages and Vero cells and selectivity of promising compounds were evaluated experimentally.
Figure 1.
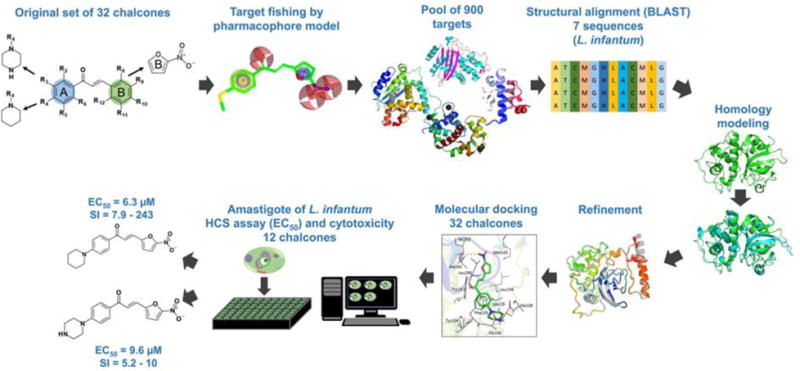
Computer-aided approach to discovery new chalcones with antileishmanial activity.
On the search of potential targets for the antileishmanial hits, we used the PharmMapper server,7,27 a database that is backed up by a large, in-house repertoire of pharmacophore information extracted from all the targets available in TargetBank, DrugBank, BindingDB, and PDTD (Potential Drug Target Database). The original dataset of 32 chalcones and chalcones-like were submitted to the web server, generating a list of targets and a maximum of 300 conformations for each ligand, which were ranked by the fit score to the pharmacophore model. These results are presented on Table S1 (Supporting Information). Then, all targets were aligned on BLAST server28. As a result, 7 sequences were identified as potential targets for L. infantum hits (Table 1), all presenting high primary sequence identity (>30%).
Table 1.
Results obtained from sequence alignment on BLAST.
| Target | Max score | Total score | Query cover | E-value | Identity |
|---|---|---|---|---|---|
| Actin | 590 | 590 | 99% | 0.0 | 70% |
| Casein Kinase II | 400 | 400 | 97% | 6.00E-143 | 62% |
| Cathepsin B | 256 | 256 | 97% | 2.00E-86 | 45% |
| Cathepsin L | 382 | 382 | 86% | 4.00E-132 | 49% |
| CKdhfr-ts | 249 | 249 | 99% | 1.00E-82 | 45% |
| GG3PD | 408 | 408 | 63% | 2.00E-147 | 85% |
| Heat stock protein 70 | 687 | 687 | 70% | 0.0 | 73% |
Dhfr-ts: Diidrofolate reductase; GG3PD: Glycosomal glyceraldehyde 3-phosphate dehydrogenase
Based on these results, homology models of these seven proteins were built on Swissmodel server29 (Table 2), by comparing target sequences with sequences of other proteins with available 3D structures, which were used as templates. The quality of the models was evaluated in PROCHECK30, and the quality of dihedral angles (phi and psi) was analyzed. Furthermore, GalaxyWEB31 was used to refine loop and terminus regions of the best template of each target. The results are presented on Table 2 and Supplementary Figures S1 (A-G). It can be observed that 89.1 to 94.7% of residues from the modeled proteins are on the most favored regions (red), 5.2 to 9.3% on the allowed regions (yellow), 0.0 to 1.6% on the generously allowed regions (beige) and just 0.0 to 1.0% on the disallowed regions (white).
Table 2.
Summary of statistics of obtained 3D models of L. infantum proteins
| Template information | PROCHECK analysis | ||||||
|---|---|---|---|---|---|---|---|
| Target Uniprot (ID) | Cov. | Seq. Id. | Temp. | MFR | AAR | GAR | DR |
| Actin (P60010) | 99% | 72% | 1YAG | 95.3% | 4.3% | 0.3% | 0.0% |
| CK2 (P68400) | 90% | 57% | 3PE2 | 93.7% | 6.0% | 0.0% | 0.4% |
| CathepsinB (Q6R7Z5) | 90% | 52% | 3MOR | 93.3% | 5.7% | 0.5% | 0.5% |
| Cathepsin L (P07711) | 62% | 41% | 1CJL | 90.4% | 9.2% | 0.0% | 0.4% |
| Dhfr-ts (A7ASX7) | 91% | 44% | 3NRR | 89.1% | 9.3% | 0.7% | 0.9% |
| G3PD (P22513) | 96% | 85% | 1K3T | 92.2% | 5.2% | 1.6% | 1.0% |
| Hsp70 (P54652) | 99% | 71% | 5FPN | 94.7% | 5.3% | 0.0% | 0.0% |
CK2: Creatine kinase 2; Cov.: coverage; Seq. Id.: Sequence Identity; Temp.: Template; MFR: Most Favored Regions; AAR: Additional Allowed Regions; GAR: Generously Allowed Regions; DR: Disallowed Regions.
The residues in the disallowed regions were located in regions far from the binding sites, and therefore, did not affect the quality of the models. Therefore, the generated homology models could be used for the estimation of the binding modes and affinity of ligands to the proteins by docking.
After building, selection, and analysis of the homology models, they were used to perform molecular docking of chalcone-like compounds. Chemical structures were carefully curated following the protocols developed by Fourches et al.32–34. Based on the results of docking (Supplemental Table S2), we have selected nine promising compounds (LabMol-69, 73, 65, 67, 70, 76, 86, 90, and 72) and potentially inactive compounds (LabMol-82, 92, and 78) as negative controls.
Twelve selected chalcone-like compounds and Amphotericin B, used as positive control, were tested against L. infantum amastigotes and differentiated THP-1 macrophages (Table 3). Three out of nine selected compounds (LabMol-65, LabMol-72, and LabMol-73) possess reasonably high activity (6.32<EC50<10.98 μM). Other six compounds were inactive. Amphotericin B exhibits EC50 of 1.9 μM. Among negative controls, LabMol-72, and LabMol-73 were expectedly inactive, while LabMol-92 has demonstrated EC50 of 9.31 μM. Among four active compounds, during cytotoxicity testing on macrophages, only LabMol-65 and Labmol-73 showed CC50>50 μM that resulted in SI > 5.2 and 7.9, respectively (see Table 3). Amphotericin B exhibits CC50 of 9.8 μM and SI of 5.2; therefore, LabMol-65 exhibited SI higher than the control and Labmol-73 showed selectivity index (SI) similar to or higher than the control. All active compounds were also tested against Vero cells26 (Table 3). LabMol-65 and Labmol-73 demonstrated SI’s in the range 55–243 and 4–10, respectively, that make them promising anti-leishmanial agents. All the details regarding conducted experiments and the complete table with biologic evaluation (Table S3) are available in Supplemental Information.
Table 3.
In vitro antileishmanial activity EC50 (μM), toxicity (CC50 μM) and selectivity index (SI) in macrophages and Vero cells of chalcones and chalcones-like.
| Code | Structure | EC50 (μM) | * CC50 (μM) | * SI | ** CC50 (μM) | ** SI |
|---|---|---|---|---|---|---|
| LabMol-69 |
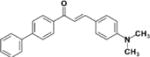
|
>50 | >50 | N.D | N.D | N.D |
| LabMol-73 |
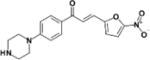
|
9.6 | >50 | >5.2 | >100 | 10 |
| LabMol-65 |
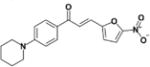
|
6.3 | >50 | >7.9 | 349 | 55 |
| LabMol-67 |
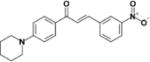
|
30.7 | >50 | >1.6 | N.D | N.D |
| LabMol-70 |
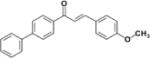
|
>50 | >50 | N.D | N.D | N.D |
| LabMol-76 |
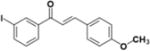
|
>50 | >50 | N.D | >100 | 2 |
| LabMol-86 |
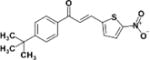
|
31.07 | 8.4 | 0.2 | >100 | 3 |
| LabMol-90 |
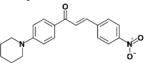
|
>50 | >50 | N.D | N.D | N.D |
| LabMol-72 |
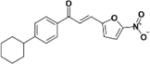
|
10.9 | 31.1 | 2.8 | >100 | 10 |
| LabMol-82 |
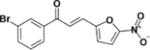
|
23.8 | 14.9 | 0.6 | 68.8 | 2.9 |
| LabMol-92 |
|
9.3 | 13.1 | 1.4 | 40.7 | 4.3 |
| LabMol-78 |
|
27.1 | 49.5 | 1.8 | >100 | 3.6 |
| Amph.B |
|
1.9 | 9.8 | >5.2 | N.D | N.D |
Macrophage,
Vero cells
Based on the experimental results, we derived the SAR rules to reveal the structural fragments responsible for anti-leishmanial activity (Figure 2). On the aryl ring B, independently of substituent positions on ring A, nitro group in position R9 and nitro-, dimethylamino-, and methoxy- groups in position R10 decrease the activity. The substitution of aryl ring B by furan and 5-nitrothiophene are also unfavorable. However, the substitution of aryl ring B by 5-nitrofuran is favorable to biological activity. Bulky groups and electron donors on R4 position of aryl ring A, e.g., piperidine, pyrrolidine, piperazine, methylthiole, imidazole, and cyclohexyl are favorable for anti-leishmanial activity, while methyl, t-buthyl, buthyl, phenyl, morpholine and halogens atoms are unfavorable. The hydrophobic substituents and halogen atoms tested in positions R2 and R3 also demonstrated negative contribution to antileishmanial activity.
Figure 2.
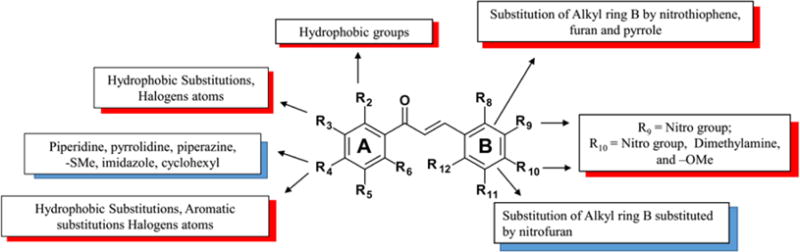
Derived SAR rules highlighting structural moieties favorable and unfavorable to the anti-leishmanial activity. Red boxes are unfavorable groups and blue boxes are favorable groups.
The results of target fishing approach, followed by homology modeling and molecular docking allowed us to rationalize the mode of action of four active compounds (LabMol-65, LabMol-72, LabMol-73, and LabMol-92). They may interact with the cysteine protease procathepsin L, demonstrating its potential for blocking the replication and differentiation of Leishmania in vitro and in vivo. These analyses revealed that the exploration of modifications on scaffolds of chalcones identified here could afford new potent leads against L. infantum and suggest that the mode of action of these compounds could be by inhibition of cysteine proteases of the parasite.
Cysteine proteases constitute an important class of enzymes responsible for virulence factors, essential to parasite survival and are potential drug targets35–37. Figures 3A and 3B show the obtained docked poses for LabMol-72 and its molecular interactions in the active site of procathepsin L. As we can see, hydrophobic interactions and the hydrogen bond are showed. The analysis of the hits in the active site cavity reveals that the hydrophobic pocket is important for interaction between four hits and procathepsin L, Trp151 plays a significant role by performing a hydrogen bond with the carboxyl group of chalcones (see SI Figures S2, S3, S4 and Table S4 on supplementary data).
Figure 3.
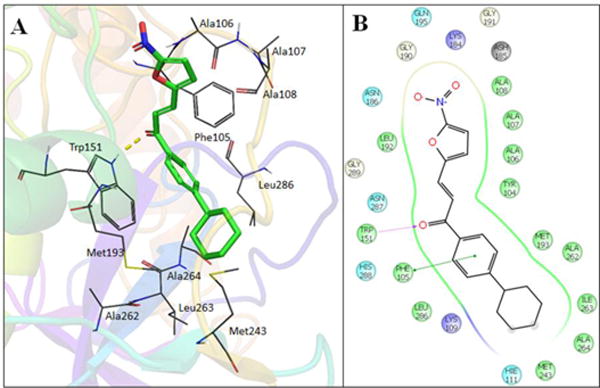
3D (A) and 2D (B) visualization of interactions of LabMol-72 within the binding site of procathepsin L, obtained by docking.
To summarize, the set of 32 recently synthesized26 chalcones and chalcone-like compounds was evaluated by computational approaches to verify their potential anti-leishmanial activity. By results of this in silico evaluation, nine potentially active and three potentially inactive compounds were experimentally tested against L. infantum. Four compounds showed EC50 < 11μM. Among them, two compounds, LabMol-65 and LabMol-73, exhibited cytotoxicity in macrophages >50 μM that resulted in better selectivity than Amphotericin B. These two compounds also demonstrated low cytotoxicity and high selectivity towards Vero cells. Based on modeling results, we suggested that activity of our compounds is caused by their interaction with cysteine proteases. We also conducted SAR analysis to derive structural recommendations useful for molecular design of new chalcones or chalcone-like compounds with antileishmanial activity. For instance, the substitution of aryl ring B by 5-nitrofuran is favorable.
The other nitrofuran analogues, nitrothiophenes, aromatic rings, pyrrole, and furan analogues were inactive against amastigotes of L. infantum (see supplementary Table S3). These results corroborates with other studies which demonstrated that chalcones5,25 and nitroheterocycle38 compounds are active against Leishmania species.
Supplementary Material
Acknowledgments
The authors thank Brazilian funding agencies, CNPq, CAPES and FAPEG for financial support and fellowships. E.M. acknowledge NIH (grant 1U01CA207160), and CNPq (grant 400760/2014-2) for partial financial support. C.H.A. is productivity fellow of CNPq. We are grateful to OpenEye Scientific Software, Inc. and ChemAxon for providing academic license of their software.
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
Supplementary Material
Supplementary data associated with this article can be found, in the online version, at https://www.journals.elsevier.com/bioorganic-and-medicinal-chemistry-letters.
References and notes
- 1.Capriles PVSZ, Baptista LPR, Guedes IA, Guimaraes ACR, Custodio FL, Alves-Ferreira M, Dardenne LE. J Mol Graph Model. 2015;55:134–147. doi: 10.1016/j.jmgm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 2.dos Santos MS, Oliveira MLV, Bernardino AMR, de Léo RM, Amaral VF, de Carvalho FT, Leon LL, Canto-Cavalheiro MM. Bioorg Med Chem Lett. 2011;21:7451–7454. doi: 10.1016/j.bmcl.2011.09.134. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization Leishmaniasis Fact sheet N° 375. 2016. [Google Scholar]
- 4.Moreno MA, Alonso A, Alcolea PJ, Abramov A, de Lacoba MG, Abendroth J, Zhang S, Edwards T, Lorimer D, Myler PJ, Larraga V. Int J Parasitol Drugs Drug Resist. 2014;4:347–354. doi: 10.1016/j.ijpddr.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogungbe IV, Erwin WR, Setzer WN. J Mol Graph Model. 2014;48:105–117. doi: 10.1016/j.jmgm.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Rognan D. Br J Pharmacol. 2007;152:38–52. doi: 10.1038/sj.bjp.0707307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu X, Ouyang S, Yu B, Liu Y, Huang K, Gong J, Zheng S, Li Z, Li H, Jiang H. Nucleic Acids Res. 2010;38:5–7. doi: 10.1093/nar/gkq300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekins S, Williams AJ, Krasowski MD, Freundlich JS. Drug Discov Today. 2011;16:298–310. doi: 10.1016/j.drudis.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 9.Erić S, Ke S, Barata T, Solmajer T, Antić Stanković J, Juranić Z, Savić V, Zloh M. Bioorganic Med Chem. 2012;20:5220–5228. doi: 10.1016/j.bmc.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 10.Wermuth CG. The Practice of Medicinal Chemistry. 2008. 3o. [Google Scholar]
- 11.Syam S, Abdelwahab SI, Al-Mamary MA, Mohan S. Molecules. 2012;17:6179–95. doi: 10.3390/molecules17066179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoki N, Muko M, Ohta E, Ohta S. J Nat Prod. 2008;71:1308–1310. doi: 10.1021/np800187f. [DOI] [PubMed] [Google Scholar]
- 13.Chen YH, Wang WH, Wang YH, Lin ZY, Wen CC, Chern CY. Molecules. 2013;18:2052–2060. doi: 10.3390/molecules18022052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahapatra DK, Bharti SK, Asati V. Eur J Med Chem. 2015;98:69–114. doi: 10.1016/j.ejmech.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Rizvi SUF, Siddiqui HL, Johns M, Detorio M, Schinazi RF. Med Chem Res. 2012;21:3741–3749. [Google Scholar]
- 16.Hans RH, Guantai EM, Lategan C, Smith PJ, Wan B, Franzblau SG, Gut J, Rosenthal PJ, Chibale K. Bioorg Med Chem Lett. 2010;20:942–4. doi: 10.1016/j.bmcl.2009.12.062. [DOI] [PubMed] [Google Scholar]
- 17.Chen M, Zhai L, Christensen SB, Theander TG, Kharazmi A. Antimicrob Agents Chemother. 2001;45:2023–2029. doi: 10.1128/AAC.45.7.2023-2029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ouattara, M.; Sissouma, D.; Koné, M. W.; Hervé, E.; Touré, S. A.; Ouattara, L. 2011, 10, 767–775.
- 19.López SN, Castelli MV, Zacchino Sa, Domínguez JN, Lobo G, Charris-Charris J, Cortés JC, Ribas JC, Devia C, Rodríguez AM, Enriz RD. Bioorg Med Chem. 2001;9:1999–2013. doi: 10.1016/s0968-0896(01)00116-x. [DOI] [PubMed] [Google Scholar]
- 20.Avila-Villarreal G, Hernández-Abreu O, Hidalgo-Figueroa S, Navarrete-Vázquez G, Escalante-Erosa F, Peña-Rodríguez LM, Villalobos-Molina R, Estrada-Soto S. Phytomedicine. 2013 doi: 10.1016/j.phymed.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto T, Yoshimura M, Yamaguchi F, Kouchi T, Tsuji R, Saito M, Obata A, Kikuchi M. Biosci Biotechnol Biochem. 2004;68:1706–1711. doi: 10.1271/bbb.68.1706. [DOI] [PubMed] [Google Scholar]
- 22.Jamal H, Ansari WH, Rizvi SJ. Fundam Clin Pharmacol. 2008;22:673–681. doi: 10.1111/j.1472-8206.2008.00639.x. [DOI] [PubMed] [Google Scholar]
- 23.Lam KW, Uddin R, Liew CY, Tham CL, Israf D a, Syahida A, Rahman MBA, Ul-Haq Z, Lajis NH. Med Chem Res. 2011;21:1953–1966. [Google Scholar]
- 24.Sato Y, He JX, Nagai H, Tani T, Akao T. Biol Pharm Bull. 2007;30:145–149. doi: 10.1248/bpb.30.145. [DOI] [PubMed] [Google Scholar]
- 25.Passalacqua TG, Torres FAE, Nogueira CT, de Almeida L, Del Cistia ML, Dos Santos MB, Regasini LO, Graminha MAS, Marchetto R, Zottis A. Bioorg Med Chem Lett. 2015;25:3564–8. doi: 10.1016/j.bmcl.2015.06.085. [DOI] [PubMed] [Google Scholar]
- 26.Gomes MN, Braga RC, Grzelak EM, Neves BJ, Muratov EN, Ma R, Klein LK, Cho S, Oliveira GR, Franzblau SG, Andrade CH. QSAR-driven Design, Synthesis and Discovery of Potent and Selective Chalcone Derivatives with Antitubercular Activity. 2017 doi: 10.1016/j.ejmech.2017.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X. PharmMapper [Google Scholar]
- 28.NCBI BLAST: Basic Local Alignment Search Tool. 2016. [Google Scholar]
- 29.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, Schmidt T, Kiefer F, Cassarino TG, Bertoni M, Bordoli L, Schwede T. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 31.Ko J, Park H, Heo L, Seok C. Nucleic Acids Res. 2012;40:294–297. doi: 10.1093/nar/gks493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fourches D, Muratov EN, Tropsha A. J Chem Inf Model. 2016 doi: 10.1021/acs.jcim.6b00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fourches D, Muratov E, Tropsha A. J Chem Inf Model. 2010;50:1189–1204. doi: 10.1021/ci100176x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fourches D, Muratov E, Tropsha A. Nat Chem Biol. 2015;11:535–535. doi: 10.1038/nchembio.1881. [DOI] [PubMed] [Google Scholar]
- 35.Caffrey CR, Steverding D. Mol Biochem Parasitol. 2009;167:12–19. doi: 10.1016/j.molbiopara.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Ascenzi P, Bocedi A, Visca P, Antonini G, Gradoni L. Biochem Biophys Res Commun. 2003;309:659–665. doi: 10.1016/j.bbrc.2003.08.015. [DOI] [PubMed] [Google Scholar]
- 37.Ascenzi P, Salvati L, Bolognesi M, Colasanti M, Polticelli F, Venturini G. Curr Protein Pept Sci. 2001;2:137–53. doi: 10.2174/1389203013381170. [DOI] [PubMed] [Google Scholar]
- 38.Petri E Silva SCS, Palace-Berl F, Tavares LC, Soares SRC, Lindoso JAL. Exp Parasitol. 2016;163:68–75. doi: 10.1016/j.exppara.2016.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


