Abstract
Genome-wide studies in Ciona often require highly purified cell populations. In this methods paper, we introduce multi-channel combinatorial Fluorescence Activated Cells Sorting (FACS) and Magnetic-Activated Cell Sorting (MACS) as two sensitive and efficient tools to isolate lineage-specific cell populations from dissociated Ciona embryos and larvae. We present isolation of trunk ventral cell (TVC) progeny as the test-case most commonly used in our laboratory. These approaches may also be applied to purify other cells populations with the proper combination of tissue-specific reporters.
Introduction
One of the greatest challenges for genome-wide studies in Ciona is to distinguish between cells and lineages of interest and the surrounding tissues, which can constitute the overwhelming majority of unsorted samples. This is crucial because most genetic elements, especially transcribed regions, show pleiotropic activity. The successful cloning of numerous enhancers for genes that are expressed exclusively in certain tissues has enabled the community to label specific lineages and tissues by using fluorescent reporters. In principle, this permits straightforward isolation of cells of interest using Fluorescence Activated Cell Sorting (FACS).
However, in practice most reporter constructs show “leaky” expression in other tissues, strongly interfering with cell isolation using a single tissue-specific fluorescent reporter. Besides, when using a combination of early and late reporter constructs for combinatorial labeling, depending upon the activation time and the number of divisions between the onsets of the early and late enhancers, only a portion of cells labeled with the early reporter will inherit and activate the late fluorescent reporter. This caveat is defined as the “mosaic expression” in electroporated Ciona embryos. Mosaic expression is inconvenient, especially when we transiently express engineered constructs with late tissue-specific enhancers. Due to the fact that only a portion of the progeny inherits the late-active plasmid, within the isolated population, a substantial number of cells fail to inherit the perturbation construct and thus remain “wild-type”. In most cases, this causes heterogeneity in experimental samples that will confound the results of whole genome assays when comparing perturbation vs. control samples.
In this methods paper, we will use trunk ventral cells purification as an example to introduce a multi-channel combinatorial FACS approach (Christiaen et al. 2008), which can overcome the obstacles described above. Trunk ventral cells (TVCs) and TVC progeny of the B7.5 lineage are rare (<0.2% of the entire cellular population) in tailbud and older embryos and larvae, but they can be labeled with B7.5-lineage-specific enhancers driving fluorescent reporters, e.g. Mesp>tagRFP (Davidson et al. 2005). However, the leaky expression of Mesp enhancer into the B-line mesenchyme cells normally results in contamination of TVC progeny samples (Fig. 1a). This issue can be avoided by using MyoD905>GFP, which is specifically expressed in mesenchyme and primary muscle cells, except the B7.5-derived anterior tail muscles (ATM), for counter-selection (Fig. 1b; Christiaen et al. 2008). In this study, we introduced a TVC-specific reporter, Hand-r>tagBFP (Fig. 1b), and used a third FACS channel to co-select the TVC progeny for two reasons. First, the extra layer of co-selection of FACS can further improve the purity of sorted TVC progeny. Second, we used Hand-r (Woznica et al. 2012; and Christiaen lab, unpublished observations) or FoxF (Beh et al. 2007) enhancer-driven constructs to express reporters and perturbation constructs (e.g. Wang et al. 2013; Razy-Krajka et al. 2014) selectively in the TVC progeny, thus avoiding precocious effects in the B7.5 lineage cells. Due to the mosaic inheritance of Hand-r and FoxF enhancer constructs, only a portion of TVC progeny labeled with Mesp>tagRFP will also express these constructs. Previous estimates suggest that 20 to 30% of the Mesp>tagRFP+ embryos can show mosaic expression of FoxF-driven reporters (Gline et al. 2015). Using the same enhancer for the perturbation plasmid to perform the co-selection avoids sorting cells that did not inherit the perturbation constructs due to mosaicism.
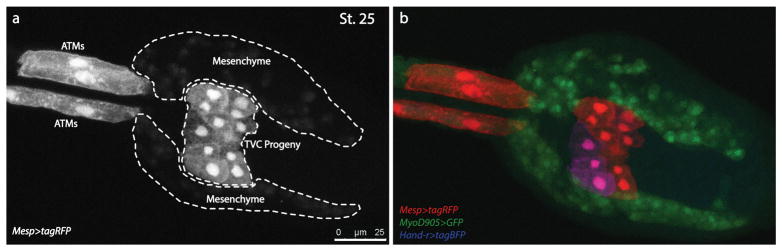
Figure 1. Ciona Late Tailbud for multi-channel combinatorial FACS.
Ciona late tailbud expresses Mesp>tagRFP, MyoD905>GFP and Hand-r>tagBFP fluorescent reporters. Embryos are fixed at St. 25 and imaged using Leica TCS SP8 X confocal microscopy. a. Isolated single channel showing the Mesp>tagRFP labelled B7.5 lineage (TVC progeny and ATMs are in grey color) and the leaky expression in mesenchyme cells (faint grey signal); b. The overlapped channels of Mesp>tagRFP, MyoD905>GFP and Hand-r>tagBFP reporters. Mesp>tagRFP labeled B7.5 lineage cells are in red. MyoD905>GFP labelled mesenchyme cells are in green. Hand-r>tagBFP reporter labels only a portion of TVC progeny due to the mosaicism. The tagRFP-tagBFP+ cells, which appear in magenta (red overlaid with blue), will be sorted for further analysis.
Although FACS is a sensitive and accurate method for purifying specific cells from the rest of embryonic cell population (Christiaen et al. 2008; Christiaen et al. 2009; Racioppi et al. 2014; Razy-Krajka et al. 2014), it requires specialized equipment and skilled operators, which are usually not available to individual laboratories, and since cells are processed through high-pressure fluids, they can easily be damaged. To circumvent these problems, affinity-based cell separation technologies are also adapted to isolating Ciona cells using membrane markers.
Magnetic-Activated Cell Sorting or MACS (Miltenyi Biotec) separates specific cells from the rest of embryonic cell population using antibody-coated superparamagnetic nanoparticles (50 nm microbeads). The antibodies are specific for particular cell surface markers, either expressed in the population of interest for positive selection, or expressed on undesired cell types for negative selection. After incubating dissociated cells expressing a trans-membrane marker with antibody-coated microbeads, the suspension is added to a separation column, which contains a matrix composed of ferromagnetic spheres. This induces the formation of a magnetic gradient, leading to microbeads-laden cells to stick to the column, while unlabeled cells flow through.
If a negative selection is performed, the flow-through is the population of interest and the bound cell-microbead complexes are discarded. If a positive selection is performed, the cells of interest are bound to the microbeads. The column is washed several times to ensure unbound or weakly-bound cells are washed away. Then the column is removed from the magnetic separator and the cell-microbead complexes are eluted.
The MACS technology permits also indirect magnetic labeling, where the cells are labeled with a primary antibody directed against a cell surface marker in a first step, then the cells are magnetically labeled with microbeads in a second step, which either bind to the primary antibody or to a molecule conjugated to the primary antibody.
MACS separation presents several advantages by comparison with FACS. It does not require an expensive dedicated instrument (although automatic platforms are available), it is generally easier to set-up than FACS, large numbers of cells can be processed simultaneously (though it is not possible to count the number of cells separated during MACS process) and it allows multiplex experiments with parallel separations from up to 24 samples in a single run.
However, one disadvantage of MACS concerns the microbeads. Most of them are only available from Miltenyi Biotec, so if there are no microbeads with the particular marker of interest available, the conjugation must be done on your own. Moreover, although combinatorial logics can, in principle, be implemented by using different membrane markers, such as human Cluster of Differentiation 4 (hCD4) and mouse Cluster of Differentiation 8 (mouse CD8); in practice, because all purifications rely on the same microbeads, this requires successive incubation and column purification, which renders these combinatorial experiments particularly impractical.
To isolate transfected cells, for example to assay CRISPR/Cas9-mediated mutagenesis rates (Stolfi et al., 2014), we have been using either glycoprotein hCD4, or mouse CD8 as cell surface markers. In our hands, hCD4 appeared to be better trafficked to the plasma membrane, usually generating more specific membrane labeling when fusion to fluorescent proteins such as GFP or mCherry (Gline et al., 2015).
Methods
Cell dissociation
Mechanical and enzymatic digestion of Ciona embryos electroporated with the combination of tissue-specific enhancers driving different fluorescent reporter releases fluorescent cells into suspension. This allows one to successfully enrich cells of interest that uniformly express a specific fluorescent reporter.
In order to proceed for cell dissociation:
Collect Ciona stage-selected embryos in borosilicate glass tubes 12x75 mm (rinse them overnight with distilled water and dry before use, in order to avoid that embryos stick to the glass).
Put the tubes on ice and let the embryos settle
Rinse the embryos three times in artificial calcium and magnesium-free sea water (CMF-ASW: 449 mM NaCl, 33 mM Na2SO4, 9 mM KCl, 2.15 mM NaHCO3, 10 mM Tris-Cl pH 8.2, 2.5 mM EGTA).
Remove the supernatant and keep the tubes on ice
Dilute the stock solution of trypsin (w/v, Sigma, T-4799, usually prepared at 2.5% in CMF-ASW) to a final concentration of 0.2% in CMF-ASW.
Add 2 mL of CMF-ASW supplemented with 0.2% trypsin and dissociate embryos by thorough pipetting (2–3 min for tailbud stage, 5–8 min for larval stage).
Block the trypsin activity by adding 2 mL of ice-cold CMF-ASW supplemented with 0.05% bovine serum albumin (BSA, Sigma, A-3311).
Keep the cell suspensions on ice and filter into 5 mL polystyrene round bottom tubes equipped with a 35μm cell strainer cap (Falcon).
Centrifuge the cell suspensions for 3 minutes at 800 rcf (relative centrifugal force) at 4°C
Remove the supernatant and rinse in CMF-ASW supplemented with 0.05% BSA.
Repeat the centrifugation and rinse steps (steps 9 and 10) at least once
Filter the cell suspensions into 5 mL polystyrene round bottom tubes equipped with a 35μm cell strainer cap before cell sorting.
Fluorescence Activated Cell Sorting (FACS)
The successful multi-channel FACS relies on the clean separation of the spectrum of fluorescent reporters that are co-expressed in the same cell population. Thus, this requires careful selection of reporters and multiple single or dual color control samples to calibrate the gate and compensation conditions. The parameters given below were determined for a BD FACSAria™ flow cytometer equipped with a 488-nm blue laser, a 561-nm yellow/green laser, a 633-nm far red laser and a 407-nm ultra violet laser.
Flow cytometry sorter gating and compensation
Four single color samples are prepared for calibration of gating and compensation on flow cytometry sorter
None-electroporated sample (no fluorescent reporter)
Single color sample 1- electroporated with MyoD905>GFP
Single color sample 2- electroporated with Mesp>tagRFP
Single color sample 3- electroporated with Hand-r>tagBFP
Three dual color samples to test the separation of channels as pair
Dual color sample 1- electroporated with MyoD905>GFP and Mesp>tagRFP
Dual color sample 2- electroporated with MyoD905>GFP and Hand-r>tagBFP
Dual color sample 3- electroporated with Mesp>tagRFP and Hand-r>tagBFP
One triple color sample electroporated with MyoD905>GFP, Mesp>tagRFP and Hand-r>tagBFP to test the separation of all the three channels
-
1
Before loading the samples into the sorter, filter the cell suspension again on the cell-strainer cap of a 5 mL round-bottom tube and gently pipette the cell suspension 3–5 times.
-
2
Load the non-electroporated sample into the sorter and count ~10,000 cells. Distinguish the well dissociated single cells from undissociated tissues and debris based on size and internal complexity which can be defined by forward scatter (FS) and side scatter (SS). Gate the single cell population as P1 (Fig. 2a).
-
3
Load the Single color sample 1 into the sorter and count ~10,000 cells. Gate the GFP+ population using 488-nm blue laser, PMT (Photomultiplier Tubes) Detector E, LP (Long Pass) Filter 505 and BP (Band Pass) Filter 530/30 for FITC (Fig. 2b).
-
4
Load the Single color sample 2 into the sorter and count ~10,000 cells. Gate the tagRFP+ population using 561-nm yellow/green laser, PMT Detector B and BP Filter 586/15 for DsRed (Fig. 2c).
-
5
Load the Single color sample 3 into the sorter and count ~10,000 cells. Gate the tagBFP+ population using 407-nm (violet) laser, PMT Detector B and BP Filter 450/50 for Pacific Blue (Fig. 2d).
-
6
Mix the Single color sample 1 and 2 by 1:1 and load the sample mixture into the sorter, and count ~10,000 cells.
-
7
Adjust the compensation, ensure clean separation of the GFP+ and tagRFP+ populations.
-
8
Repeat the same steps using mixtures of single color samples 1 and 3, and 2 and 3, adjust the compensation to ensure the clean separation of GFP+ vs. tagBFP+ populations and tagRFP+ vs tagBFP+ populations.
-
9
Mix the Single color samples 1, 2 and 3 in 1:1:1 proportions. Load the sample mixture into the sorter and count ~10,000 cells, making sure the GFP+, tagRFP+ and tagBFP+ cell populations are still well separated from each other.
Load the Dual color sample 1 and count ~10,000 cells. Use the GFP+ and tagRFP+ gate, which was defined by single color sample to identify the cell population with only GFP expression, only tagRFP expression and with both GFP and tagRFP expression. Gate the GFP-tagRFP+ population.
-
10
Repeat the same steps for 2-color samples 2 and 3, define the gates for GFP-tagBFP+ and tagRFP-tagBFP+ populations.
Note: The calibration of gating (step1-step10) is recommended every time before sorting. However, the BD FACSAria™ flow cytometer has been proved to be consistent and repeatable across daily operations. Therefore, the calibration of gating might be skipped when sorting the same type of samples frequently in a short period, such as ONE month.
Figure 2. FACS gating strategy for TVC progeny isolation.
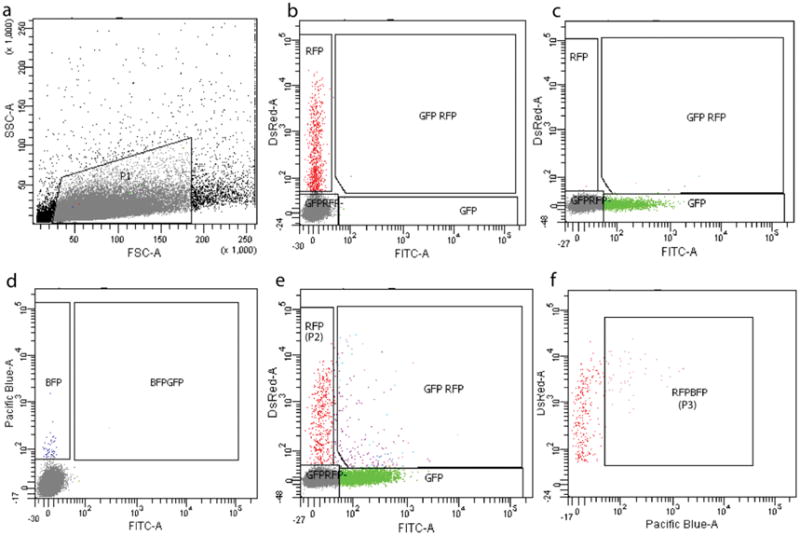
Samples electroporated with blank buffer, Mesp>tagRFP, MyoD905>GFP and Hand-r>tagBFP fluorescent reporters are used to gate the TVC progeny populations. a. Gate P1 for single cell population based off the FS and SS from the blank sample. b. Gate the tagRFP+ cell population using the sample electroporated with Mesp>tagRFP. c. Gate the GFP+ cell population using the sample electroporated with MyoD905>GFP. d. Gate the tagBFP+ cell population using the sample electroporated with Hand-r>tagBFP. e. Define the cells only have tagRFP as P2 in the sample co-electroporated with Mesp>tagRFP, MyoD905>GFP and Hand-r>tagBFP. f. Analyze P2 with gates set for tagRFP and tagBFP, define the tagRFP-tagBFP+ cell population as P3. P3 is TVC progeny that we sort for downstream genome-wide analysis.
Load the triple color sample and count ~10,000 cells.
-
11
Analyze the sample with the lasers and filters for FITC and DsRed, select the population with only tagRFP signal, mark it as P2 (Fig. 2e).
-
12
Analyze the P2 population with the lasers and filters for DsRed and Pacific Blue. Select the cell population that falls into the tagRFP-tagBFP+ gate and give it the P3 identification (Fig. 2f).
In our experiments, this P3 population consists of TVC progeny which co-express tagRFP and tagBFP (co-selection) but without the expression of GFP (counter selection of mesenchyme cells with leaky Mesp>tagRFP and Hand-r>tagBFP expression). These cells only represent 0.1%–0.2% of the P1 population from the whole embryo.
Magnet Activated Cell Sorting (MACS)
After the last step of cell dissociation from Ciona embryos protocol:
Transfer each cell suspension into 2 mL non-adhesive tubes.
Centrifuge for 3 minutes at 800 rcf at 4°C.
Discard supernatant and resuspend cells in 90 μL CMF-ASW supplemented with 0.05% BSA.
Add 10 μL of superparamagnetic microbeads coupled with the anti-human CD4 antibody (Miltenyi Biotec) to select for positive hCD4 cells.
Combine each sample into one 2 mL non-adhesive tube to have a total of 20 μL microbeads in a final volume of 200 μL.
Incubate samples for 1 hour on agitation at 4°C.
Place the MS columns inside the permanent magnet OctoMACs separator.
Equilibrate the MS columns with 500μL of CMF-ASW. Note that it is important to degas all the buffers, for example by filtration using a vacuum system, just prior to using them, otherwise microbubbles can interfere with the process and cause hCD4-negative cells to be retained in the column.
Assemble pre-separation filters (70 μm; Miltenyi Biotec) onto the MS columns (to remove larger debris for a smooth flow through of the columns and an efficient magnetic hold of the hCD4+ cells labeled with the microbeads).
Add 1 mL CMF-ASW supplemented with 0.05% BSA to the samples (after incubation to rinse the cell suspension).
Centrifuge for 3 minutes at 1000 rcf at 4°C.
Suspend each sample in 500 μL CMF-ASW supplemented with 0.05% BSA.
Load samples onto the pre-separation filters over the MS columns.
Collect flow through in 2 mL non-adhesive tubes as hCD4-negative cells.
Wash with 500 μL CMF-ASW supplemented with 0.05% BSA and collect in the same tube.
Repeat this step 2 more times.
Remove each MS column from the OctoMACS separator and place above another 2 mL non-adhesive tube.
Add 1 mL CMF-ASW supplemented with 0.05% BSA to the top of the column.
Gently flush out with a plunger supplied by the kit, on ice.
Collect flow through containing positive hCD4 cells.
Centrifuge for 3 minutes at 1000 rcf at 4°C.
RNA Extraction and Cell Purity Check
From typical FACS experiments, 200 to 2000 cells are collected from each sample, for RNA extraction if transcriptome profiling by RNA-seq is the desired outcome.
Both RNAqueous®-Micro Total RNA Isolation Kit (Thermo Fisher Scientific, Catalog number: AM1931) and AGENCOURT® RNACLEAN® XP (Beckman Coulter, Part Number: A63987) have shown good efficiency for extraction and purifying total RNA from the small amounts of sorted Ciona cells.
The quality and quantity of total RNA are checked by 2200 TapeStation system (Agilent), using High Sensitivity RNA ScreenTape (5067–5579) with High Sensitivity RNA ScreenTape Sample Buffer (5067–5580) and High Sensitivity RNA ScreenTape Ladder (5067–5581). The range of total RNA input is 500 – 10,000 pg/μL, and the total RNA integrity is estimated using the software RNA Integrity Number or RIN. RNA samples pass the quality control if RIN>8 and then will be used for downstream cDNA synthesis, and high-throughput sequencing. As an alternative, especially when the input total RNA concentration is low (extracted from less than 400 cells, for example), Agilent 2100 Bioanalyzer also serves well for quality and quantity check with Agilent RNA 6000 Pico Kit (5067–1511), which has the quantitative range from 50 to 5000 pg/μl.
Checking sample purity is indispensable following either FACS- and MACS-based purification. When sorting B7.5-lineage-derived trunk ventral cells, the main contaminations are normally contributed by the mesenchyme and tail muscle. Thus, we performed semi-quantitative PCR (real time PCR will be even better) using primers designed for TVC specific genes (e.g. Hand-r), mesenchymal cell specific gene (e.g. Twist1/2) and tail muscle specific gene (e.g. Mox) to check the purity of TVC progeny after sorting. Our recent single cell RNA-seq experiments also provided evidence that multi-channel FACS can enrich the purity of TVC progeny up to 85% out of the sorted cells (Wang et al., in preparation).
Acknowledgments
We are grateful to Pui-Leng Ip, Ken Birnbaum and the personnel of GenCore at NYU for their expert assistance with the FACS, ScreenTape, and high-throughput sequencing. We thank Nicole Kaplan for sharing the image of Mesp>tagRFP, MyoD905>GFP and Hand-r>tagBFP labelled Ciona late tailbud. Research in the laboratory of L.C. is supported by R01 awards HL108643 and GM096032 from the NIH/NHLBI and NIH/NIGMS, respectively; and by grant 15CVD01 from the Leducq Foundation. C.R. has been supported by a long-term fellowship ALTF 1608-2014 from EMBO.
References
- Beh Jeni, Shi Weiyang, Levine Mike, Davidson Brad, Christiaen Lionel. FoxF Is Essential for FGF-Induced Migration of Heart Progenitor Cells in the Ascidian Ciona Intestinalis. Development. 2007;134(18):3297–3305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- Christiaen Lionel, Davidson Brad, Kawashima Takeshi, Powell Weston, Nolla Hector, Vranizan Karen, Levine Michael. The Transcription/migration Interface in Heart Precursors of Ciona Intestinalis. Science. 2008;320(5881):1349–52. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- Christiaen Lionel, Wagner Eileen, Shi Weiyang, Levine Michael. Isolation of Individual Cells and Tissues from Electroporated Sea Squirt (Ciona) Embryos by Fluorescence-Activated Cell Sorting (FACS) Cold Spring Harbor Protocols. 2009;2009(12):db.prot5349. doi: 10.1101/pdb.prot5349. [DOI] [PubMed] [Google Scholar]
- Davidson Brad, Shi Weiyang, Levine Michael. Uncoupling Heart Cell Specification and Migration in the Simple Chordate Ciona Intestinalis. Development. 2005;132(21):4811–18. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- Gline Stephanie, Kaplan Nicole, Bernadskaya Yelena, Abdu Yusuff, Christiaen Lionel. Surrounding Tissues Canalize Motile Cardiopharyngeal Progenitors towards Collective Polarity and Directed Migration. Development. 2015;142(3):544–54. doi: 10.1242/dev.115444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racioppi Claudia, Kamal Ashwani K, Razy-Krajka Florian, Gambardella Gennaro, Zanetti Laura, di Bernardo Diego, Sanges Remo, Christiaen Lionel A, Ristoratore Filomena. Fibroblast Growth Factor Signalling Controls Nervous System Patterning and Pigment Cell Formation in Ciona Intestinalis. Nature Communications. 2014 Sep;5:4830. doi: 10.1038/ncomms5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razy-Krajka Florian, Lam Karen, Wang Wei, Stolfi Alberto, Joly Marine, Bonneau Richard, Christiaen Lionel. Collier/OLF/EBF-Dependent Transcriptional Dynamics Control Pharyngeal Muscle Specification from Primed Cardiopharyngeal Progenitors. Developmental Cell. 2014;29(3):263–76. doi: 10.1016/j.devcel.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Wei, Razy-Krajka Florian, Siu Eric, Ketcham Alexandra, Christiaen Lionel. NK4 Antagonizes Tbx1/10 to Promote Cardiac versus Pharyngeal Muscle Fate in the Ascidian Second Heart Field. PLoS Biology. 2013;11(12):e1001725. doi: 10.1371/journal.pbio.1001725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woznica Arielle, Haeussler Maximilian, Starobinska Ella, Jemmett Jessica, Li Younan, Mount David, Davidson Brad. Initial Deployment of the Cardiogenic Gene Regulatory Network in the Basal Chordate, Ciona Intestinalis. Developmental Biology. 2012;368(1):127–39. doi: 10.1016/j.ydbio.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]