SUMMARY
The mechanisms how the mammalian airway detects invading viral pathogens to trigger protective innate neutrophilic inflammation are incompletely understood. We observe that innate activation of the NFκB/RelA transcription factor indirectly activates atypical BRD4 histone acetyltransferase (HAT) activity, RNA Polymerase II (Pol II) phosphorylation, and secretion of neutrophilic chemokines. To study this pathway in vivo, we develop a conditional knockout of RelA in the distal airway epithelial cell; these animals have reduced mucosal BRD4/Pol II activation and neutrophilic inflammation to viral patterns. To further understand the role of BRD4 in vivo, two potent, highly selective small-molecule BRD4 inhibitors were developed. These well-tolerated inhibitors disrupt BRD4 complex with Pol II and histones, completely blocking inducible epithelial chemokine production and neutrophilia. We conclude RelA-BRD4 signaling in distal tracheobronchiolar epithelial cells mediates acute inflammation in response to luminal viral patterns. These potent BRD4 antagonists are versatile pharmacological tools for investigating BRD4 functions in vivo.
INTRODUCTION
Approximately 384 million people worldwide are affected by chronic obstructive pulmonary disease (COPD) a heterogeneous, environmentally caused lung disease where inflammation accelerates irreversible remodelling of the airways (Rabe et al., 2007). COPD is an age-related disease that typically starting beings at middle-age, increasing in prevalence, severity and mortality thereafter (Centers for Disease and Prevention, 2008). Consequently, COPD is the fourth largest cause of death globally, and the third largest cause in the United States. The course of COPD is punctuated by intermittent, acute exacerbations of obstructive symptoms. Exacerbations reduce exercise capacity, quality of life and increase health care costs (Donaldson et al., 2005; Seemungal et al., 1998).
Respiratory RNA viral infections are responsible for over half of the acute decompensations in patients with COPD (Wedzicha, 2004). Viral pathogen-associated patterns (PAMPs), notably double-stranded (ds) RNA produced by active infection from these ubiquitous viruses, trigger leukocytic inflammation, mucous production, and airway hyper-reactivity. PAMPs are detected in the airway by membrane-associated Toll-like receptor (TLR)-3 and intracellular retinoic acid inducible gene (RIG)-I, two receptors that trigger transcription factors signalling the innate immune response (Alexopoulou et al., 2001; Bertolusso et al., 2014). Of these, NFκB plays a major role in innate inflammation, controlling the expression of inflammatory chemokines as well as the mucosal IFNs (Brasier et al., 2011; Tian et al., 2017). NFκB activation involves RelA release from cytoplasmic inhibitors coupled with a nuclear oxidative stress-induced serine phosphorylation, promoting RelA binding to bromodomain-containing protein 4 (BRD4) (Brasier et al., 2011; Tian et al., 2017). Through sequence-specific binding, NFκB recruits BRD4 to latent innate genes promoting their rapid expression by a process of transcriptional elongation (Nowak et al., 2008; Tian et al., 2013). In this manner, the NFκB-BRD4 complex binds and directly activates immediate-early inflammatory CXC chemokines, and indirectly activates mucosal type I and III interferons (IFNs) via the IFN regulatory factor (IRF)-RIG-I pattern recognition receptor “cross-talk pathway” (Fang et al., 2015). Upon secretion, these cytokines activate airway dendritic cells and innate lymphocytes to initiate the pulmonary adaptive immune response (Holt and Strickland, 2010).
An unresolved question is the identity of the major sentinel cell of the airway responsible for detecting viral PAMPs. Airway epithelial cells dynamically respond to viral patterns in a cell-type specific pattern by synthesizing and secreting over 400 proteins including CXC and CC-type chemokines, type I and III IFNs, and IFN stimulated genes (ISGs) (Zhao et al., 2017). Similarly, resident alveolar macrophages synthesize cytokine mediators that play roles in initial anti-viral defense and inflammation (Kolli et al., 2014). Recent work has also implicated the role of group 2 innate lymphoid cells in the host response to viruses (Peebles et al., 2001). To provide greater insight into the role of the epithelial cell as a sentinel cell responding to viral PAMPs, we developed a conditional knockout (CKO) mouse selectively deleting RelA in tracheo-bronchiolar cells and challenged them with poly(I:C), a viral pattern that simulates acute RNA virus infections in COPD (Harris et al., 2013; Stowell et al., 2009). We observed that poly(I:C) induces mucosal BRD4 (HAT)-dependent histone acetyltransferase (HAT) activity on Histone H3K122 and BRD4-dependent phosphorylation of Ser2 on the RNA Pol II carboxy terminal domain (CTD). Interestingly, both of these activities were blocked by RelA silencing in cellulo and in vivo. To selectively study the functional role of BRD4 in vivo, we utilized a structure-based design approach to develop two BRD4-selective inhibitors that bind to the BRD4 bromodomains (BDs) with nanomolar affinity. BRD4 inhibition also completely blocked poly(I:C)-induced inflammatory gene programs in cellulo and airway inflammation and neutrophil recruitment in vivo. Our studies indicate that the tracheo-bronchiolar epithelial cell is an innate sensor to viral patterns and validate BRD4 as a rational target to prevent virus-induced lung inflammation.
RESULTS
NFκB/RelA mediates TLR3 induced neutrophil chemokine expression in bronchiolar cells
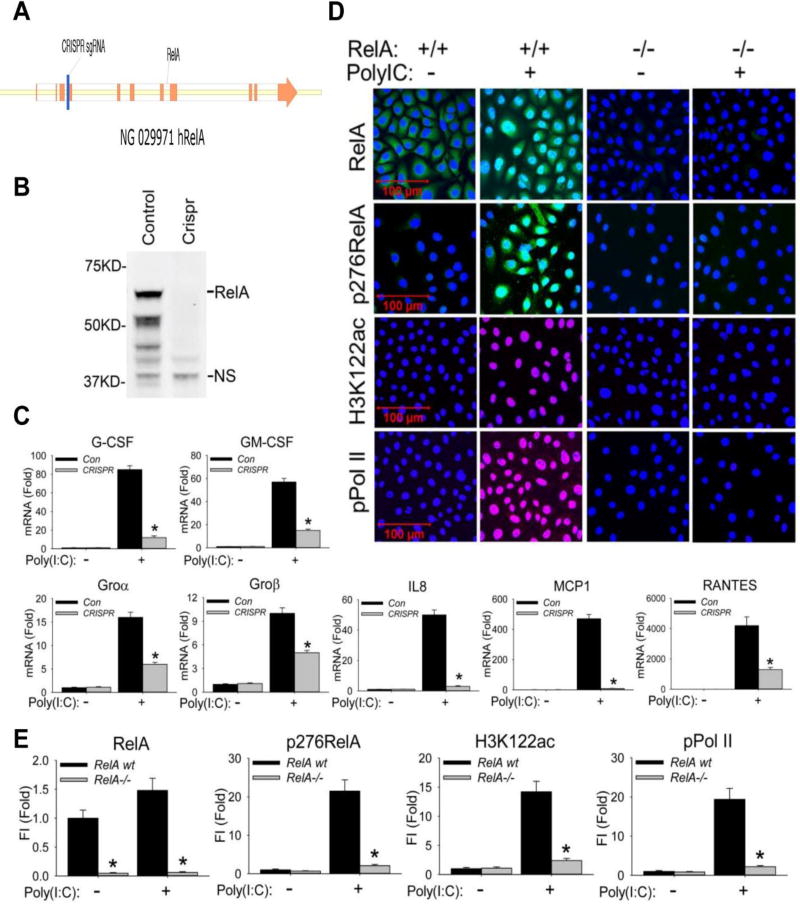
To extend our earlier studies that NFκB-BRD4 signaling mediates the RIG-I-initiated anti-viral innate response (Fang et al., 2015; Tian et al., 2017), we evaluated the role of NFκB/RelA in TLR3-mediated activation of neutrophilic chemoattractant secretion. This model used telomerase-immortalized human small airway epithelial cells (hSAECs), differentiated epithelial cells that exhibit similar innate responses as primary SAECs whose stable phenotype enables genomic manipulation. hSAECs permit study of signalling pathways relevant to those in vivo (Tian et al., 2017; Zhao et al., 2017). To establish stable RelA-deficient hSAECs, we used CRISPR/Cas9 genome editing to introduce a frameshift mutation in Exon 4 of RelA (Figure 1A), resulting in a complete absence of the 65 kDa RelA by Western immunoblot (Figure 1B). Wild type (RelA+/+) or RelA−/− hSAECs were stimulated with poly(I:C), and neutrophilic chemokine expression was quantified. In RelA+/+ cells, poly(I:C) induced an 85-fold induction of CSF3/G-CSF mRNA after 6 h of stimulation; this induction was reduced to less than 12-fold in the RelA−/− hSAECs (Figure 1C). Similarly, the 57-fold induction of CSF2/GM-CSF, the 16-fold induction of CXCL1/Groα, the 10-fold induction of CXCL2/Groβ, the 50-fold induction of CXCL8/IL8, the 470-fold induction of CCL2/MCP1 and the 4200-fold induction of CCL5/RANTES were all significantly reduced by RelA silencing (Figure 1C). These data indicated that the neutrophilic chemokine response in hSAECs was RelA-dependent.
Figure 1. RelA mediates poly(I:C) stimulation induced BRD4 activation.
(A). RelA deletion was performed by targeting a 20 nt seed sequence in Exon 4 of the RelA gene, introducing a stop codon that induces nonsense-mediated RNA decay. The seed sequence is in blue; exons are indicated in orange. (B). Western blot of RelA expression in wild type (control) and CRISPR/Cas9 genome edited cells. NS, nonspecific band. (C). Wild type (control) and CRISPR/Cas9 genome edited hSAECs were challenged with poly(I:C) (10 µg/ml) for 0 and 6 h. Expression of CSF3/G-CSF, CSF2/GM-CSF, CXCL1/Groα, CXCL2/Groβ, CXCL8/IL8, CCL2/MCP1, and CCL5/RANTES was quantified by Q-RT-PCR. Shown is the fold-change in mRNA abundance normalized to cyclophilin (PPIA). *: p < 0.01 compared to control hSAECs. n=3. (D). Cells were stimulated with poly(I:C). After fixation, cells were stained with primary abs of RelA, phospho-Ser 276 RelA, H3K122ac, and phospho-Ser 2 CTD RNA Pol II. Secondary detection was Alexa 488-(green, for RelA and p276 RelA), 568-(red, for pPol II), and 647-(deep red, for H3K122ac) conjugated goat anti-rabbit IgG respectively. Nuclei were counterstained with DAPI (blue). Images were acquired by confocal microscopy at 63× magnification. (E). Quantifications of total fluorescence intensities is shown as fold changes compared to WT hSAECs. * p<0.01, n = 5. FI: relative total fluorescence intensity.
RelA is required for poly(I:C)-induced RNA Pol II phosphorylation and BRD4 histone acetyl transferase activity
We earlier found that intracellular RSV replication promoted the formation of a ternary RelA·BRD4·CDK9 complex (Brasier et al., 2011; Tian et al., 2017). RelA+/+ or RelA−/− hSAECs were stimulated with extracellular poly(I:C) and analysed by immunofluorescence confocal microscopy (IFCM). Poly(I:C) stimulation induced RelA nuclear translocation in 88 % of RelA+/+ cells and induced phospho-Ser 276 RelA. Both signals were abolished by RelA silencing, confirming knockdown and staining specificity (Figures 1D and 1E).
Our previous work showed that RIG-I was required for RSV induction of BRD4 HAT activity forming acetylated H3 on Lys (K) 122 (Tian et al., 2017), a post translational modification that destabilizes nucleosomes, promoting transcriptional elongation by enabling RNA Pol II to traverse nucleosome-rich coding sequences (Devaiah et al., 2016). We found that poly(I:C) produced a 14.5-fold increase of nuclear H3K122 Ac that was also inhibited in the RelA-deficient hSAECs (Figures 1D and 1E).
Besides its HAT activity forming acetylated H3K122 (Devaiah et al., 2016), BRD4 is also an atypical kinase that phosphorylates Ser2 CTD RNA Pol II (Devaiah et al., 2012). We found that poly(I:C) treatment induced a 20-fold increase in phospho-Ser 2 CTD RNA Pol II formation in wild-type cells, an activation that was silenced in the RelA-depleted cells (Figure 1D and 1E). These data indicate that RelA is required for global activation of atypical BRD4 HAT activity and phosphorylation of Ser 2 in the RNA Pol II CTD by extracellular poly(I:C).
RelA is required for BRD4 HAT activity and RNA Pol II phosphorylation in response to TNFα
We next asked whether RelA mediated BRD4 HAT activity and phospho-Ser 2 CTD RNA Pol II formation were inducible in a stimulus-independent mechanism by other innate activators. We examined the effects of TNFα, a monokine that activates RelA via TNFRI (Tian et al., 2005), that produces nuclear translocation in 90% of cells (Supplemental Figure S1). TNFα stimulation induced a 4-fold increase in H3K122 Ac and a 15-fold increase in phospho-Ser2 CTD RNA Pol II formation that was also dependent on RelA (Supplemental Figure S1). Similarly, TNFα-induced expression of CSF3/G-CSF, CSF2/GM-CSF, CXCL1/Groα, CXCL2/Groβ, CXCL8/IL8, and CCL2/MCP1 chemokines were blocked by RelA silencing (Supplemental Figure S1). Together these data indicate that RelA is required for global inducible regulation of atypical BRD4 HAT activity and RNA Pol II kinase activity in response to viral and cytokine innate activators.
BRD4 HAT activity is required for formation of phospho-Ser 2 CTD RNA Pol II formation and acetylation of H3K122 as well as transcriptional activation
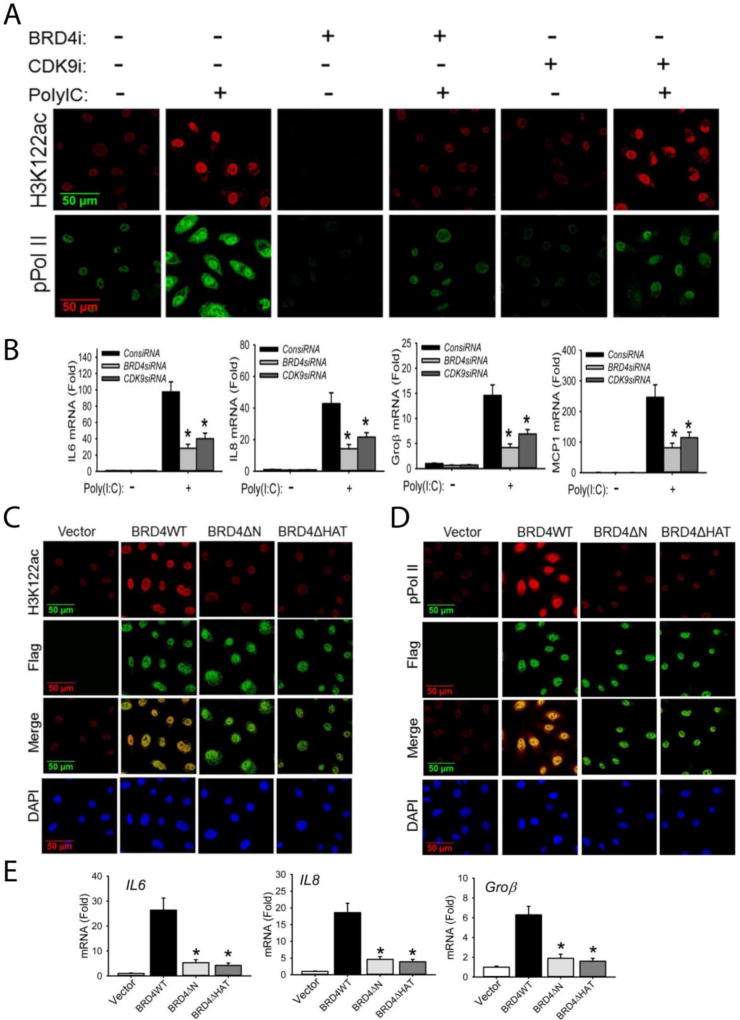
BRD4 is an RNA Pol II serine kinase that functions in cooperation with CDK9 (Devaiah and Singer, 2012). To examine the individual contributions of endogenous BRD4 and CDK9 in poly(I:C)-induced acetylation of H3K122 and formation of phospho-Ser 2 CTD RNA Pol II, we depleted endogenous BRD4 or CDK9 separately using siRNA mediated transfection. We observed that BRD4 silencing blocked both poly(I:C)-induced induction of H3K122 Ac and phospho-Ser 2 CTD RNA Pol II formation by IFCM (Figure 2A). Conversely, CDK9 silencing inhibited phospho-Ser2 CTD RNA Pol II activation to a similar extent as seen by BRD4 silencing, but did not affect induction of H3K122 Ac (Figure 2A). Both BRD4 and CDK9 silencing reduced the expression of RelA-dependent genes IL6, IL8, Groβ, and MCP1 (Figure 2B).
Figure 2. BRD4 is required for acetylation of H3K122 and cooperates with CDK9 for inducible phosphorylation of the RNA Pol II CTD.
(A). hSAECs were transfected with scrambled siRNA (control), BRD4-specific siRNA, or CDK9-specific siRNA (100 nM). 48 h later, cells were stimulated with extracellular poly(I:C) for 4 h. After fixation, cells were stained with anti-H3K122ac and anti-phospho-Ser 2 CTD RNA Pol II Abs as indicated. Secondary detection was Alexa 568-(red, for H3K122ac) and 488-(green, for pPol II) conjugated goat anti-rabbit IgGs. Images were acquired by confocal microscopy at 63× magnification. (B). Expression of IL6, IL8, Groβ, and MCP1 genes from the same experiment were quantified by Q-RT-PCR. Shown is the fold-change in mRNA abundance normalized to cyclophilin (PPIA). *: p < 0.01 compared to poly(I:C) only. Data are the means ± S.D. from n=3 experiments. (C). hSAECs were transfected with 4 µg pCMV2 empty vector (control), BRD4WT, BRD4 kinase mutant (BRD4ΔN) or BRD4 HAT mutant (BRD4ΔHAT) expression vectors. 24 h later, cells were fixed and stained with anti-H3K122 Ac rabbit Ab and anti-FLAG M2 mouse Ab. Secondary detection was Alexa 568-conjugated goat anti-rabbit IgG (red, for H3K122ac) and 488-(green, for Flag) conjugated goat anti-mouse IgG respectively. Nuclei were counterstained and images acquired as in (A). (D). Transient transfectants were stained with anti-phospho-Ser 2 CTD RNA Pol II rabbit Ab and anti-FLAG M2 mouse Ab. Secondary detection, nuclear counterstaining and image acquisition are as in (C). (E).Expression of IL6, IL8, and Groβ were quantified by Q-RT-PCR. *: p < 0.01 compared to BRD4WT transfectants. Data are the means ± S.D. from n=3 experiments.
To directly examine the role of BRD4 kinase or HAT domains in regulating transcription, previously characterized Flag epitope-tagged BRD4 wild type (WT) or mutants were transfected into hSEACs. The BRD4ΔN mutation deletes both the NH2 terminal kinase domain and the acetyl-CoA binding domains, whereas the BRD4ΔHAT mutation destroys its HAT activity (Devaiah et al., 2016). Consistent with the earlier observations (Devaiah et al., 2016), BRD4 WT overexpression itself significantly increased the formation of H3K122 Ac and phospho-Ser 2 CTD RNA Pol II in the Flag-expressing cells (Figures 2C,D). Correspondingly, BRD4 WT overexpression also activated of RelA-dependent IL6, IL8, and Groβ gene expression (Figure 2E). By contrast, expression of either BRD4ΔN or BRD4ΔHAT interfered with H3K122 Ac in the Flag-expressing cells (Figure 2C). Similarly, inducible formation of phospho-Ser 2 RNA Pol II CTD was blocked as was induction of the RelA-dependent genes (Figures 2D,E). We conclude that both BRD4 kinase and HAT activities are required for formation of H3K122 Ac, phospho-Ser 2 CTD RNA Pol II and RelA-dependent transcriptional activation.
Conditional knockout (CKO) of RelA in Secretoglobin-expressing epithelial cells
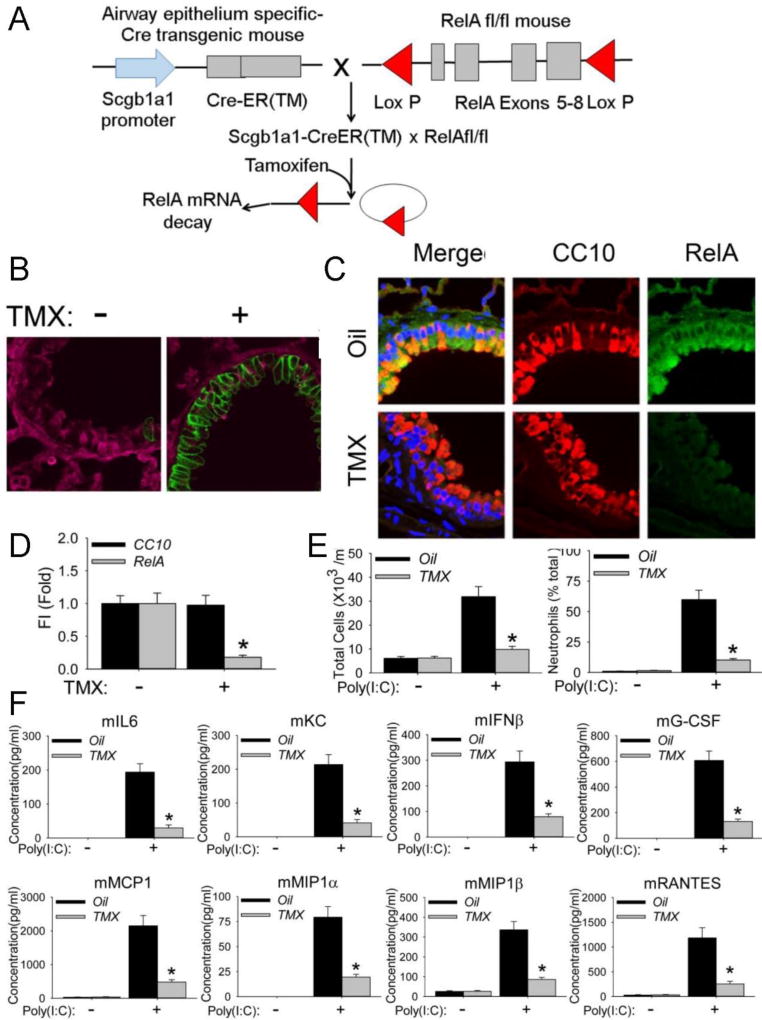
Recognizing that unscheduled RelA activity affects normal lung branching (Benjamin et al., 2010), we used generated an epithelial-specific conditional knockout by crossing our novel NH2 terminal RelAfl/fl mouse (Ijaz et al., 2016) with an epithelial-specific Secretoglobin Family 1A Member 1 (Scgb1a1) promoter-driven inducible Cre recombinase-estrogen receptor fusion protein (CreER™) transgenic mouse (Figure 3A). Scgb1a1-CreER™ expresses Cre selectively in the basal cells of the tracheal and bronchiolar epithelium (Rawlins et al., 2009). This inducible RelACKO enables us to disrupt NFκB signalling activated by either the canonical (IκBα) or the cross-talk (RelA-NFκB2) pathways (Liu et al., 2008).
Figure 3. Conditional knockout of RelA in non-ciliated epithelial Clara cells.
(A). Strategy for generating conditional knockout of RelA in non-ciliated epithelial Clara cells. RelA fl/fl conditional knockout (CKO) were crossed with an epithelial-specific Secretoglobin Family 1A Member 1 (Scgb1a1) promoter-driven inducible Cre recombinase-estrogen receptor fusion protein [CreER(™)] transgenic mouse in the C57BL6/J background. Scgb1a1-CreER(™) × RelA fl/fl mice were injected (i.p.) with oil or Tamoxifen (TMX) 10 d for epithelial RelA knockout. (B).The Scgb1a1-CreER™ × RelAfl/fl mice crossed with the dimeric tomato (mT)/membrane EGFP (mG) mouse and exposed to vehicle oil or TMX for 10 d; 2 weeks later the lungs were analysed by IFCM. Note the membrane localized EGFP (green signal) was observed in the TMX-treated mice. (C). Validation of epithelial RelA conditional knockout. Co-staining of RelA (in green color) and epithelium-specific marker Scgb1a (in red color) were performed using sections of paraffin-embedded lung of mice were treated with oil (control) or TMX for 10 d. (D) Quantifications of total fluorescence intensities Scgb1a1 (CC10) and RelA staining. * p<0.01, n = 5. (E) Total cells and neutrophils count in the bronchoalveolar lavage fluid (BALF), expressed as total number of cells X 103/ml (left) and percentage of neutrophils (right). #, p<0.01 compared with Oil-treated mice (n=5). (F). BALF cytokine secretion was measured using multiplex ELISA (n=5 mice per group). Protein concentration (in pg/ml) is shown. *, p<0.01, n=5.
Confirmation of RelACKO in the Scgb1a1-expressing bronchiolar epithelium
We evaluated the effectiveness of the bronchiolar epithelial RelACKO on acute poly(I:C)-induced airway inflammation. Scgb1a1-CreER™ × RelAfl/fl mice were treated with oil (control) or tamoxifen (TMX) to excise RelA. To confirm selective recombination in the bronchiolar epithelium, we crossed the Scgb1a1-CreER™ × RelAfl/fl mice with the double-fluorescent dimeric tomato (mT)/membrane EGFP (mG) mouse (Muzumdar et al., 2007). In mice harbouring the mT/mG alleles, the presence of active Cre leads to excision of the tandem dimer tomato reporter and expression of a membrane-localized EGFP. The Scgb1a1-CreER™ × RelAfl/fl× mT/mG mice were then exposed to oil or TMX for 10 d; 2 weeks later the lungs were analysed by IFCM. In the oil-treated mice, uniform staining of the tomato reporter was observed throughout the bronchioles, identified as airspaces lined by single layered epithelial cells without surrounding tracheal cartilage (Figure 3B). By contrast, EGFP fluorescence was observed in the epithelial layer of the TMX-treated mice, confirming functional Cre expression (Figure 3B). RelA expression was significantly reduced in these cells after TMX treatment (Figures 3C and 3D).
Bronchiolar epithelial RelACKO blocks poly(I:C)-induced leukocytic inflammation
To determine the role of the epithlium as a sentinel cell detecting viral luminal PAMPs, we challenged oil- or TMX-treated Scgb1a1-CreER™ × RelAfl/fl with intranasal poly(I:C). Poly(I:C) increased the leukocyte number ~ 5-times greater than that of PBS challenge control in oil-treated mice, an induction that was effectively inhibited by TMX treatment (Figure 3E). Also, the percentage of BALF neutrophils increased from 1% to 56.3% in oil-treated mice exposed to poly(I:C), and this induction was significantly reduced to 9.7% in TMX-treated mice (Figure 3E).
Correspondingly, bronchiolar RelACKO had a dramatic effect on poly(I:C)-induced BALF chemokine expression. In comparison to PBS-treated mice, where IL6 protein abundance was below the lower limit of detection, poly(I:C) induced IL6 to 193 pg/ml, an increase largely reduced by TMX (Figure 3F). Similar poly(I:C)-induced expression patterns were seen for KC, IFNβ, CCL2/MCP1, G-CSF, MIP1α, MIP1β, and CCL5/RANTES. In the TMX-treated animals, poly(I:C)-induced values were all significantly reduced (p<0.01, Figure 3F). We confirmed that these reductions in chemokine expression were mediated by inhibition of mRNA expression (Supplementary Figure S2). In addition to the effects on neutrophilic chemokines, we recently described RelA to be a master regulator of mucosal IFN response through a cross-talk pathway (Fang et al., 2015; Tian et al., 2017). Here, IRF7 is induced in a RelA-dependent manner; this newly formed IRF7 binds and activates the RIG-I promoter, sustaining IFNβ production. We confirmed that this pathway is also activated by poly(I:C) and is blocked by the RelACKO in bronchiolar epithelium (Supplemental Figure S3).
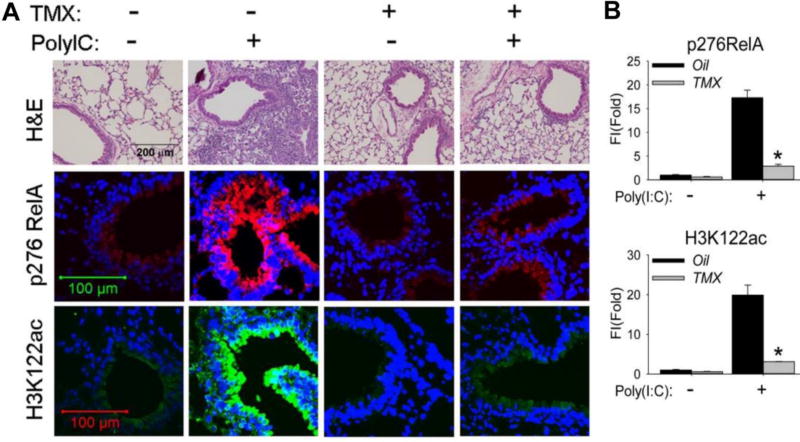
In the oil-treated animals, poly(I:C) stimulation produced a marked infiltration of neutrophils into the alveolar spaces and peribronchial regions (Figure 4A). By contrast, leukocytic inflammation was completely blocked in the TMX-treated RelACKO. We next determined whether poly(I:C) induced BRD4 HAT activity. In oil-treated mice challenged with PBS, only faint phospho-Ser 276 RelA signals were produced; poly(I:C) stimulation highly induced phospho-Ser 276 RelA in the bronchiolar epithelium of the oil-treated mice(Figure 4A, and quantitated in Figure 4B). By contrast, the phospho-Ser 276 RelA staining was absent in TMX-treated RelACKO, confirming staining specificity.
Figure 4. Bronchiolar RelA is required for poly(I:C) induced inflammation in vivo.
Scgb1a1/CC10 -Cre X RelACKO mice were pretreated with oil (control) or TMX 21 d prior to experimentation. For poly(I:C) challenge, mice were administered either 50 µL of PBS or 50 µL PBS + 100 µg poly(I:C) via the i.n. route. (A). Hematoxylin and Eosin (H&E) staining (upper panel), immunofluorescence staining of phospho-276 RelA, (middle panel, in red color) and immunofluorescence staining of H3K122 Ac (lower panel, in green color) were performed on paraffin-embedded lung sections. (B). Quantifications of total fluorescence intensities on immunofluorescence staining of phospho-276 RelA and H3K122 Ac. * p<0.01, n = 5.
Examination of the staining for H3K122 Ac showed a similar pattern of induction as that observed for the phospho-Ser 276 RelA, with faint levels observed in the PBS-treated wild type airways, and robust 21-fold induction of staining in the bronchiolar epithelium in response to poly(I:C) (Figures 4A,B). Strikingly, poly(I:C)-induced activation of H3K122 Ac was effectively blocked by TMX treatment of the RelACKO.
Collectively, all of these data provide compelling evidence that activation of bronchiolar RelA is required for BRD4 HAT activity and poly(I:C)-induced airway inflammation. We next sought to advance the use of small molecule probes of BRD4 activity using small molecule probes.
BRD4 expression is required for induction of the RelA-dependent gene network
First, to confirm that BRD4 expression mediates activation of poly(I:C)-induced gene regulatory networks, we observed the robust, 120-fold increase in IL6 mRNA by poly(I:C) was reduced to less than 35-fold in BRD4 siRNA-silenced cells (Supplemental Figure S4). Similarly, BRD4 siRNA silencing effectively blocked poly(I:C)-induced expression of CXCL2/Groβ, CXCL8/IL8, CCL2/MCP1, and CCL5/RANTES mRNAs (Supplemental Figure S4).
BRD4 bromodomain (BD) interactions are required for induction of the RelA-dependent gene network
To determine whether disruption of BRD4-acetyl Lys interactions produced similar results as those produced by BRD4 depletion, we tested a panel of chemically unrelated BRD4 BD inhibitors on poly(I:C)-induced neutrophilic chemokine expression. The inhibitors tested included: the diazobenzene MS436 (Zhang et al., 2013); the amide JQ1 analog, CPI203 (Filippakopoulos et al., 2010); the apabetalone RVX-208 (Picaud et al., 2013) and the dimethylsoxazole OXFBD2 (Hewings et al., 2011). All produced a profound inhibition of poly(I:C)-induced IL6, CXCL8/IL8, and CXCL2/Groβ mRNA expression (Supplemental Figure S4). These significant reductions in gene expression were confirmed by measurement of cytokine secretion in to the cell culture supernatant (Supplemental Figure S4). We conclude that BRD4 BD interactions are required for poly (I:C)-induced expression of RelA-dependent gene networks and neutrophilic chemokine secretion, making this an attractive target for small molecule inhibition.
Development of selective BRD4 inhibitors targeting the BD domain
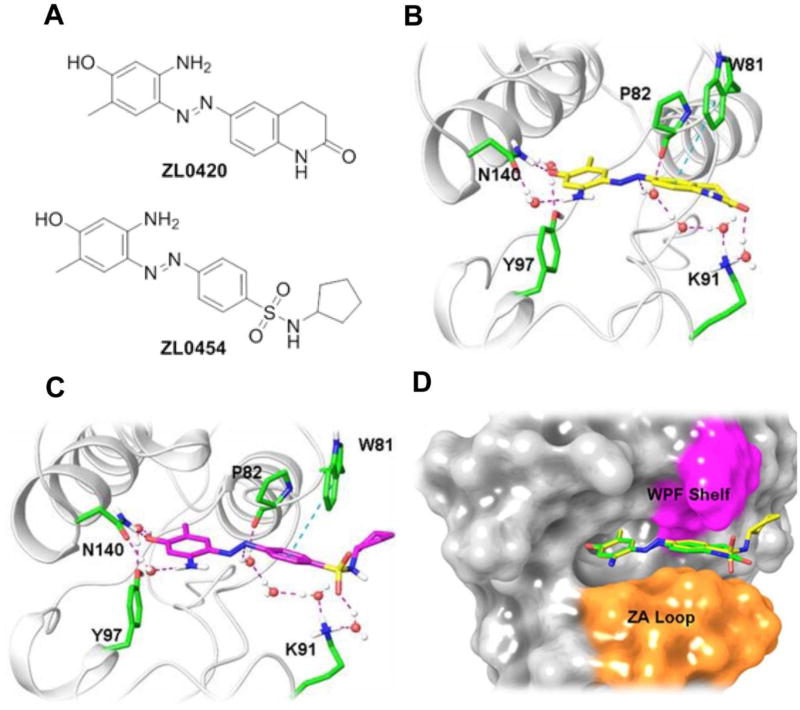
We therefore sought to designe and synthesize BRD4 inhibitors with improved selectivity and drug-like properties. The KAc binding pocket in the BRD4 BD is anchored by the conserved residue Asn140 and Tyr97 and surrounded by a hydrophobic tripeptide tryptophan-proline-phenylalanine (“WPF”) shelf and linking domain “ZA” loop. New BRD4 inhibitors ZL0420 and ZL0454 (Figure 5A) were designed by taking advantage of a substituted amino phenol moiety as the polar head of our ligand, diazene as the linker and dihydroquinolin-2(1H)-one or N-cyclopentylbenzenesulfonamide as the binding tails to more efficiently fit into the BRD4 BD1 domain. As depicted in Figures 5B and 5C, docking studies demonstrated that these two inhibitors fit into the BRD4 BD1 in a similar binding mode. Both compounds interact with Asn140 directly through the hydroxyl group of their phenol ring and form hydrogen bonds with Tyr97 mediated by a water molecule. In addition, the phenyl ring of both molecules on their tails can form a T shape π-π interaction with Trp81, while the diazene linker interacts with Lys91 mediated by water molecules. These two inhibitors are sandwiched by the WPF shelf and residues from ZA loop (Figure 5D), thereby allowing strong interactions of the ligand producing high binding affinity to the BRD4 BD.
Figure 5. Design and structures of novel BRD4 inhibitors ZL0420 and ZL0454.
(A) Chemical structures of novel BRD4 inhibitors ZL0420 and ZL0454. (B). Binding mode of ZL0420 (yellow) with BRD4 BD1 in ribbon representation. The highlighted residues are Asn140 (N140), Tyr97 (Y97), Pro82 (P82), Trp81 (W81) and Lys91 (K91). (C). Binding mode of ZL0454 (magenta) with BRD4 BD1 in ribbon representation. (D). Superimposition of ZL0420 (green) and ZL0454 (yellow) docked into BRD4 BD1 in the form of surface representation. WPF (Trp81-Pro82-Phe83) shelf is highlighted in magenta and part of ZA loop is colored orange. Asn, Asparagine; Tyr, Tyrosine; Pro, Proline; Trp, Tryptophan; Lys, Lysine.
ZL lead compounds are potent and highly specific BRD4 inhibitors
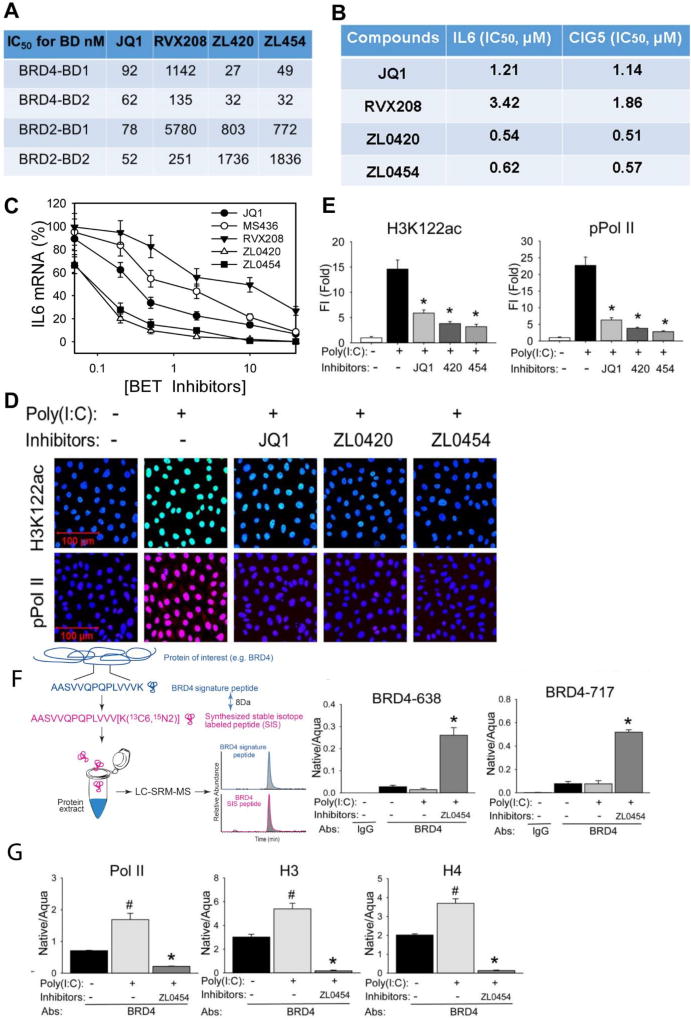
Despite significant effort invested in the development of small molecule BRD4 inhibitors, none have been yet reported that are highly selective for BRD4 over other members of the BET family (Liu et al., 2017). To this end, by utilizing a structure-based drug design for the KAc binding pocket, ZL-0420 and ZL-0454, containing a dihydroquinolin-2(1H)-one or N-cyclopentyl-benzenesulfonamide binding tail, were synthesized. Specificity for the BRD4 BDs was determined by TR-FRET measurements vs the BDs from the closely related homolog, BRD2. Strikingly, ZL-0420 and -0454 display nanomolar binding affinities for BRD4, while exhibiting approximately 30~60-fold selectivity over BRD2 BDs (Figure 6A).
Figure 6. ZL lead compounds are potent and highly specific BRD4 inhibitors.
(A). Specificity of BRD4 inhibitors to BRD4 BDs was determined using TR-FRET assays. Data shown as the IC50 values of tested BRD4 inhibitors to the BRD2 and BRD4 BDs separately. (B). IC50 values of ZL lead compounds. hSAECs were pretreated with indicated series of BRD4 inhibitors from 0.01 nM to 100 µM final concentrations overnight prior to poly(I:C) stimulation (10 µg/ml for 4 h). The IC50 values of each inhibitor were estimated by poly(I:C) induced expression of innate immune genes IL6 and CIG5. (C). BRD4 inhibitors block IL6 expression. Shown as % inhibition of JQ1, MS436, RVX208, ZL0420, and ZL0454 inhibitors on poly(I:C)-induced IL6 expression. The results are from three independent experiments (n=3). (D) hSAECs were preincubated with ZL0420, ZL0454, and JQ1 overnight respectively before poly(I:C) stimulation. After fixation, cells were stained with anti-H3K122ac and phospho-Ser 2 CTD RNA Pol II Abs. (E). Quantifications of total fluorescence intensities on images of immunostaining of H3K122ac and phospho-Ser 2 CTDRNA Pol II in hSAECs. * p<0.01, n = 5. (F). Left, schematic diagram of the IP-SID-SRM-MS assay. Stable isotope labelled internal control peptides are used to quantify target proteins in a triple quadrupole MS. Right, SID-SRM-MS assay for two BRD4 peptides (BRD4-638 peptide and 717 peptide). (G). IP-SID-SRM-MS for BRD4 associated Pol II, H3, and H4 proteins were normalized by the input protein concentration and presented as native/aqua values. *: p < 0.01 compared to poly(I:C) only and #: p < 0.01 compared to mock treatment, n=3.
ZL lead compounds block poly(I:C) induced chemokine expression and BRD4 HAT activity in vitro
We evaluated the effectiveness of the two ZL lead compounds for inhibiting poly(I:C)-induced innate immune response genes. hSAECs were pretreated with a series of BRD4 inhibitors from 0.01 nM to 100 µM (final concentration) for 24 h, stimulated with poly(I:C) (10 µg/ml, 4 h) and innate gene expression of IL6 and RSAD2/CIG5 determined by Q-RT-PCR. These data showed that both ZL compounds have a submicromolar IC50 for inhibiting poly(I:C) induced expression of innate signalling and were more potent than either JQ1 (Filippakopoulos et al., 2010) and RVX-208 (Picaud et al., 2013) reference compounds (Figures 6B and 6C). Also, both ZL0420 and ZL0454 effectively blocked poly(I:C)-induced activation of H3K122Ac and formation of phospho-Ser2 CTD RNA Pol II (Figure 6D, quantitated in Figure 6E). This selectivity profile indicates that our new inhibitors have a much higher BRD4 specificity than available reference molecules.
ZL lead compounds disrupt BRD4 binding to Pol II and histones
To determine the mechanism of inhibition, we measured the effect of ZL0454 on BRD4 binding to Pol II and histones. Soluble nuclear proteins were prepared from hSAECs pretreated with ZL0454 for 24 h and subsequently stimulated with poly(I:C). The BRD4 complex was captured by IP with anti-rabbit BRD4 Ab and the complex was analysed for the presence of BRD4, Pol II, H3, and H4 proteins quantified by stable isotope dilution (SID)-selected reaction monitoring (SRM)-MS, normalized by the input protein concentration (Tian et al., 2013; Zhao and Brasier, 2013). A schematic of the SRM assays is shown in Figure 6F. BRD4 abundance was measured using two independent proteotypic peptides, BRD4-638 and -717 (Supplemental Table 2). Compared to control IgG IPs, where the BRD4 abundance was completely undetectable, BRD4 was significantly enriched in the samples IPed with anti-BRD4 Ab (Figure 6F). We detected a strong increase of BRD4 signals in the IPed samples of ZL0454 pretreated hSAECs, indicating an increase of soluble fraction of BRD4 in the nucleoplasm released from the insoluble chromatin (Figure 6G). More importantly, poly(I:C) stimulation increased the association of BRD4 with Pol II, H3, and H4 and the effect of poly (I:C) was completely blocked with ZL0454 pretreatment (Figure 6G). We therefore conclude that ZL BD inhibitors disrupt BRD4 binding to Pol II and histones, releasing it from chromatin into the soluble fraction of the nucleoplasm.
In vivo effect of ZL0420 and ZL0454 on poly(I:C) induced airway inflammation
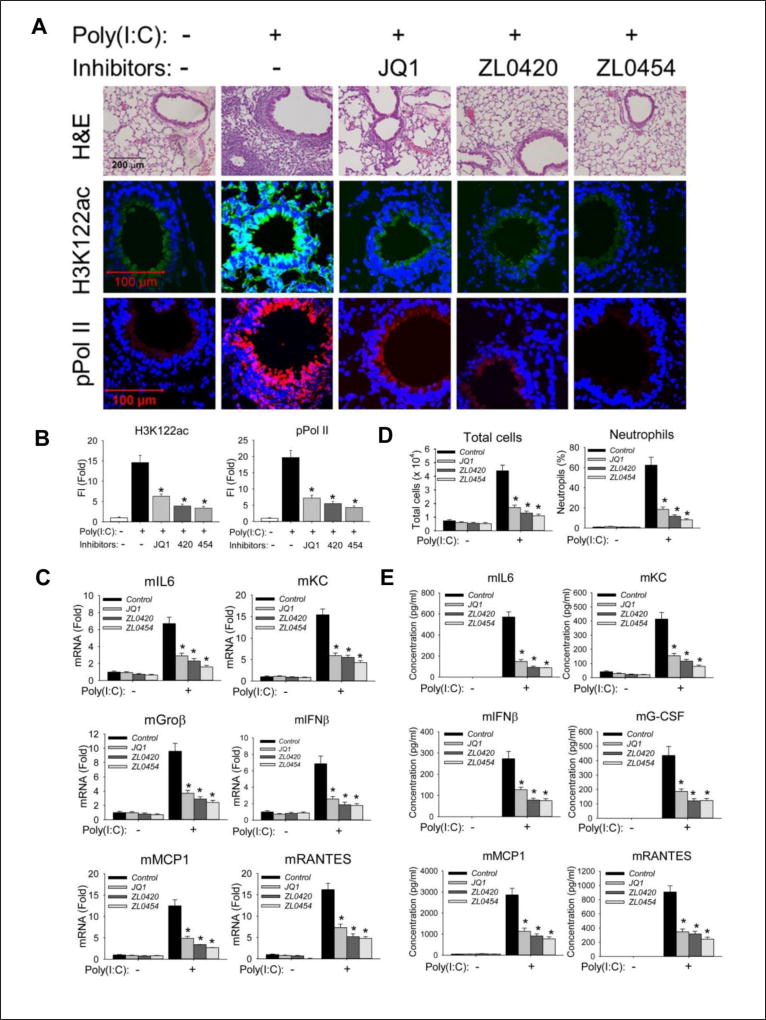
We evaluated the effectiveness of the lead inhibitors (ZL-0420, -0454) in blocking acute poly(I:C)-induced airway inflammation in vivo. Histologically, we observed that poly(I:C) induced a profound neutrophilic inflammation around the small and medium sized airways 24 h after poly(I:C) stimulation that was completely inhibited by ZL inhibitor administration (Figure 7A, upper panel). ZL inhibitors also effectively blocked poly(I:C)-induced activation of H3K122 Ac, a selective marker of the activated BRD4 HAT and formation of phospho-Ser2 CTD RNA Pol II (Figure 7A, quantitated in Figure 7B). Furthermore, ZL inhibitors prevented poly(I:C)-induced mRNA expression of the NFκB/RelA-dependent genes IL6, KC, CXCL2/Groβ, CCL2/MCP1, and CCL5/RANTES as well as IFNβ in the mouse lung tissue (Figure 7C). Meanwhile, ZL inhibitors also induced HEXIM1 expression, indicating that these compounds were interacted with BRD4 in the mouse lung tissue (Supplemental Figure S5) (Fiskus et al., 2014).
Figure 7. BRD4 inhibitors block poly(I:C) induced inflammation and neutrophilia in vivo.
C57BL6/J mice were pre-treated (IP) with ZL compounds before intranasal (IN) poly(I:C) exposure. (A). Hematoxylin and Eosin (H&E) staining (upper panel), immunofluorescence staining of H3K122ac (middle panel, in green color) and phospho-Ser 2 CTD RNA Pol II (lower panel, in red color) were performed on paraffin-embedded lung sections. (B). Quantifications of total fluorescence intensities of immunofluorescence staining of H3K122ac and phospho-Ser 2 CTD RNA Pol II. * p<0.01, n = 5. (C). Total lung RNA was isolated for measurement of mKC, mGroβ, mIL6, mIFNβ, mMCP1, and mRANTES mRNA abundance by Q-RT-PCR. m, mouse. *, p<0.01 compared to PBS only treated mice, n=5. (D). Total cells and neutrophils in the BALF, expressed as total number of cells X 104/ml (left) and percentage of neutrophils (right). (E). BALF cytokine abundance was measured using multiplex ELISA. *, p<0.01, n=5.
The number of leukocytes in BALF of poly(I:C)-treated mice increased about 6-times compared to that of PBS-treated mice, an induction completely inhibited by treatment with either ZL compound (Figure 7D, left panel). Also, we observed the percentage of BALF neutrophils that increased from 1% to 62.4% in PBS-treated mice exposed to poly(I:C), was reduced to 11.7% and 8.2%, respectively, in ZL0420 and ZL0454 treated mice (Figure 7D right panel). Meanwhile, we also observed that ZL inhibitors effectively suppress poly(I:C)-induced inflammatory cytokine secretion in the BALF (Figure 7E). Collectively, these findings provide strong evidence that BRD4 is required for poly(I:C)-induced inflammation, and that the ZL inhibitors are efficacious in vivo.
DISCUSSION
Viral infections are responsible for unscheduled hospital visits, prolonged recovery time and accelerated loss of pulmonary function in patients with COPD (Donaldson et al., 2002; Seemungal et al., 1998). One mechanism is that viruses promote exacerbations by triggering host inflammatory responses resulting in cellular infiltration, mucous production, airway hyper-reactivity and remodelling (Wedzicha, 2004). Using poly(I:C) as a prototypical viral PAMP that mimics the inflammatory exacerbations of COPD (Harris et al., 2013), we examined the role of the Scgb1a1-expressing tracheo-bronchiolar epithelial cell as an innate sentinel cell, and specifically, the role of NFκB-BRD4 pathway in mediating airway inflammation. Our findings that selective depletion of RelA in tracheal and bronchiolar epithelial cells prevented BRD4 activation and poly(I:C) induced expression of inflammatory cytokines and IFNs, indicate that epithelial cells are a major sensor of luminal poly(I:C). Two highly selective BRD4 small molecular antagonists disrupt the BRD4 complex and consequently block viral inflammatory gene programs in vitro and airway inflammation in vivo. Together, these data indicate that the bronchiolar epithelial NFκB-BRD4 complex plays a primary role in acute neutrophilic response to viral molecular patterns.
The pulmonary mucosal innate defense is coordinated by a complex interaction between resident epithelial cells, alveolar macrophages, innate lymphoid cells and other innate leukocytes (Holt and Strickland, 2010). Epithelial cells are a dynamically responsive and structurally diverse population of cells that play essential roles in host defense by forming a semi-impermeable barrier, clearing inhaled microparticles by the ciliary escalator, secreting anti-viral mucins and expressing anti-viral cytokines (Lambrecht and Hammad, 2012). Despite this understanding, the relative contributions of epithelial vs other sentinel cells of the airways (macrophages, innate epithelial lymphocytes, NK cells and others) has not been definitely established. Here we demonstrate that a Scgb1a1/CC10- expressing lineage of epithelial cells represents the major sensor of viral patterns in the airway lumen. Scgb1a1 is selectively expressed in distal bronchial and bronchiolar epithelial Clara cells, a dynamic population of basal (nonciliated) epithelial cells that play important roles in xenobiotic metabolism and as stem cell precursors to repopulate the postnatal airway after injury(Rawlins et al., 2009).
Our studies extend the function of the Scgb1a1-expressing Clara cell as an innate sensor and mediator of anti-viral response. It is interesting that diverse phenotypes of airway epithelial cells- express the TLR3 pattern recognition receptors on their luminal surface and respond to extracellular dsRNA in contrast to neutrophils which lack TLR3 expression(Hayashi et al., 2003). Consequently neutrophil recruitment is an indirect process, primarily mediated by epithelial secretion of neutrophilic chemokines. Given this observation, why would depletion of NFκB/RelA selectively in these tracheobronchial cell population have such a dramatic effect on airway inflammation? Once explanation may be provided by our systems-level proteomics studies that have shown cell-type differences in innate cytokine production in response to RSV infection. Here, we observed that, although a significant amount of proteins were shared between tracheal and bronchiolar cells, the bronchiolar cells selectively secreted TSLP, CCL20 and IL6, proteins that promote Th2 and mucin expression important in the pathogenesis of RSV-induced lower respiratory tract infections (Zhao et al., 2017). These subtle differences in chemokine expression patterns by different epithelial cell types are therefore immunologically significant.
Our experimental approach using a TMX-inducible conditional RelACKO in the Scgb1a1 expressing cell is distinct from several studies that have used the Scgb1a1 promoter to inhibit the canonical NFκB pathway in airway inflammation and fibrosis in dust mite allergen exposure (Tully et al., 2013). Expression of the non-degradable IκBα ”super-repressor” selectively inhibits canonical pathway activation, but does not affect the cross-talk pathway (Brasier, 2006). It is important to recognize that NFκB activation by viral patterns involves both the canonical and cross-talk pathways (Liu et al., 2008); our RelACKO strategy interferes with both.
NFκB mediates rapid genomic responses by recruiting transcriptional elongation complexes to immediate early genes (Brasier et al., 2011). Viral PAMPs induce ROS production that interfaces with DNA damage response pathways to produce Ser phosphorylation of RelA, a post-translational modification coupled with lysine acetylation that promotes complex formation with BRD4 (Brasier et al., 2011; Zou et al., 2014). BRD4 is a acetylated lysine reader containing intrinsic kinase activity (Devaiah et al., 2012) that bridges RelA with CDK9 of the positive transcriptional elongation complex (Jang et al., 2005). Phosphorylation on Ser2 of RNA Pol II CTD is a key regulatory step in the activation of NFκB-dependent genes by transcriptional elongation. Our findings that global Ser2 CTD RNA Pol II activation requires both activated RelA as well as BRD4 suggests that the RelA·BRD4 complex may be a global genome regulatory complex. Our data demonstrating that BRD4 is required for serine phosphorylation of RNA Pol II is consistent with earlier findings showing BRD4 mediates innate signalling by cytokines (Huang et al., 2009) and RNA viruses (Tian et al., 2017).
In addition, BRD4 is an atypical HAT producing H3 K122 Ac, a modification that reduces histone side charge and enables RNA Pol II to transcribe through GC rich regions of innate immune response genes, an independent effect on transcriptional elongation (Devaiah et al., 2016). Our data shows that BRD4 HAT activity is upregulated by activated RelA downstream of diverse innate stimuli. Our transient transfection studies, interpreted with the effects of our selective BRD4 BD inhibitors both suggest that the H3K122 acetyltransferase requires BRD4 HAT activity. We note previous work that have shown H3K122 acetylation is independent of p300/CBP acetyltransferases (Devaiah et al., 2016). Because RelA lacks any known enzymatic activity, the RelA-dependent activation of BRD4 HAT must be indirect. Our previous experimental and computational work has identified 900 proteins that interact with RelA that modify its activity (Li et al., 2014; Zhang et al., 2017), the identification of those responsible for activating BRD4 HAT will require further work.
Because of its diverse roles in inflammation, cancer and fibrosis, development of BRD4 inhibitors have been the subject of intense focus (Liu et al., 2017). In this study, we have applied structure-based drug design to advance the pharmacopeia of BRD4 inhibitors. Our small molecule inhibitors demonstrate enhanced potency and selectivity for both BDs of BRD4 but not for the closely related BRD2 isoform. More detailed structure-function relationship studies will be required to understand this BD preference. BRD4 forms protein-protein interactions via the BDs (Huang et al., 2009) and its COOH extra-terminal domain (Jonkers and Lis, 2015). Our recent unbiased proteomics studies demonstrate that BRD4 is a hub signalling protein that complexes with the AP2 adapter, SWI/SNF, and DNA directed RNA Pol II complex (Zhang et al., 2017). Here we demonstrate the mechanism of inhibition of BD-directed inhibitors. Affinity purification-SID-SRM studies demonstrate that the BRD4 inhibitors not only disrupt BRD4 from its tight association with the chromatin, and histone H3/H4 interaction, but also disrupt BRD4 binding with RNA Pol II. Earlier work has shown that the BRD4 BD is required for interaction with acetylated RelA (Zou et al., 2014). Further studies are in progress to systematically identify what components of the BRD4 interactome require binding to the BD.
The ZL compound forms extensive and strong interactions with the BRD4 BD1 including direct and multiple H2O-mediated indirect hydrogen bond networks, π- π interactions and hydrophobic contacts. By contrast, very limited interactions were observed BRD2 BD1. In molecular docking experiments, the ZL NH2 was rotated where it formed a single hydrogen bond with Pro98. No direct or indirect hydrogen bonds were seen. Hydrophobic contacts were observed with Asn156. In this conformation, the hydrophobic tail of ZL was away from the WPF shelf. Such differences in key interactions may explain the selectivity of ZL compound for BRD4 over other relevant proteins BRD2, BRD3, and BRDT.
Using these unique molecular probes, we are able to elucidate the biological roles of BRD4 in vivo. The effect of our selective inhibitors to block H3K122 Ac and inflammation validate BRD4 as a target for treatment of acute lung inflammation mediated by viral PAMPs triggering the RIG-I and the TLR3 pathway. Recently, we have discovered the important role of the RelA-BRD4 pathway in controlling growth factor-induced mesenchymal transition, airway remodelling and airway fibrosis (Tian et al., 2017; Tian et al., 2016). Further work will be needed to determine whether BRD4 inhibitors are effective in reducing airway remodelling as a consequence of repetitive viral exposures.
EXPERIMENTAL PROCEDURES
A complete description of experimental procedures is found in the Supplemental Files
Efficacy of BRD4 inhibitors in acute poly(I:C) induced airway inflammation
Animal experiments were performed according to the NIH Guide for Care and Use of Experimental Animals and approved by the University of Texas Medical Branch (UTMB) Animal Care and Use Committee (approval no. 1312058A). Male C57BL6/J mice (18 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME) and housed under pathogen-free conditions with food and water ad libitum. C57BL/6 mice were pre-treated in the absence or presence of the indicated BRD4 inhibitor [50 mg/kg body weight, via the intraperitoneal (i. p.) route] one day prior to poly(I:C) stimulation. The next day, mice were given another dose of BRD4 inhibitor immediately followed by intranasal (i.n.) administration of phosphate-buffered saline (PBS, 50 µL) or poly(I:C) (100 µg dissolved in 50 µL PBS). One day later, the mice were euthanized. For bronchoalveolar lavage (BAL), the trachea was cannulated and 1 ml of PBS was introduced by syringe. The BAL fluid was analysed for total and differential cell counts and quantified for secreted cytokines/chemokines using multiplex immunoassays (BioPlex). Half of each lung was fixed with 10% (v/v) neutral buffered formalin for 3 d processed into paraffin blocks, and cut into 5-µm sections for H&E staining. The other half of each lung was immediately frozen in liquid N2 and pulverized. One ml of Tri Reagent was added, followed by the extraction of total RNA as per the manufacturer’s directions. The total RNA was reverse transcribed and gene expression quantified using Q-RT-PCR (Tian et al., 2017).
Evaluation of airway inflammation
Lungs were perfused twice with 0.75 mL of sterile PBS (pH 7.4) to obtain the BALF. Total cell counts were determined by trypan blue staining 50 µL of BALF and counting viable cells using a hemocytometer. Differential cell counts were performed on cytocentrifuge preparations (Cytospin 3; Thermo Shandon, Pittsburgh, Pa) stained with Wright-Giemsa. A total of 300 cells were counted per sample using light microscopy. Formalin-fixed lungs were embedded in paraffin, sectioned at a 4 µm thickness, and stained with hematoxylin and eosin or Masson's trichrome. Microscopy was performed on a NIKON Eclipse Ti System (Tian et al., 2016).
Statistical analysis
One-way analysis of variance (ANOVA) was performed for time differences, followed by Tukey's post hoc test to determine significance. Mann-Whitney tests were used for nonparametric data. All data subjected to statistical analysis are the mean ± S.D. A P value of <0.05 was considered significant.
Supplementary Material
Highlights.
Innate stimuli activate NFκB/RelA, whose binding indirectly induces BRD4 HAT and kinase activity.
Activity of the RelA·BRD4 complex controls neutrophil chemokine production.
RelA·BRD4 signaling in Scgb1a1-expressing basal cells of the distal epithelium is required for airway inflammation.
Highly selective, small-molecule inhibitors of BRD4 reduce neutrophilic airway inflammation.
In Brief.
Tian et al. demonstrate that viral patterns activate RelA to complex with BRD4 and indirectly activate its enzymatic activity in distal trachea-bronchiolar cells to induce acute neutrophilic airway inflammation. Two highly selective, small-molecule inhibitors of the BRD4 BD were developed. These disrupt BRD4 complex formation, HAT activity and neutrophilia in vivo.
Acknowledgments
We thank Dr. Dinah Singer for sharing BRD4 expression plasmids. Core laboratory support was provided by the UTMB Transgenic mouse facility, Immunohistochemistry Core, and Optical Imaging Core. This work was supported, in part, by NIH grants NIAID AI062885 (ARB), UL1TR001439 (ARB), NIEHS ES006676 (ARB, BT), NSF grant DMS-1361411/DMS-1361318 (ARB), UTMB Technology Commercialization Program (ARB, BT, JZ &ER), and Sanofi Innovation Awards (iAwards) (ARB, JZ, BT, &ER).
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, two tables, and can be found online with this article.
AUTHOR CONTRIBUTIONS
Conceptualization, BT, ER, JZ, ARB; Methodology, BT, ZL, JZ, ARB; Resources, JZ, ARB; Writing-Original Draft, BT, ARB, ZL, JZ; Reviewing and Editing BT, ARB, ZL, YZ, JZ, ER; Funding acquisition-ARB, BT, JZ, ER.
References
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-[kappa]B by Toll-like receptor 3. Nature (London) 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- Benjamin JT, Carver BJ, Plosa EJ, Yamamoto Y, Miller JD, Liu JH, van der Meer R, Blackwell TS, Prince LS. NF-kappaB activation limits airway branching through inhibition of Sp1-mediated fibroblast growth factor-10 expression. Journal of immunology (Baltimore, Md : 1950) 2010;185:4896–4903. doi: 10.4049/jimmunol.1001857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolusso R, Tian B, Zhao Y, Vergara LA, Sabree A, Iwanaszko M, Lipniacki T, Brasier A, Kimmel M. Dynamic Cross Talk Model Of The Epithelial Innate Immune Response To Double-Stranded Rna Stimulation: Coordinated Dynamics Emerging From Cell-Level Noise. PLoS ONE. 2014;9:e93396. doi: 10.1371/journal.pone.0093396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasier AR. The NF-kappaB regulatory network. Cardiovasc Toxicol. 2006;6:111–130. doi: 10.1385/ct:6:2:111. [DOI] [PubMed] [Google Scholar]
- Brasier AR, Tian B, Jamaluddin M, Kalita MK, Garofalo RP, Lu M. RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection. J Virol. 2011;85:11752–11769. doi: 10.1128/JVI.05360-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease, C., and Prevention. Deaths from chronic obstructive pulmonary disease--United States, 2000–2005. MMWR Morb Mortal Wkly Rep. 2008;57:1229–1232. [PubMed] [Google Scholar]
- Devaiah BN, Case-Borden C, Gegonne A, Hsu CH, Chen Q, Meerzaman D, Dey A, Ozato K, Singer DS. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol. 2016;23:540–548. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, Ozato K, Sims RJ, 3rd, Singer DS. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A. 2012;109:6927–6932. doi: 10.1073/pnas.1120422109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah BN, Singer DS. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem. 2012;287:38755–38766. doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GC, Seemungal TA, Bhowmik A, Wedzicha JA. Relationship between exacerbation frequency and lung function decline in chronic obstructive pulmonary disease. Thorax. 2002;57:847–852. doi: 10.1136/thorax.57.10.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GC, Seemungal TA, Patel IS, Bhowmik A, Wilkinson TM, Hurst JR, Maccallum PK, Wedzicha JA. Airway and systemic inflammation and decline in lung function in patients with COPD. Chest. 2005;128:1995–2004. doi: 10.1378/chest.128.4.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Choudhary S, Tian B, Boldogh I, Yang C, Ivanciuc T, Ma Y, Garofalo RP, Brasier AR. Ataxia Telangiectasia Mutated Kinase Mediates NF-kappaB Serine 276 Phosphorylation and Interferon Expression via the IRF7-RIG-I Amplification Loop in Paramyxovirus Infection. J Virol. 2015;89:2628–2642. doi: 10.1128/JVI.02458-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskus W, Sharma S, Qi J, Shah B, Devaraj SG, Leveque C, Portier BP, Iyer S, Bradner JE, Bhalla KN. BET protein antagonist JQ1 is synergistically lethal with FLT3 tyrosine kinase inhibitor (TKI) and overcomes resistance to FLT3-TKI in AML cells expressing FLT-ITD. Mol Cancer Ther. 2014;13:2315–2327. doi: 10.1158/1535-7163.MCT-14-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P, Sridhar S, Peng R, Phillips JE, Cohn RG, Burns L, Woods J, Ramanujam M, Loubeau M, Tyagi G, et al. Double-stranded RNA induces molecular and inflammatory signatures that are directly relevant to COPD. Mucosal immunology. 2013;6:474–484. doi: 10.1038/mi.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi F, Means TK, Luster AD. Toll-like receptors stimulate human neutrophil function. Blood. 2003;102:2660–2669. doi: 10.1182/blood-2003-04-1078. [DOI] [PubMed] [Google Scholar]
- Hewings DS, Wang M, Philpott M, Fedorov O, Uttarkar S, Filippakopoulos P, Picaud S, Vuppusetty C, Marsden B, Knapp S, et al. 3,5-dimethylisoxazoles act as acetyl-lysine-mimetic bromodomain ligands. J Med Chem. 2011;54:6761–6770. doi: 10.1021/jm200640v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt PG, Strickland DH. Interactions between innate and adaptive immunity in asthma pathogenesis: New perspectives from studies on acute exacerbations. The Journal of allergy and clinical immunology. 2010;125:963–972. doi: 10.1016/j.jaci.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Huang B, Yang XD, Zhou MM, Ozato K, Chen Lf. Brd4 Coactivates Transcriptional Activation of NF-{kappa}B via Specific Binding to Acetylated RelA. Molecular and Cellular Biology. 2009;29:1375–1387. doi: 10.1128/MCB.01365-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ijaz T, Wakamiya M, Sun H, Recinos A, 3rd, Tilton RG, Brasier AR. Generation and characterization of a novel transgenic mouse harboring conditional nuclear factor-kappa B/RelA knockout alleles. BMC Dev Biol. 2016;16:32. doi: 10.1186/s12861-016-0135-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The Bromodomain Protein Brd4 Is a Positive Regulatory Component of P-TEFb and Stimulates RNA Polymerase II-Dependent Transcription. Molecular Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167–177. doi: 10.1038/nrm3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolli D, Gupta MB, Sbrana E, Velayutham TS, Chao H, Casola A, Garofalo R. Alveolar Macrophages Contribute to the Pathogenesis of hMPV Infection While Protecting Against RSV Infection. American Journal of Respiratory Cell & Molecular Biology. 2014 doi: 10.1165/rcmb.2013-0414OC. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrecht BN, Hammad H. The airway epithelium in asthma. Nat Med. 2012;18:684–692. doi: 10.1038/nm.2737. [DOI] [PubMed] [Google Scholar]
- Li X, Zhao Y, Tian B, Jamaluddin M, Mitra A, Yang J, Rowicka M, Brasier AR, Kudlicki A. Modulation of Gene Expression Regulated by the Transcription Factor NF-kappaB/RelA. J Biol Chem. 2014;289:11927–11944. doi: 10.1074/jbc.M113.539965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li K, Garofalo RP, Brasier AR. Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-kappa B2 complexes via a novel retinoic acid-inducible gene-I{middle dot}NF-kappa B-inducing kinase signaling pathway. J Biol Chem. 2008;283:23169–23178. doi: 10.1074/jbc.M802729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wang P, Chen H, Wold E, Tian B, Brasier AR, Zhou J. Drug Discovery Targeting Bromodomain-Containing Protein 4 (BRD4) Journal of Medical Chemistry. 2017;60:4533–4558. doi: 10.1021/acs.jmedchem.6b01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nowak DE, Tian B, Jamaluddin M, Boldogh I, Vergara LA, Choudhary S, Brasier AR. RelA Ser276 Phosphorylation Is Required for Activation of a Subset of NF-{kappa}B-Dependent Genes by Recruiting Cyclin-Dependent Kinase 9/Cyclin T1 Complexes. Molecular and Cellular Biology. 2008;28:3623–3638. doi: 10.1128/MCB.01152-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles JRS, Hashimoto K, Collins RD, Jarzecka K, Furlong J, Mitchell DB, Sheller JR, Graham BS. Immune Interaction between Respiratory Syncytial Virus Infection and Allergen Sensitization Critically Depends on Timing of Challenges. The Journal of Infectious Diseases. 2001;184:1374–1379. doi: 10.1086/324429. [DOI] [PubMed] [Google Scholar]
- Picaud S, Wells C, Felletar I, Brotherton D, Martin S, Savitsky P, Diez-Dacal B, Philpott M, Bountra C, Lingard H, et al. RVX-208, an inhibitor of BET transcriptional regulators with selectivity for the second bromodomain. Proc Natl Acad Sci U S A. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van Weel C, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemungal TA, Donaldson GC, Paul EA, Bestall JC, Jeffries DJ, Wedzicha JA. Effect of exacerbation on quality of life in patients with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 1998;157:1418–1422. doi: 10.1164/ajrccm.157.5.9709032. [DOI] [PubMed] [Google Scholar]
- Stowell NC, Seideman J, Raymond HA, Smalley KA, Lamb RJ, Egenolf DD, Bugelski PJ, Murray LA, Marsters PA, Bunting RA, et al. Long-term activation of TLR3 by poly(I:C) induces inflammation and impairs lung function in mice. Respir Res. 2009;10:43. doi: 10.1186/1465-9921-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Nowak DE, Jamaluddin M, Wang S, Brasier AR. Identification of Direct Genomic Targets Downstream of the Nuclear Factor-{kappa}B Transcription Factor Mediating Tumor Necrosis Factor Signaling. Journal of Biological Chemistry. 2005;280:17435–17448. doi: 10.1074/jbc.M500437200. [DOI] [PubMed] [Google Scholar]
- Tian B, Yang J, Zhao Y, Ivanciuc T, Sun H, Garofalo RP, Brasier AR. Bromodomain Containing 4 (BRD4) Couples NFκB/RelA With Airway Inflammation And The IRF-RIG-I Amplification Loop In Respiratory Syncytial Virus Infection. Journal of Virology. 2017;91 doi: 10.1128/JVI.00007-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Zhao Y, Kalita M, Edeh CB, Paessler S, Casola A, Teng MN, Garofalo RP, Brasier AR. CDK9-dependent transcriptional elongation in the innate interferon-stimulated gene response to respiratory syncytial virus infection in airway epithelial cells. J Virol. 2013;87:7075–7092. doi: 10.1128/JVI.03399-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Zhao Y, Sun H, Zhang Y, Yang J, Brasier AR. BRD4 Mediates NFkB-dependent Epithelial-Mesenchymal Transition and Pulmonary Fibrosis via Transcriptional Elongation. The American Journal of Physiology-Lung Cellular and Molecular Physiology. 2016;311:L1183–L1201. doi: 10.1152/ajplung.00224.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully JE, Hoffman SM, Lahue KG, Nolin JD, Anathy V, Lundblad LKA, Daphtary N, Aliyeva M, Black KE, Dixon AE, et al. Epithelial NF-κB Orchestrates House Dust Mite–Induced Airway Inflammation, Hyperresponsiveness, and Fibrotic Remodeling. The Journal of Immunology. 2013 doi: 10.4049/jimmunol.1301329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedzicha JA. Role of Viruses in Exacerbations of Chronic Obstructive Pulmonary Disease. Proceedings of the American Thoracic Society. 2004;1:115–120. doi: 10.1513/pats.2306030. [DOI] [PubMed] [Google Scholar]
- Zhang G, Plotnikov AN, Rusinova E, Shen T, Morohashi K, Joshua J, Zeng L, Mujtaba S, Ohlmeyer M, Zhou M-M. Structure-Guided Design of Potent Diazobenzene Inhibitors for the BET Bromodomains. Journal of Medicinal Chemistry. 2013;56:9251–9264. doi: 10.1021/jm401334s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sun H, Zhang J, Brasier AR, Zhao Y. Quantitative Assessment of the Effects of Trypsin Digestion Methods on Affinity Purification-Mass Spectrometry-based Protein-Protein Interaction Analysis. J Proteome Res. 2017;16:3068–3082. doi: 10.1021/acs.jproteome.7b00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Brasier AR. Applications Of Selected Reaction Monitoring (SRM)-Mass Spectrometry (MS) For Quantitative Measurement Of Signaling Pathways. Methods. 2013 doi: 10.1016/j.ymeth.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Jamaluddin M, Zhang Y, Sun H, Ivanciuc T, Garofalo RP, Brasier AR. Systematic Analysis of Cell-Type Differences in the Epithelial Secretome Reveals Insights into the Pathogenesis of Respiratory Syncytial Virus-Induced Lower Respiratory Tract Infections. Journal of immunology (Baltimore, Md : 1950) 2017;198:3345–3364. doi: 10.4049/jimmunol.1601291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z, Huang B, Wu X, Zhang H, Qi J, Bradner J, Nair S, Chen LF. Brd4 maintains constitutively active NF-kappaB in cancer cells by binding to acetylated RelA. Oncogene. 2014;33:2395–2404. doi: 10.1038/onc.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.