Abstract
Functional tissue regeneration is required for restoration of normal organ homeostasis after severe injury. While some organs, such as the intestine, harbor active stem cells throughout homeostasis and regeneration1, more quiescent organs like the lung often contain facultative progenitor cells which are recruited after injury to participate in regeneration2,3. Here we show that a Wnt-responsive alveolar epithelial progenitor (AEP) lineage within the alveolar type 2 (AT2) cell population acts as a major facultative progenitor cell in the distal lung. AEPs are a stable lineage during alveolar homeostasis but expand rapidly to regenerate a large proportion of the alveolar epithelium after acute lung injury. AEPs exhibit a distinct transcriptome, epigenome, and functional phenotype with specific responsiveness to Wnt and Fgf signaling. In distinction to other proposed lung progenitor cells, human AEPs (hAEPs) can be directly isolated via expression of the conserved cell surface marker TM4SF1, and hAEPs act as functional human alveolar epithelial progenitor cells in 3D organoids. Together, our results identify the AEP lineage as an evolutionarily conserved alveolar progenitor and a new target for human lung regeneration strategies.
We previously showed that Wnt signaling, evidenced by Axin2 expression, plays an important role in development of both surfactant-producing AT2 cells and alveolar type 1 (AT1) cells that form the gas exchange surface of the lung alveolus4. In the adult lung, Axin2+ Wnt-responsive epithelial cells, identified with Axin2creERT2:TdT:R26REYFP mice, are restricted to the alveolar region and express the AT2 cell marker Sftpc (Fig. 1A–D, Extended Data Fig.1A–E). Few Axin2+ cells express AT1 markers, including Hopx (Fig. 1E, Extended Data Fig.1K–L). These Axin2+ AT2 cells, hereafter referred to as AEPs, comprise approximately 20% of adult AT2 cells (Fig. 1F). AEPs express the same level of AT2 marker genes as other AT2 (Extended Data Fig. 1F) with enriched expression of Wnt targets (Extended Data Fig. 1G). We performed one-, three-, and nine-month lineage tracing using Axin2creERT2:TdT:R26REYFP mice to define AEP dynamics during adult homeostasis (Fig. 1A). AEPs are remarkably stable, with only a small increase in the number of AEP-marked cells after nine months (Fig. 1G and Extended Data Fig. 2A–C). In contrast to alveologenesis4 (Extended Data Fig. 3), few non-Axin2+ AT2 become AEPs during homeostasis (Fig. 1H).
Figure 1. Identification of an Axin2+ alveolar epithelial progenitor (AEP) in the adult lung that regenerates a substantial percentage of the alveolar epithelium.
(A) Schematic of Axin2creERT2:TdT:R26REYFP mice. EYFP is detected by an anti-GFP antibody. Lineage tracing experimental design is as indicated. (B–D) Axin2 marks a subset of AT2 cells. Unmarked = white arrowheads. AEP-marked = yellow arrowheads. D shows orthogonal view of C. (E) Hopx+ AT1 cells are not marked by EYFP. (F) Approximately 20% of AT2 cells express Axin2. (G–H) Epithelial Wnt responsiveness is stable for up to 9 months. The majority of the AEP lineage remains Axin2TdTomato+, while some AEP progeny lose Axin2TdTomato+ expression. Very few Sftpc+/Axin2− cells gain Axin2TdTomato+ expression. Red arrow indicates an Axin2+ mesenchymal cell. (I) Influenza-induced lung injury results in regionalized alveolar damage: minimal (Zone 1), mild (Zone 2), severe (Zone 3), or complete (Zone 4). (J–L) AEP-generated Sftpc+ cells (J–K) and Hopx+ AT1 cells (L) expand in Zones 2 and 3. (M) Ki67+ AEPs preferentially re-enter the cell cycle in areas of regeneration. (N) AEPs can self-renew (YFP+/RFP+) while regenerating a significant number of AT2 cells (YFP+/RFP−), but very few non-AEP cells acquire Axin2 expression (YFP−/RFP+). (O) A region of regenerated lung epithelium near a persistent Krt5+ pod. Black line shows border of Krt5+ pod. Yellow dotted line indicates region of regeneration. (P–Q) A large number of new AEP-derived AT1 and AT2 cells are found within 3 alveolar units (regenerated Zone 3) of Krt5+ pods. N=5 (M,N), N=6 (F–H, O–P), or N=10 (others) animals from 2 (G–H, O–P) or 3 (others) individual experiments. Statistics are representative of all biological replicates. Plots are centered on mean with bars indicating SD. *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001 by two-tailed T-test (E, P–Q) or ANOVA with adjustment for multiple comparison testing (others). Scale bars: B=100μm, C–E, G, J, O=50μm.
To assess AEPs dynamics in lung injury, we used H1N1 influenza virus to injure adult lungs, which causes a spatially heterogeneous injury, similar to human influenza infection5. We defined four regions of injury severity: Zone 1 - no morphological changes, Zone 2 - minor injury with mild interstitial thickening, Zone 3 - significant injury, and Zone 4 - total alveolar destruction (Fig. 1I). We utilized this spatially specific response to analyze the contribution of AEPs to lung regeneration.
Recent studies have shown that Sox2-derived, Krt5+ epithelial cells migrate to damaged distal lung regions to recreate an epithelial barrier6–10. We observe Krt5+ epithelium specifically in Zone 4 after influenza infection (Extended Data Fig. 4A–D,F), but lineage tracing demonstrates no Krt5+ cells are derived from AEPs (Extended Data Fig. 4G). Moreover, AEPs express minimal levels of Krt5 or Sox2 RNA and no detectable protein (Extended Data Figs. 1F and 4E), demonstrating that AEPs and Krt5+ cells derive from distinct lineages. In Zone 4, Sftpc+ and Krt5+/Sftpc+ cells are very rare (Extended Data Fig. 4I), confirming prior reports that the Krt5+ lineage cells do not efficiently regenerate Sftpc+ cells7, except following forced Wnt activation9.
One month after influenza injury, AEPs and their progeny are present at homeostatic levels in Zone 1. However, in Zones 2 and 3, the number of AT2 cells expands significantly (Extended Data Fig. 4H)11,12, with a large increase in the percentage of AT1 and AT2 cells arising from the AEP lineage (Fig. 1J–L; Extended Data Figs. 2D–I and 4J–L). This robust labeling is independent of the timing of tamoxifen injection prior to influenza infection (Extended Data Fig. 5G–I). Notably, in Zone 2 and Zone 3 the AEP lineage shows a significant and specific increase in proliferation (Fig. 1M, Extended Data Fig. 2K–O). Three months after injury, within 300 microns of a persistent Krt5+ pod, a majority of AT2 cells and many AT1 cells in regenerated alveoli are derived from the AEP lineage (Fig. 1O–Q). IHC and FACS analysis after influenza injury demonstrate that AEPs self-renew to maintain the AEP lineage and generate a large number of new lineage-traced alveolar epithelial progeny (Fig. 1N, Extended Data Figs. 2J and 5A–E). Importantly, few non-AEP AT2s acquire the AEP phenotype even in the setting of significant lung injury (Fig. 1N, Extended Data Fig. 5E).
AEPs exhibit a distinct gene expression profile enriched in lung developmental genes (Fig. 2A–D), including the key genes Fgfr2, Nkx2.1, Id2, Etv4, Etv5, and Foxa1 (Extended Data Fig. 6, Table S1). Furthermore, ATAC-seq analysis (Extended Data Fig. 7) revealed a dramatic difference in AEPs versus AT2s, with more than 40% of the genome containing differential open chromatin (Fig. 2A). While many regions of common open chromatin are found near housekeeping genes, regions of AEP-enriched open chromatin are found near lung development genes (Extended Data Fig. 7C). DNA binding site motif analysis shows that AEP-enriched chromatin contains binding sites for AEP-enriched transcription factors of the Klf, Six, Sox, Nkx2, and Elf/Ets families (Extended Data Fig. 7D–E), all known regulators of progenitor cell behavior13–17. Moreover, a group of primed cell cycle regulators near AEP-enriched open chromatin were dynamically regulated in AEPs two weeks after influenza infection (Fig. 2B–D and Extended Data Fig. 6E–G)18–21.
Figure 2. AEPs possess a distinct transcriptome and chromatin architecture enriched for cell cycle and progenitor cell pathways.
(A) RNA-seq and ATAC-seq was performed on AEPs and AT2s. PCA plot of the top 500 most variable genes showing that AEPs segregate into a distinct population from AT2. By ATAC-seq, more than 40% of the genome was differentially accessible in AEPs (16.2%) or AT2 cells (24.1%). (B) ATAC-seq heatmap showing genome wide regions of differential open chromatin peaks in AT2s versus AEPs. (C) AEP-enriched ATAC-seq peaks compared to RNA-seq expression shows that a majority of genes associated with AEP open chromatin are not differentially expressed but primed for rapid activation. (D) A subset of these primed genes are associated with cell cycle activation. Full details on the experimental design and statistical methods for these analyses are available in the Methods.
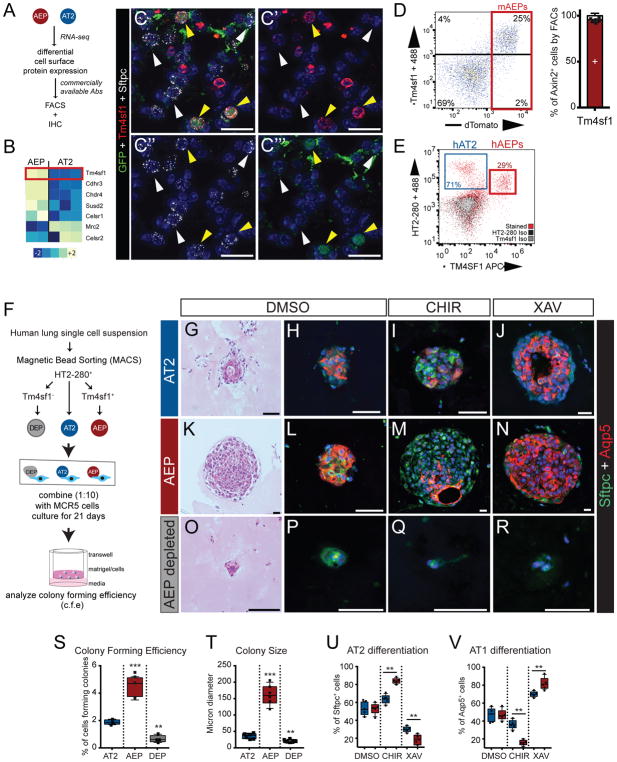
To isolate human AEPs, we identified cell surface markers enriched in mouse AEPs (mAEPs) (Fig. 3A). These studies identified the epithelial cancer stem cell membrane protein Tm4sf122,23 as a marker for mAEPs (Fig. 3B and Extended Data Fig. 8A–C). IHC and FACS analysis demonstrates that Tm4sf1 marks approximately 20% of labeled mAT2 cells and more than 90% of mAEPs (Fig. 3C and D, Extended Data Fig. 8A). Using a combination of a human TM4SF1 antibody (Extended Data Fig. 8D) and human AT2 (hAT2) specific HTII-280 antibody24 (Extended Data Fig. 8B,E–H), we were able to identify a distinct subset of HTII-280+/TM4SF1+/EPCAM+ putative hAEPs in normal human lung. These hAEPs comprise approximately 29% of the hAT2 population (Fig. 3E) and express SFTPC but not KRT5 or SOX2 mRNA (Table S2).
Figure 3. Identification of Tm4sf1 as an AEP-specific cell surface marker capable of isolating functional human AEPs.
(A and B) AEP enriched cell surface proteins with an available antibody. Tm4sf1 is highlighted. (C) Tm4sf1 is expressed in mAEPs. (D) FACS analysis (of N=6 individual animals, see Extended Data Fig. 8) demonstrating that Tm4sf1 correlates strongly with Axin2TdTomato expression. (E) A human anti-TM4SF1 antibody marks a subset of human HTII-280+ AT2s, which are putative human AEPs (hAEPs). (F) Schematic diagram of human lung alveolar organoid assay using either total hAT2 cells (HTII-280+), hAEPs (HTII-280+/TM4SF1+), or AEP-depleted hAT2 cells (HTII-280+/TM4SF1−). Indicated cultures were treated with CHIR or XAV to modulate Wnt signaling. (G–J) The complete hAT2 population generates alveolar organoids and responds to Wnt activation by increasing AT2 cell differentiation or to Wnt inhibition by increasing AT1 cell differentiation. (K–N) hAEPs generate more and larger organoids that respond more robustly to Wnt modulation. (O–R) Depletion (DEP) of TM4SF1+ cells from hAT2s results in a loss of alveolar organoid formation and Wnt responsiveness. (S–V) Quantification of colony forming efficiency (S), colony size (T), AT2 (U) and AT1 (V) cell differentiation. N=4 individual human organoid experiments. Statistics are inclusive of all biological replicates. **=p<0.01, ***=p<0.001 by ANOVA with adjustment for multiple comparison testing. Plots are centered on mean with bars indicating SD. Scale bar C=50μm, F–Q=25μm.
Using clonal alveolar organoid assays25, both mAEP and hAEPs form more and larger organoids containing AT1 and AT2 cells but no SOX2 or KRT5+ cells (Extended Data Fig. 8I–J), and demonstrate increased responsiveness to Wnt modulation compared to AT2 cells (Fig. 3F–N, Extended Data Fig. 9). Importantly, depletion of TM4SF1+ cells from the hAT2 population leads to a dramatic loss of organoid formation (Fig. 3O–S). Notably, Wnt inhibition promoted AT1 cell differentiation and Wnt activation promoted AT2 formation in both mouse and human organoids but not hAEP-depleted organoids (Fig. 3O–R, U–V, Extended Data Fig. 9O–P). These data suggest that TM4SF1+/HTII-280+ hAEPs are the functional equivalent of mAEPs.
RNA-seq analysis demonstrated that a large proportion of hAEP-enriched genes (35.6%) were evolutionarily conserved with mAEPs including key progenitor cell regulators (Fig. 4A–B and Extended Data Fig. 10A–B). In particular, mAEPs and hAEPs are both enriched for Wnt pathway targets including AXIN2 and FGFR2, the primary receptor for Fgf7 and Fgf10 (Fig. 4C, Extended Data Fig.10K, Table S2)26–32. DNA binding motif analysis shows that LEF/TCF binding sites are enriched in open chromatin near conserved AEP genes and β-catenin bound to some of these genomic regions (Extended Data Fig. 10C–E), supporting the evolutionarily conserved Wnt-responsiveness of AEPs. Importantly, treatment of both mAEP and hAEPs with Fgf7 or Fgf10 ligand resulted in substantial increases in colony size and colony-forming efficiency, while mAT2 and hAEP-depleted hAT2 cells exhibited a diminished response (Fig. 4D–W, Extended Data Fig. 10F–Q).
Figure 4. AEPs display an evolutionarily conserved response to Wnt and Fgf signaling.
(A) hAEPs exhibit a distinct transcriptome enriched for Wnt responsiveness. (B) More than one-third of hAEP-enriched genes are shared with mAEPs. (C) Volcano plot of 15,628 genes tested using limma shows extensive overlap between up-regulated genes in mAEPs and hAEPs. FGFR2 is indicated. (D–W) Alveolar organoid assays show mAEPs (D–M) and hAEPs (N–W) display a significant increase in colony formation and size upon FGF7 treatment. Additional data is shown in Extended Data Fig 10. N=4 individual organoid experiments. Statistics are inclusive of all biological replicates. **=p<0.01, ***=p<0.001 by ANOVA with adjustment for multiple comparison testing. Plots are centered on mean with bars indicating SD. Scale bars=25μm.
Our data reveal that AEPs are a major lineage contributing to functional alveolar epithelial regeneration by producing a significant plurality of both AT2 and AT1 cells after injury. In contrast, Sox2-derived Krt5+ cells migrate from the proximal airway after acute lung injury, preventing loss of the epithelial barrier 6–9. AEPs and Krt5+ cells likely act in concert, with Krt5+ cells acting rapidly to prevent immediate loss of epithelial barrier while AEPs simultaneously regenerate functional alveoli. AEPs respond robustly to both Wnt and Fgf signals, with Wnt signaling a key factor in modulating the AT2 to AT1 transition4, and Fgfr2 activation promoting AT2 cell proliferation (Extended Data Fig 10J). Importantly, the conservation and accessibility of both mAEPs and hAEPs provides an opportunity for mechanistic studies to elucidate human lung progenitor cell biology and development of new treatments for acute and chronic lung diseases.
METHODS
Ethical Compliance
All animal studies were performed under guidance of the University of Pennsylvania Institutional Animal Care and Use Committee in accordance with institutional and regulatory guidelines. This study utilized cells derived from de-identified non-utilized lungs donated for organ transplantation via an established protocol (PROPEL, approved by University of Pennsylvania Institutional Review Board) with informed consent in accordance with institutional and NIH procedures. All patient information was removed prior to use. This use does not meet the current NIH definition of human subject research, but all institutional procedures required for human subject research were followed throughout the reported experiments.
Animals and Cre recombinase induction
The generation and genotyping of the Axin2CreERT2-TdTomato mouse line generated in our laboratory has been previously described4. The SftpcCreERT2 mouse line was a generous gift of Hal Chapman at the University of California at San Francisco and their genotyping and generation have been previously described34. Hopx3FlagGFP mice35 were a generous gift of Rajan Jain and Jonathan Epstein at the University of Pennsylvania and are available at Jackson Laboratories. The R26REYFP mice are available at Jackson Laboratories. All animal studies were performed under guidance of the University of Pennsylvania Institutional Animal Care and Use Committee. Animals were maintained on a mixed CD-1 and C57BL/6 background. For induction of all Cre recombinase models, tamoxifen (Sigma) was dissolved in 100% ethanol and diluted with corn oil (Sigma) to produce a 10% ethanol:tamoxifen:corn oil mixture at 20 mg/mL. 6–8 week old mice were injected intraperitoneally (IP) with 200μg/gm on 3–5 consecutive days to induce recombination. All lineage tracing experiments represent a minimum of N=6 animals in all groups to allow for effective statistical evaluation. QPCR experiments represent a minimum of N=3 animals in all groups. Animal experiments were performed on both male and female animals in all conditions, and animals were chosen at random from the cohort but not formally randomized. Blinding for experimental condition was not possible due to the nature of the injury experiments.
Influenza Lung Injury
PR8 H1N1 influenza was a generous gift of Dr. John Wherry at the University of Pennsylvania. Recombination for lineage tracing was performed using 3 daily tamoxifen injections 7 or 28 days prior to viral infection. For infection, virus was diluted in PBS and a dose of 0.3 LD50 was administered via intranasal instillation. Following infection, animals were weighted and monitored daily for 14–28 days and animals which lost >30% of starting weight or were moribund were sacrificed humanely. Post-influenza RNA was obtained at 14d post infection, and lung regeneration was analyzed from tissue collected from animals 28 days to 3 months after infection. FACS data was generated from influenza- infected and uninfected animals using the same protocols; please see below. Regionalized lung injury was assessed via histology, and adjacent sections where utilized for all immunostaining and quantification.
Histology
At the time of tissue harvest mice were euthanized by CO2 inhalation. The chest cavity was exposed and the lungs cleared of blood by perfusion with cold PBS via the right ventricle. Lungs were inflated with 2% paraformaldehyde under constant pressure of 30 cm water and allowed to fix overnight. Tissue was then dehydrated, paraffin embedded, and sectioned. Hematoxylin and eosin staining was performed to examine morphology, and to score regions based on the severity of injury. Immunohistochemistry was used to detect protein expression using the following antibodies on paraffin sections: GFP (chicken, Aves, GFP-1020, 1:500), GFP (goat, Abcam, ab5450, 1:100), RFP (rabbit, Rockland, 600-901-379, 1:250), Scgb1a1 (goat, Santa Cruz, sc-9772, 1:20), Tubb4 (mouse, BioGenex, MU178-UC, 1:20), Sftpc (rabbit, Millipore, ABC99, 1:250), Sftpc (goat, Santa Cruz, sc-7750, 1:50), Pdpn (mouse, Hybridoma Bank, Clone 8.1.1, 1:50), Aqp5 (rabbit, Abcam, ab92320, 1:100), and Ki67 (rabbit, Abcam, clone SP6, ab16667, 1:50), anti-mouse Tm4sf1 (rabbit, LSBiosciences, B7077, 1:500).
Alveolar Epithelial Cell Number and Lineage Imaging and Quantification
Following immunostaining for alveolar epithelial lineages and proliferation, images were captured using a Nikon Eclipse Ni wide field microscope or a Leica TCS SP8 confocal microscope. We captured images containing at least eight individual 1μm optical sections. Z-stacks were obtained from at least five random areas of each histological zone in a minimum of N=5 animals. All images were processed with ImageJ software. Cell counts were performed using the Cell Counter plug-in for ImageJ. Cells were counted in at least three different areas of each histological injury zone for each mouse, to obtain a total count of >1000 cells counted for each condition. Only true confocal images were used for quantification. For image presentation, both confocal images and images obtained with automatic deconvolution algorithms in Nikon Elements software are presented, with source as noted.
Lung Alveolar Epithelial Cell Isolation and FACS analysis
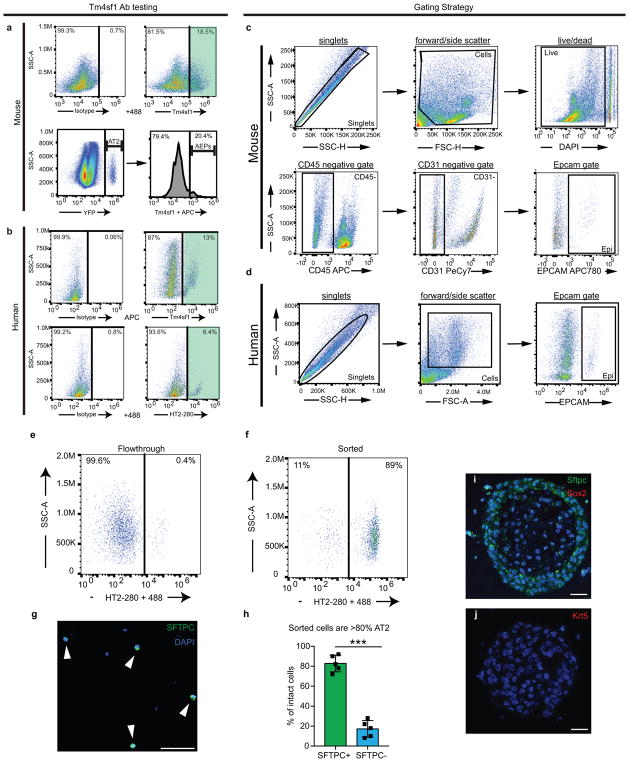
Mouse
Lungs from Axin2CreERT2-TdTomato animals were harvested at 6–8 weeks of age and processed into a single cell suspension using dispase, collagenase I, and DNase as previously described36,37. EPCAM+ Axin2+ cells (TdTomato+) were identified via FACS sorting as previously described37. The total AT2 population (Sftpc+ AT2s) was isolated from lungs of 6–8 week old SftpcCreERT2:R26REYFP animals 5 days after induction with 200μg/gm tamoxifen. EYFP+ cells were then isolated via FACS sorting as previously described37. For sorting and quantification, the following antibodies were used: Pdpn-eFluor660 (eBioscience, Clone 8.1.1, 1:100) EpCAM-APC (eBioscience, Clone G8.8, 1:200), EpCAM-eFluor488 (eBioscience, Clone G8.8, 1:200), CD31-PeCy7 (eBioscience, Clone 390, 1:200), CD45-PeCy7 (eBioscience, Clone 30-F11, 1:200). Two anti-mouse Tm4sf1 antibodies were used to ensure specificity: Sheep anti-mouse Tm4sf1 (R&D systems, AF7514, 1:10) and Sheep IgG isotype control (R&D systems, 5-001-A, 1:10) with anti-Sheep 488 secondary (Abcam, ab150177, 1:50) or Rabbit anti-mouse Tm4sf1 (LS Biosciences, B7077, 1:25) and Rabbit IgG isotype control (LS Biosciences, LS-C109221, 1:25) with Donkey anti-rabbit 488 secondary (Life Technologies, A212016, 1:200).
Human
Samples of normal, de-identified human lungs were obtained from non-utilized lungs donated for organ transplantation via an established protocol (PROPEL, approved by University of Pennsylvania Institutional Review Board) with informed consent in accordance with institutional procedures. A 2x2cm piece of distal lung tissue was obtained, pleura and large airways were carefully dissected away, and tissue was processed into a single cell suspension using the same combination of dispase, collagenase I, and DNase used for mouse lungs. A Miltenyi gentleMACS dissociator was used for mincing and incubation for 35min at 37°C. Cells were washed, passed over 70μM and 40μM filters, and RBCs were lysed with ACK lysis buffer. After a single cell suspension was obtained, cells were analyzed by FACS or sorted using the MACS multisort kit, MACs LS columns, and the following antibodies: EPCAM-PE (BD, mouse, Clone 1B7, 1:50), HT2-280 (mouse IgM, a gift of Leland Dobbs, UCSF, 1:50), TM4SF1-APC (mouse, R&D Systems, Clone 877621, 1:100), Mouse IgG1-APC isotype control (R&D systems, 1C002A, 1:100), anti-APC microbeads (Miltenyi, 130-090-855, 1:20), anti-mouse IgM microbeads (Miltenyi, 130-047-302, 1:20). The full protocol for digestion and sorting of human lung epithelial cells, and their propogation as alveolar organoids, has been made available via the Nature Protocol Exchange36.
Ex Vivo Alveolar Organoids
Clonal alveolar organoid assays were performed as described previously with some modifications from the original protocol4,11,25,37 In brief, 5 × 103 epithelial cells (AT2 or AEP for mouse, HT2-280+, HT2-280/TM4SF1+, HT2-280/TM4SF1- for human) were isolated as described above and mixed with 5 x104 lung fibroblasts (isolated from adult wild type mice as previously described37 for mouse, MRC5 cells (ATCC CCL-171, tested negative for mycobacterial contamination, at no greater than passage 10) for human. Cells were then suspended in a 1:1 mixture of SAGM media (Lonza, with all additives except epinephrine) and growth factor-reduced, phenol-free Matrigel (Corning). 90μl of the cell/media/matrigel mixture was then aliquoted into individual 24 well cell culture inserts and allowed to solidify at 37°C. SAGM was then placed into each well of the 24-well plate. The Rock inhibitor Y27632 (Sigma) was included in the media for the first two days. After two days of culture, Y276632 was removed and ligand treatments of organoids were performed using the following reagents at the indicated concentrations: Wnt3a 200ng/ml (R&D systems), Fgf7 50ng/ml (R&D Systems), Fgf10 50ng/ml (R&D Systems), XAV939 10μM (Sigma), CHIR99021 1μm (Fisher). DMSO was used a control. The media was changed every 48 hours, and fresh ligands were included at each media change. After 21 days of culture, organoids were fixed in 2% paraformaldehyde, embedded in Histogel (Richard-Allen), dehydrated, paraffin embedded, and sectioned and immunostained as described above.
RNA-seq analysis
Cells were sorted using the protocols above into Trizol LS (Life Technologies). For mouse, 6 individual animals were sorted and pooled into 2 individual pools for Axin2+ cells and 3 individual pools for Sftpc+ cells. For human, 3 individual patients were sorted separately and prepared for sequencing individually. RNA was then extracted using a combination of the Trizol protocol and MinElute RNA Cleanup Kit (Qiagen). RNA integrity was confirmed via Bionanalyzer evaluation and samples with RIN>8.5 were chosen for library preparation. Library prep was conducted using Illumina truSeq stranded mRNA kit followed by the Nugen Ovation amplification kit. Fastq files were assessed for quality control using the FastQC program. Fastq files were aligned against the mouse reference genome (mm9) or human reference (hg19/hGRC37) genome using the STAR aligner 38. Duplicate reads were flag using the MarkDuplicates program from Picard tools. Per gene read counts for Ensembl (v67) gene annotations for the mouse samples or Ensembl (v75) for the human were computed using the R package Rsubread with duplicate reads removed. Gene counts represented as counts per million (CPM) were first nominalized using trimmed mean of M-values method in the R package edgeR and genes with 25% of samples with a CPM < 1 were removed and deemed low expressed. This data was transformed using the VOOM function from the limma R package39. Differential gene expression was performed using a linear model with the limma package. Given the small sample size of the experiment, we employed the empirical Bayes procedure as implemented in limma to adjust the linear fit and calculate P values. P values were adjusted for multiple comparisons using Benjamini-Hochberg procedure. For the human data a paired analysis was employed using the patient as a blocking variable. Heatmaps and PCA plots were generated in R. Gene Ontology enrichment analysis was performed using the ToppGene Suite (http://toppgene.cchmc.org/)40.
ATAC-seq analysis
Individual ATAC-seq libraries were generated from sorted Axin2+ and Sftpc+ AT2s as above using previously published methods41. In brief, 5x104 cells were sorted into PBS, washed, and lysed to obtain nuclei. Nuclei were exposed to Tn5 transposase (Illumina), and fractionated DNA was used for amplification and library preparation. Libraries were then purified and paired-end sequenced. Following sequencing, Fastq files were aligned against the mouse reference genome (mm9) using the STAR aligner38, with default parameters plus options to suppress the matching of spliced reads (‘--outFilterMatchNminOverLread 0.4 --outFilterScoreMinOverLread 0.4’). Duplicate reads were flagged using the MarkDuplicates program from Picard tools and removed using samtools. MACS2 was used to call peaks with the following options ‘--nomodel --shift -100 --extsize 200’ 42. Differential ATAC-seq peaks were determined using the bdgdiff command from MACS2 and default options. Peaks were filtered to have a MACS2 log10 likelihood ratio score > 10 and within -50kb and +10kb of the transcription start site of Ensembl 67 protein coding genes. ATAC-seq enrichment heatmaps were created using deepTools243.
Motif Analysis
The intersection of gene promoter regions (−5kb,+600bp, Ensembl v67) with identified ATAC-seq peaks was performed using Bedtools. Fasta file of genome sequence (mm9) of promoter ATAC-seq peaks was created using bedtools and scanned for a TCF/LEF motifs using FIMO44. Motif enrichment analysis was performed using the findMotifsGenome.pl program in the HOMER software suite45, with the peak search size option set to 50bp.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed using the High Sensitivity ChIP Kit (Abcam) with 3 μg of anti-β-catenin (Santa Cruz sc-7963) or anti-IgG1 isotype control (Santa Cruz sc-3877). In brief, 1x105 Axin2+ or Sftpc+ AT2s were sorted into SAGM (Lonza), whole chromatin was prepared, chromatin was cross-linked and sonicated using a Covaris sonicator to an optimal size of 300bp, and chromatin was immunoprecipitated using the antibodies above following the Abcam protocol. Library quality was confirmed via Bioanalyzer, and enrichment of genomic DNA was assessed via QPCR comparing β-catenin IP vs IgG control for each cell type. QPCR data represents N=2 individual immunoprecipitation experiments and was performed in triplicate.
Statistical Analysis
Statistical analysis was performed in Prism for Mac and R. A two-tailed Student’s t-test was used for the comparison between two experimental groups. For experiments with more than 2 groups, an ANOVA was performed followed by planned contrasts, with pairwise comparisons and P value adjustments for multiple comparisons were performed using Dunnett HSD. Generation of odds ratios for distribution of ATAC regions near genes was evaluated using Fisher’s exact test and contingency table analysis. Statistical data was considered significant if P < 0.05. Center values of all plots represent means, and error bars represent standard deviations, with the exception of error bars for odds ratios which represent confidence intervals.
Data Availability
Sequencing (ATAC-seq and RNA-seq) data generated during this study has been deposited in the GEO database with the primary accession GSE97055 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97055). All upregulated and downregulated genes identified during the described RNA-seq experiments are found in Supplemental Table 1 (mouse data) and Supplemental Table 2 (human data). Source data underlying all plots in all figures is available per Nature policies. The detailed protocol for the cell isolation and propogation of human AEPs has been made available on Nature Protocol Exchange36. All other datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
Extended Data
Extended Data Figure 1. Location of Axin2+ epithelial cells within the adult mouse lung.
(A) Low power view of the lung showing that E-cadherin+ Axin2+ epithelial cells are found only in the alveolar region and not the airway of the lung. (B–C) IHC for ciliated (B) and secretory (C) markers shows no evidence of Axin2-lineaged labeled cells co-expressing either of these markers. (D–E) Quantification of the location of Axin2+ epithelial cell distribution in the lung. (F) QPCR showing that Axin2+ AEPs and AT2 cells express similar levels of AT2 markers and other lung epithelial cell markers. AEPs express slightly higher levels of Abca3. (G) AEPs express increased levels of Wnt signaling pathway components and targets by QPCR. (H–J) Cytopsins and quantification demonstrating that the majority of sorted Axin2+ epithelial cells are Sftpc+. (K–L) FACS analysis of Axin2tdTomato+/HopxEYFP animals demonstrating that few Axin2+ epithelial cells express Hopx, consistent with the IHC data shown in Figure 1. Data in this figure represent N=3 (K–L), N=4 (E–G) N=4 (D, H–J), or N=10 (all other data) animals from 3 individual experiments. Statistics are representative of all biological replicates. All data is shown as centered on mean with bars indicating standard deviation. *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001 by two-tailed T-test (F–G) or ANOVA with preplanned pairwise comparisons and adjustment for multiple comparison testing (D). Scale bars A–C 100μm, H–I 25μm.
Extended Data Figure 2. Characterization of Axin2+ Wnt responsive cells in the adult lung.
(A) 3 month lineage tracing shows a stable population of AEPs and progeny in the alveolar epithelium. (B–C) Quantification of AT1 and AT2 cells labeled by the AEP lineage mark at homeostasis. 7d lineage tracing data is re-plotted here for comparison to later time points. (D–I) Lower power (D–F) and higher power (G–I) images showing expansion of AEPs in a regional fashion 1 month after influenza injury. Dotted white line in F shows the edge of a Krt5+ pod, with a dearth of AEP-lineage labeled cells. G–I show additional channels of the same fields as Fig 1IJ. (J) Representative FACS plot showing expansion of AEP-lineage labeled epithelial cells following influenza. Quantification found in Fig 1N. (K–O) Comparison of AT2 and AEP Ki67+ expression following influenza. Ki67+ AEPs re-enter the cell cycle almost exclusively compared to AT2s in areas of regeneration. Data shown in this figure represent N=5 (J–O), N=6 (A–C), or N=10 (D–I) independent animals from 3 individual experiments, except for the 9 month lineage tracing which was performed in two separate experiments. Statistics are representative of all biological replicates. All data is shown as centered on mean with bars indicating standard deviation. *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001 by ANOVA with preplanned pairwise comparisons and adjustment for multiple comparison testing. Scale bar=50μm.
Extended Data Figure 3. In contrast to adult lung homeostasis, the Wnt response in the alveolar epithelium during alveologenesis is dynamic.
(A) Schematic of lineage labeling procedure to assess Wnt-responsive epithelium during alveologenesis. (B) Epithelial cells were identified by FACS as Epcam+ CD45− CD31−. Cells were then gated for TdTomato and EYFP expression as shown. (C) Quantification of Wnt responsiveness in the alveolar epithelium over a 1 day or 3 week lineage trace. (D) Model of directionality and magnitude of AT2 and AEP transitions. During alveologenesis, AT2 and AEP fates are somewhat fluid, though the AEP population narrows during this period of lung development. During adult homeostasis, few if any AT2 cells take on the AEP fate (see Figure 2). After injury, AEPs expand to create AT2 cells, but even after injury very few AT2 cells adopt the AEP fate. Data shown in this figure represents N=3 animals. Statistics are representative of all biological replicates. C is centered on mean with bars indicating standard error of the mean.
Extended Data Figure 4. AEPs are a distinct lineage compared to Sox2-derived Krt5+ cells and are capable of generating AT1 cells.
(A–D) AEPs and Krt5+ cells inhabit distinct regions of the regenerating mouse lung. (A) Overview of a region surrounding a Krt5+ pod. (B) In regions of mild injury AEPs and AEP lineage marked AT2 cells predominate, with no Krt5+ cells seen. (C) At the border of Zone 4 areas of alveolar destruction, AEPs are observed regenerating AT2 cells. (D) Krt5+ cells are distinct from AEPs and never bear the AEP lineage mark. (E) AEP-lineage cells do not expression Krt5+ or Sox2+ protein at baseline, in distinction to previously reported lineages. Arrows represent probable AEPs by morphology. (F) Krt5+ cells predominate in Zone 4 regions, while AEPs are not present in these regions. (G) Quantification demonstrating that Krt5+ cells are never marked with the AEP lineage mark. (H) AT2 populations expand dramatically after influenza injury except in Zone 4. (I) Krt5+ cells rarely express Sftpc in Zone 4 regions. (J–L). One month after influenza injury, AEPs give rise to a small number of Hopx+ AT1 cells, predominantly in Zone 2 of mild injury. Zone 3 (L) has very few AEP derived Hopx+ cells, which may be due to a lag in AT1 regeneration from AEPs in this more severely affected region. Data shown in this figure represent N=6 (A–G,I) or N=10 (H, J–K) independent animals across 3 individual experiments. Statistics are representative of all biological replicates. All data is shown as centered on mean with bars indicating standard deviation. **=p<0.01, ***=p<0.001, by ANOVA with preplanned pairwise comparisons and adjustment for multiple comparison testing. Scale bars: A=200μm, B–D,J–L,=50μm.
Extended Data Figure 5. Wnt signaling in the alveolar epithelium is largely stable following influenza infection and AEP lineage labeling is not affected by tamoxifen perdurance.
(A) FACS gating strategy used for all post-influenza FACS experiments in Figure 1, Extended Data Figure 2, and this figure. (B–C) FACS analysis demonstrates that Axin2tdTom intensity is mildly decreased in the epithelium at 7d and 14d time points following influenza infection. (D) In regions of more mild lung injury, most lineage labeled AT2 cells are EYFP+ and tdTomato-, suggesting these are progeny of AEPs. (E) In Zone 3, we detect a mix of EYFP+/tdTomato+ AEPs (red arrowheads) and EYFP+/tdTomato- AEP progeny (yellow arrowheads) among the AT2 cell population. (F) Experimental design of lineage tracing experiment in the second half of this figure, with longer incubation time following tamoxifen treatment than the data presented in A–E and the main Figures. (G–H) Confocal imaging demonstrating lineage labeling of AT2 cells with AEP lineage mark 28d after influenza-mediated injury. (I) Quantification of lineage labeled AT2 cells in multiple regions of lung injury. Representative 7d lineage data for comparison is reproduced from Figure 2. Data shown in this figure represent N=4 (A–C) or N=5 (D–I) independent animals across 2 different experiments. Statistics are representative of all biological replicates. All data is was analyzed with ANOVA followed by preplanned pairwise comparisons and adjustment for multiple comparison testing and shown centered on mean with bars indicating standard deviation with **=p<0.01. Scale bars = 50μm.
Extended Data Figure 6. Transcriptome analysis of AEPs versus AT2s and activation of cell cycle genes in AEPs after influenza injury.
(A) Volcano plot of 14,618 genes tested using a linear model using the R package limma showing the distinct differences in gene expression in AEPs versus AT2s. Notable lung progenitor developmental signaling and transcription factors are indicated. (B) GO analysis of top 500 most differentially expressed genes showing the enrichment of categories related to lung development and morphogenesis in AEPs. (C) Heatmaps of two of the AEP-enriched GO categories with important regulators of lung progenitor cell biology indicated. (D) QPCR confirming up-regulation of a subset of important regulators of lung progenitor biology in AEPs. (E) AT2 and AEP open chromatin is found near a distinct sets of genes involved in the cell cycle. (F) Schematic of analysis of changes in expression of AEP primed genes after influenza infection. (G) A subset of primed cell cycle regulators in AEPs show expression changes following influenza infection. QPCR data is from N=4 animals from 2 separate infections. All data is shown as centered on mean with bars indicating standard deviation. Statistics are representative of all biological replicates. *= p<0.05, **=p<0.01 by two-tailed T-test.
Extended Data Figure 7. ATAC-seq reveals distinct differences in open chromatin architecture in AEPs versus AT2s.
(A) ATAC-seq peaks in both AT2 and AEP cells are similar to previously described mouse lung genome wide DNase hypersensitivity profiling. (B) AT2 and AEP ATAC peaks are distributed similarly, predominantly within intergenic regions and introns. (C) GO enrichment analysis of nearest neighbor genes near AT2 vs AEP vs common peaks shows that common peaks are enriched for general cellular housekeeping roles while AT2 open chromatin is enriched near genes associated with exocytosis and cell differentiation. In contrast, AEP peaks are enriched near genes associated with lung development processes. (D–E) Examination of the genes associated with open chromatin in AEPs reveals a strong enrichment for lung endoderm progenitor cell associated transcription factors including members of the Klf, Six, Sox, Nkx2, and Elf/Ets families in AEPs. In contrast, AT2 cell open chromatin is associated with a unique set of transcriptional regulators including members of the NfI and Cebp families which are known to regulate AT2 cell surfactant genes. Detailed methods for ATAC analysis are reported in the Materials and Methods.
Extended Data Figure 8. The combination of HT2-280 and TM4SF1 antibodies are capable of identifying AEPs in human lung.
(A) Top of panel shows isotype and active antibody gates for Sheep anti-mouse Tm4sf1 FACS. The lower panel shows that the Tm4sf1 antibody detects approximately 20% of SftpcCreER;EYFP labeled AT2 cells. (B) Isotype and active antibody gates for human HT2-280 (AT2 marker) antibody and TM4SF1 antibody. (C–D) An example of the FACS gating strategy used to generate the data shown in Figure 3. (E–F) Selection for HT2-280 strongly enriches for hAT2 cells. (G–H) The majority of isolated HT2-280 cells express SFTPC protein by cytospin. (I–J) Human AEPs in organoid culture do not express KRT5 or SOX2 protein at detectable levels. Each FACS panel shown in A–F shows gates from one individual animal or patient and is representative of N=6 independent animals across 2 individual experiments or N=4 human patients. Isotype staining was done 3 times to confirm specificity. Statistics are representative of all biological replicates. Statistics in H are calculated with two-tailed T-test and displayed as mean with bars showing standard deviation. Scale bars = 25μm (D) or 50μm (I–J).
Extended Data Figure 9. Mouse AEPs generate more alveolar organoids which are restricted from AT1 cell differentiation by Wnt signaling.
(A) Schematic of mouse alveolar organoid culture method. Sftpc+ mAT2s (B–D, H–J) and mAEPs (E–G, K–M) were isolated from the indicated mouse lines and cultured in alveolar organoid assays. AT2s (B) and AEPs (E) both form alveolar organoids, while AEPs generating more and larger organoids. Activation of Wnt signaling using CHIR99021 does not increase the organoid forming efficiency of either AT2s (C) or AEPs (F) but does increase the number of Sftpc+ cells in treated organoids (I, L, O). Inhibition of Wnt signaling using XAV939 increases the number and size of alveolar organoids (D, G, N, Q), decreases the number of Sftpc+ AT2 cells, and increases the number of Aqp5+ AT1 cells (J, M, P). For all parameters tests, AEPs respond more dramatically to Wnt modulation than AT2s. Data shown in this figure represent N=12 wells across from N=4 individual animals in each group across 3 individual experiments. Quantitative counting shown for cell differentiation (O,P) represents counting of N>400 organoids from N=4 animals. All data was analyzed with ANOVA followed by preplanned pairwise comparisons and adjustment for multiple comparison testing and shown centered on mean with bars indicating standard deviation with *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001. Statistics are representative of all biological replicates. Scale bars: H–M=50μm.
Extended Data Figure 10. Combination ATAC-seq and RNA-seq emphasizes the Wnt- and FGF-responsive nature of AEPs and identifies several novel AEP-enriched direct Wnt target genes.
(A) Schematic representation of the design of the human RNA-seq experiments. (B) GO Term analysis of the top 300 hAEP enriched genes shows enrichment of several categories associated with lung progenitor cell function, similar to what was observed in mAEP. (C) Evaluation of the chromatin accessibility in the mouse genome near common AEP enriched genes demonstrates a significant overrepresentation of Tcf binding sites, particularly in 5kb immediate upstream putative regulatory regions. Full details of enrichment analysis are found in the Methods. (D) Schematic depiction of areas of AEP-enriched open chromatin near selected AEP-enriched genes. Peak height represents coverage of the indicated genomic region in the ATAC library, while the number indicates the fold enrichment in the indicated peak. (E) ChIP QPCR on AEP vs AT2 chromatin demonstrates Ctnnb1 antibody binding at the differentially accessible genomic regions near Etv4, Sftpa, Lamp3, and Gpr116 in AEP cells, indicating that these genes are direct Wnt targets. Data is shown as mean with individual data points showing summary data from 2 independent ChIP experiments with multiple technical replicates. (F–J) Fgfr2 activation in mAEPs drives increased proliferation and formation of larger organoids. Quantification in (E). Correlate to Figure 4. (K) RNAscope showing enriched expression of Fgfr2 (red) in lineage labeled AEPs. (L–Q) Similar to Fgf7, Fgf10 treatment drives increased colony forming efficiency in both mAEP (L–P) and hAEP (Q). Data shown in F–J,L–Q represent a minimum of N=12 wells across 2 individual experiments. Statistics are representative of all biological replicates. Data was analyzed with ANOVA followed by preplanned pairwise comparisons and adjustment for multiple comparison testing and shown centered on mean with bars indicating standard deviation with *=p<0.05, **=p<0.01, ***=p<0.001, ****=p<0.0001.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (T32-HL007586 to W.J.Z; T32-HL007915, K12-HD043245 to D.B.F., T32-HL007843 to J.A.Z., and HL110942, HL087825, HL132999, HL129478, HL134745 to EEM). The authors thank the Flow Cytometry Core Laboratory of Children’s Hospital of Philadelphia and the CVI Histology Core, Next Generation Sequencing Core, and CDB Microscopy Core at the University of Pennsylvania for extensive technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS
W.J.Z, D.B.F, J.A.Z, F.A., S.Z., and J.K performed the experiments. W.J.Z., D.B.F, J.A.Z, M.P.M., and E.E.M. analyzed the data. E.C. provided access to human samples and assisted W.J.Z. with all human experiments. E.E.M. supervised the project. W.J.Z. wrote the first draft of the manuscript. All authors contributed to the writing of the final manuscript.
The authors declare no competing financial interests.
References
- 1.Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 2.Stanger BZ. Probing hepatocyte heterogeneity. Cell Res. 2015;25:1181–1182. doi: 10.1038/cr.2015.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afelik S, Rovira M. Pancreatic beta-cell regeneration: Facultative or dedicated progenitors? Mol Cell Endocrinol. 2017;445:85–94. doi: 10.1016/j.mce.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 4.Frank DB, et al. Emergence of a Wave of Wnt Signaling that Regulates Lung Alveologenesis by Controlling Epithelial Self-Renewal and Differentiation. Cell Rep. 2016;17:2312–2325. doi: 10.1016/j.celrep.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Töpfer L, et al. Influenza A (H1N1) vs non-H1N1 ARDS: analysis of clinical course. J Crit Care. 2014;29:340–346. doi: 10.1016/j.jcrc.2013.12.013. papers3://publication/doi/10.1016/j.jcrc.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Kumar PA, et al. Distal airway stem cells yield alveoli in vitro and during lung regeneration following H1N1 influenza infection. Cell. 2011;147:525–538. doi: 10.1016/j.cell.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaughan AE, et al. Lineage-negative progenitors mobilize to regenerate lung epithelium after major injury. Nature. 2015;517:621–625. doi: 10.1038/nature14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zuo W, et al. p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature. 2015;517:616–620. doi: 10.1038/nature13903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xi Y, et al. Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol. 2017;19:904–914. doi: 10.1038/ncb3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ray S, et al. Rare SOX2+ Airway Progenitor Cells Generate KRT5+ Cells that Repopulate Damaged Alveolar Parenchyma following Influenza Virus Infection. Stem Cell Reports. 2016;7:817–825. doi: 10.1016/j.stemcr.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barkauskas CE, et al. Type 2 alveolar cells are stem cells in adult lung. J Clin Invest. 2013;123:3025–3036. doi: 10.1172/JCI68782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rock JR, et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108:E1475–1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Hashash AH, et al. Six1 transcription factor is critical for coordination of epithelial, mesenchymal and vascular morphogenesis in the mammalian lung. Dev Biol. 2011;353:242–258. doi: 10.1016/j.ydbio.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herriges JC, et al. FGF-Regulated ETV Transcription Factors Control FGF-SHH Feedback Loop in Lung Branching. Dev Cell. 2015;35:322–332. doi: 10.1016/j.devcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kherrouche Z, et al. PEA3 transcription factors are downstream effectors of Met signaling involved in migration and invasiveness of Met-addicted tumor cells. Mol Oncol. 2015;9:1852–1867. doi: 10.1016/j.molonc.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rockich BE, et al. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci U S A. 2013;110:E4456–4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan H, et al. Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development. 2008;135:2563–2572. doi: 10.1242/dev.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogunovic M, et al. Origin of the Lamina Propria Dendritic Cell Network. Immunity. 2009;31:513–525. doi: 10.1016/j.immuni.2009.08.010. papers3://publication/doi/10.1016/j.immuni.2009.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu L, et al. MAT1-modulated CAK activity regulates cell cycle G(1) exit. Mol Cell Biol. 2001;21:260–270. doi: 10.1128/MCB.21.1.260-270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin D, et al. Constitutive expression of B-myb can bypass p53-induced Waf1/Cip1–mediated G1 arrest. Proc Natl Acad Sci U S A. 1994;91:10079–10083. doi: 10.1073/pnas.91.21.10079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt-Edelkraut U, Daniel G, Hoffmann A, Spengler D. Zac1 regulates cell cycle arrest in neuronal progenitors via Tcf4. Mol Cell Biol. 2014;34:1020–1030. doi: 10.1128/MCB.01195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marken JS, Schieven GL, Hellstrom I, Hellstrom KE, Aruffo A. Cloning and expression of the tumor-associated antigen L6. Proc Natl Acad Sci U S A. 1992;89:3503–3507. doi: 10.1073/pnas.89.8.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H, et al. Multi-organ Site Metastatic Reactivation Mediated by Non-canonical Discoidin Domain Receptor 1 Signaling. Cell. 2016;166:47–62. doi: 10.1016/j.cell.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez RF, Allen L, Gonzales L, Ballard PL, Dobbs LG. HTII-280, a Biomarker Specific to the Apical Plasma Membrane of Human Lung Alveolar Type II Cells. Journal of Histochemistry and Cytochemistry. 2010;58:891–901. doi: 10.1369/jhc.2010.956433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barkauskas CE, et al. Lung organoids: current uses and future promise. Development. 2017;144:986–997. doi: 10.1242/dev.140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu W, et al. Wnt/beta-catenin signaling acts upstream of N-myc, BMP4, and FGF signaling to regulate proximal-distal patterning in the lung. Dev Biol. 2005;283:226–239. doi: 10.1016/j.ydbio.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X, et al. Receptor specificity of the fibroblast growth factor family. The complete mammalian FGF family. J Biol Chem. 2006;281:15694–15700. doi: 10.1074/jbc.M601252200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yano T, et al. KGF regulates pulmonary epithelial proliferation and surfactant protein gene expression in adult rat lung. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1146–1158. doi: 10.1152/ajplung.2000.279.6.L1146. [DOI] [PubMed] [Google Scholar]
- 29.Yano T, Deterding RR, Simonet WS, Shannon JM, Mason RJ. Keratinocyte growth factor reduces lung damage due to acid instillation in rats. Am J Respir Cell Mol Biol. 1996;15:433–442. doi: 10.1165/ajrcmb.15.4.8879176. [DOI] [PubMed] [Google Scholar]
- 30.Panos RJ, Rubin JS, Csaky KG, Aaronson SA, Mason RJ. Keratinocyte growth factor and hepatocyte growth factor/scatter factor are heparin-binding growth factors for alveolar type II cells in fibroblast-conditioned medium. J Clin Invest. 1993;92:969–977. doi: 10.1172/JCI116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quantius J, et al. Influenza Virus Infects Epithelial Stem/Progenitor Cells of the Distal Lung: Impact on Fgfr2b-Driven Epithelial Repair. PLoS Pathog. 2016;12:e1005544. doi: 10.1371/journal.ppat.1005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolaidis NM, et al. Mitogenic stimulation accelerates influenza-induced mortality by increasing susceptibility of alveolar type II cells to infection. Proc Natl Acad Sci U S A. 2017;114:E6613–E6622. doi: 10.1073/pnas.1621172114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chapman HA, et al. Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest. 2011;121:2855–2862. doi: 10.1172/JCI57673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takeda N, et al. Hopx expression defines a subset of multipotent hair follicle stem cells and a progenitor population primed to give rise to K6+ niche cells. Development. 2013;140:1655–1664. doi: 10.1242/dev.093005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zacharias WJ, Morrisey EE. Isolation and culture of human alveolar epithelial progenitor cells. Protocol Exchange. 2018 doi: 10.1038/protex.2018.015. [DOI] [Google Scholar]
- 37.Peng T, et al. Hedgehog actively maintains adult lung quiescence and regulates repair and regeneration. Nature. 2015;526:578–582. doi: 10.1038/nature14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol. 2015;109:21 29 21–29. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing (ATAC-seq and RNA-seq) data generated during this study has been deposited in the GEO database with the primary accession GSE97055 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97055). All upregulated and downregulated genes identified during the described RNA-seq experiments are found in Supplemental Table 1 (mouse data) and Supplemental Table 2 (human data). Source data underlying all plots in all figures is available per Nature policies. The detailed protocol for the cell isolation and propogation of human AEPs has been made available on Nature Protocol Exchange36. All other datasets generated during and/or analyzed during the current study are available from the corresponding author on request.