Abstract
We describe TissueInfo, a knowledge-based method for the high-throughput identification of tissue expression profiles and tissue specificity. TissueInfo defines a set of tissue information calculations that can be computed for large numbers of genes, expressed sequence tags (ESTs) or proteins. Tissue information records that result from the TissueInfo calculations are used to generate tables suitable for data mining and for the selection of genes according to a given expression profile or specificity. When benchmarked against a test set of 116 proteins and literature information, TissueInfo was found to be accurate for 69% of identified tissue specificities and for 80% of expression profiles. The accuracy of the identifications can be increased if query sequences for which little information is available from dbEST are ignored. Thus, with 80% coverage, TissueInfo achieves an accuracy of 76% for specificity and 89% for expression. For the same set of proteins, the curated tissue specificity offered in SWISS-PROT was accurate in 78% of cases. TissueInfo can be useful for the selection of clones for custom microarrays, selection of training sets for ab initio identification of tissue information, gene discovery and genome-wide predictions. Further information about the program can be found at http://icb.mssm.edu/tissueinfo.
INTRODUCTION
Expressed sequence tags (ESTs) derive from libraries of expressed genes made from specific tissues, organs or cell types. Clones are selected randomly from these libraries and their cDNA inserts are sequenced, either randomly from anywhere in the cDNA or more generally from the 3′- or 5′-ends. ESTs from the 3′-end frequently correspond to the 3′-untranslated region (3′-UTR) of the gene (1). ESTs are usually short (∼400 bp) (2) and, because they are subjected to unedited single pass sequencing, they can incorporate a high error rate (∼2–3%; 2,3).
EST sequencing projects escalated in the early 1980s when it was recognized that short stretches of DNA sequence from cDNAs could be used to identify genes (4). In 1992, a central repository for EST sequences, called dbEST, was established at NCBI (5). The biggest contributors to the human and mouse EST sequencing projects have been the WashingtonU Merck EST sequencing project (6) and the project funded by the Howard Hughes Medical Institute (7), respectively. ESTs now currently sample more eukaryotic genes than any other data source (8).
The information available from dbEST has been used primarily for the identification of new genes, physical map construction, identification of disease-causing mutations and other polymorphisms, and annotation of genomic sequence (e.g. finding splice sites) (3,9–11). They have also been used to determine expression profiles of genes (12,13), compare expression patterns in different tissues or disease states (11) and in microarray studies (14,15).
As ESTs have been primarily used to identify new genes, it has become redundant to sequence repetitively the more commonly expressed genes, which can comprise as much as 50–65% of the total mRNA mass (16). Therefore, different methods of eliminating, or significantly reducing, highly expressed transcripts have been developed (1). The most common of these is normalization (16,17), where the frequency of finding rare messages in a normalized library is increased to a frequency similar to that of finding the more common mRNAs. Normalization does not result in the complete removal of members of gene families (6). Libraries can also be subtracted (i.e. made more specific to a particular tissue or to remove clones that have already been sampled) (16) or enriched (i.e. clones selected for a particular feature) (18). Because of the different ways of preparing EST libraries, the quantitative distribution of tissue expression becomes harder to determine and therefore the frequency of representation of a gene in dbEST should not be used to predict its expression level (19).
However, the presence in a tissue-specific library of an EST attributed to a certain gene implies that the given gene is expressed in that specific tissue (or set of tissues). This reasoning can be used to calculate not only general expression profiles, but also to identify genes that are specifically expressed in one particular tissue or organ. In 1998, Vasmatzis et al. used EST data to predict genes specific to the human prostate (20). Of 15 predicted genes, four were already known to be prostate specific and three were experimentally verified to be novel prostate-specific genes. Four more predictions could not be experimentally confirmed, as there was no hybridization of the cDNA probes in either RNA dot blots or northern blots. The authors also reported that PSSPP, a gene previously believed to be expressed specifically in the prostate, is also found to be expressed in lung and trachea (which was both predicted and experimentally verified).
In this paper, we generalize and extend the approach pioneered by Vasmatzis et al. to calculate, for any query sequence, the tissue expression profile and specificity to any tissue type represented in dbEST. We present a new method (TissueInfo) that makes possible the high-throughput identification of tissue distribution of ESTs, mRNAs and protein sequences. To evaluate the approach that TissueInfo automates, we compared the tissue expression profiles obtained for 116 proteins to their SWISS-PROT entry annotations and to a subset of the literature referenced by SWISS-PROT (21). This evaluation helps highlight drawbacks of this approach but shows that TissueInfo can identify tissue specificity or expression with ~75% accuracy.
MATERIALS AND METHODS
TissueInfo: a tissue expression identification pipeline
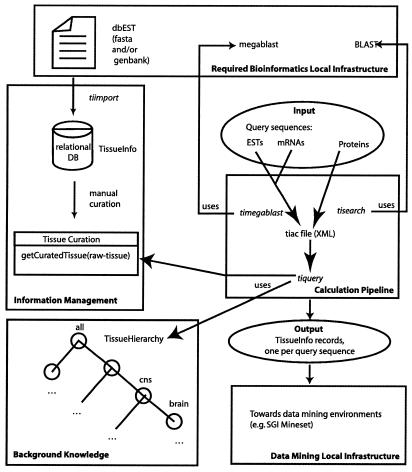
The main components of TissueInfo are illustrated in Figure 1. These include the following. (i) The Calculation Pipeline determines which ESTs are to be used to calculate the tissue expression profile of a query sequence. The selected ESTs are said to match the query sequence. The set of matching ESTs is then used to retrieve information about the set of tissues and/or organs from which their libraries have been prepared. This is achieved by using the Information Management component. (ii) Information Management involves the importation of data from the public dbEST section of GenBank. For each EST in dbEST, TissueInfo attempts to extract the following triplet: accession code of the EST; tissue or organ from which the EST library has been made; organism from which the tissue or organ has been collected. Raw data are stored as imported from dbEST, in a relational database, which provides the indexing capabilities required for quick querying, and enables scheduled updates of any new information in dbEST. Data cleaning is performed by a manual curation that removes natural language variations, aliases and typographical errors from the tissue description, which are frequent in the raw data. (iii) Background Knowledge is provided about the tissues. We defined tissue hierarchies (described later) to provide TissueInfo with the knowledge that ‘cerebral cortex’ is a part of ‘brain’, such that genes found to be expressed in a library made from cerebral cortex are considered to be expressed in brain for the purpose of tissue distribution calculations. The default tissue hierarchy provided with TissueInfo describes more than 165 tissues and their relationships.
Figure 1.
The components of TissueInfo are illustrated and their relation to a local infrastructure. Arrows represent either a flow of data or relationships among components that provide or use services. Information Management imports raw data from dbEST, stores it in a relational database and supports the curation of such data. Curated information is made available as objects empowered with methods necessary to implement the algorithms described in Table 1. See the text for a description of the other components. TissueInfo requires access to dbEST, MegaBLAST and BLAST, which must be provided by the local bioinformatics infrastructure. TissueInfo records are generated for each query sequence going through the pipeline. A suitable data mining environment helps navigate data sets produced when large numbers of query sequences are annotated.
Calculation Pipeline
ESTs that match the query sequence. Depending on the sequence type of the query sequence, one of two approaches is taken to identify ESTs that match the query sequence: (i) protein query sequences are searched against dbEST using BLAST (22); (ii) nucleic acid query sequences are searched against dbEST using MegaBLAST (23). Both approaches yield a list of high scoring pairs (HSPs) that match the query sequence over a given length, with a certain proportion of mismatches. Because ESTs result from a single pass sequencing run, complete sequence identity cannot be assumed with the query sequence. HSPs are considered to match the query sequence if the length of their alignment is longer than min_length and the sequence identity between the query sequence and the HSP is ≥ (100% – sequencing_error). An EST is considered to match the query sequence if at least one of its HSPs matches the query sequence. In typical cases, min_length is set to 100 bp and sequencing_error to 5%. Due to the occasional presence of introns in pre-spliced mRNAs, these query sequences can be treated slightly differently. ESTs that span over two exons may prevent HSPs from reaching min_length. In these cases, lengths of HSPs from the same EST are summed before min_length is tested. Each HSP is still required to pass the sequence identity test independently to prevent low sequence identity regions from acting as a bridge between two short segments of high sequence identity.
This procedure identifies ESTs that are likely to be derived from the mRNA of the gene which encodes the query sequence. The second step implemented by the Calculation Pipeline retrieves the tissue information associated with each matching EST.
Querying TissueInfo. The set of ESTs that match each query sequence is considered in turn. Accession numbers of matching ESTs are used to retrieve the tissue information that is stored in dbEST with most entries. The direct consultation of dbEST for each EST would be prohibitively slow. In addition, the raw data available in dbEST are unsuitable for direct calculations. Therefore, we designed the Information Management component of TissueInfo to accelerate this retrieval, clean the data and transform it into a machine-friendly representation of the tissue information. Details of this process are provided in the Information Management section.
Once the tissue information is obtained as illustrated in Figure 1, the Calculation Pipeline performs a series of calculations. Each calculation uses the tissue information with some optional parameters and returns a single value. The simplest calculation, isExpressedIn(tissue), determines if tissue appears in the tissue information associated with the query sequence. Therefore, isExpressedIn(tissue) returns true when tissue is an element of the set of matching tissues, otherwise false. Another calculation, mostExpressedIn() returns the tissue in which the query sequence is found expressed most often.
Algorithms can be formally defined for each calculation applied to the tissue information. The calculations that we devised are described in the Algorithm section of this paper. Names of the calculations have been chosen such that they allude to the biological concepts that they were designed to calculate. This makes the data analysis a more intuitive process, but should not hide the fact that each result is obtained through an objective, algorithmic process, making large-scale analysis of tissue distribution information possible.
Information Management
Importing raw data from dbEST. TissueInfo relies on a local bioinformatics infrastructure to provide GenBank EST files. Each file is parsed to retrieve the organism and tissue type for every EST accession number. The organism name is extracted from the line beginning with ‘ORGANISM’. The tissue type is retrieved from the line containing ‘TAG_TISSUE’, ‘tissue_type’ or ‘/Note: ‘Organ’’, in that order of preference. This procedure extracts a triplet for each EST (accession number, tissue and organism), where tissue can be a tissue, organ or cell type. For a number of dbEST entries (∼30%), the tissue data cannot be identified using this automated approach. Manual inspection of these records shows that the tissue data are often found embedded in the Note field and thus cannot be extracted accurately by conventional parsing approaches. In these cases, no data are stored in TissueInfo and subsequent queries from the Calculation Pipeline will result in no tissue information being returned (defined as being ‘null’).
Tissue curation: curating the tissue information. There is no set method for describing libraries or describing the tissue type from which the EST library was made. Many libraries from the same tissue are labeled differently. The library information can also contain spelling mistakes. For example, ‘kkin’ is described as coming from primary fibroblasts, but as there is no published data, it is difficult to know what the tissue type actually is, and it has therefore been assumed to be ‘skin’; a more straightforward example is ‘sanal mucosa’, which is a simple misspelling of ‘nasal mucosa’. These differences may appear subtle but could introduce serious algorithmic complications if tolerated in the input data. Therefore we opted to clean raw tissue data to correct for these variations.
In addition to spelling mistakes, the tissue data found in dbEST present additional challenges to computational analysis: ‘whole brain’, ‘brain’, ‘left brain’, ‘corpora quadrigemina’ all represent the ‘brain’ tissue in part or its entirety. The Calculation Pipeline needs a quick way to identify that isExpressedIn(brain) equals true for any raw tissue data synonymous with ‘brain’.
EST libraries can also be derived from pools of tissues or whole organisms. In this case, the tissue data play a special role, as they do not identify the tissue in which the gene is expressed. Any EST that is labeled as coming from a whole body library or from an unspecified tissue, such as ‘whole embryo’, ‘Mixed’, ‘pooled organs’ or ‘Cell line’, is annotated as being non-specific. We label this special tissue information ‘all’.
Finally, ESTs derived from a mixture of specified tissues, such as ‘brain,liver,kidney,lung,heart,spleen’, are annotated as coming from each of those tissues; plant tissues are also flagged as such.
As stated previously, TissueInfo stores the raw tissue data as imported from dbEST. The process of curation consists of reading each raw tissue type to associate it with a curated tissue. For curation convenience, we defined a text-based language to represent curated tissues. Sentences of this language include ‘+brain’, ‘!all’, ‘+brain, +liver, +kidney, +lung, +heart, +spleen’ or ‘+leaf, !plant’.
Background Knowledge. The Background Knowledge component provides the Calculation Pipeline with a mechanism to determine when a tissue is part of an organ or larger tissue. Many entries in dbEST are annotated as coming from very specific tissues, such as the hypothalamus or the corpus striatum, as well as more general tissue types, such as the brain. For example, to determine whether a query sequence is expressed in brain, ESTs that are annotated as coming from a brain library, as well as those annotated as coming from more specific libraries such as hypothalamus and hippocampus libraries, must all be taken into account. To facilitate this process, we have created a Tissue Hierarchy. This enables us to construct queries for very specific tissue types, as well as the more general tissues, without losing any data. For example, a query sequence is determined to be expressed in the hippocampus if it matches any ESTs that come from a hippocampus library. Similarly, a query sequence is determined to be expressed in the eye if it matches any ESTs that come from an eye library or any library created from an eye-specific tissue type (e.g. retina, cornea, iris or lens) or from even more specific libraries such as pigmented retinal epithelium libraries. The level of detail in our hierarchy is determined by the histological detail given in the library annotations in dbEST.
We define Background Knowledge as a Tissue Hierarchy where each node is a tissue and where links are made between tissues when the parent tissue contains the child tissue. The root of the hierarchy is the special ‘all’ tissue.
Algorithms
Table 1 presents the algorithms of the TissueInfo calculations. These calculations provide an objective way to obtain answers to the following biological questions. Is the query sequence expressed in a given tissue? Does the query sequence show specific tissue expression? Is the query sequence specific to a given tissue? In which tissue is the query sequence most expressed? What are all the tissues in which the query sequence is found to be expressed? Is the query sequence expressed in plants?
Table 1. Calculations supported by TissueInfo.
| Calculation |
Algorithm |
| numTotalHits() | Total number of ESTs that match the query sequence. |
| numAll() | Total number of ESTs that are labeled as coming from the special tissue ‘all’. |
| tissueSummary() | Gives a set of all the tissues that the query sequence is shown to be expressed in. Card(tissueSummary()) is the number of elements of the set. |
| numExpressedIn(tissue) | The number of ESTs that match the query sequence which are expressed in tissue. An EST is labeled as being expressed in tissue if the library that the EST derives from is, or is a subset of, tissue as defined by the Tissue Hierarchy. |
| isExpressedIn(tissue) | Tests if the query sequence is expressed in tissue. If numExpressedIn(tissue) > 0 then TRUE, else FALSE. |
| mostExpressedIn() | Gives the tissue type in which the query sequence is predominantly expressed. If the query sequence is equally expressed in two or more tissues, the first one is returned. |
| isSpecificTo(tissue) | Tests if the query sequence is expressed predominantly in tissue. If numExpressedIn(tissue)/numTotalHits() > α then TRUE, else FALSE. |
| isTissueSpecific() | Tests if the query sequence is tissue specific in its expression. A query sequence is assumed to be tissue specific if it is expressed in no more than two tissues, and no more than [100 × (1–α)]% of the tissues in its tissue information are labeled as ‘all’. If (numAll()/numTotalHits() > (1–α) or card(tissueSummary()) > 2 then FALSE, else TRUE. |
| nullCount() | Gives the number of hits for the query sequence that do not have organism and tissue information. |
| numPlant() | Gives the number of hits for the query sequence that come from plant tissue. |
The α parameter controls the stringency of the calculations. For example, if α = 0.95, then isSpecificTo(brain) is TRUE only when the tissue information for the query sequence contains at least 95% of curated tissues that contain brain.
Implementation
Triplets imported from dbEST are stored in a relational database. This makes possible concurrent updates to the database and provides support for integrity constraints. The content of the relational database can be dumped periodically to flat files.
The Calculation Pipeline consists of timegablast and tisearch. These two Java applications filter the output of BLAST or MegaBLAST to extract ESTs that match the query sequence. Both applications share the same XML output file format so successive steps in the pipeline are independent of the path used to calculate the matches. A third Java application, tiquery, reads the XML tiac file (TissueInfo Accession Codes) and queries the Information Management component for raw tissue data. Two Information Management implementations have been developed. The first one uses a JDBC driver to connect to the central TissueInfo relational database. A caching mechanism is implemented to speed up repetitive queries. A second implementation reads a flat file representation of the TissueInfo raw data and holds it entirely in memory for the duration of the queries. This is the faster alternative but requires >300 Mb of memory to hold mouse tissue information.
The approach we discuss here is suitable for the high-throughput identification of tissue information. We generated TissueInfo records for the NIA 15K Mouse cDNA Clone Set (24) in 8 h on one SGI R10000 processor with 512 Mb of memory. The query included about 150 TissueInfo calculations for each of the 14 400 clones that had at least one hit in dbEST. The total number of hits in dbEST was 588 491.
RESULTS AND DISCUSSION
Method evaluation
In order to evaluate the accuracy of TissueInfo in identifying the tissue distribution of genes, we compared calculated tissue expression profiles to information extracted from SWISS-PROT and the literature.
Test set construction. A subset of SWISS-PROT entries have a Tissue field, which indicates the tissues from which the researchers cloned their sequence (in the References section), and includes a Tissue Specificity field in the Comments section. To evaluate the accuracy of our method, we assembled a test set of 116 proteins from four tissues (brain, liver, kidney and pancreas). These were extracted from SWISS-PROT release 39.14 according to the following criteria: the SWISS-PROT entry included a Tissue field in the reference section and a Tissue Specificity field in the Comments section. Both the Tissue field and the Tissue Specificity field had to contain one of the tissues under analysis. This selection process generated a set of proteins likely to be tissue specific to brain, liver, kidney or pancreas.
Identification parameters. The set of proteins was processed with tisearch and tiquery with raw data obtained from dbEST, as available from the NCBI web site on March 26, 2001. Parameters for tisearch were set as follows: min_length = 50, sequencing_error = 5%. The α parameter for tiquery was set to 0.95.
Evaluation criteria. The tissue profiles thus generated were compared with the information given in the Tissue Specificity field of each SWISS-PROT entry analyzed. The abstracts for every paper mentioned in SWISS-PROT for that protein entry were also read. The specificity of the prediction was marked ‘correct’ if our prediction matched either the literature or SWISS-PROT (L/SW) annotation (with the literature review taking precedence) or if neither the L/SW nor our prediction was tissue specific. The prediction was marked ‘agree’ if the L/SW was tissue specific and our prediction was not, but the tissue in which the query sequence was predicted to be most expressed was consistent with that tissue specificity. Finally, it was marked ‘wrong’ if (i) both L/SW and our prediction were tissue specific but the tissues were different; or (ii) if L/SW was tissue specific and our prediction was not and the tissue in which the protein was most expressed did not match the L/SW tissue specificity; or (iii) if the EST matches were all ‘null’. The expression of a protein was marked as: ‘correct’ if the prediction had most of the tissues mentioned by L/SW or if neither L/SW nor our prediction were tissue specific (but not necessarily identical); ‘wrong’ if L/SW was tissue specific but our prediction was not or if most of the L/SW tissues were not mentioned in the prediction. Any proteins that had no hits in dbEST are marked ‘no hits’.
Evaluation results. Evaluation results are summarized in Table 2. The complete results, including the annotations from SWISS-PROT and the literature, can be accessed at http://icb.mssm.ed u/tiss ueinfo. When the proteins labeled ‘no hits’ are not taken into account, TissueInfo finds the ‘correct’ or ‘agree’ tissue specificity in 69% of cases. When expression is assessed the accuracy reaches 80%.
Table 2. Summary of evaluation results.
| Tissue | Correct |
|
Agree |
|
Wrong |
|
No hits | Total |
| |
Spec |
Exp |
Spec |
Exp |
Spec |
Exp |
|
|
| Brain | 22 | 44 | 17 | 0 | 18 | 13 | 5 | 62 |
| Liver | 9 | 28 | 15 | 0 | 6 | 2 | 0 | 30 |
| Kidney | 2 | 6 | 2 | 0 | 5 | 3 | 3 | 11 |
| Pancreas | 4 | 7 | 2 | 0 | 4 | 3 | 2 | 12 |
| Total | 37 | 85 | 36 | 0 | 33 | 21 | 10 | 116 |
| Percent total | 32 | 73 | 31 | 0 | 28 | 18 | 9 | 100 |
| Percent without no hits | 35 | 80 | 34 | 0 | 31 | 20 | 100 |
Spec and Exp indicate the number of agreements between the TissueInfo predictions and the literature search for tissue specificity and tissue expression, respectively. See text for the construction of this test set and at http://icb.mssm.edu/tissueinfo for the evaluation criteria.
Furthermore, as shown in Figure 2, the accuracy of the identification of the tissue expression profile for a particular query sequence correlates with the amount of information available in TissueInfo. Calculations made for proteins found to have less than three hits with information in TissueInfo are unreliable. These calculations represent 20% of the data set. Therefore, when these are removed and the method is re-evaluated at 80% coverage the accuracy reaches 76% for specificity and 89% for expression.
Figure 2.
Reliability of predictions. A summary of the number of times the ‘correct’ or ‘agree’ tissue specificity or tissue expression is found when predictions are compared to SWISS-PROT and the literature. First line, accuracy of specificity prediction. Second line, accuracy of expression prediction. Each column represents a protein, an X indicates an incorrect prediction and a period represents a ‘correct’ or ‘agree’ prediction. (See text for evaluation details.) Third and following lines, adjusted number of hits, i.e. number of hits for which information is available in TissueInfo. The adjusted number of hits is given reading downwards at every point. Predictions are sorted according to increasing adjusted hit number.
It should be noted that this evaluation was carried out with protein sequences, which are less likely to match ESTs that overlap the 3′-UTR of the gene than mRNAs. Since UTR regions of the gene are less conserved than the coding sequence, searches with mRNA will probably generate more hits than the numbers reported here.
Most common pitfalls. An analysis of the incorrect results revealed that TissueInfo is least successful in those cases where highly homologous proteins have different tissue specificities. In these cases, it is not possible to assign one gene unambiguously to a set of ESTs because sequencing errors are of the order of magnitude of sequence variations found among close homologs. Therefore, ESTs for several closely homologous genes can contribute to the calculated tissue distribution of the query sequence and cause errors. The finding of Vasmatzis et al. (20) that some supposedly prostate-specific genes were found expressed in other libraries could be a consequence of the close homolog problem.
Tissue-dependent splice forms of the same gene, used as independent query sequences, are likely to result in the same tissue information prediction, unless ESTs can be matched to the sequence in the regions that are spliced out of some variants. Because this is unlikely, we do not expect TissueInfo to differentiate very well between splice forms.
TissueInfo belongs to the category of knowledge-based prediction methods and exhibits the usual problems associated with this class of methods. In practical terms, the most important problem is that some genes are expressed at low levels in any tissue and therefore are not represented in dbEST. For these genes, prediction records simply list no match. As the number of normalized libraries grows, this problem will be alleviated for some tissues, but other tissues or cell lines will be likely to remain under-represented in dbEST (for example, human taste buds).
TissueInfo can complement curated databases. In the process of our evaluation, we found SWISS-PROT to be misleading, inaccurate or inconsistent with the literature in its annotation of 22% of the proteins considered. (Details for these 25 cases are provided at http://icb.mssm.edu/tissueinfo.) Given this substantial number of curation issues and considering that SWISS-PROT represents tissue information as a text field difficult to parse automatically, TissueInfo appears to be a useful complement to the information that can be extracted from today’s curated databases.
Referring to the primary literature was sometimes necessary to provide an objective benchmark for TissueInfo. This source of information also has drawbacks. (i) Tissue information about one gene is scattered in several papers. These articles are difficult to identify. Genes are often named inconsistently from one article to another and the gene accession codes are of limited use in identifying articles. Retrieval of the information in the primary literature is therefore a manual task that does not scale well for large numbers of genes. (ii) Experimental evidence for tissue expression profiles is provided by northern blot analysis or dot blots. Each article usually reports blots made with a collection of about 10 tissues. This set represents only a fraction of the tissues in which the gene is potentially expressed. The primary literature is therefore an incomplete source of information about tissue expression and specificity.
Large-scale analysis of tissue information. TissueInfo can be used to identify the tissue expression profiles and specificities of thousands of genes or ESTs at once. Analysis of such amounts of data is greatly facilitated by the careful organization of the calculated records. Thus, we store all calculation results in a tabular format. Each query sequence is represented as a row in the output and the results of TissueInfo calculations requested by the user are stored in columns. Table 3 shows the records for three proteins and seven calculations.
Table 3. Sample records for three proteins.
| AC |
No. of hits |
specificTo(hypothalamus) |
specificTo(brain) |
specificTo(muscle) |
specificTo(liver) |
mostExpressedIn |
tissueSpecific |
| P1223 | 12 | TRUE | TRUE | FALSE | FALSE | hypothalamus | TRUE |
| P3483 | 50 | FALSE | FALSE | FALSE | FALSE | kidney | FALSE |
| P9273 | 0 | FALSE | FALSE | FALSE | FALSE | FALSE |
Seven calculations have been performed on each protein. Details of the algorithms behind these calculations are described in Table 1. Note that the third protein has no hit in dbEST.
We have used the SGI Mineset data mining environment to analyze these records. Operations such as filtering can be performed interactively to select genes that are expressed in a combination of tissues, are specific to others, are expressed in plants, etc. The record format is also ideally suited for transfer to a relational database and the selection of subsets of genes using SQL queries.
Finally, TissueInfo can also produce textual reports that output the original dbEST information so that any user who wishes to investigate small sets of genes selected during the data mining process can follow up on them, verify the accuracy of TissueInfo records or investigate ‘null’ hits. The reports therefore link tissue expression calculations to the raw data.
Relation to other tissue information resources. TissueInfo is unique in its ability to process any query sequence to identify its tissue expression profile, if it finds any matching ESTs in dbEST. We also introduce the notion of tissue information calculations and evaluate the accuracy of our calculations. Several other groups have developed resources designed to organize expression data and enhance the information content of the raw data provided by dbEST.
The TIGR Gene Indices (25) are assemblies of ESTs (and annotated genes from GenBank) into clusters called Tentative Consensus sequences (TCs). Each TC references ESTs used for its construction together with the raw tissue data provided by dbEST. This makes possible query of the TIGR Gene Indices to extract TCs found to be expressed in a given tissue. STACK (26,27) provides another collection of EST assemblies, which can be queried with a tissue hierarchy (similar but less extensive than the one offered in TissueInfo). The Unigene system (2) also provides assemblies (but not consensus sequences) that include both gene and EST sequences, and uses the information from the ESTs to determine the tissues in which that gene is expressed. EST library names are not curated. BodyMap (12) takes a different approach. All the BodyMap data are locally produced, from non-normalized libraries, so that the abundance of clones from each library are representative of the abundance of the expressed mRNA transcripts (28). This makes BodyMap ideally suited for quantitative analysis, where differential expression in different tissues or disease states can be detected by the variation in the numbers of mRNAs (29). BodyMap therefore makes possible quantitative analyses of expression profiles that are not possible with dbEST (or consequently with TissueInfo), but is limited to a somewhat smaller collection of ESTs (18 998 gene sequences for human, 16 772 GS for mouse). BodyMap is also the only resource that can specify tissues where genes are not expressed.
TissueInfo is similar in concept to the Serial Analysis of Gene Expression (SAGE) method (30). In contrast to SAGE, which relies on tags of 9–12 bp from the 3′-UTR to uniquely identify a gene, TissueInfo can use any part of the full-length sequence of a gene or protein to determine its expression profile. For instance, TissueInfo can identify the expression profiles of proteins, as shown by SWISS-PROT, whereas SAGE requires the sequence of the 3′-UTR of the gene. Using full-length ESTs as tags helps lessen ambiguity problems encountered with SAGE when one given tag matches several genes. SAGE analysis is usually performed on non-normalized libraries and therefore allows quantitative analysis.
Several problems with the data, identified in the current work, include typographical errors, the presence of synonyms and the need for Background Knowledge. These have not been addressed by TIGR, which also does not curate raw tissue information. It is therefore difficult to compare TissueInfo with TIGR. STACK uses Background Knowledge to a more limited extent than TissueInfo to enhance tissue information queries, but tissue specificity cannot be determined. TIGR, STACK and BodyMap allow the user to search the databases with their own sequence, but if the query sequence is not an exact match to any ‘contig’ in their databases (allowing for sequencing error), it is impossible to make any tissue expression profile predictions for that query sequence.
CONCLUSION
We have designed a computational approach to automate the identification of tissue information. In contrast to previous approaches, TissueInfo makes possible (i) the calculation of this information for large numbers of genes, proteins or expressed sequence tags, and (ii) analysis of the resulting data to identify genes that exhibit specific expression patterns.
Evaluation of the accuracy of this identification method is limited by the amount of experimentally verified information directly available for computational comparisons. Today’s databases rarely describe the tissue expression or tissue specificity of genes. SWISS-PROT is an exception but was found to reveal two major problems. First, the tissue specificity information contained in SWISS-PROT is represented as text. This complicates the automatic extraction of sets of proteins experimentally shown to be specific to a given tissue. Second, some entries were found to disagree with the primary literature referenced by the SWISS-PROT entry. When compared to the primary literature, TissueInfo correctly identifies the tissue specificity of 69% of the genes in the test set, and 80% when expression is considered (excluding genes with no matches in dbEST). Accuracy can be raised to 76% (specificity) and 89% (expression) at a coverage level of 80%. The TissueInfo approach should therefore be most useful in performing the following high-throughput tasks.
Selection of clones for cDNA microarrays
The selection of clones specific to a given tissue is the first step in the design of custom microarray chips. Custom microarrays can be cheaper to produce than general purpose arrays and show a better signal-to-noise ratio. As illustrated by Rockett et al. (31), the traditional selection of genes for a single, tissue-focused microarray is a lengthy process. TissueInfo can be applied routinely to filter collections of cDNA clones for a given tissue.
Ab initio prediction of tissue information
Tissue-specific gene expression is controlled by regulatory sequences. Many regulatory sequences are still unknown, but should in principle be identifiable with ab initio methods (32). Ab initio methods are required to decipher expression patterns of genes with low expression. Development of such methods has probably been hindered by the lack of training and test data sets. We expect that TissueInfo can be used to generate such data sets by grouping genes according to their expression profiles and to foster the development of ab initio prediction methods.
Gene discovery
TissueInfo can be integrated into gene discovery pipelines to supplement them with the ability to calculate tissue expression profiles and specificity for candidate genes. As shown in our recent identification of the Sac sensory receptor gene candidate (33), prediction of restricted tissue expression or other specific expression profiles can contribute to the identification of a gene candidate.
Genome-wide predictions
The performance of our algorithms and implementations makes possible the calculation of tissue expression profiles for whole genomes. This can produce a very valuable resource for identification of tissue-specific gene candidates and can complement experimental approaches recently developed for such a purpose (34). Similarly, when quantities of tissue information at the scale of a genome become available, statistical estimates of the gene expression of tissues can be obtained. Analysis of these data should produce important insights into the mechanisms of cell differentiation and tissue development.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Harel Weinstein for reviewing the manuscript and continuous support during the development of this work. Our special thanks go to Marianna Max and Charles Mobbs for introducing us to the biological problems that motivated this work. We thank Dr Sherman Kupfer for his help with the physiological aspects of Tissue Hierarchy and TogetherSoft for free academic access to their Together software design and development platform. This work was supported by the Institute for Computational Biomedicine of the Mount Sinai School of Medicine.
References
- 1 Wolfsberg T.G. and Landsman,D. (1997) A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res., 25, 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 3.Marra M.A., Hillier,L. and Waterston,R.H. (1998) Expressed sequence tags—ESTablishing bridges between genomes. Trends Genet., 14, 4–7. [DOI] [PubMed] [Google Scholar]
- 4.Putney S.D., Herlihy,W.C. and Schimmel,P. (1983) A new troponin T and cDNA clones for 13 different muscle proteins, found by shotgun sequencing. Nature, 302, 718–721. [DOI] [PubMed] [Google Scholar]
- 5.Boguski M.S., Lowe,T.M. and Tolstoshev,C.M. (1993) dbEST—database for ‘expressed sequence tags’. Nature Genet., 4, 332–333. [DOI] [PubMed] [Google Scholar]
- 6.Hillier L.D., Lennon,G., Becker,M., Bonaldo,M.F., Chiapelli,B., Chissoe,S., Dietrich,N., DuBuque,T., Favello,A., Gish,W., Hawkins,M., Hultman,M., Kucaba,T., Lacy,M., Le,M., Le,N., Mardis,E., Moore,B., Morris,M., Parsons,J., Prange,C., Rifkin,L., Rohlfing,T., Schellenberg,K., Marra,M. et al. (1996) Generation and analysis of 280,000 human expressed sequence tags. Genome Res., 6, 807–828. [DOI] [PubMed] [Google Scholar]
- 7.Marra M., Hillier,L., Kucaba,T., Allen,M., Barstead,R., Beck,C., Blistain,A., Bonaldo,M., Bowers,Y., Bowles,L., Cardenas,M., Chamberlain,A., Chappell,J., Clifton,S., Favello,A., Geisel,S., Gibbons,M., Harvey,N., Hill,F., Jackson,Y., Kohn,S., Lennon,G., Mardis,E., Martin,J., Waterston,R. et al. (1999) An encyclopedia of mouse genes. Nature Genet., 21, 191–194. [DOI] [PubMed] [Google Scholar]
- 8.Parsons J.D. and Rodriguez-Tomé,P. (2000) JESAM: CORBA software components to create and publish EST alignments and clusters. Bioinformatics, 16, 313–325. [DOI] [PubMed] [Google Scholar]
- 9.Gerhold D. and Caskey,C.T. (1996) It’s the genes! EST access to human genome content. Bioessays, 18, 973–981. [DOI] [PubMed] [Google Scholar]
- 10.Schultz J., Doerks,T., Ponting,C.P., Copley,R.R. and Bork,P. (2000) More than 1,000 putative new human signalling proteins revealed by EST data mining. Nature Genet., 25, 201–204. [DOI] [PubMed] [Google Scholar]
- 11.Gill R.W. and Sanseau,P. (2000) Rapid in silico cloning of genes using expressed sequence tags (ESTs). Biotechnol. Annu. Rev., 5, 25–44. [DOI] [PubMed] [Google Scholar]
- 12.Okubo K. and Matsubara,K. (1997) Complementary DNA sequence (EST) collections and the expression information of the human genome. FEBS Lett., 403, 225–229. [DOI] [PubMed] [Google Scholar]
- 13.Colantuoni C., Purcell,A.E., Bouton,C.M. and Pevsner,J. (2000) High throughput analysis of gene expression in the human brain. J. Neurosci. Res., 59, 1–10. [DOI] [PubMed] [Google Scholar]
- 14.Carulli J.P., Artinger,M., Swain,P.M., Root,C.D., Chee,L., Tulig,C., Guerin,J., Osborne,M., Stein,G., Lian,J. and Lomedico,P.T. (1998) High throughput analysis of differential gene expression. J. Cell. Biochem. Suppl., 31, 286–296. [DOI] [PubMed] [Google Scholar]
- 15.Khan J., Saal,L.H., Bittner,M.L., Chen,Y., Trent,J.M. and Meltzer,P.S. (1999) Expression profiling in cancer using cDNA microarrays. Electrophoresis, 20, 223–229. [DOI] [PubMed] [Google Scholar]
- 16.Bonaldo M.F., Lennon,G. and Soares,M.B. (1996) Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res., 6, 791–806. [DOI] [PubMed] [Google Scholar]
- 17.Soares M.B., Bonaldo,M.F., Jelene,P., Su,L., Lawton,L. and Efstratiadis,A. (1994) Construction and characterization of a normalized cDNA library. Proc. Natl Acad. Sci. USA, 91, 9228–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki Y., Yoshitomo-Nakagawa,K., Maruyama,K., Suyama,A. and Sugano,S. (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene, 200, 149–156. [DOI] [PubMed] [Google Scholar]
- 19.Bains W. (1996) Virtually sequenced: the next genomic generation. Nature Biotechnol., 14, 711–713. [DOI] [PubMed] [Google Scholar]
- 20.Vasmatzis G., Essand,M., Brinkmann,U., Lee,B. and Pastan,I. (1998) Discovery of three genes specifically expressed in human prostate by expressed sequence tag database analysis. Proc. Natl Acad. Sci. USA, 95, 300–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z., Schwartz,S., Wagner,L. and Miller,W. (2000) A greedy algorithm for aligning DNA sequences. J. Comput. Biol., 7, 203–214. [DOI] [PubMed] [Google Scholar]
- 24.Kargul G., Dudekula,D., Qian,Y., Lim,M., Jaradat,S., Tanaka,T., Carter,M. and Ko,M. (2001) Verification and initial annotation of the NIA mouse 15K cDNA clone set. Nature Genet., 28, 17–18. [DOI] [PubMed] [Google Scholar]
- 25.Quackenbush J., Cho,J., Lee,D., Liang,F., Holt,I., Karamycheva,S., Parvizi,B., Pertea,G., Sultana,R. and White,J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res., 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christoffels A., van Gelder,A., Greyling,G., Miller,R., Hide,T. and Hide,W. (2001) STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucleic Acids Res., 29, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller R.T., Christoffels,A.G., Gopalakrishnan,C., Burke,J., Ptitsyn,A.A., Broveak,T.R. and Hide,W.A. (1999) A comprehensive approach to clustering of expressed human gene sequence: the sequence tag alignment and consensus knowledge base. Genome Res., 9, 1143–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okubo K., Hori,N., Matoba,R., Niiyama,T., Fukushima,A., Kojima,Y. and Matsubara,K. (1992) Large scale cDNA sequencing for analysis of quantitative and qualitative aspects of gene expression. Nature Genet., 2, 173–179. [DOI] [PubMed] [Google Scholar]
- 29.Audic S. and Claverie,J.M. (1997) The significance of digital gene expression profiles. Genome Res., 7, 986–995. [DOI] [PubMed] [Google Scholar]
- 30.Velculescu V.E., Zhang,L., Vogelstein,B. and Kinzler,K.W. (1995) Serial analysis of gene expression. Science, 270, 484–487. [DOI] [PubMed] [Google Scholar]
- 31.Rockett J.C., Luft,J.C., Garges,J.B., Krawetz,S.A., Hughes,M.R., Kirn,K.H., Oudes,A.J. and Dix,D.J. (2001) Development of a 950-gene DNA array for examining gene expression patterns in mouse testis. Genome Biol., 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Editorial. (2001) The human genome: what next? Nature Neurosci., 4, 217. [DOI] [PubMed] [Google Scholar]
- 33.Max M., Shanker,Y.G., Huang,L., Rong,M., Liu,Z., Campagne,F., Weinstein,H., Damak,S. and Margolskee,R.F. (2001) Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nature Genet., 28, 58–63. [DOI] [PubMed] [Google Scholar]
- 34.Shoemaker D.D., Schadt,E.E., Armour,C.D., He,Y.D., Garrett-Engele,P., McDonagh,P.D., Loerch,P.M., Leonardson,A., Lum,P.Y., Cavet,G., Wu,L.F., Altschuler,S.J., Edwards,S., King,J., Tsang,J.S., Schimmack,G., Schelter,J.M., Koch,J., Ziman,M., Marton,M.J., Li,B., Cundiff,P., Ward,T., Castle,J., Krolewski,M., Meyer,M.R., Mao,M., Burchard,J., Kidd,M.J., Dai,H., Phillips,J.W., Linsley,P.S., Stoughton,R., Scherer,S. and Boguski,M.S. (2001) Experimental annotation of the human genome using microarray technology. Nature, 409, 922–927. [DOI] [PubMed] [Google Scholar]