Abstract
Multiple myeloma (MM) is a heterogeneous disease with a poor prognosis. Circulating microRNAs (miRNAs) have shown potential as non-invasive prognostic biomarkers for heterogeneous diseases. miR-20a has been shown involved in various human cancers, and the phosphatase and tensin homolog/phosphoinositide 3-kinase/protein kinase B (PTEN/P13K/Akt) signaling pathway plays a key role in cell proliferation, migration and apoptosis. Here, we investigated the effect of miR-20a on the PTEN/PI3K/Akt signaling pathway during MM cell proliferation, migration and apoptosis. Reverse transcription quantitative polymerase chain reaction was applied to detect miR-20a expression in plasma from 30 MM patients and MM cell lines. CCK-8 assays, Transwell assays, Annexin V/PI double-staining and western blotting were performed to examine the protein expressions of PTEN, PI3K and Akt during cellullar proliferation, migration, cycling, and apoptosis. Significant upregulation of miR-20a and deregulation of PTEN were observed in MM cells. We also identified PTEN as a downstream target gene of miR-20a, which bound to the 3′-untranslated region of PTEN. Overexpression of miR-20a was associated with decreased PTEN expression, and treatment with miR-20a inhibitors decreased cell proliferation, migration and clonogenicity and reduced the protein expressions of PI3K and p-Akt but increased PTEN protein expression compared with blank and negative control groups. Taken together, these results showed that inhibition of miR-20a suppresses MM progression by modulating the PTEN/PI3K/Akt signaling pathway. These findings suggest that miR-20a may be a novel molecular therapeutic target for the treatment of MM.
Keywords: microR-20a, multiple myeloma, metastasis, phosphatase and tensin homolog, cell proliferation, cell apoptosis, cell migration
Introduction
Multiple myeloma (MM) is a clonal B cell malignancy which is characterized by the proliferation of plasma cells (PCs) within the bone marrow (BM). MM is the second most common hematologic malignancy, but it remains incurable with survival duration ranging from a few months to over 10 years (1,2). Although treatment strategies have evolved from traditional chemotherapy and autologous hematopoietic stem cell trans-plantation to novel targeted drug therapy, patient outcomes have not improved (3), and the 5-year survival rate remains only 30–40%, mainly due to the development of drug resistance (4). Therefore, it is important to elucidate the mechanisms regulating the malignant behavior of MM and identify key genes contributing to cancer progression for improving patient prognosis. One hot topic of chromosomal abnormalities and other types of genetic or epigenetic alterations might contribute to microRNA (miRNA) deregulation in cancer progression (5–7).
miRNAs, a newly discovered class of endogenous, small, non-coding RNAs, regulate gene expression by base pairing to partially or fully complementary sites in the 3′-untranslated regions (3′-UTRs) of target mRNAs (8). miRNAs are involved in various biological processes, including cellular growth, development, metabolism, apoptosis and development (9). Increasing evidence has demonstrated that differential miRNAs (miR-21, miR-155, miR-17-92, and miR-125a-5p) have key roles in MM, suggesting the importance of miRNAs in MM progression (8,10,11). Deregulated miRNAs can act as oncogenes or tumor suppressors upregulated or downregulated in cancer cells (12–14). miR-20a has an important in various human cancers, including gastric cancer (15), colorectal cancer (16), non-small cell lung cancer (17) and cervical cancer (18). Importantly, significant expression of miR-20 has been found in myeloma (19,20). Several studies have suggested that the dysregulation of miRNAs initiates the activation of the phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling pathway, which is involved in the progression of breast cancer, bladder cancer and non-small-cell lung cancer (21–24). However, the effects of the PTEN/PI3K/Akt signaling pathway and miRNA dysregulation on MM pathogenesis and progression remain unknown. Here, we reported for the first time a miRNA that directly regulated PTEN: miR-20a acts as a negative regulator of PTEN in MM cell lines. Downregulation of miR-20a expression inhibited MM cell growth in vivo. Furthermore, PTEN was identified as a downstream target gene of miR-20a, which bound to the 3′-UTR of PTEN. Our study provides new insights into the function of miR-20a during development and suggests that this miRNA has a role in tumorigenesis. The overall aim of this study was to investigate the effects of miR-20a on the cell proliferation, migration and invasion of MM cells.
Materials and methods
Study subjects and sample collection
The specimens in this study were obtained from 30 patients who were diagnosed according to the NCCN clinical practice guidelines for MM at the First Affiliated Hospital of Nanchang University between October 2015 and May 2017 (25). We obtained plasma samples from 30 patients diagnosed with MM. All patients who were receiving chemotherapy and/or biotherapy were excluded and those with other types of malignant tumors were also eliminated. To investigate miR-20a expression in the different genetic subtypes of MM, 30 cases were cytogenetically classified using FISH (Abbott Diagnostics, Berkshire, UK). Cytogenetic abnormalities involving 13q deletions and immunoglobulin heavy-chain gene rearrangements were investigated. The patient details were shown in Tables I and II. 8 plasma samples from healthy people were used as control in the validation set. Additionally, venous blood was collected in EDTA tubes (BD Biosciences, Franklin Lakes, NJ, USA). The plasma was transferred to a fresh tube and stored at −80°C after freezing with liquid nitrogen. The study was approved by the Local Research Ethics Committee, and written informed consent was obtained from all study subjects.
Table I.
Summary of clinical details of multiple myeloma patients used for quantitative polymerase chain reaction analysis of plasma samples (n=30).
| Clinical characteristics | n (%) |
|---|---|
| Sex | |
| Male | 21 (70) |
| Female | 9 (30) |
| Age range | 35–75 |
| Mean | 56.5 |
| Durie salmon stage | |
| I | 8 (26.67) |
| II | 5 (16.67) |
| III | 17 (56.67) |
| Karyotype | |
| t(4:14) | 10 (33.33) |
| t(11:14) | 5 (3.33) |
| t(14:16) | 3 (10) |
| del(13q) as a unique abnormality | 7 (23.33) |
| Normal FISH | 5 (16.67) |
FISH, fluorescence in situ hybridization.
Cell lines and RNA extraction
Human MM cell lines (MM1S, U266, and RPMI-8226) and normal plasma cells (nPCs) were cultured in RPMI-1640 (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 5% CO2 atmosphere. When cells reached 80% confluency, cells at the logarithmic growth phase were collected.
RNA isolation and quantitative RT-PCR
Total RNA was extracted from human plasma and human MM cell lines using the mirVana Paris RNA Isolation kit (Ambion, Austin, TX, USA) following the manufacture's instructions for liquid samples. The concentration and purity of extracted RNA were measured at 260 and 280 nm optical densities. cDNA was synthesized from RNAs using gene specific primers (Applied Biosystems, Beijing, China) with the M-MLV Reverse Transcriptase kit (GeneCopoeia, Rockville, MD, USA) according to the manufacturer's instructions. To determine miR-20a expression levels, real-time PCR was performed with SYBR Green (Takara, Osaka, Japan). β-actin was used as internal control. PCR conditions were as follows: 95°C for 5 min, 95°C for 45 sec, 55°C for 15 sec, 72°C for 50 sec, for 40 cycles. The primers used in PCR were as follows: miR-20a forward, 5′-TGCGCTAAAGTGCTTATAGTGC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGTATT-3′; PTEN forward, 5′-TTGTGGCAACAGCTGAATCTGCAGTTGGCTAAGAGAGGTT-3′ and reverse, 5′-ATGTAGCAAAACCCTTCGGAAACCTCTCTTAGCCAACTGC-3′; β-actin forward, 5′-AGCGAGCATCCCCCAAAGTT-3′ and reverse, 5′-GGGCACGAAGGCTCATCATT-3′. Samples were analyzed in triplicate and gene expression was quantified by normalizing target gene expression to that of the internal control using the 2−ΔΔCt formula.
Plasmid construction and transfection
The 5′-flanking regions of the pre-miR-20a-5p and 3′-UTR sequences of PTEN [the mutated (mu) PTEN 3′-UTR was designated as PTEN] containing the putative miR-20a target sites were isolated using specific PCR primers. For luciferase assays, the PTEN 3′-UTR sequence was inserted into the pGL3 luciferase reporter vector (Promega Corporation, Madison, WI, USA). Target sites were mutated using the QuickChange Site-Directed Mutagenesis (Agilent Technologies, Inc., Santa Clara, CA, USA). The miR-20a NC (5′-CAGUACUUUUGUGUAGUACAA-3′) and the miR-20a inhibitor (5′-CUACCUGCACUAUAAGCACUUUA-3′) were synthesized by Genepharma (Shanghai, China). Cell transfection was performed using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions then cells were added and cultured in complete medium. Cells in the logarithmic growth phase were seeded in 12-well plates at a density of 1×105 cells, and divided into three groups: miR-20a inhibitor group (transfected with anti-miR-20a), scramble group (transfected with small interfering RNA negative control) and blank group (untransfected cells). Three triplicates were set for each group. The medium was replaced with normal medium 6 h after transfection. DMEM containing 10% FBS and G418 was added for selection 24 h later, and clones were obtained after two weeks. The selected clones were cultured for the subsequent experiments.
Cell proliferation analysis
For cell proliferation assays, cells were plated 96-well plate (1,500 per well) and examined 48 h after transfection using the Cell Counting Kit-8 (CCK-8) (Dojindo Laboratories, Shanghai, China). Optical density (OD) values were determined at a wavelength of 450 nm. The plates were then read on a micro plate reader using a test wave length of 450 nm. For survival rate assay, the number of viable cells were counted using Trypan blue (Solarbio, Beijing, China) on Countstar Cell Counter (Inno-Alliance Biotech, Inc., Wilmington, NC, USA) according to the manufacturer's instructions. All assays were performed in triplicate.
Cell migration and invasion assays
The cells were collected and suspended 72 h after transfection for the cell migration assays. Then, the cells were inoculated into the Transwell upper chamber and placed in a 5% CO2 incubator at 37°C for 48 h. The cells that failed to penetrate the upper chamber were removed, and the membrane was fixed in 95% ethanol for 15–20 min and then soaked with water. The membrane was tinted with crystal violet for 10 min, soaked again in water, photographed and observed under a high magnification microscope, followed by cell counting on the back of the membrane. Five high-power fields were randomly chosen, and the number of cells penetrating through the polycarbonate membrane was used to evaluate migration ability. For the invasion assay, Matrigel matrix was dissolved at 4°C overnight and diluted with 1:3 serum-free DMEM medium. A total volume of 30 µl of Matrigel was added to the Transwell upper chamber until it covered the bottom of the upper chamber. The cell suspension was added to the upper chamber, and 0.5 ml of DMEM containing 10% FBS was added to the lower chamber of the 24-well plate. The number of cells penetrating through the Matrigel was determined to assess the cell invasive ability.
Luciferase reporter analysis
For luciferase reporter experiments, the 3′-UTR segments of PTEN predicted to interact with miR-20a were amplified by PCR and inserted into pGL3 vector immediately downstream from the stop codon of luciferase (Promega Corporation). The mutated (mu) PTEN 3′-UTR without miR-20a binding sites was designated as PTEN 3′-UTR-mu (forward, 5′-TTGTGGCAACAGCTGAATCTGCAGTTGGCTAAGAGAGGTT-3′ and reverse, 5′-ATGTAGCAAAACCCTTCGGAAACCTCTCTTAGCCAACTGC-3′). To construct the luciferase reporter vector, 293T cells were cotransfected in 24-well plates with 0.4 mg of the firefly luciferase report vector and 0.08 mg of the control vector containing Renilla luciferase, pRL-TK (Promega Corporation), as well as with 100 nM miR-20a mimics, inhibitor or control miRNA. Luminescence was detected using the Dual-Luciferase Reporter Assay System (Promega Corporation) according to the instructions 48 h after transfection. Data were normalized to the Renilla luminescence and presented relative to control miRNA transfected group.
Apoptosis analysis
The cells in each group were collected at 24, 48 and 72 h after transfection, and cold PBS was used to wash cells for three times. The cells were resuspended with 500 µl pre-cooled binding buffer, and the concentration was adjusted to 5×106 ml. A total of 100 µl of the cell suspension was added to flow cytometry tubes and 5 µl of Annexin V-fluorescein isothiocyanate (Beyotime Institute of Biotechnology, Shanghai, China) was added. After mixture, samples were incubated at room temperature in the dark for 15 min, and 5 µl of 10 mg/l propidium iodide (PI) dye (Beyotime Institute of Biotechnology) was added 5 min prior to the measurements. Samples were immediately analyzed with a FacSort without wash or fixation. Cell Quest FCS 3.0 software (BD Biosciences) was used for data analysis. Each sample was repeated three times.
Western blotting
Whole cell protein extracts were obtained from MM cell lines. Cell lysates were loaded and polyacrylamide gel electrophoresis separated. Proteins were transferred by Trans-Blot Turbo Transfer Starter System (Bio-Rad Laboratories, Inc., Hercules, CA, USA) for 7 min. After protein transfer, the membranes were blotted with the primary antibodies. The cells in the logarithmic growth phase with 80% confluency were collected, and radio immunoprecipitation assay lysis buffer containing protease inhibitors (Beyotime Institute of Biotechnology, Haimen, China) was used for conventional extraction of total cellular protein. A bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology) was used for protein quantification. An equal amount of protein was separated by running on 10% SDS gel electrophoresis under denaturing and nonreducing conditions and then transferred to a nitrocellulose membrane. 5% skim milk was added to block the membrane in a sealed container for 1 h. The membranes were incubated overnight at 4°C using the following antibodies: anti-PTEN-7 (1:300), anti-p13k (1:1,000), anti-p-p13k (1:500), anti-AKT (1:500) and anti-p-AKT (1:500) (all from Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Anti-β-actin (1:200; Abcam, Cambridge, MA, USA) was used as a loading control.
Statistical analysis
Each experiment was performed at least three times and values are reported as mean ± SD. Differences between two groups were evaluated by a Student's t-test, and differences among multiple groups were evaluated by one-way analysis of variance and a Newman-Keuls post-hoc test using SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA). P-values <0.05 were considered to indicate a statistically significant difference.
Results
miR-20a was overexpressed in plasma from MM patients and MM cell lines
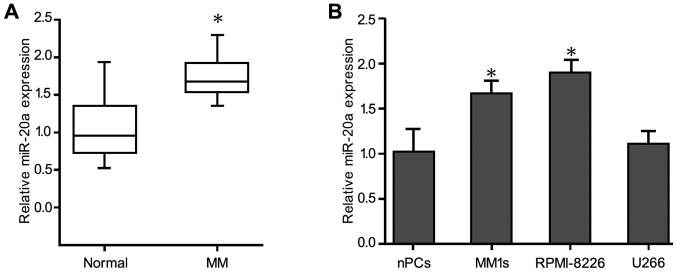
Real-time quantitative PCR revealed that miR-20a was significantly upregulated in 30 plasma samples from MM patients compared with healthy control subjects (Fig. 1A). The levels of miR-20a expression in the MM patients and the control subjects were 1.70±0.02 and 0.94±0.03, respectively, and miR-20a expression in the MM patients was significantly higher than that in the normal individuals (P<0.05). Consistent with these data, miR-20a expression in the U266, MM1 sec, and RPMI-8226 MM cell lines, was significantly increased compared with the normal healthy bone marrow-derived plasma cells (Fig. 1B). In addition, miR-20a expression in stage III patients (1.80±0.31) was significantly higher than that in stage I/II patients (1.63±0.03) (P<0.05) (Table I). However, miR-20a expression was not associated with the patient age, sex or karyotype (P>0.05) (Table II).
Figure 1.
Determination of miR-20a expression in human MM cell lines and clinical samples via RT-qPCR. (A) RT-qPCR detection of miR-20a in plasma cells derived from bone marrow of healthy donors (normal) and MM patients. (B) Expression of miR-20a in the MM cell lines MM.1S, U266 and RPMI-8226 compared with nPCs. Each sample was analyzed in triplicate and error bars represent standard deviation. *P<0.05 vs. nPCs. nPCS, normal plasma cells; MM, multiple myeloma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, microRNA.
Table II.
Relationship between the miR-20a expression and the clinical pathological characteristics of patients with multiple myeloma.
| Characteristics | Cases, % | miR-20a expression | P-value |
|---|---|---|---|
| Age, years | 0.053 | ||
| ≤50 | 14 | 1.67±0.03 | |
| >50 | 16 | 1.73±0.04 | |
| Sex | 0.112 | ||
| Male | 21 | 1.68±0.05 | |
| Female | 9 | 1.77±0.05 | |
| Durie-salmon stage | <0.05 (P=0.004) | ||
| I/II phase | 13 | 1.63±0.03 | |
| III phase | 17 | 1.80±0.31 | |
| Extramedullary infiltration | 0.413 | ||
| Yes | 13 | 1.68±0.03 | |
| No | 17 | 1.72±0.04 | |
| Karyotype | 0.282 | ||
| Normal FISH | 12 | 1.74±0.03 | |
| Abnormal FISH | 18 | 1.68±0.03 |
FISH, fluorescence in situ hybridization; miR, microRNA.
Effect of miR-20a expression on cellular growth, migration, invasion and apoptosis
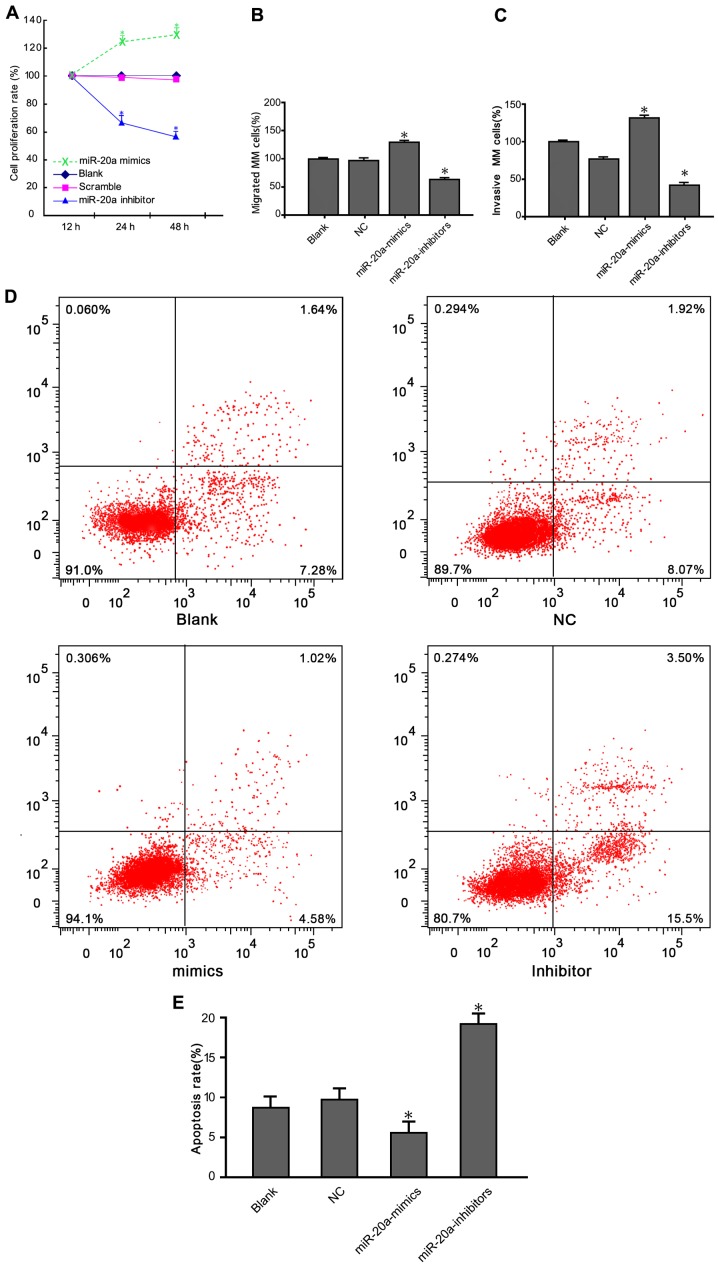
CCK-8 assay results showed that the viability of U266 and RPMI-8226 cells was reduced following transfection with a miR-20a inhibitor (Fig. 2A). Cell growth in the miR-20a inhibitor group was significantly decreased compared with blank and negative control (NC) groups (P<0.05). We next investigated whether treatment with the miR-20a inhibitor inhibited MM cell migration using a Transwell assay. Consistent with the CCK-8 assays results, cells migration rate transfected with the miR-20a inhibitor was reduced (Fig. 2B). The number of migrated cells in the miR-20a inhibitor group was significantly lower than that in the blank and NC groups (P<0.05). Cell invasion assay showed that the number of cells penetrating through the Matrigel into the back of the Transwell membrane was significantly lower in the treatment group compared with the blank and NC groups (Fig. 2C) (P<0.05). Moreover, apoptosis rates for the cells in the blank group, NC group, miR-20a mimics group and miR-20a-inhibitor group was 8.92, 9.99, 5.62 and 19.9% 48 h after transfection, respectively (Fig. 2D). Compared with the two control groups, the apoptotic rate in the miR-20a-inhibitor group was significantly increased (P<0.05) (Fig. 2E).
Figure 2.
Regulatory effects of miR-20a inhibitor on MM cell proliferation, migration, invasion and apoptosis. (A) The CCK-8 assay showed that RPMI-8226 cells transfected with miR-20a inhibitor grew slower than cells transfected with NC and blank cells. (B) Transwell migration assays of RPMI-8226 cells were performed after transfection with miR-20a inhibitor, NC and blank cells. Cells transfected with miR-20a inhibitor exhibited significantly lower motility than control-treated cells and blank. (C) The invasion ability of RPMI-8226 cells after transfection in each group. (D) Cell cycle analysis. Data represent means ± standard deviation (n=3). (E) The apoptosis rates of the transfected cells in each group. *P<0.05 compared with the blank and NC groups. miR, microRNA; NC, negative control; MM, multiple myeloma.
PTEN as a target gene of miR-20a
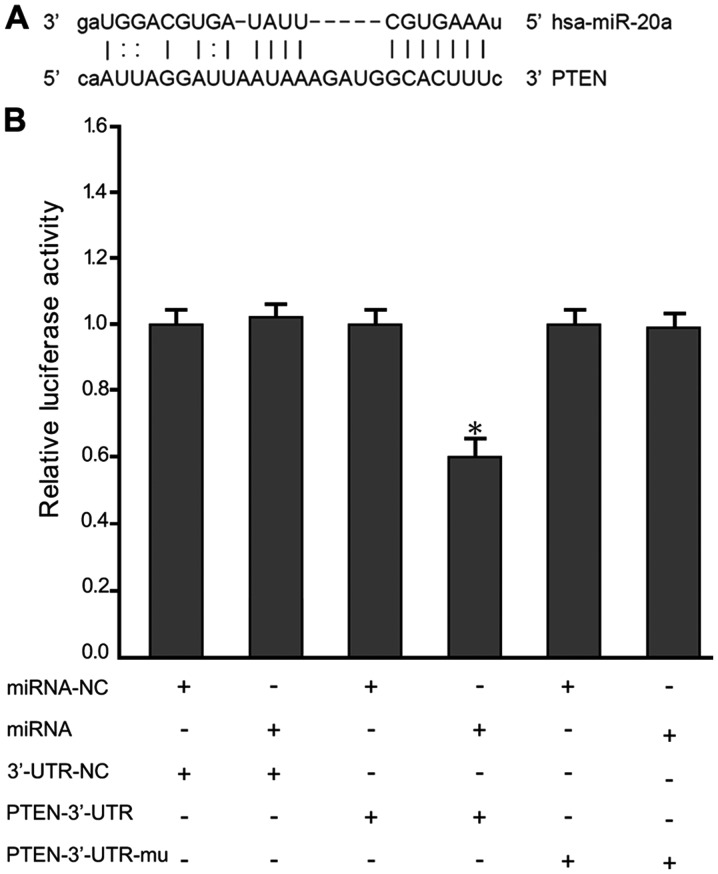
To further explore the mechanisms that miR-20a regulated MM cell growth and metastasis, we identified candidate targets of miR-20a using the TargetScan program. Among the identified genes, we chose to further investigate PTEN (Fig. 3A). To determine whether miR-20a binds to the 3′-UTR of PTEN, we utilized a luciferase reporter vector containing 3′-UTR of PTEN. As expected, miR-20a directly bound to the 3′-UTR and remarkably reduced the vector's luciferase activity. In contrast, cells with mutant PTEN 3′-UTRs displayed much higher luciferase activity (Fig. 3B).
Figure 3.
miR-20a targets PTEN by binding to its 3′-UTR. (A) Image of miR-20a bound to the PTEN 3′-UTR. (B) The relative luciferase activity detected by dual-luciferase reporter gene activity assay. *P<0.05 in miRNA/PTEN-3′-UTR-mu vs. miRNA-NC/3′-UTR-NC, miRNA/3′-UTR-NC, miRNA-NC/PTEN-3′-UTR and miRNA-NC/PTEN-3′-UTR. miR, microRNA; PTEN, phosphatase tensin homolog; NC, negative control; UTR, untranslated region.
PTEN, PI3K, AKT and p-AKT protein expression
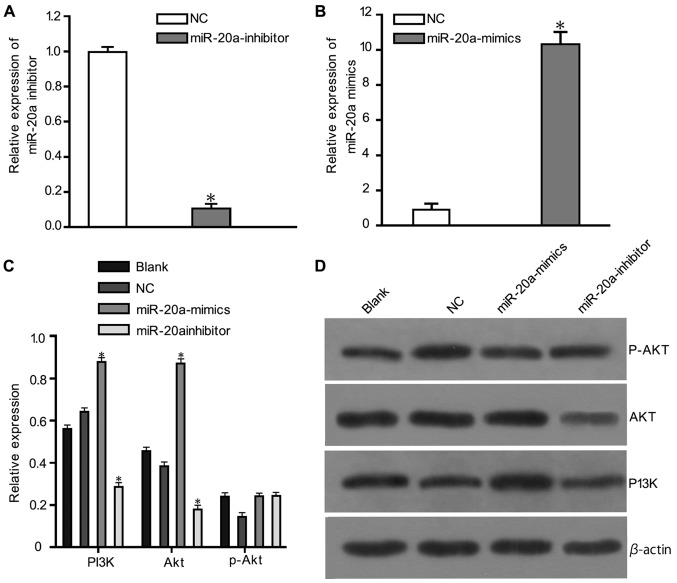
The relative expression of miR-20a and inhibitor was shown in Fig. 4A and B. Western blotting results (Fig. 4) showed that PTEN expression in the miR-20a-mimic group was downregulated compared with the blank and NC groups (P<0.05). p-PI3K and p-Akt expression levels were also markedly downregulated in the miR-20a-mimics group compared with the blank and NC groups (P<0.05). There were no statistically significant differences in the protein expression between the blank and NC groups.
Figure 4.
The relative expression of miR-20a, inhibitor, and protein levels of PTEN, PI3K, Akt and p-Akt detected by western blotting. (A) The relative expression of miR-20a inhibitor. (B) The relative expression of miR-20a. (C) Diagram of the protein bands in each group detected by western blotting. (D) Histogram of the protein expression levels in each group; *P<0.05 compared with the blank and NC groups. NC, negative control; PTEN, phosphatase and tensin homolog; PI3K, phosphoinositide 3-kinase; AKT, protein kinase B; p-Akt, phosphorylated Akt.
Discussion
Increasing studies focusing the molecular biology of cancer have revealed that the PTEN/PI3K/AKT pathway is an important oncogenic pathway that is frequently activated during tumorigenesis, and plays a crucial role in the growth, proliferation, migration, and invasion of malignant cells (26,27). MM is a plasma cell disorder with a relatively high incidence rate among hematological malignancies. Despite significant progress in elucidating the biology of MM and identifying treatment options, the disease remains incurable, highlighting the urgent need for digging novel and effective therapies (28). As miRNAs have important functions in many cancers, therapeutic modulation of miRNAs might be a valuable strategy for cancer study. The overall aim of current study was to explore the roles of miR-20a and the PTEN/PI3K/AKT signaling pathway in the proliferation, migration and invasion of MM cells. To the best of our knowledge, this was the first examination has been reported in the present day.
Initially, we demonstrated that miR-20a was upregulated in plasma from MM patients and MM cells. Notably, miR-20a, a member of the miR-17-92 cluster, has been shown to function as an oncomir in many human cancers. A study performed by Wang et al demonstrated that miR-20a is highly expressed in brainstem gliomas and potentially associated with gliomagenesis (29). Li et al have reported that miR-20a expression was remarkably increased in the breast cancer of patients (30). Moreover, previous research has also shown that high levels of miR-20a are associated with shortened progression-free survival (PFS) (31). In the current study, miR-20a expression in plasma was significantly higher in stage III MM patients than stage I/II patients, suggesting a key role for this miRNA in survival prediction. Considering endogenously expression of miR-20a in stage III MM patients, we chose to utilize RPMI-8226 cells for subsequent loss-of-function studies. Our results showed a marked reduction in RPMI-8226 cell viability after transfection of with a miR-20a inhibitor. The transfected cells also showed inhibited migration, however, apoptosis was promoted. These data suggests that the upregulation of miR-20a in MM is related to the development and progression of the disease.
Recent research has shown that activation of PI3K/AKT reduces the p21Cip1 levels and increases CCND1 expression (32,33), which regulates cell cycle progression through the G1 phase. In addition, activated PI3K can catalyze 3,4-phosphatidylinositol trisphosphate phosphorylation and subsequently activate the protein kinase AKT to promote cell growth and proliferation (26). PTEN, a tumor suppressor gene, was confirmed as a target of miR-20a by a luciferase assay. Notably, one prior study speculated that miR-20a plays an oncogenic role in hepatocellular carcinoma, at least partially by negatively regulating PTEN (27). In the present study, we found that PTEN expression decreased after overexpression of miR-20a, suggesting that PTEN is regulated by miR-20a. These data were further confirmed by a luciferase activity assay. Our results suggest that PTEN is a direct target of miR-20a. The PTEN/PI3K/AKT signaling pathway is known to have important roles in regulating biological processes including cell growth and proliferation, metabolism, and apoptosis, which makes the pathway an attractive candidate for drug discovery (34). Chen et al reported that PTEN and activated AKT are associated with intrahepatic metastasis, tumor grade, and a high proliferation index. Furthermore the PI3K/PTEN/AKT pathway is a diagnostic and prognostic indicator of invasion and metastasis hepatocellular carcinoma as well as a therapeutic target for this cancer (35). Our current correlation analyses of miR-20a, PTEN, PI3K and Akt proteins expression also confirmed the relationships existing among these proteins.
In conclusion, we report that miR-20a regulates the proliferation and migration of MM cells by modulating the PTEN/PI3K/AKT signaling pathway. Meanwhile, this pathway may serve as a novel therapeutic target for MM. Unraveling the relationship between miR-20a and the PTEN/PI3K/Akt signaling pathway may help identify new molecular markers and potential therapeutic targets that will provide a theoretical basis for future studies. The effects of miR-20a on MM cell proliferation, migration and invasion should be confirmed using animal models in future study. Performing such experiment will bring us closer to the goal of developing new genetic and therapeutic strategies for treating MM.
Acknowledgements
International Collaboration Fund from National Science and Technology Committee of China (no. 2011DFA32820). Innovation fund project in Jiangxi Province (no. YC2016-B018). The National Natural Science Fund Project (no. 81460037).
References
- 1.Sonneveld P, Avet-Loiseau H, Lonial S, Usmani S, Siegel D, Anderson KC, Chng WJ, Moreau P, Attal M, Kyle RA, et al. Treatment of multiple myeloma with high-risk cytogenetics: A consensus of the international myeloma working group. Blood. 2016;127:2955–2962. doi: 10.1182/blood-2016-01-631200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdi J, Chen G, Chang H. Erratum: Drug resistance in multiple myeloma: Latest findings and new concepts on molecular mechanisms. Oncotarget. 2015;6:7364. doi: 10.18632/oncotarget.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimura N, Hideshima T, Anderson KC. Novel therapeutic strategies for multiple myeloma. Exp Hematol. 2015;43:732–741. doi: 10.1016/j.exphem.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar SK, Lee JH, Lahuerta JJ, Morgan G, Richardson PG, Crowley J, Haessler J, Feather J, Hoering A, Moreau P, et al. Risk of progression and survival in multiple myeloma relapsing after therapy with IMiDs and bortezomib: A multicenter international myeloma working group study. Leukemia. 2012;26:149–157. doi: 10.1038/leu.2012.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, Sales G, Deliliers GL, Bicciato S, Lombardi L, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–e26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene. 2006;25:6202–6210. doi: 10.1038/sj.onc.1209910. [DOI] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA-cancer connection: The beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 8.Luo X, Gu J, Zhu R, Feng M, Zhu X, Li Y, Fei J. Integrative analysis of differential miRNA and functional study of miR-21 by seed-targeting inhibition in multiple myeloma cells in response to berberine. BMC Syst Biol. 2014;8:82. doi: 10.1186/1752-0509-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seckinger A, Meißner T, Moreaux J, Benes V, Hillengass J, Castoldi M, Zimmermann J, Ho AD, Jauch A, Goldschmidt H, et al. miRNAs in multiple myeloma-a survival relevant complex regulator of gene expression. Oncotarget. 2015;6:39165–39183. doi: 10.18632/oncotarget.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leotta M, Biamonte L, Raimondi L, Ronchetti D, Di Martino MT, Botta C, Leone E, Pitari MR, Neri A, Giordano A, et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells. J Cell Physiol. 2014;229:2106–2116. doi: 10.1002/jcp.24669. [DOI] [PubMed] [Google Scholar]
- 11.Chi J, Ballabio E, Chen XH, Kušec R, Taylor S, Hay D, Tramonti D, Saunders NJ, Littlewood T, Pezzella F, et al. MicroRNA expression in multiple myeloma is associated with genetic subtype, isotype and survival. Biol Direct. 2011;6:23. doi: 10.1186/1745-6150-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esquela-Kerscher A. The lin-4 microRNA: The ultimate micromanager. Cell Cycle. 2014;13:1060–1061. doi: 10.4161/cc.28384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33:1126–1133. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzon R, Marcucci G. Potential of microRNAs for cancer diagnostics, prognostication and therapy. Curr Opin Oncol. 2012;24:655–659. doi: 10.1097/CCO.0b013e328358522c. [DOI] [PubMed] [Google Scholar]
- 15.Yang R, Fu Y, Zeng Y, Xiang M, Yin Y, Li L, Xu H, Zhong J, Zeng X. Serum miR-20a is a promising biomarker for gastric cancer. Biomed Rep. 2017;6:429–434. doi: 10.3892/br.2017.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang G, Chen X, Cai Y, Wang X, Xing C. miR-20a-directed regulation of BID is associated with the TRAIL sensitivity in colorectal cancer. Oncol Rep. 2017;37:571–578. doi: 10.3892/or.2016.5278. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Mao F, Shen T, Luo Q, Ding Z, Qian L, Huang J. Plasma miR-145, miR-20a, miR-21 and miR-223 as novel biomarkers for screening early-stage non-small cell lung cancer. Oncol Lett. 2017;13:669–676. doi: 10.3892/ol.2016.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiong Y, Sun F, Dong P, Watari H, Yue J, Yu MF, Lan CY, Wang Y, Ma ZB. iASPP induces EMT and cisplatin resistance in human cervical cancer through miR-20a-FBXL5/BTG3 signaling. J Exp Clin Cancer Res. 2017;36:48. doi: 10.1186/s13046-017-0520-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng J, Thakur A, Zhang S, Dong Y, Wang X, Yuan R, Zhang K, Guo X. Expressions of miR-181a and miR-20a in RPMI8226 cell line and their potential as biomarkers for multiple myeloma. Tumour Biol. 2015;36:8545–8552. doi: 10.1007/s13277-015-3600-2. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Corrigan-Cummins M, Barber EA, Saleh LM, Zingone A, Ghafoor A, Costello R, Zhang Y, Kurlander RJ, Korde N, et al. Aberrant levels of miRNAs in bone marrow microenvironment and peripheral blood of myeloma patients and disease progression. J Mol Diagn. 2015;17:669–678. doi: 10.1016/j.jmoldx.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo M, Tan X, Mu L, Luo Y, Li R, Deng X, Chen N, Ren M, Li Y, Wang L, et al. MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci Rep. 2017;7:43427. doi: 10.1038/srep43427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu MH, Yang L, Liu XJ, Nie ZY, Luo JM. Targeted suppression of miRNA-21 inhibit K562 cells growth through PTEN-PI3K/AKT signaling pathway. Zhonghua Xue Ye Xue Za Zhi. 2016;37:982–986. doi: 10.3760/cma.j.issn.0253-2727.2016.11.011. (In Chinese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang X, Cheng Y, Li P, Tao J, Deng X, Zhang X, Gu M, Lu Q, Yin C. A lentiviral sponge for miRNA-21 diminishes aerobic glycolysis in bladder cancer T24 cells via the PTEN/PI3K/AKT/mTOR axis. Tumour Biol. 2015;36:383–91. doi: 10.1007/s13277-014-2617-2. [DOI] [PubMed] [Google Scholar]
- 24.Wang F, Li L, Chen Z, Zhu M, Gu Y. MicroRNA-214 acts as a potential oncogene in breast cancer by targeting the PTEN-PI3K/Akt signaling pathway. Int J Mol Med. 2016;37:1421–1428. doi: 10.3892/ijmm.2016.2518. [DOI] [PubMed] [Google Scholar]
- 25.Anderson KC. Progress and paradigms in multiple myeloma. Clin Cancer Res. 2016;22:5419–5427. doi: 10.1158/1078-0432.CCR-16-0625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z, Fang S, Di Y, Ying W, Tan Y, Gu W. Modulation of NF-κB/miR-21/PTEN pathway sensitizes non-small cell lung cancer to cisplatin. PLoS One. 2015;10:e0121547. doi: 10.1371/journal.pone.0121547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, Zheng L, Ding Y, Li Q, Wang R, Liu T, Sun Q, Yang H, Peng S, Wang W, Chen L. MiR-20a induces cell radioresistance by activating the PTEN/PI3K/Akt signaling pathway in hepatocellular carcinoma. Int J Radiat Oncol Biol Phys. 2015;92:1132–1140. doi: 10.1016/j.ijrobp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Bates SE. Multiple myeloma: Multiplying therapies. Clin Cancer Res. 2016;22:5418. doi: 10.1158/1078-0432.CCR-16-0624. [DOI] [PubMed] [Google Scholar]
- 29.Wang X, Zhang H, Zhang A, Han L, Wang K, Liu R, Yang S, Pu P, Shen C, Kang O, Yu C. Upregulation of miR-20a and miR-106b is involved in the acquisition of malignancy of pediatric brainstem gliomas. Oncol Rep. 2012;28:1293–1300. doi: 10.3892/or.2012.1927. [DOI] [PubMed] [Google Scholar]
- 30.Li S, Qiang Q, Shan H, Shi M, Gan G, Ma F, Chen B. miR-20a and miR-20b negatively regulate autophagy by targeting RB1CC1/FIP200 in breast cancer cells. Life Sci. 2016;147:143–152. doi: 10.1016/j.lfs.2016.01.044. [DOI] [PubMed] [Google Scholar]
- 31.Gao X, Zhang R, Qu X, Zhao M, Zhang S, Wu H, Jianyong L, Chen L. MiR-15a, miR-16-1 and miR-17-92 cluster expression are linked to poor prognosis in multiple myeloma. Leuk Res. 2012;36:1505–1509. doi: 10.1016/j.leukres.2012.08.021. [DOI] [PubMed] [Google Scholar]
- 32.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 33.Roy SK, Srivastava RK, Shankar S. Inhibition of PI3K/AKT and MAPK/ERK pathways causes activation of FOXO transcription factor, leading to cell cycle arrest and apoptosis in pancreatic cancer. J Mol Signal. 2010;5:10. doi: 10.1186/1750-2187-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8:187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 35.Chen JS, Wang Q, Fu XH, Huang XH, Chen XL, Cao LQ, Chen LZ, Tan HX, Li W, Bi J, Zhang LJ. Involvement of PI3K/PTEN/AKT/mTOR pathway in invasion and metastasis in hepatocellular carcinoma: Association with MMP-9. Hepatol Res. 2009;39:177–186. doi: 10.1111/j.1872-034X.2008.00449.x. [DOI] [PubMed] [Google Scholar]