Abstract
The interactions of self-complementary oligonucleotides with a group of metal-mediated DNA-binding drugs, including chromomycin A3, mithramycin and the novel compound UK-1, were examined via electrospray ionization quadrupole ion trap mass spectrometry. Both chromomycin and mithramycin were shown to bind preferentially to GC-rich oligonucleotide duplexes in a 2:1 drug:metal ratio, while UK-1 was shown to bind in a 1:1 drug:metal stoichiometric ratio without a strong sequence preference. These trends were observed in the presence of Co2+, Ni2+ and Zn2+, with the exception that chromomycin–Zn2+ complexes were not readily observed. The binding stoichiometries as well as the sequence specificities are in agreement with literature reports for solution studies. Binding selectivities and stabilities of the complexes were also probed using electrospray ionization mass spectrometry. Both of the GC-rich oligomers 5′-GCGCGC-3′ and 5′-GCGCATGCGC-3′ exhibited a binding preference for chromomycin over mithramycin in the presence of Co2+ and Ni2+. Energy-variable collisionally activated dissociation of the complexes was employed to determine the stabilities of the complexes. The relative metal-dependent binding energies were Ni2+ > Zn2+ > Co2+ for UK-1–oligomer complexes and Ni2+ > Co2+ for both mithramycin and chromomycin complexes.
INTRODUCTION
The mechanism of action of many antitumor and antibiotic pharmaceutical agents involves the formation of non-covalent complexes with strands of DNA as a prelude to DNA cleavage or inhibition of DNA-associated enzymes (1). It is crucial from a drug design point of view to accurately assess the mode of binding (via the minor groove, intercalation or mixed mode binding), the sequence specificity (for either AT- or GC-rich regions of DNA) and the stoichiometry of the bound complex in order to fully understand how a given drug works and to design more effective or more specific drug candidates. Complicating matters, some of these DNA-binding drugs exert their effects only in the presence of a metal cation, such as Mg2+ or Zn2+. Thus issues such as the identity and size (ionic radius) of the metal ion, electronic effects and the stoichiometry of the drug–metal ion complex also become important for elucidating structure–function relationships and for directing the design of future drug candidates.
Electrospray ionization mass spectrometry (ESI-MS) has been shown to be effective for the study of non-covalently bound complexes (2,3). Because electrospray ionization is a soft ionization technique, strongly bound non-covalent complexes can survive the electrospray process and be transported intact from solution to the gas phase. Recently, several reports have described the application of ESI-MS for evaluating the non-covalent binding of drugs to DNA (4–16), RNA (17–20) and peptides (21–25). These studies have primarily involved well-studied model binding drugs, including the classic minor groove binders distamycin and Hoechst 33258 and intercalators such as actinomycin. Information regarding binding stoichiometries, relative binding affinities and stabilities of the non-covalent complexes were rapidly and easily obtained from the mass spectra, making ESI-MS a promising method for the evaluation of these types of complexes.
For example, Gabelica et al. (9) observed the preferential formation of 1:2 duplex:distamycin A complexes from an equimolar solution of a dodecamer oligonucleotide and distamycin A. However, exclusively 1:1 complexes were observed with netropsin and berenil and the same dodecamer. These binding stoichiometries parallel those found in solution. In addition, several groups have performed competition experiments, either adding two drugs to a solution containing one oligonucleotide or adding two oligonucleotides to a solution containing one drug. The relative intensities of the complexes in the mass spectra were then used to evaluate the relative binding affinities of the drugs (8–10). Collisionally activated dissociation of oligonucleotide duplex–drug complexes has been used to estimate the stabilities of the complexes in the gas phase (11–12). Energy-resolved dissociation curves of a given duplex or duplex–drug complex, analogous to DNA melting curves in solution, have been constructed. Complexes that require more collision energy to dissociate are considered more stable, similar to DNA duplexes in solution that have higher melting temperatures.
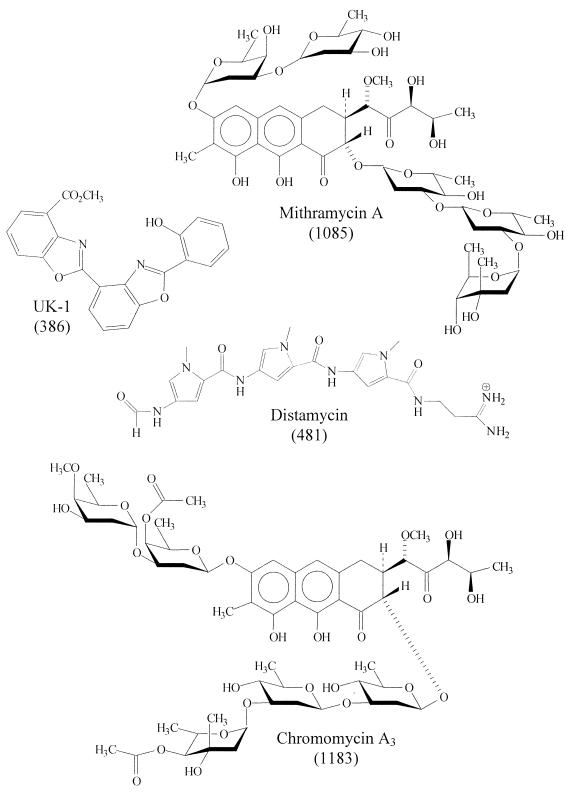
The goal of the present study was to evaluate for the first time the metal ion-dependent DNA binding of a group of antitumor agents via ESI-MS. It was recently reported that the novel cytotoxic natural product UK-1, a metabolite produced by Streptomyces sp. 517-02, binds Mg2+ and Zn2+ and binds to DNA in a Mg2+ ion-dependent fashion (26). Similarly, the antitumor agents mithramycin A and chromomycin A3 are known to interact with DNA in the presence of metal ions (27–33). It has been reported that both chromomycin and mithramycin bind preferentially to GC-rich sites in DNA and that a divalent metal cation of radius <0.85 Å (i.e. especially Mg2+, Zn2+ and Ni2+) is necessary for binding (27,32). Both chromomycin and mithramycin reportedly bind as 2:1 drug:metal complexes to the minor groove of DNA, with association constants of ∼106–107 M–1 (27–33). The binding abilities of these three drugs, shown in Figure 1, to self-complementary oligodeoxynucleotides, were assessed in the presence and absence of different metal cations. In addition, complexation of the classic minor groove binder distamycin was also studied for comparison, as it does not require metal ions for binding. As shown in this report, ESI-MS offers a rapid way to screen the metal-binding abilities of drugs and to further elucidate binding of the metal–drug complexes to DNA.
Figure 1.
Structures of DNA-binding drugs (molecular weight).
MATERIALS AND METHODS
Self-complementary oligodeoxynucleotides were obtained from TriLink BioTechnologies (San Diego, CA), custom synthesized on the 1.0 µmol scale with purification by HPLC in an ammonium acetate buffer. The DNA-binding drugs distamycin, chromomycin A3 and mithramycin A were purchased from Sigma Chemical Co. (St Louis, MO), while UK-1 was prepared as previously reported (34). The metal salts CoBr2, NiBr2 and ZnBr2 were purchased from Aldrich Chemical (Milwaukee, WI) and Mg(NO3)2·6H2O came from EM Science (Gibbstown, NJ). All compounds were used without further purification.
Stock oligonucleotide solutions (445 µM single strand) were prepared in 1.0 M ammonium acetate at nominally pH 6.5. The oligonucleotide solutions were annealed (if necessary) by heating to 90°C and cooling slowly to room temperature over 2–3 h (9,10). The oligonucleotides were then diluted to 25 µM with water and methanol, with the resulting electrospray solution being 75% 75 mM ammonium acetate in water and 25% methanol. However, solutions containing UK-1 were ultimately 67% methanol due to the low solubility of UK-1 in water. Oligonucleotide:drug:metal mixtures were generally prepared for analysis in 1:1:1 ratios, except for specific experiments in which the ratio of drug to metal was varied to assess this factor. When mixtures of the drugs and metals with the oligonucleotides were prepared, mixtures of the drugs and metals were prepared first and allowed to mix for ∼5 min. The oligonucleotide solution was then added, the final mixture allowed to mix for ∼5 min and the resulting three component mixture was analyzed by mass spectrometry.
All experiments were performed with a Finnigan LCQ Duo mass spectrometer equipped with an electrospray interface (ThermoFinnigan, San Jose, CA). The negative ion electrospray mode was used for all samples, with an electrospray voltage of 4.0 kV. Solutions containing the oligomers with or without the drugs and metals were delivered to the interface via a Harvard syringe pump (Holliston, MA) at 3.0 µl/min. The heated capillary was kept at 85°C to minimize premature dissociation of the double-stranded oligonucleotide complexes (9,10). The base pressure of the ion trap was ∼1 × 10–5 torr. Collisionally activated dissociation experiments were performed on selected complexes. The precursor ion was isolated and spectra were obtained as a function of increasing collision energy applied to the ion trap. The energy used is reported as a percentage of 5 V0-p.
RESULTS AND DISCUSSION
A variety of experiments were undertaken to evaluate the ability to detect complexes and differentiate non-specific complex formation by ESI-MS. Several oligomers, varying in both size and sequence, an array of metal ions, varying in both size and electronic nature, and four different drugs, both metal-dependent and non-metal-dependent DNA binders, were examined with the aim of providing evidence for binding selectivity in the ESI-MS experiments. Moreover, solution mixtures in which either the metal, the drug or the oligomer was omitted were evaluated to further confirm the importance of each constituent on formation and detection of the targeted complexes by ESI-MS. Experiments in which capillary temperature, pH and concentrations of constituents in solution were varied were also undertaken in the same vein, as discussed in the following sections.
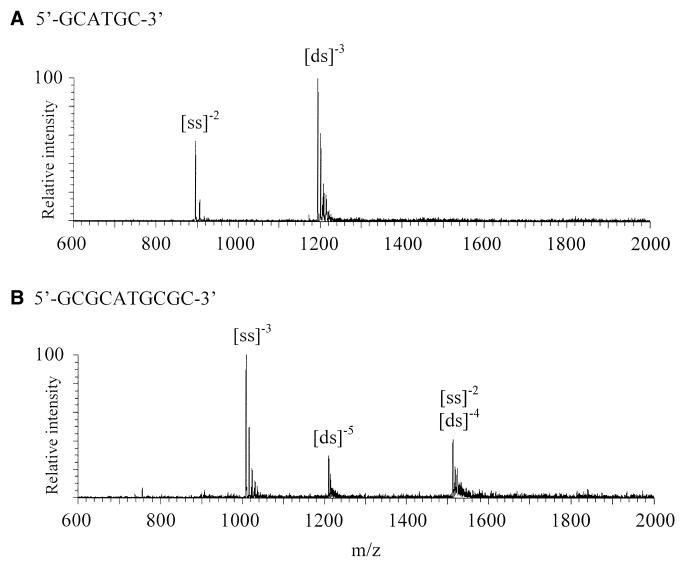
As reported previously, mass spectral analysis of self-complementary oligonucleotides is relatively straightforward and generally yields simple spectra with all ions originating from one species (9,10). Thus, a series of 6mer and 10mer self-complementary oligonucleotides varying in sequence from GC-rich to AT-rich were employed in the present study. These included 5′-GCGCGC-3′, 5′-GCATGC-3′, 5′-GAATTC-3′, 5′-GCGCATGCGC-3′, 5′-GCGAATTCGC-3′ and 5′-GCAAATTTGC-3′. Figure 2 shows example ESI mass spectra obtained from the 6mer 5′-GCATGC-3′ (Fig. 2A) and the 10mer 5′-GCGCATGCGC-3′ (Fig. 2B); both were unannealed. As shown, the spectra are uncluttered and yield only ions from both single-stranded (ss) and double-stranded (ds) oligonucleotides (as multiply deprotonated species) in both cases. Annealing was not necessary for the formation of duplex ions for the GC-rich oligomers, but it aided duplex formation for the more AT-rich oligomers.
Figure 2.
Electrospray spectra of unannealed oligomers (A) 5′-GCATGC-3′ and (B) 5′-GCGCATGCGC-3′.
One drawback to the use of self-complementary oligonucleotides is that the mass-to-charge ratio for double-stranded even charge states is the same as for single-stranded ions with half the charge, thus duplex formation cannot be unequivocally observed (10). For example, the (ss)–2 and (ds)–4 ions of 5′-GCGCATGCGC-3′ shown in Figure 2B both occur at m/z 1513.4. Sometimes it is possible to confirm the presence of a double-stranded complex in an ambiguous ion cluster by examining the sodium adducts, but this is difficult in practice when the mass resolution may be poor or desolvation incomplete. Additionally, fragment ions originating from these species cannot be identified unambiguously. However, odd charge state duplex ions are unambiguous because their mass-to-charge ratios do not overlap with those of the single-stranded ions and as such are of special interest because our primary concern is with the complexation of drugs to duplex (not single-stranded) DNA.
Several other oligonucleotides were examined in this study in order to verify the generality of duplex formation and potential production of complexes incorporating a drug and metal ion. These other oligonucleotides included a random sequence oligomer, 5′-GGCACG-3′, and two complementary oligomers, 5′-GGGGGG-3′ and 5′-CCCCCC-3′. Electrospray of the random sequence oligomer produced primarily doubly deprotonated single-stranded species, however, a small amount of double-stranded oligonucleotide, (ds)–3, was present. While this oligomer is not self-complementary, there is a self-complementary 5′-GC-3′:3′-CG-5′ region near the ends of the oligomer, which may be enough for some duplex formation. It is also possible that the duplex ion is indicative of some non-specific interaction. For 5′-CCCCCC-3′ duplexes were not formed and 5′-GGGGGG-3′ suffered from solubility problems that prevented conclusive investigation.
Drug–metal complexation in the absence of the oligonucleotides was also examined. For example, the mass spectrum obtained in the positive electrospray mode for an equimolar solution of UK-1 and ZnBr2 revealed only one complex, corresponding to [UK1 – H + Zn2+]+ (data not shown). Similar complexes were observed with Ni2+ and Co2+, supporting the behavior observed in solution whereby UK-1 binds divalent cations as a phenolate species (26). The complexation behaviors are different for chromomycin and mithramycin, as both singly and doubly charged drug–metal complexes of the type [drug + Na]+, [drug – H + M2+]+ and [2 × drug + M2+ + 44]2+, where M = Ni or Co, were observed.
Metal-dependent complexation of drugs with oligonucleotides
The metal-dependent complexation of chromomycin and mithramycin, two well-studied DNA-binding drugs, was evaluated prior to experiments with UK-1, the novel cytotoxic natural product. An array of metals, including Ni2+, Co2+, Zn2+, Mg2+, Ca2+ and Mn2+, and the six oligomers listed in Table 1 were used for this facet of the study. The ionic radii of the metals used are (in Å): Ni2+, 0.69; Zn2+, 0.74; Co2+, 0.72; Mg2+, 0.66; Ca2+, 0.99; Mn2+, 0.80 (35). In solution it has been reported that chromomycin binds as a metal-bound dimer (with a divalent cation having a radius of <0.85 Å forming the center of the dimer) preferentially to GC-rich DNA (30). Ca2+ (ionic radius 0.99 Å) is known not to promote DNA binding (32).
Table 1. Complexation of mithramycin and chromomycin to oligomers.
| Oligomer |
Ni2+ |
Co2+ |
Zn2+ |
| 5′-GCGCGC-3′ | M and C | M and C | M |
| 5′-GCATGC-3′ | C | C | None |
| 5′-GAATTC-3′ | None | None | None |
| 5′-GCGCATGCGC-3′ | M and C | M and C | M |
| 5′-GCGAATTCGC-3′ | M and C | M and C | M |
| 5′-GCAAATTTGC-3′ | None | None | None |
M indicates that complexes of the type [ds + 2 × mithramycin + M2+]–4 formed with the 6mers or [ds + 2 × mithramycin + M2+]–5 formed with the 10mers; C indicates that complexes of the type [ds + 2 × chromomycin + M2+]–4 formed with the 6mers or [ds + 2 × chromomycin + M2+]–5 formed with the 10mers; None indicates that no oligomer–drug–metal complexes were observed.
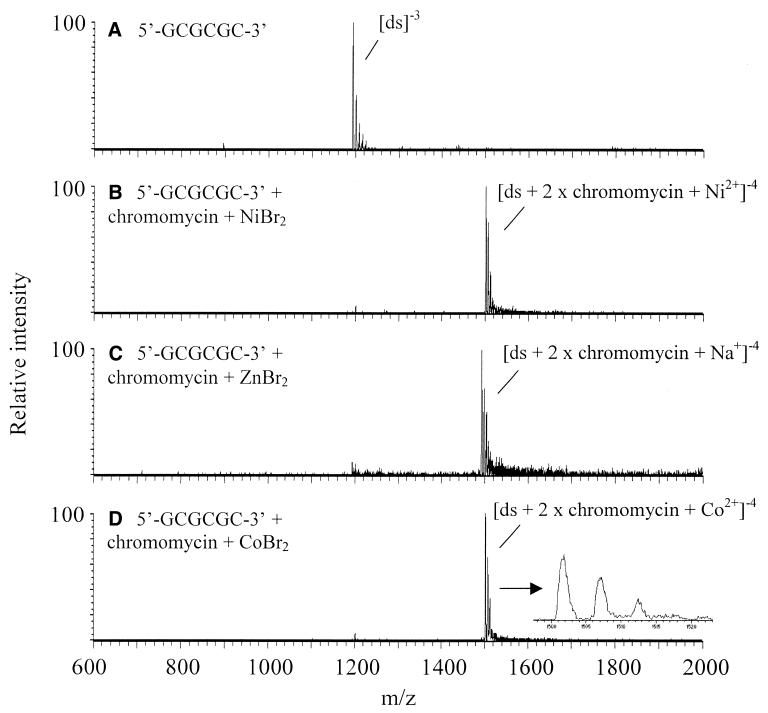
Examples of spectra obtained for chromomycin interacting with 5′-GCGCGC-3′ in the presence of Ni2+, Zn2+ and Co2+ metal ions are shown in Figure 3, and a summary of the ESI-MS complexation results is shown in Table 1. In the case of the Ni2+ and Co2+ solutions, complexes of the type [ds + 2 × chromomycin + metal2+]–4 form from equimolar solutions of all three components (chromomycin, 6mer and metal) for the four most GC-rich oligomers. (Due to the presence of the metal ions, these complexes can be unambiguously identified as double-stranded species, even though they have an even charge state.) It should be noted that the sites of deprotonation in the complex are not readily determined. Thus it is unknown whether none, one or both of the chromomycin molecules is deprotonated in the complex, although it is likely that at least one chromomycin is deprotonated to facilitate binding with the metal ion. It is unclear why the charge state of the double-stranded oligomer changes from –3 to –4 upon complexation with the chromomycin–metal dimer. Again, the site of the additional deprotonation is not known. It is possible that because the chromomycin–metal dimer has a mass that is ∼70% of the mass of the double-stranded oligomer, the resulting drug–metal–oligomer complex is large enough to stabilize another negative charge in the gas phase. Thus the complex with an additional group deprotonated (likely a phosphate group from the oligonucleotide) in a basic solution could survive the electrospray process and be observed in the gas phase preferentially as the –4 charge state instead of –3.
Figure 3.
Spectra of 5′-GCGCGC-3′ (A) and complexes with chromomycin and various metals (B–D). The inset in (D) is a close-up of the (ds + 2 × chromomycin + Co2+)–4 complex at m/z 1501 showing the sodium adduct peaks.
The complexes observed in these spectra with Ni2+ and Co2+ are in good agreement with what is known to occur in solution. The main complex evident for the analogous Zn2+ solution is [ds + 2 × chromomycin + Na+]–4. There is evidence of a [ds + 2 × chromomycin + Zn2+]–4 complex in the spectrum, but it was not unambiguously identified from the sodium adducts associated with the main ion, [ds + 2 × chromomycin + Na+]–4. Thus it appears under these conditions that both cobalt and nickel promote binding to a greater extent than zinc.
Interestingly, the complex [ds + 2 × chromomycin + Na+]–4 also appeared upon ESI of the solution containing chromomycin and 5′-GCGCGC-3′ in the presence of Mg2+, Ca2+ and Mn2+ or in the absence of any added metal. The spectra for all of these solutions look very similar to those shown in Figure 3C, where the complexes are abundant and are the only ones visible. Mg2+ is known to promote binding between chromomycin and DNA in solution (27,32), thus [ds + 2 × chromomycin + Mg2+] was expected to be present in the mass spectra. Mn2+ and Zn2+ have ionic radii of 0.80 and 0.74 Å (34), so they could promote binding as well, but complexes containing these metals were not confirmed in the ESI mass spectra. The ionic radius of Na+ is larger than that typically thought to encourage chromomycin dimer formation (0.97 Å) (34), however, there is clearly significant sodium contamination present in the solutions, as evidenced by the sodium adducts associated with the oligonucleotides. The ubiquitous presence of sodium, especially in the commercially available drugs, likely accounts for these dominant sodium complexes. Despite many attempts to remove sodium from the system (rinsing the transfer lines and all glassware thoroughly, restricting the use of the tubing to one user, flushing the system prior to beginning experiments, etc.), trace amounts were always present. Thus, when the [ds + 2 × chromomycin + Na+]–4 complex emerged as the prominent ion from a given solution, the metal in that solution was considered to be a poor promoter of binding, thus providing a qualitative way to evaluate the impact of different metals on complexation.
Mithramycin and chromomycin have very similar structures (see Fig. 1) and exhibit similar binding behaviors, which is evident in the ESI-MS results obtained for mithramycin with the metals Co2+, Ni2+ and Zn2+ and the oligonucleotides 5′-GCGCGC-3′, 5′-GCATGC-3′, 5′-GAATTC-3′, 5′-GCGCATGCGC-3′, 5′-GCGAATTCGC-3′ and 5′-GCAAATTTGC-3′ (see Table 1). The most notable exception occurred for the Zn2+ solutions, in which binding between mithramycin and three of the oligomers was evident, while Zn2+ did not promote substantial binding for chromomycin. For the Mg2+, Ca2+ and Mn2+ solutions, in addition to solutions containing no metal ions, complexes of the type [ds + 2 × mithramycin + Na+]–4 were observed, analogous to those described above for the chromomycin solutions.
Solutions containing the GC-rich 6mer 5′-GCGCGC-3′ and Ni2+, Zn2+ and Co2+ produced abundant complexes of the type [ds + 2 × mithramycin + metal2+]–4, analogous to what was observed for chromomycin. In contrast, very little binding was observed for mixtures containing either mithramycin or chromomycin with a 6mer incorporating one AT sequence, 5′-GCATGC-3′, and no binding was observed with 5′-GAATTC-3′. Similarly, for the 10mers the binding preferences for both chromomycin and mithramycin were 5′-GCGCATGCGC-3′ > 5′-GCGAATTCGC-3′ > 5′-GCAAATTTGC-3′ with Co2+, Zn2+ and Ni2+, again with Zn2+ producing complexes with mithramycin but not chromomycin. In these cases, complexes of the type [ds + 2 × drug + metal2+]–5 were observed. Measurable complexes were detected for both 5′-GCGCATGCGC-3′ and 5′-GCGAATTCGC-3′, while no complexes were observed for either chromomycin or mithramycin with the AT-rich 10mer 5′-GCAAATTTCG-3′. When the random sequence oligomer 5′-GGCACG-3′ was mixed with chromomycin and NiBr2, an insubstantial amount of the complex [ds + 2 × chromomycin + Ni2+]–4 was observed. Due to the low intensity, further evaluation of this complex was unsuccessful. These results clearly indicate a binding preference for GC-rich duplex oligonucleotides, regardless of the metal ion present, which is in agreement with the solution binding preferences (27–33). Moreover, the results point to a preference for complexation of Ni2+, Co2+ or Na+ , as opposed to Mg2+, Ca2+, Mn2+ or Zn2+.
Concentration effects
Solutions with varying ratios of oligomer, drug and metal were examined using 5′-GCGCGC-3′, chromomycin and NiBr2 to evaluate how changes in the solution conditions affect the abundance, types and stoichiometry of complexes observed by electrospray. Even when the metal was present at one-tenth the drug and oligomer concentrations (while keeping the drug and oligomer at 2.5 × 10–5 M), abundant complexes of the type [ds + 2 × chromomycin + Ni2+]–4 were observed. In addition, the [ds + 2 × chromomycin + Na+]–4 complex was also present, as already noted for cases when no metal was added to the oligomer and drug solution. As the ratio of oligomer:chromomycin:metal was increased from 1:1:0.1 to 1:1:0.5, the [ds + 2 × chromomycin + Na+]–4 complex disappeared and only the oligomer–chromomycin–nickel complex was observed, similar to that noted for the 1:1:1 solutions (as shown in Fig. 3B). When a 10-fold excess of metal was used, the complex was still abundant, but an additional ion corresponding to [ds + 2 × chromomycin + 2 × Ni2+]–4 was observed, at about the same intensity as the complex containing one metal ion (data not shown). It should be noted that the complex with two metal ions, two drug molecules and the duplex oligomer cannot be unequivocally assigned as a duplex, because it has the same m/z as a single-stranded complex containing one drug and one metal ion. However, the sodium adducts suggest there is some double-stranded component. In addition, it is likely that in the presence of such a large excess of Ni2+ in the solution, complexes containing more than one nickel ion could form.
There were fewer differences in the mass spectra as the concentration of chromomycin was varied with respect to the oligomer and NiBr2. Most notable was the absence of any chromomycin-containing complexes when the concentration of chromomycin was one-tenth that of the oligomer, while abundant duplex oligomer complexes, [ds]–3, were visible, as well as a small amount of single-stranded oligomer, [ss]–2. At equimolar concentrations of oligomer, drug and Ni2+ and at 1:2:1 concentrations, abundant [ds + 2 × chromomycin + Ni2+]–4 ions were observed. These studies support the idea that changes made to the solution conditions are reflected in the electrospray mass spectra.
Complexation of UK-1
Like mithramycin and chromomycin, the novel natural product UK-1 has been shown to exhibit metal ion-mediated binding to DNA (26). However, unlike the two previously discussed drugs, it has been shown to bind metal ions with an overall 1:1 ligand:metal ratio and to bind DNA in the presence of Mg2+ with an ∼10-fold lower association constant than the corresponding (mithramycin)2Mg2+ or (chromomycin)2Mg2+ species. In addition, no sequence specificity was reported (26).
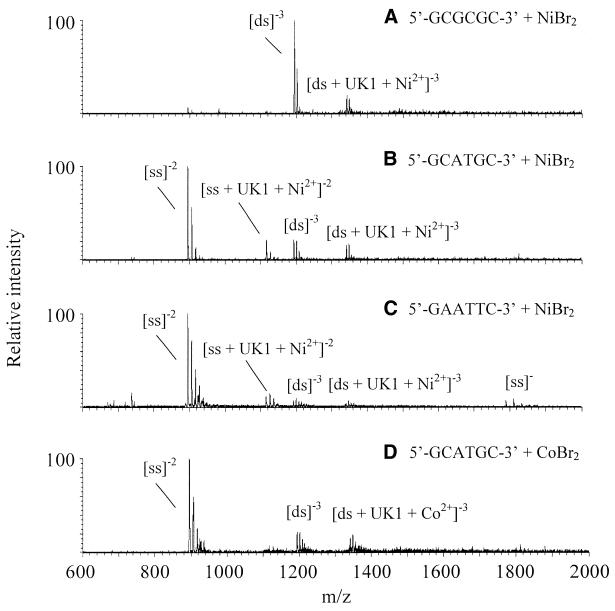
The electrospray mass spectra again parallel what is known about UK-1 in solution. Mass spectra obtained from equimolar solutions of UK-1 and NiBr2 with each of the three 6mers studied, 5′-GCGCGC-3′, 5′-GCATGC-3′ and 5′-GAATTC-3′, are shown in Figure 4A–C and the results are summarized in Table 2. Complexes of the type [ds + UK-1 + Ni2+]–3 are observed for each 6mer. Overall, the ions corresponding to oligonucleotide–drug–metal complexes are much lower in intensity compared to the complexes shown in Figure 3 for chromomycin with 5′-GCGCGC-3′. This is consistent with UK-1 having a 10-fold lower association constant for DNA binding than chromomycin.
Figure 4.
Electrospray spectra of equimolar solutions of oligomer with UK-1 and transition metal salts.
Table 2. Complexation of UK-1 to oligomers.
| Oligomer |
Ni2+ |
Co2+ |
Zn2+ |
| 5′-GCGCGC-3′ | U | U | U |
| 5′-GCATGC-3′ | U | U | U |
| 5′-GAATTC-3′ | U | U | U |
U indicates that complexes of the type [ds + UK-1 + M2+]–3 formed with the 6mers.
The spectra for 5′-GCATGC-3′ and 5′-GAATTC-3′ mixed with UK-1 are dominated by ions corresponding to the uncomplexed single-stranded oligomer in the –2 charge state. The dominance of the single-stranded ion for 5′-GCATGC-3′ in ionspray mass spectrometry has previously been observed and attributed to the gas phase instability of the duplex, as the duplex was shown to be stable in solution (7). The relatively short chain length and the inclusion of an AT sequence in the oligomer (which forms fewer hydrogen bonds in a duplex than a corresponding GC sequence) contribute to the relative instability. The same issues also affect 5′-GAATTC-3′. However, signals corresponding to (ds)–3 and complexed [ds + UK-1 + M2+]–3 are unequivocally observed for both 5′-GCATGC-3′ and 5′-GAATTC-3′, but are of much smaller intensity than [ss]–2. Because [ds + UK-1 + M2+]–3 complexes were observed for all three oligomers examined over a range of GC- and AT-rich sequences (in contrast to chromomycin and mithramycin, where no complexes were observed with oligomers containing primarily AT base pairs), no sequence specificity is indicated. Due to the disparity between the abundances of single-stranded versus double-stranded oligomers in the gas phase compared to solution, no assignments of binding preferences were made based on the relative intensities of the complexes observed. Moreover, a variety of solutions containing the longer duplexes and UK-1 were also examined by ESI-MS. Complexes containing the duplexes and UK-1 could not be readily identified, suggesting that these complexes are less stable or that UK-1 is disrupting some of the duplex interactions.
Interestingly, nickel appears to promote binding to ssDNA. The mass spectra recorded for both 5′-GCATGC-3′ and 5′-GAATTC-3′ with nickel and UK-1 show evidence of [ss + UK-1 + Ni2+]–2 ions (Fig. 4B and C, respectively). The analogous spectrum for 5′-GCATGC-3′ and Co2+, shown in Figure 4D, reveals only negligible single-stranded–drug–metal complexes. Note that for this metal-binding drug, complexes of the type [ds + UK-1 + Na+]–3, similar to those observed in the mithramycin and chromomycin experiments, were not observed upon analysis of Mg2+/oligomer or Ca2+/oligomer solutions or in the absence of any added metal. In fact, in the absence of any metal or with the addition of Mg2+, no complexes involving the oligomers and UK-1 were detected.
Complexation of distamycin
In order to examine the influence of a metal on the DNA binding of a non-metal-mediated binder, the classic minor groove binder distamycin was examined. An equimolar solution of 5′-GAATTC-3′ and distamycin produced complexes in which distamycin was bound to both single-stranded and double-stranded AT-rich DNA (data not shown). The addition of NiBr2 in a 1:1:1 ratio with the oligomer and drug produced no noticeable effect on the binding of distamycin and no metal-containing complexes were detected. Thus, non-specific binding of metal to drug or metal to oligonucleotide was not observed.
Temperature and pH effects
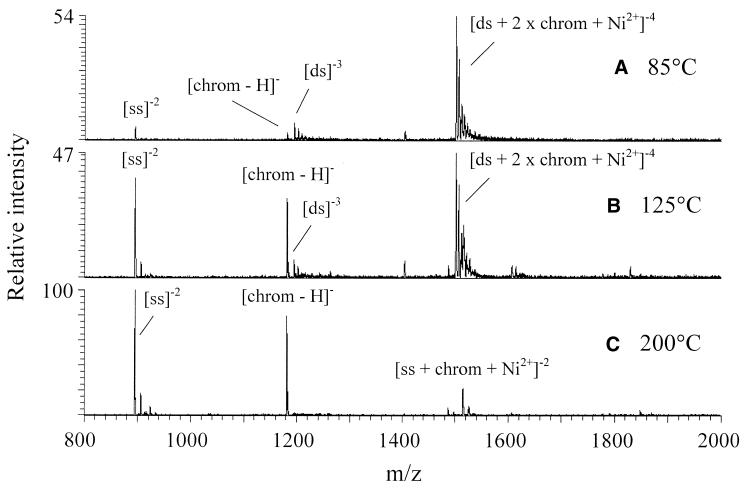
The effect of the electrospray process, specifically transport of the ions through the heated capillary for desolvation, on the structure of these non-covalent complexes was further evaluated. It has been shown that having the heated capillary temperature too high has a deleterious effect on the formation of non-covalent complexes (10–11). The effect of increasing the heated capillary temperature on products resulting from an equimolar solution of 5′-GCGCGC-3′, chromomycin and Ni2+ is shown in Figure 5. At 85°C, the [ds + 2 × chromomycin + Ni2+]–4 complex is the dominant ion (Fig. 5A). At 125°C, the ion corresponding to the complex has decreased and is about equal in intensity to ions corresponding to the single-stranded oligomer, [ss]–2, and deprotonated chromomycin (Fig. 5B). At 200°C, no duplex complexes are visible (Fig. 5C). These spectra are consistent with the dissociation of a non-covalent complex, as increasing the temperature does not have a destructive effect on either the chromomycin ion or the single-stranded oligomer ion.
Figure 5.
Effect of heated capillary temperature on the spectra of an equimolar solution of 5′-GCGCGC-3′, chromomycin and NiBr2.
The influence of the pH of the solution was also evaluated. The solutions previously discussed were made slightly basic with the addition of ammonium acetate in order to promote deprotonation of the drugs and subsequent binding of the deprotonated drugs to the positively charged metal ions. In addition, a slightly basic solution encourages deprotonation of the oligomers and enhances detection by negative electrospray. When the pH of the solution was changed to nominally ∼5 with the addition of a small amount of acetic acid, no complexes were observed and the signal intensity and spectral quality were greatly diminished.
Binding selectivities
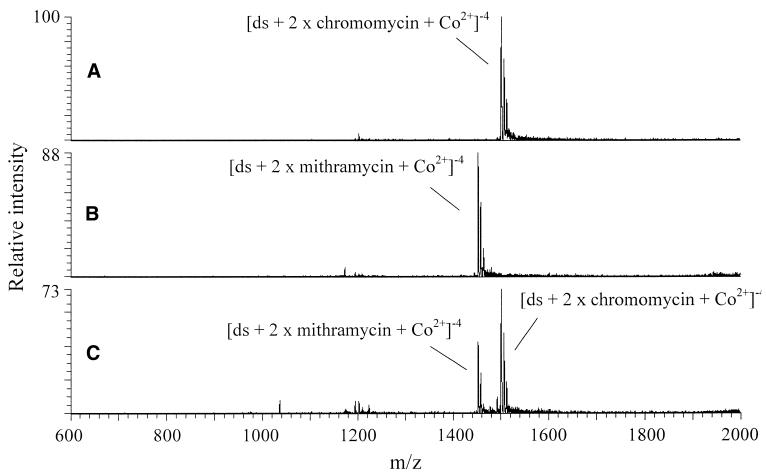
Competition experiments were undertaken with chromomycin, mithramycin and the two oligomers to which they bind the best, 5′-GCGCGC-3′ and 5′-GCGCATGCGC-3′, in order to determine the drug binding selectivity of the oligomers. Figure 6 shows the resulting mass spectra for the competition experiment with 5′-GCGCGC-3′ in the presence of Co2+. Figure 6A and B shows the spectra for individual equimolar solutions of the oligomer, CoBr2 and either chromomycin or mithramycin, respectively. As shown, the relative intensities of the two [ds + 2 × drug + Co2+]–4 complexes are within 10%. However, in the 2:1:1:2 solution of oligomer:chromomycin:mithramycin:CoBr2 shown in Figure 6C, the chromomycin complex is ∼60% of the signal, while the mithramycin complex is ∼40%. As both of these antibiotics are similar in size, structure and binding mode, minimal differences in ionization efficiencies are expected. Thus the difference in mass spectral intensities suggests that the 5′-GCGCGC-3′ oligomer has a binding preference for chromomycin over mithramycin. Similar results were obtained for Ni2+ experiments, suggesting that the structures of the complexes are similar regardless of the identity of the metal ion. The results for the GC-rich 10mer were also similar, showing a binding preference for chromomycin over mithramycin for both Co2+ and Ni2+. As binding likely involves the GC-rich sites, it is reasonable that binding is similar for these oligomers and that they show the same preference for chromomycin over mithramycin.
Figure 6.
Binding selectivity of 5′-GCGCGC-3′ for chromomycin and mithramycin with CoBr2. (A) 5′-GCGCGC-3′ + chromomycin + CoBr2, 1:1:1. (B) 5′-GCGCGC-3′ + mithramycin + CoBr2, 1:1:1. (C) 5′-GCGCGC-3′ + chromomycin + mithramycin + CoBr2, 2:1:1:2.
Competition experiments involving an oligomer, one drug and two different metal ions were difficult to undertake directly, due to the limited resolution of the LCQ and the small mass range separating the metal ions of interest. One such experiment, using 5′-GCGCGC-3′, mithramycin, Ni2+ and Zn2+, was attempted using the high resolution mode of the ion trap (ZoomScan). This mode was necessary to distinguish the [ds + 2 × mithramycin + M2+]–4 ions containing Ni2+ (m/z 1452.2) from Zn2+ (m/z 1453.6). However, due to the loss in sensitivity that accompanies use of the ZoomScan mode, the results were inconclusive. Competition experiments could not be undertaken with two different oligomers with the same mass (or masses within 1–2 a.m.u. on multiply charged complexes) for the same reason of limited resolution.
Stabilities of complexes
Energy-variable collisionally activated dissociation curves were obtained for some of the drug–metal–oligomer complexes to evaluate their relative stabilities. In this method, a complex is isolated and subjected to collisional activation in which the collision energy is varied to change the energy deposition. Following the procedure described by Wan et al. (11), E values were obtained, defined as the collision energy required to dissociate half of the precursor complexes (analogous to the melting temperature, Tm, of DNA in solution). Complexes with higher E values are considered more stable than complexes with lower E values. As Wan et al. (11) and Gabelica et al. (12) reported previously, in order to reflect the differences due to the stabilities of the complexes, only species with the same charge state should be compared to each other. In the gas phase when two identical complexes of different charge states are compared, dissociation of the one with the higher charge state results in a lower E value, indicating that the complex is less stable. With no solvent to stabilize the charges, this is expected due to the greater magnitude of Coulombic repulsions for the complexes with higher charge states. Thus, in order to compare relative stabilities attributed to the nature of the complexes themselves, only complexes with the same charge state were compared to each other.
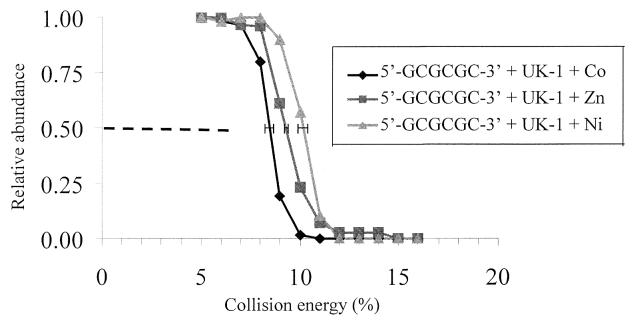
An example of the resulting dissociation curves is shown for the [ds + UK-1 + metal2+]–3 complexes incorporating 5′-GCGCGC-3′ in Figure 7. E values of 10.1 ± 0.2% (505 mV), 9.3 ± 0.1% (465 mV) and 8.4 ± 0.2% (420 mV) were observed for the nickel, zinc and cobalt complexes, respectively. This indicates that the stabilities of complexes formed with UK-1 and 5′-GCGCGC-3′ are in the order nickel > zinc > cobalt. The same trend was observed for two other oligomers, 5′-GCATGC-3′ and 5′-GCGCATGCGC-3′. As the ionic radii of the metal ions are roughly the same (∼0.7 Å), this may point to a metal-dependent electronic effect on the overall stabilities of the complexes. Similar results were obtained for [ds + 2 × chromomycin + metal2+]–3 complexes with 5′-GCGCGC-3′ (data not shown), where the E values for Co2+ and Ni2+ were 9.7 ± 0.2% (485 mV) and 10.6 ± 0.2% (530 mV), respectively. The Zn2+ complexes could not be evaluated for chromomycin because a complex could not be unequivocally identified as Zn2+-containing. The results for the mithramycin complexes were comparable to those obtained for the chromomycin complexes, except that the E values for the mithramycin were more compressed and thus less differentiated than those for the chromomycin complexes. The E values for the mithramycin complexes were 9.8 ± 0.2% for Co2+, 10.3 ± 0.2% for Zn2+ and 10.4 ± 0.2% for Ni2+.
Figure 7.
Collisionally activated dissociation of 5′-GCGCGC-3′ complexes: [ds + UK-1 + M2+]–3. Each curve represents the average of three trials. The error bars at relative abundance = 0.50 show the standard deviations for three independent determinations of E for each complex.
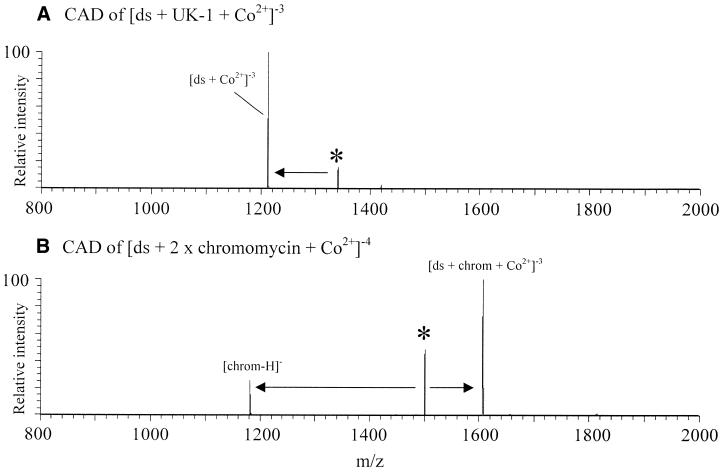
For the energy-resolved collisionally activated dissociation experiments described above, dissociation of the (ds + UK-1 + metal2+)–3 complexes resulted in loss of the neutral UK-1 molecule, with the double-stranded oligomer retaining the metal ion (Fig. 8A). Dissociation of [ds + 2 × (chromomycin or mithramycin) + metal2+]–4 complexes resulted in loss of (chromomycin)– or (mithramycin)– (deprotonated drug), with the double-stranded oligomer retaining one drug molecule and the metal ion and a net charge of –3 (Fig. 8B). These dissociation patterns are similar to those found by Gabelica et al. (9) for the dissociation of the minor groove binders Hoechst 33258 and netropsin from a non-self-complementary duplex of 5′-GGGGATATGGGG-3′ and 5′-CCCCATATCCCC-3′. They found the –5 charge state duplex–Hoechst 33258 complex dissociated ‘into the duplex and the single strands, with release of the neutral drug’ (9). The analogous complex with netropsin dissociated at the same collision energy into the single strands with netropsin remaining attached to the G-rich strand. No covalent bond cleavage was reported for these complexes.
Figure 8.
Collisionally activated dissociation of complexes of 5′-GCGCGC-3′ and CoBr2 with either (A) UK-1 or (B) chromomycin.
These dissociation patterns are similar to those found by Wan et al. (11) for the ruthenium-containing drug Ru(II)12S4dppz interacting with duplex 5′-ATATAT-3′ (11). In that case, the drug–duplex complex (–3 charge state) dissociated into two single-stranded ions of –2 and –1 charge state with loss of the drug compound. This was interpreted as indicating that the Ru2+-containing drug does not bind in the minor groove, as their analysis of the dissociation of duplex oligomer complexes containing classic minor groove binders (including distamycin and Hoechst 33258) showed that the primary dissociation pattern was preferential cleavage of covalent bonds (11). Given these conflicting results reported by Gabelica et al. (9) and Wan et al. (11) concerning the dissociation patterns of classic minor groove binders, it is difficult to comment at this time on what the dissociation patterns of the metal ion-mediated DNA-binding drug complexes indicate about their modes of binding, i.e. whether it can be determined if these ligands are minor groove binders based on their dissociation patterns. Further investigation is required.
CONCLUSIONS
ESI-MS has been shown to be useful for the analysis of metal ion-mediated drug–DNA binding, especially for the determination of binding stoichiometries. The promise of this method, shown by others for mixtures of well-studied drugs and oligomers, has been extended here to multi-component mixtures of drugs with more specific requirements for binding (i.e. the presence of a particular metal ion) and more complex binding modes (not purely minor groove binding or intercalation). Although sodium contamination limited the ability to examine the binding interactions of some metals with known biological activity (i.e. Mg2+), specific, non-covalent binding mediated by other metal ions was observed by this method, and the results were in agreement with the solution binding behavior previously reported for these drugs. In addition, ESI-MS offers significant advantages over traditional solution techniques such as visible and fluorescence spectrophotometry and circular dichroism by enabling absolute binding stoichiometries (through the analysis of bound complex molecular weights) to be determined directly. The minimal sample consumption should also be an advantage relative to other conventional methods for studying DNA–drug complexation.
At present, this method cannot establish the mode of binding nor the binding location of the DNA-binding drugs. It is clear from the current conflicting reports in the literature that further study is required before the method may be reliably extended towards that goal. Additional comparisons of ESI-MS results to those obtained by conventional methods will reveal whether ESI-MS can be used to resolve these questions.
The results obtained by the ESI-MS method show that both chromomycin and mithramycin bind as metal ion-bound dimers preferentially to GC-rich oligomers, in agreement with reported solution binding behaviors. UK-1 was shown to bind in a 1:1 drug:metal ratio to oligomers with no dramatic sequence specificity observed. The intensities of the UK-1 complexes were much lower than those of either the chromomycin or mithramycin complexes, suggesting that UK-1 has a lower association constant, in agreement with what is reported in the literature. Additionally, differences in binding behaviors and stabilities based on metal ion composition were able to be rapidly assessed. Thus, this method may prove useful for the initial screening of novel metal ion-mediated DNA-binding drugs to select the drug/metal combination that exhibits the desired binding behavior.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by the Welch Foundation (F-1155) and the National Science Foundation (CHE-9820755).
References
- 1.Chabrer B.A., Allegra,C.J., Curt,C.A. and Calabresi,P. (1996) Chemotherapy of Neoplastic Diseases. In Hardman,J.G., Limbird,L.E., Molinoff,P.B. and Ruddon,R.W. (eds), Goodman and Gilman’s The Pharmacological Basis of Therapeutics, 9th Edn. McGraw-Hill, New York, NY, pp. 1233–1287.
- 2.Cole R.B. (ed.) (1997) Electrospray Ionization Mass Spectrometry: Fundamentals, Instrumentation & Applications. John Wiley & Sons, New York, NY.
- 3.Schalley C.A. (2000) Supramolecular chemistry goes gas phase: the mass spectrometric examination of noncovalent interactions in host-guest chemistry and molecular recognition. Int. J. Mass Spectrom., 194, 11–39. [Google Scholar]
- 4.Gale D.C., Goodlett,D.R., Light-Wahl,K.J. and Smith,R.D. (1994) Observation of duplex DNA-drug noncovalent complexes by electrospray ionization mass spectrometry. J. Am. Chem. Soc., 116, 6027–6028. [DOI] [PubMed] [Google Scholar]
- 5.Hsieh Y.L., Li,Y.-T., Henion,J.D. and Ganem,B. (1994) Studies of non-covalent interactions of actinomycin D with single-stranded oligodeoxynucleotides by ion spray mass spectrometry and tandem mass spectrometry. Biol. Mass Spectrom., 23, 272–276. [DOI] [PubMed] [Google Scholar]
- 6.Gale D.C. and Smith,R.D. (1995) Characterization of noncovalent complexes formed between minor groove binding molecules and duplex DNA by electrospray ionization-mass spectrometry. J. Am. Soc. Mass Spectrom., 6, 1154–1164. [DOI] [PubMed] [Google Scholar]
- 7.Triolo A., Arcamone,F.M., Raffaelli,A. and Salvadori,P. (1997) Non-covalent complexes between DNA-binding drugs and double-stranded deoxyoligonucleotides: a study by ionspray mass spectrometry. J. Mass Spectrom., 32, 1186–1194. [DOI] [PubMed] [Google Scholar]
- 8.Kapur A., Beck,J.L. and Sheil,M.M. (1999) Observation of daunomycin and nogalamycin complexes with duplex DNA using electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom., 13, 2489–2497. [DOI] [PubMed] [Google Scholar]
- 9.Gabelica V., De Pauw,E. and Rosu,F. (1999) Interaction between antitumor drugs and a double-stranded oligonucleotide studied by electrospray ionization mass spectrometry. J. Mass Spectrom., 34, 1328–1337. [DOI] [PubMed] [Google Scholar]
- 10.Wan K.X., Shibue,T. and Gross,M.L. (2000) Non-covalent complexes between DNA-binding drugs and double-stranded oligodeoxynucleotides: a study by ESI ion-trap mass spectrometry. J. Am. Chem. Soc., 122, 300–307. [DOI] [PubMed] [Google Scholar]
- 11.Wan K.X., Gross,M.L. and Shibue,T. (2000) Gas-phase stability of double-stranded oligodeoxynucleotides and their noncovalent complexes with DNA-binding drugs as revealed by collisional activation in an ion trap. J. Am. Soc. Mass Spectrom., 11, 450–457. [DOI] [PubMed] [Google Scholar]
- 12.Gabelica V., Rosu,F., Houssier,C. and De Pauw,E. (2000) Gas phase thermal denaturation of an oligonucleotide duplex and its complexes with minor groove binders. Rapid Commun. Mass Spectrom., 14, 464–467. [DOI] [PubMed] [Google Scholar]
- 13.Iannitti-Tito P., Weimann,A., Wickham,G. and Sheil,M.M. (2000) Structural analysis of drug-DNA adducts by tandem mass spectrometry. Analyst, 125, 627–634. [DOI] [PubMed] [Google Scholar]
- 14.Carte N., Legendre,F., Leize,E., Potier,N., Reeder,F., Chottard,J.-C. and Van Dorsselaer,A. (2000) Determination by electrospray mass spectrometry of the outersphere association constants of DNA/platinum complexes using 20-mer oligonucleotides and ((Pt(NH3)4)2+, 2 Cl–) or ((Pt(py)4)2+, 2 Cl–). Anal. Biochem., 284, 77–86. [DOI] [PubMed] [Google Scholar]
- 15.Harsch A., Marzilli,L.A., Bunt,R.C., Stubbe,J. and Vouros,P. (2000) Accurate and rapid modeling of iron–bleomycin-induced DNA damage using tethered duplex oligonucleotides and electrospray ionization ion trap mass spectrometric analysis. Nucleic Acids Res., 28, 1978–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffey R.H., Greig,M.J., An,H., Sasmor,H. and Manalili,S. (1999) Targeted site-specific gas-phase cleavage of oligonucleotides. Application in mass spectrometry-based identification of ligand binding sites. J. Am. Chem. Soc., 121, 474–475. [Google Scholar]
- 17.Griffey R.H., Hofstadler,S.A., Sannes-Lowery,K.A., Ecker,D.J. and Crooke,S.T. (1999) Determinants of aminoglycoside-binding specificity for rRNA by using mass spectrometry. Proc. Natl Acad. Sci. USA, 96, 10129–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sannes-Lowery K.A., Mei,H.-Y. and Loo,J.A. (1999) Studying aminoglycoside antibiotic binding to HIV-1 TAR RNA by electrospray ionization mass spectrometry. Int. J. Mass Spectrom., 193, 115–122. [Google Scholar]
- 19.Sannes-Lowery K.A., Drader,J.J., Griffey,R.H. and Hofstadler,S.A. (2000) Fourier transform ion cyclotron resonance mass spectrometry as a high throughput affinity screen to identify RNA binding ligands. Trends Anal. Chem., 19, 481–491. [Google Scholar]
- 20.Griffey R.H., Sannes-Lowery,K.A., Drader,J.J., Venkratraman,M., Swayze,E.E. and Hofstadler,S.A. (2000) Characterization of low-affinity complexes between RNA and small molecules using electrospray ionization mass spectrometry. J. Am. Chem. Soc., 122, 9933–9938. [Google Scholar]
- 21.Jorgensen T.J.D., Roepstorff,P. and Heck,A.J.R. (1998) Direct determination of solution binding constants for noncovalent complexes between bacterial cell wall peptide analogs and vancomycin group antibiotics by electrospray ionization mass spectrometry. Anal. Chem., 70, 4427–4432. [Google Scholar]
- 22.Ganguly A.K., Pramanik,B.N., Huang,E.C., Liberles,S., Heimark,L., Liu,Y.H., Tsarbopoulos,A., Doll,R.J., Taveras,A.G., Remiszewski,S., Snow,M.E., Wang,Y.S., Vibulbhan,B., Cesarz,D., Brown,J.E., del Rosario,J., James,L., Kirschmeier,P. and Girijavallabhan,V. (1997) Detection and structural characterization of Ras oncoprotein-inhibitors complexes by electrospray mass spectrometry. Bioorg. Med. Chem., 5, 817–820. [DOI] [PubMed] [Google Scholar]
- 23.Lim H.-K., Hsieh,Y.L., Ganem,B. and Henion,J. (1995) Recognition of cell-wall peptide ligands by vancomycin group antibiotics: studies using ion spray mass spectrometry. J. Mass Spectrom., 30, 708–714. [Google Scholar]
- 24.Ganem B., Li,Y.-T. and Henion,J.D. (1991) Detection of noncovalent receptor-ligand complexes by mass spectrometry. J. Am. Chem. Soc., 113, 6294–6296. [Google Scholar]
- 25.Hamdan M., Curcuruto,O. and Di Modugno,E. (1995) Investigation of complexes between some glycopeptide antibiotics and bacterial cell-wall analogs by electrospray- and capillary zone electrophoresis/electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom., 9, 883–887. [DOI] [PubMed] [Google Scholar]
- 26.Reynolds M.B., DeLuca,M.R. and Kerwin,S.M. (1999) The novel bis(benzoxazole) cytotoxic natural product UK-1 is a magnesium ion-dependent DNA binding agent and inhibitor of human topoisomerase II. Bioorg. Chem., 27, 326–337. [Google Scholar]
- 27.Demicheli C., Albertini,J.-P. and Garnier-Suillerot,A. (1991) Interaction of mithramycin with DNA: evidence that mithramycin binds to DNA as a dimer in a right-handed screw conformation. Eur. J. Biochem., 198, 333–338. [DOI] [PubMed] [Google Scholar]
- 28.Hardenbol P. and Van Dyke,M.W. (1992) In vitro inhibition of c-myc transcription by mithramycin. Biochem. Biophys. Res. Commun., 185, 553–558. [DOI] [PubMed] [Google Scholar]
- 29.Aich P. and Dasgupta,D. (1995) Role of magnesium ion in mithramycin-DNA interaction: binding of mithramycin-Mg2+ complexes with DNA. Biochemistry, 34, 1376–1385. [DOI] [PubMed] [Google Scholar]
- 30.Silva D.J., Goodnow,R.,Jr and Kahne,D. (1993) The sugars in chromomycin A3 stabilize the Mg2+-dimer complex. Biochemistry, 32, 463–471. [DOI] [PubMed] [Google Scholar]
- 31.Gambari R., Feriotto,G., Rutigliano,C., Bianchi,N. and Mischiati,C. (2000) Biospecific interaction analysis (BIA) of low-molecular weight DNA-binding drugs. J. Pharmacol. Exp. Ther., 294, 370–377. [PubMed] [Google Scholar]
- 32.Itzhaki L., Weinberger,S., Livnah,N. and Berman,E. (1990) A unique binding cavity for divalent cations in the DNA-metal-chromomycin A3 complex. Biopolymers, 29, 481–489. [DOI] [PubMed] [Google Scholar]
- 33.Gao X. and Patel,D.J. (1990) Chromomycin dimer-DNA oligomer complexes. Sequence selectivity and divalent cation specificity. Biochemistry, 29, 10940–10956. [DOI] [PubMed] [Google Scholar]
- 34.DeLuca M.R. and Kerwin,S.M. (1997) The total synthesis of UK-1. Tetrahedron Lett., 38, 199–202. [Google Scholar]
- 35.Weast R.C. (ed.) (1988) CRC Handbook of Chemistry and Physics, 1st Student Edn. CRC Press, Boca Raton, FL, F-105.