Abstract
The mutation of the Parkin gene is a cause of familial Parkinson’s disease. A growing body of evidence suggests that Parkin also functions as a tumor suppressor. Parkin is an ubiquitin E3 ligase, and plays important roles in a variety of cellular processes implicated in tumorigenesis, including cell cycle, cell proliferation, apoptosis, metastasis, mitophagy and metabolic reprogramming. Here we review the role and mechanism of Parkin in cancer.
Keywords: Parkin, Parkinson’s disease, Cancer, E3 ubiquitin ligase, Tumor suppressor
Background
Parkinson’s disease (PD), the second most common neurodegenerative disorder after Alzheimer’s disease, affects 1–2% of the general population [1–3]. PD is characterized by the progressive loss of dopaminergic neurons in the substantia nigra [1–3]. Mutations of the Parkin gene (PARK2) have been linked to autosomal recessive juvenile PD (ARJPD), one of the most common familial forms of PD [3, 4]. Approximately 50% of the individuals with ARJPD carry PARK2 mutations [3, 5]. Parkin dysfunction has also been implicated, albeit to a lesser degree, in the more common sporadic form of PD, as well as other neurodegenerative diseases, including Alzheimer’s disease and amyotrophic lateral sclerosis [2, 6, 7].
PARK2 encodes a 465-residue protein comprising an N-terminal ubiquitin-like domain (Ubl), a cysteine-rich RING0 domain, and two C-terminal RING domains (RING1, RING2) separated by an “in-between RING” (IBR) domain. All these domains appear to be functionally important, since mutations in all domains have been identified in PD patients [3, 8]. Similar to many other proteins containing RING-domains, Parkin functions as an E3 ubiquitin ligase. It ubiquitinates various proteins to regulate a variety of cellular processes, including mitochondrial homeostasis, anti-oxidative stress and mitophagy (mitochondria-specific autophagy), and such actions are thought to explain at least partly how Parkin prevents PD [2, 6, 9, 10]. PARK2 mutations could reduce its ability to ubiquitinate substrates such as CDCrel-1, Pael receptor, α-synuclein and synphilin-1, leading to their toxic build-up in the brain, which in turn causes PD [10–15]. Parkin regulates mitophagy to clear damaged mitochondria, thus preventing the accumulation of reactive oxygen species (ROS) and limiting oxidative damages in cells [9, 10, 16].
A growing body of evidence suggests that Parkin also functions as a tumor suppressor. In this review, we summarize recent advances on the tumor suppressive function of Parkin and its underlying mechanisms.
Parkin is a tumor suppressor
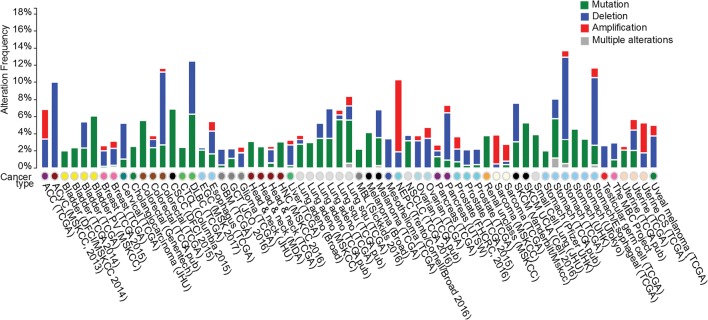
Parkin is ubiquitously expressed but predominantly in the brain. PARK2 is localized to human chromosome 6q25-27, a region frequently lost in cancers [17]. Loss of PARK2 heterozygosity and copy number has been observed in breast, lung, colorectal, and ovarian cancers [17–20]. Mutations of the Parkin gene have been reported in many types of cancers, although the frequency of these mutations appears to be relatively low [21, 22]. For instance, analysis of the datasets from cBioportal (http://www.cbioportal.org) [23] indicates that the Parkin gene is mutated in < 1% of breast cancer, 2–5% of colorectal cancer, ~ 5% of lung squamous cell carcinoma, and ~ 5% of gastric cancer [22, 24] (Fig. 1). Most Parkin gene mutations linked to cancer are missense mutations, with > 10% involving frameshifts or truncations [22]. Many missense mutations in cancer, such as T173A, T240M and P294S, impair E3 ubiquitin ligase activity and the tumor suppressive function of Parkin [24, 25]. In addition to mutations within the PARK2 coding sequence, levels of PARK2 mRNA and protein are frequently down-regulated in various types of cancers [17–20, 24]. Loss of PARK2 heterozygosity and copy number contributes to this down-regulation; hypermethylation of the PARK2 promoter may also be involved in certain cancers such as leukemia and colorectal cancer [26, 27].
Fig. 1.
Parkin alterations in human cancers. Summary of Parkin alterations associated with different cancers, based on the datasets in cBioPortal. Deletions, mutations and amplifications are depicted in different colors
Parkin undergoes various types of post-translational modifications that modulate its level and activity [28]. For instance, PINK1 phosphorylates Parkin to activate its E3 ubiquitin ligase activity, and recruits cytosolic Parkin to damaged mitochondria [9, 16, 29, 30]. Cyclin-dependent kinase (CDK) 5 and CK-1 phosphorylate Parkin, affecting its solubility without altering its activity [31, 32]. In contrast, c-Abl phosphorylates Parkin at Tyr 143 and inactivates it [33]. Sumoylation of Parkin promotes its nuclear localization and auto-ubiquitination [34], neddylation increases its activity [35], and S-nitrosylation reduces its activity [36]. In the brain, many of these post-translational modifications modulate Parkin’s neuroprotective function. It remains unclear whether these post-translational modifications are altered in cancer cells and contribute to the observed reduction in Parkin expression or activity in cancer.
Many different Parkin knockout mouse models have been established to study the role of Parkin in PD [37, 38]. However, most strains of Parkin knockout mice fail to model PD pathophysiology or display the selective loss of dopaminergic neurons characteristic of human PD [37, 38]. Instead, experiments with some of these Parkin knockout mice support the idea that Parkin is a potential tumor suppressor gene. Parkin knockout mice lacking exon 3 of PARK2 show enhanced hepatocyte proliferation and develop macroscopic hepatic tumors that resemble human hepatocellular carcinoma, indicating that Parkin-deficient mice are susceptible to spontaneous tumorigenesis [39]. Parkin+/− Apc+/min mice show higher incidence of intestinal adenomas and earlier onset of all adenoma stages (monocryptal, oligocryptal, and established) than Parkin+/+ Apc+/min mice [27]. Work from our laboratory has shown that Parkin−/− mice are more susceptible than Parkin+/+ mice to γ-irradiation-induced tumorigenesis, although the resulting tumor spectrum (mainly lymphomas) is similar in the two strains [40].
Mechanisms of Parkin-mediated tumor suppression
Exactly how Parkin may suppress tumor formation is poorly understood. Mechanisms proposed in the literature are discussed below.
Cell cycle and proliferation
Parkin has been reported to play an important role in inhibiting cell cycle progression (Fig. 2). Parkin ubiquitinates and degrades cyclin E [41, 42], which in turn binds to CDK2 to promote the transition from G1 to S phase of the cell cycle. Parkin also ubiquitinates and degrades cyclin D, a regulator of CDK4/6 [43]. In this way, Parkin induces G1/S cell cycle arrest and inhibits cell proliferation. Parkin also induces expression of Myt1, which phosphorylates CDK1 to inhibit CDK1 activity and cause G2/M cell cycle arrest in HeLa cells treated with TNF-α [44].
Fig. 2.
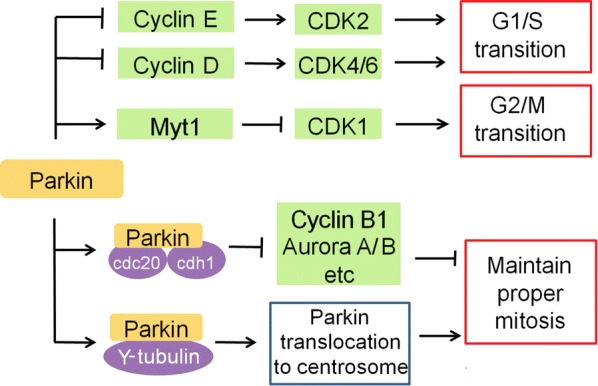
Parkin regulates cell cycle and mitosis. → : promote;
 : inhibit
: inhibit
In addition to regulating cell cycle, Parkin also regulates mitosis. Parkin interacts with anaphase-promoting complex/cyclosome (APC/C) co-activators Cdc20 and Cdh1 to mediate the degradation of several key mitotic regulators, including cyclin B1 and Aurora A/B [45]. Parkin deficiency results in overexpression of cyclin B1, Aurora A/B and other mitotic regulators, which leads to mitotic defects, genomic instability and tumorigenesis [45], suggesting that the Parkin-Cdc20/Cdh1 complex is important to ensure proper mitosis. In cells treated with the proteasome inhibitor lactacystin, Parkin forms a complex with γ-tubulin and then is recruited to the centrosome through a microtubule-dependent mechanism [46], where it may assist in mitotic spindle formation. Parkin deficiency induces spindle multipolarity and misorientation as well as multinucleation, promoting tumorigenesis [47]. These results suggest that Parkin may help suppress tumor formation by ensuring proper mitosis.
Apoptosis
In contrast to promoting survival of neurons [48, 49], Parkin appears to promote apoptosis of cancer cells (Fig. 3). Parkin has been reported to promote apoptosis induced by mitochondrial depolarization [50]. In response to mitochondrial depolarization caused by CCCP treatment, Parkin promotes ubiquitination and degradation of the Bcl-2 family member Mcl-1, which opens the Bax/Bak channel and thereby sensitizes cells to apoptosis. This Parkin-dependent apoptosis requires PINK1 and can be blocked by knockdown of Bax and Bak [50]. Restoration of Parkin expression in cervical cancer HeLa cells acts via a poorly understood mechanism to reduce levels of survivin, an inhibitor of apoptosis (IAP) family member that inhibits caspase, thereby sensitizing the cells to TNF-α-induced apoptosis [51]. Parkin sensitizes breast cancer MCF7 cells to apoptosis induced by microtubule-stabilizing drugs such as paclitaxel; Parkin binds to the outer surface of microtubules and increases paclitaxel–microtubule interaction [52]. Parkin also promotes apoptosis induced by HDAC inhibitors in hepatocellular carcinoma cells via a poorly understood mechanism [53]. In mouse hepatocytes, Parkin induces apoptosis by transcriptionally down-regulating follistatin, which antagonizes the pro-apoptotic cytokine activin [54, 55]. Conversely, Parkin deficiency in Parkin−/− mouse hepatocytes up-regulates follistatin to inhibit apoptosis induced by chemotherapeutic agents such as cisplatin, doxorubicin and etoposide [39]. These results suggest that Parkin normally functions to keep follistatin expression at a low level and thereby limit hepatocyte proliferation, which may explain the elevated occurrence of hepatic tumor development in Parkin−/− mice [39].
Fig. 3.
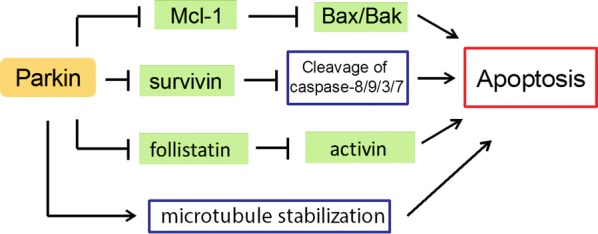
Parkin promotes apoptosis in cancer
Migration, invasion and metastasis
Metastasis is a major cause of cancer-related death. Parkin expression is significantly lower in tumors with lymph node metastases than in tumors without such metastases in the case of clear-cell renal cell carcinoma [56], pancreatic cancer and nasopharyngeal carcinoma [47, 57]. Work from our laboratory has shown that Parkin is frequently down-regulated in breast cancer, and that lower Parkin expression correlates with worse distant metastasis-free survival [24]. These results suggest that Parkin plays an important role in suppressing metastasis.
One mechanism through which Parkin may help suppress metastasis is by ubiquitinating HIF-1α and triggering its degradation, thereby inhibiting the migration and invasion of breast cancer cells [24] (Fig. 4). Stabilization and accumulation of HIF-1α in cancer cells promote metastasis [58, 59], and HIF-1α expression correlates inversely with Parkin expression in breast cancer specimens. Given that HIF-1α regulates, in addition to metastasis, several other cellular processes, including cell survival, metabolic reprogramming, and angiogenesis [58, 59], Parkin may exert its tumor suppressive function by inhibiting these processes, which should be addressed in future studies.
Fig. 4.
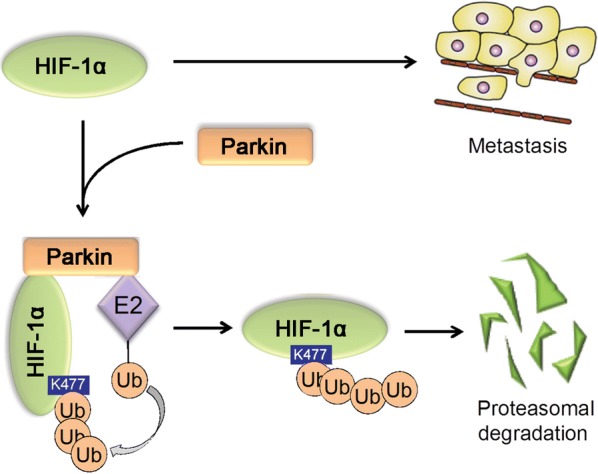
Parkin inhibits cancer metastasis by ubiquitinating HIF-1α
Metabolic reprogramming
Metabolic reprogramming is a hallmark of cancers and plays a key role in cancer progression by driving cancer cell proliferation, survival and metastasis [60–64]. Metabolic reprogramming in cancer can be driven by the inactivation of tumor suppressors such as p53 and PTEN, or activation of oncoproteins such as HIF-1α, Myc and PI3K [60–63, 65]. The most well characterized metabolic change in cancers is enhanced glycolysis, also known as the Warburg effect [60–63, 65]. Work from our group has shown that p53, which is known to suppress glycolysis [61, 65–68], could transcriptionally activate Parkin expression by binding to the p53-responsive elements in PARK2 [40]. Parkin suppresses glycolysis and promotes mitochondrial oxidative phosphorylation. How Parkin inhibits glycolysis is poorly understood. One possibility is that Parkin-mediated ubiquitination and degradation of HIF-1α prevents HIF-1α from transcriptionally activating the proteins involved in glycolysis [24] (Fig. 5). Similarly, another group reported that p53 transcriptionally activates Parkin in glioma [69]. The same group reported that Parkin, independently of its E3 ligase activity, transcriptionally represses p53 expression in mouse and human brains affected by ARJPD, suggesting that p53 can form a negative feedback loop with Parkin in the brain [70]. Further studies are needed to clarify whether this negative feedback loop exists in tissues outside the brain and in tumors.
Fig. 5.
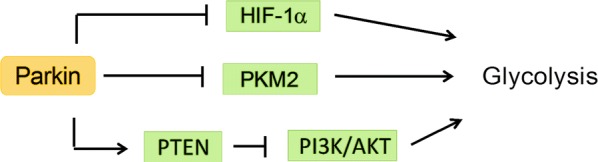
Parkin inhibits glycolysis in cancer
Similar to p53, PTEN is involved in Parkin-mediated metabolic regulation. Parkin deficiency promotes PTEN inactivation through S-nitrosylation and ubiquitination, which activates PI3K/AKT signaling in cancer cells [71] (Fig. 5). PI3K/AKT signaling drives metabolic reprogramming in cancer cells, including promotion of glycolysis [60–64].
Parkin interacts with PKM2, a glycolytic enzyme that is frequently overexpressed in cancer, and catalyzes ubiquitin conjugation to PKM2 [72]. This ubiquitination inhibits PKM2 activity without destabilizing the protein. Loss of Parkin function in cancer cells enhances PKM2 enzymatic activity, thus promoting glycolysis [72] (Fig. 5).
These studies suggest that one mechanism by which Parkin suppresses tumorigenesis is by inhibiting glycolysis.
Mitophagy
Parkin-mediated neuroprotection involves regulation of mitophagy [9, 10, 16]. In response to mitochondrial damage, the serine/threonine kinase PINK1 is stabilized and accumulates on the outer membrane of the damaged mitochondria, where it phosphorylates Parkin and thereby activates its E3 ubiquitin ligase activity and recruits it from cytosol to damaged mitochondria [9, 16]. Parkin ubiquitinates outer mitochondrial membrane proteins, triggering mitophagy that selectively clears damaged mitochondria [9, 16]. Thus, PINK1 and Parkin work together to prevent PD by eliminating the accumulation of damaged mitochondria, ROS and mitochondrial DNA mutations [9, 10, 16].
Consistent with the idea that Parkin is important for both mitophagy regulation and tumor suppression, mitophagy defects have been linked with cancer development [73, 74]. In parallel, BNIP3 and Nix (also named BNIP3L) induce mitophagy independently of Parkin and suppress growth of certain types of tumors [10, 73, 74]. BNIP3 is silenced in liver, lung, pancreatic and colorectal cancers [75–77], and this loss leads to mitophagy defects, increases glycolysis and ROS production, and promotes growth and metastasis of breast cancer cells in mouse models [78, 79]. Similarly, knockdown of Nix promotes tumorigenesis in mouse models [80]. These studies suggest that loss of mitophagy mediated by Parkin or BNIP3/Nix contributes to the progression of certain types of cancers.
Analogously to the dual role of autophagy in cancer, mitophagy may promote or suppress tumorigenesis depending on the cellular circumstances [10, 73, 74]. Future studies should examine the role of mitophagy in the development and progression of different types of cancers.
Conclusions and perspectives
Increasing evidence from cell culture systems, xenograft tumor models, Parkin knockout mouse models, and clinical studies suggests that Parkin is an important tumor suppressor. How the protein suppresses tumor growth is poorly understood, with many questions requiring further study. One question is how Parkin inhibits apoptosis and promotes survival in neuronal cells, yet promotes apoptosis in various types of cancer cells and mouse hepatocytes. One possible explanation is brain-specific regulation of p53 by Parkin. Induction of apoptosis is an important mechanism by which p53 exerts its tumor suppressive function in cancers [81, 82]. Interestingly, p53 activation has been observed in the brain of PD patients as well as mouse models for PD mice, suggesting that p53 activation and p53-mediated apoptosis contribute to PD [83–87]. Parkin inhibits p53 activation in the brain [70]. Accordingly, inhibition of p53-mediated apoptosis could be an important mechanism that contributes to the neuroprotective effect of Parkin. It remains unclear whether Parkin affects p53 activation or p53-mediated apoptosis in cancer cells. Future work should address this question.
Another unanswered question is why Parkin is down-regulated in many types of cancers. This cannot be explained solely on the basis of mutations, loss of heterozygosity or copy number, and promoter hypermethylation. Future studies should examine whether the transcriptional regulation of PARK2, epigenetic modifications of the gene, or post-translational modifications of Parkin (e.g., include phosphorylation, ubiquitination, sumoylation, neddylation, and S-nitrosylation) are altered in cancer [22, 28].
The third question is whether and how the anti-cancer activity of PD relates to the function of Parkin in PD. Many epidemiological studies have indicated an association between PD and reduced risk of prostate, lung, bladder, stomach, uterine, and colorectal cancers, and increased risk of melanoma, brain and breast cancers [88–91]. The association between PD and cancer risk appears to be complex and may be linked to factors such as ethnicity: PD among Asians in one study was linked to increased risk of brain, kidney, uterine, stomach and lung cancers [92]. Future studies should examine why PD shows opposite associations with different types of cancers, or even opposite associations with the same cancer in different patient populations. Future studies should also examine the potential role of Parkin gene mutations in mediating the association between PD and cancer risk. Such analyses will need to take genetic, epigenetic and environmental factors into account.
The role of Parkin in PD is well established, and its role as a tumor suppressor has recently emerged. How Parkin protects against PD and cancer is poorly understood. Future studies in this area may lead to novel therapeutic targets and strategies for both diseases.
Authors’ contributions
All authors contributed substantially to the writing of this review. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by grants to Z.F. from the NIH (R01CA227912) and Rutgers Cancer Institute of New Jersey Pilot Award, and by grants to W.H. from the NIH (R01CA160558, R01CA203965) and Ellison Medical Foundation.
Footnotes
Juan Liu and Cen Zhang contributed equally to this work
Contributor Information
Juan Liu, Email: jl1530@cinj.rutgers.edu.
Cen Zhang, Email: zhangce@cinj.rutgers.edu.
Wenwei Hu, Email: wh221@cinj.rutgers.edu.
Zhaohui Feng, Email: fengzh@cinj.rutgers.edu.
References
- 1.Tanner CM, Goldman SM. Epidemiology of Parkinson’s disease. Neurol Clin. 1996;14(2):317–335. doi: 10.1016/S0733-8619(05)70259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson TM, Dawson VL. The role of parkin in familial and sporadic Parkinson’s disease. Mov Disord. 2010;25(Suppl 1):S32–S39. doi: 10.1002/mds.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein C, Westenberger A. Genetics of Parkinson’s disease. Cold Spring Harb Perspect Med. 2012;2(1):a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsumine H, Saito M, Shimoda-Matsubayashi S, Tanaka H, Ishikawa A, Nakagawa-Hattori Y, et al. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2-27. Am J Hum Genet. 1997;60(3):588–596. [PMC free article] [PubMed] [Google Scholar]
- 5.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 6.Zhang CW, Hang L, Yao TP, Lim KL. Parkin regulation and neurodegenerative disorders. Front Aging Neurosci. 2015;7:248. doi: 10.3389/fnagi.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilcher H. Parkin implicated in sporadic Parkinson’s disease. Lancet Neurol. 2005;4(12):798. doi: 10.1016/S1474-4422(05)70237-4. [DOI] [PubMed] [Google Scholar]
- 8.Trempe JF, Sauve V, Grenier K, Seirafi M, Tang MY, Menade M, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340(6139):1451–1455. doi: 10.1126/science.1237908. [DOI] [PubMed] [Google Scholar]
- 9.Pickrell AM, Youle RJ. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron. 2015;85(2):257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardini JP, Lazarou M, Dewson G. Parkin and mitophagy in cancer. Oncogene. 2017;36(10):1315–1327. doi: 10.1038/onc.2016.302. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin—protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci USA. 2000;97(24):13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105(7):891–902. doi: 10.1016/S0092-8674(01)00407-X. [DOI] [PubMed] [Google Scholar]
- 13.Chung KK, Zhang Y, Lim KL, Tanaka Y, Huang H, Gao J, et al. Parkin ubiquitinates the alpha-synuclein-interacting protein, synphilin-1: implications for Lewy-body formation in Parkinson disease. Nat Med. 2001;7(10):1144–1150. doi: 10.1038/nm1001-1144. [DOI] [PubMed] [Google Scholar]
- 14.Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson’s disease. Science. 2001;293(5528):263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 15.Shimura H, Hattori N, Kubo S, Mizuno Y, Asakawa S, Minoshima S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25(3):302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 16.McWilliams TG, Muqit MM. PINK1 and Parkin: emerging themes in mitochondrial homeostasis. Curr Opin Cell Biol. 2017;45:83–91. doi: 10.1016/j.ceb.2017.03.013. [DOI] [PubMed] [Google Scholar]
- 17.Cesari R, Martin ES, Calin GA, Pentimalli F, Bichi R, McAdams H, et al. Parkin, a gene implicated in autosomal recessive juvenile parkinsonism, is a candidate tumor suppressor gene on chromosome 6q25-q27. Proc Natl Acad Sci USA. 2003;100(10):5956–5961. doi: 10.1073/pnas.0931262100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denison SR, Wang F, Becker NA, Schule B, Kock N, Phillips LA, et al. Alterations in the common fragile site gene Parkin in ovarian and other cancers. Oncogene. 2003;22(51):8370–8378. doi: 10.1038/sj.onc.1207072. [DOI] [PubMed] [Google Scholar]
- 19.Picchio MC, Martin ES, Cesari R, Calin GA, Yendamuri S, Kuroki T, et al. Alterations of the tumor suppressor gene Parkin in non-small cell lung cancer. Clin Cancer Res. 2004;10(8):2720–2724. doi: 10.1158/1078-0432.CCR-03-0086. [DOI] [PubMed] [Google Scholar]
- 20.Tay SP, Yeo CW, Chai C, Chua PJ, Tan HM, Ang AX, et al. Parkin enhances the expression of cyclin-dependent kinase 6 and negatively regulates the proliferation of breast cancer cells. J Biol Chem. 2010;285(38):29231–29238. doi: 10.1074/jbc.M110.108241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Je EM, Yoo NJ, Lee SH. Somatic mutation of PARK2 tumor suppressor gene is not common in common solid cancers. Pathol Oncol Res. 2013;19(3):393–395. doi: 10.1007/s12253-012-9591-z. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, Lin DC, Yin D, Koeffler HP. An emerging role of PARK2 in cancer. J Mol Med. 2014;92(1):31–42. doi: 10.1007/s00109-013-1107-0. [DOI] [PubMed] [Google Scholar]
- 23.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J, Zhang C, Zhao Y, Yue X, Wu H, Huang S, et al. Parkin targets HIF-1alpha for ubiquitination and degradation to inhibit breast tumor progression. Nat commun. 2017;8(1):1823. doi: 10.1038/s41467-017-01947-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin DC, Xu L, Chen Y, Yan H, Hazawa M, Doan N, et al. Genomic and functional analysis of the E3 Ligase PARK2 in glioma. Cancer Res. 2015;75(9):1815–1827. doi: 10.1158/0008-5472.CAN-14-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agirre X, Roman-Gomez J, Vazquez I, Jimenez-Velasco A, Garate L, Montiel-Duarte C, et al. Abnormal methylation of the common PARK2 and PACRG promoter is associated with downregulation of gene expression in acute lymphoblastic leukemia and chronic myeloid leukemia. Int J Cancer. 2006;118(8):1945–1953. doi: 10.1002/ijc.21584. [DOI] [PubMed] [Google Scholar]
- 27.Poulogiannis G, McIntyre RE, Dimitriadi M, Apps JR, Wilson CH, Ichimura K, et al. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc Natl Acad Sci USA. 2010;107(34):15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakraborty J, Basso V, Ziviani E. Post translational modification of Parkin. Biol Direct. 2017;12(1):6. doi: 10.1186/s13062-017-0176-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Park J, Kim S, Song S, Kwon SK, Lee SH, et al. PINK1 controls mitochondrial localization of Parkin through direct phosphorylation. Biochem Biophys Res Commun. 2008;377(3):975–980. doi: 10.1016/j.bbrc.2008.10.104. [DOI] [PubMed] [Google Scholar]
- 30.Kondapalli C, Kazlauskaite A, Zhang N, Woodroof HI, Campbell DG, Gourlay R, et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2012;2(5):120080. doi: 10.1098/rsob.120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Avraham E, Rott R, Liani E, Szargel R, Engelender S. Phosphorylation of Parkin by the cyclin-dependent kinase 5 at the linker region modulates its ubiquitin-ligase activity and aggregation. J Biol Chem. 2007;282(17):12842–12850. doi: 10.1074/jbc.M608243200. [DOI] [PubMed] [Google Scholar]
- 32.de la Rubio Torre E, Luzon-Toro B, Forte-Lago I, Minguez-Castellanos A, Ferrer I, Hilfiker S. Combined kinase inhibition modulates parkin inactivation. Hum Mol Genet. 2009;18(5):809–823. doi: 10.1093/hmg/ddn407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ko HS, Lee Y, Shin JH, Karuppagounder SS, Gadad BS, Koleske AJ, et al. Phosphorylation by the c-Abl protein tyrosine kinase inhibits parkin’s ubiquitination and protective function. Proc Natl Acad Sci USA. 2010;107(38):16691–16696. doi: 10.1073/pnas.1006083107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Um JW, Chung KC. Functional modulation of parkin through physical interaction with SUMO-1. J Neurosci Res. 2006;84(7):1543–1554. doi: 10.1002/jnr.21041. [DOI] [PubMed] [Google Scholar]
- 35.Um JW, Han KA, Im E, Oh Y, Lee K, Chung KC. Neddylation positively regulates the ubiquitin E3 ligase activity of Parkin. J Neurosci Res. 2012;90(5):1030–1042. doi: 10.1002/jnr.22828. [DOI] [PubMed] [Google Scholar]
- 36.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, et al. S-nitrosylation of parkin regulates ubiquitination and compromises Parkin’s protective function. Science. 2004;304(5675):1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 37.Lee Y, Dawson VL, Dawson TM. Animal models of Parkinson’s disease: vertebrate genetics. Cold Spring Harb Perspect Med. 2012;2(10):a009324. doi: 10.1101/cshperspect.a009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perez FA, Palmiter RD. Parkin-deficient mice are not a robust model of parkinsonism. Proc Natl Acad Sci USA. 2005;102(6):2174–2179. doi: 10.1073/pnas.0409598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujiwara M, Marusawa H, Wang HQ, Iwai A, Ikeuchi K, Imai Y, et al. Parkin as a tumor suppressor gene for hepatocellular carcinoma. Oncogene. 2008;27(46):6002–6011. doi: 10.1038/onc.2008.199. [DOI] [PubMed] [Google Scholar]
- 40.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, et al. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci USA. 2011;108(39):16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Veeriah S, Taylor BS, Meng S, Fang F, Yilmaz E, Vivanco I, et al. Somatic mutations of the Parkinson’s disease-associated gene PARK2 in glioblastoma and other human malignancies. Nat Genet. 2010;42(1):77–82. doi: 10.1038/ng.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ikeuchi K, Marusawa H, Fujiwara M, Matsumoto Y, Endo Y, Watanabe T, et al. Attenuation of proteolysis-mediated cyclin E regulation by alternatively spliced Parkin in human colorectal cancers. Int J Cancer. 2009;125(9):2029–2035. doi: 10.1002/ijc.24565. [DOI] [PubMed] [Google Scholar]
- 43.Yeo CW, Ng FS, Chai C, Tan JM, Koh GR, Chong YK, et al. Parkin pathway activation mitigates glioma cell proliferation and predicts patient survival. Cancer Res. 2012;72(10):2543–2553. doi: 10.1158/0008-5472.CAN-11-3060. [DOI] [PubMed] [Google Scholar]
- 44.Lee MH, Cho Y, Jung BC, Kim SH, Kang YW, Pan CH, et al. Parkin induces G2/M cell cycle arrest in TNF-alpha-treated HeLa cells. Biochem Biophys Res Commun. 2015;464(1):63–69. doi: 10.1016/j.bbrc.2015.05.101. [DOI] [PubMed] [Google Scholar]
- 45.Lee SB, Kim JJ, Nam HJ, Gao B, Yin P, Qin B, et al. Parkin regulates mitosis and genomic stability through Cdc20/Cdh1. Mol Cell. 2015;60(1):21–34. doi: 10.1016/j.molcel.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao J, Ren Y, Jiang Q, Feng J. Parkin is recruited to the centrosome in response to inhibition of proteasomes. J Cell Sci. 2003;116(Pt 19):4011–4019. doi: 10.1242/jcs.00700. [DOI] [PubMed] [Google Scholar]
- 47.Sun X, Liu M, Hao J, Li D, Luo Y, Wang X, et al. Parkin deficiency contributes to pancreatic tumorigenesis by inducing spindle multipolarity and misorientation. Cell Cycle. 2013;12(7):1133–1141. doi: 10.4161/cc.24215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seirafi M, Kozlov G, Gehring K. Parkin structure and function. FEBS J. 2015;282(11):2076–2088. doi: 10.1111/febs.13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hang L, Thundyil J, Lim KL. Mitochondrial dysfunction and Parkinson disease: a Parkin-AMPK alliance in neuroprotection. Ann N Y Acad Sci. 2015;1350:37–47. doi: 10.1111/nyas.12820. [DOI] [PubMed] [Google Scholar]
- 50.Carroll RG, Hollville E, Martin SJ. Parkin sensitizes toward apoptosis induced by mitochondrial depolarization through promoting degradation of Mcl-1. Cell Rep. 2014;9(4):1538–1553. doi: 10.1016/j.celrep.2014.10.046. [DOI] [PubMed] [Google Scholar]
- 51.Lee K, Lee MH, Kang YW, Rhee KJ, Kim TU, Kim YS. Parkin induces apoptotic cell death in TNF-alpha-treated cervical cancer cells. BMB Rep. 2012;45(9):526–531. doi: 10.5483/BMBRep.2012.45.9.104. [DOI] [PubMed] [Google Scholar]
- 52.Wang H, Liu B, Zhang C, Peng G, Liu M, Li D, et al. Parkin regulates paclitaxel sensitivity in breast cancer via a microtubule-dependent mechanism. J Pathol. 2009;218(1):76–85. doi: 10.1002/path.2512. [DOI] [PubMed] [Google Scholar]
- 53.Wang F, Denison S, Lai JP, Philips LA, Montoya D, Kock N, et al. Parkin gene alterations in hepatocellular carcinoma. Genes Chromosom Cancer. 2004;40(2):85–96. doi: 10.1002/gcc.20020. [DOI] [PubMed] [Google Scholar]
- 54.Chen YG, Lui HM, Lin SL, Lee JM, Ying SY. Regulation of cell proliferation, apoptosis, and carcinogenesis by activin. Exp Biol Med. 2002;227(2):75–87. doi: 10.1177/153537020222700201. [DOI] [PubMed] [Google Scholar]
- 55.Harrison CA, Gray PC, Vale WW, Robertson DM. Antagonists of activin signaling: mechanisms and potential biological applications. Trends Endocrinol Metab. 2005;16(2):73–78. doi: 10.1016/j.tem.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 56.Toma MI, Wuttig D, Kaiser S, Herr A, Weber T, Zastrow S, et al. PARK2 and PACRG are commonly downregulated in clear-cell renal cell carcinoma and are associated with aggressive disease and poor clinical outcome. Genes Chromosomes Cancer. 2013;52(3):265–273. doi: 10.1002/gcc.22026. [DOI] [PubMed] [Google Scholar]
- 57.Ni H, Zhou Z, Jiang B, Yuan X, Cao X, Huang G, et al. Inactivation of parkin by promoter methylation correlated with lymph node metastasis and genomic instability in nasopharyngeal carcinoma. Tumour Biol. 2017;39(3):1010428317695025. doi: 10.1177/1010428317695025. [DOI] [PubMed] [Google Scholar]
- 58.LaGory EL, Giaccia AJ. The ever-expanding role of HIF in tumour and stromal biology. Nat Cell Biol. 2016;18(4):356–365. doi: 10.1038/ncb3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones RG, Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev. 2009;23(5):537–548. doi: 10.1101/gad.1756509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic regulation by p53 family members. Cell Metab. 2013;18(5):617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 63.DeNicola GM, Cantley LC. Cancer’s fuel choice: new flavors for a picky eater. Mol Cell. 2015;60(4):514–523. doi: 10.1016/j.molcel.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 2010;20(7):427–434. doi: 10.1016/j.tcb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, Zhang C, Hu W, Feng Z. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett. 2015;356(2 Pt A):197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang SJ, Gu W. To be, or not to be: functional dilemma of p53 metabolic regulation. Curr Opin Oncol. 2014;26(1):78–85. doi: 10.1097/CCO.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 69.Viotti J, Duplan E, Caillava C, Condat J, Goiran T, Giordano C, et al. Glioma tumor grade correlates with parkin depletion in mutant p53-linked tumors and results from loss of function of p53 transcriptional activity. Oncogene. 2014;33(14):1764–1775. doi: 10.1038/onc.2013.124. [DOI] [PubMed] [Google Scholar]
- 70.da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, et al. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson’s disease. Nat Cell Biol. 2009;11(11):1370–1375. doi: 10.1038/ncb1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gupta A, Anjomani-Virmouni S, Koundouros N, Dimitriadi M, Choo-Wing R, Valle A, et al. PARK2 depletion connects energy and oxidative stress to PI3K/Akt activation via PTEN S-nitrosylation. Mol cell. 2017;65(6):999–1013. doi: 10.1016/j.molcel.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu K, Li F, Han H, Chen Y, Mao Z, Luo J, et al. Parkin regulates the activity of pyruvate kinase M2. J Biol Chem. 2016;291(19):10307–10317. doi: 10.1074/jbc.M115.703066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kulikov AV, Luchkina EA, Gogvadze V, Zhivotovsky B. Mitophagy: link to cancer development and therapy. Biochem Biophys Res Commun. 2017;482(3):432–439. doi: 10.1016/j.bbrc.2016.10.088. [DOI] [PubMed] [Google Scholar]
- 74.Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4. doi: 10.1186/s40170-015-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Okami J, Simeone DM, Logsdon CD. Silencing of the hypoxia-inducible cell death protein BNIP3 in pancreatic cancer. Can Res. 2004;64(15):5338–5346. doi: 10.1158/0008-5472.CAN-04-0089. [DOI] [PubMed] [Google Scholar]
- 76.Murai M, Toyota M, Suzuki H, Satoh A, Sasaki Y, Akino K, et al. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin Can Res. 2005;11(3):1021–1027. [PubMed] [Google Scholar]
- 77.Koop EA, van Laar T, van Wichen DF, de Weger RA, Wall E, van Diest PJ. Expression of BNIP3 in invasive breast cancer: correlations with the hypoxic response and clinicopathological features. BMC Cancer. 2009;9:175. doi: 10.1186/1471-2407-9-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manka D, Spicer Z, Millhorn DE. Bcl-2/adenovirus E1B 19 kDa interacting protein-3 knockdown enables growth of breast cancer metastases in the lung, liver, and bone. Can Res. 2005;65(24):11689–11693. doi: 10.1158/0008-5472.CAN-05-3091. [DOI] [PubMed] [Google Scholar]
- 79.Chourasia AH, Tracy K, Frankenberger C, Boland ML, Sharifi MN, Drake LE, et al. Mitophagy defects arising from BNip3 loss promote mammary tumor progression to metastasis. EMBO Rep. 2015;16(9):1145–1163. doi: 10.15252/embr.201540759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, et al. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6(6):597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 81.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 82.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13(6):1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 83.Nair VD, McNaught KS, Gonzalez-Maeso J, Sealfon SC, Olanow CW. p53 mediates non-transcriptional cell death in dopaminergic cells in response to proteasome inhibition. J Biol Chem. 2006;281(51):39550–39560. doi: 10.1074/jbc.M603950200. [DOI] [PubMed] [Google Scholar]
- 84.Mogi M, Kondo T, Mizuno Y, Nagatsu T. p53 protein, interferon-gamma, and NF-kappaB levels are elevated in the parkinsonian brain. Neurosci Lett. 2007;414(1):94–97. doi: 10.1016/j.neulet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 85.Lee SJ, Kim DC, Choi BH, Ha H, Kim KT. Regulation of p53 by activated protein kinase C-delta during nitric oxide-induced dopaminergic cell death. J Biol Chem. 2006;281(4):2215–2224. doi: 10.1074/jbc.M509509200. [DOI] [PubMed] [Google Scholar]
- 86.Martin LJ, Pan Y, Price AC, Sterling W, Copeland NG, Jenkins NA, et al. Parkinson’s disease alpha-synuclein transgenic mice develop neuronal mitochondrial degeneration and cell death. J Neurosci. 2006;26(1):41–50. doi: 10.1523/JNEUROSCI.4308-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chang JR, Ghafouri M, Mukerjee R, Bagashev A, Chabrashvili T, Sawaya BE. Role of p53 in neurodegenerative diseases. Neurodegener Dis. 2012;9(2):68–80. doi: 10.1159/000329999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wirdefeldt K, Adami HO, Cole P, Trichopoulos D, Mandel J. Epidemiology and etiology of Parkinson’s disease: a review of the evidence. Eur J Epidemiol. 2011;26(Suppl 1):S1–S58. doi: 10.1007/s10654-011-9581-6. [DOI] [PubMed] [Google Scholar]
- 89.Bajaj A, Driver JA, Schernhammer ES. Parkinson’s disease and cancer risk: a systematic review and meta-analysis. Cancer Causes Control. 2010;21(5):697–707. doi: 10.1007/s10552-009-9497-6. [DOI] [PubMed] [Google Scholar]
- 90.Kareus SA, Figueroa KP, Cannon-Albright LA, Pulst SM. Shared predispositions of parkinsonism and cancer: a population-based pedigree-linked study. Arch Neurol. 2012;69(12):1572–1577. doi: 10.1001/archneurol.2012.2261. [DOI] [PubMed] [Google Scholar]
- 91.Catala-Lopez F, Suarez-Pinilla M, Suarez-Pinilla P, Valderas JM, Gomez-Beneyto M, Martinez S, et al. Inverse and direct cancer comorbidity in people with central nervous system disorders: a meta-analysis of cancer incidence in 577,013 participants of 50 observational studies. Psychother Psychosom. 2014;83(2):89–105. doi: 10.1159/000356498. [DOI] [PubMed] [Google Scholar]
- 92.Lin PY, Chang SN, Hsiao TH, Huang BT, Lin CH, Yang PC. Association between Parkinson disease and risk of cancer in Taiwan. JAMA Oncol. 2015;1(5):633–640. doi: 10.1001/jamaoncol.2015.1752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.