ABSTRACT
Strains of Rickettsia rickettsii, the tick-borne agent of Rocky Mountain spotted fever, vary considerably in virulence. Genomic comparisons of R. rickettsii strains have identified a relatively small number of genes divergent in an avirulent strain. Among these is one annotated as Rickettsia ankyrin repeat protein 2 (RARP-2). Homologs of RARP-2 are present in all strains of R. rickettsii, but the protein in the avirulent strain Iowa contains a large internal deletion relative to the virulent Sheila Smith strain. RARP-2 is secreted in a type IV secretion system-dependent manner and exposed to the host cell cytosol. RARP-2 of Sheila Smith colocalizes with multilamellar membranous structures bearing markers of the endoplasmic reticulum (ER), whereas the Iowa protein shows no colocalization with host cell organelles and evidence of proteolytic degradation is detected. Overexpression of Sheila Smith RARP-2 in R. rickettsii Iowa converts this avirulent strain’s typically nonlytic or opaque plaque type to a lytic plaque phenotype similar to that of the virulent Sheila Smith strain. Mutation of a predicted proteolytic active site of Sheila Smith RARP-2 abolished the lytic plaque phenotype but did not eliminate association with host membrane. RARP-2 is thus a type IV secreted effector and released from the rickettsiae into the host cytosol to modulate host processes during infection. Overexpression of Sheila Smith RARP-2 did not, however, restore the virulence of the Iowa strain in a guinea pig model, likely due to the multifactorial nature of rickettsial virulence.
KEYWORDS: ankyrin repeat, genome, rickettsia, type IV secretion, virulence
IMPORTANCE
Members of the genus Rickettsia are obligate intracellular bacteria that exhibit a range of virulence from harmless endosymbionts of arthropods to the etiologic agents of severe disease. Despite the growing number of available genomes, little is known regarding virulence determinants of rickettsiae. Here, we have characterized an ankyrin repeat-containing protein, RARP-2, which differs between a highly virulent and an avirulent strain of R. rickettsii, the agent of Rocky Mountain spotted fever. RARP-2 is secreted by a type IV secretion system into the cytosol of the host cell, where it interacts with and manipulates the structure of the endoplasmic reticulum. RARP-2 from the avirulent strain is truncated by the loss of seven of 10 ankyrin repeat units but, although secreted, fails to alter ER structure. Recognition of those rickettsial factors associated with virulence will facilitate understanding of regional and strain-specific variation in severity of disease.
INTRODUCTION
Rickettsia spp. are arthropod-borne Gram-negative, obligate intracellular parasites of their eukaryotic hosts. Members of the genus Rickettsia are classified into three monophyletic groups: the spotted fever group (SFG), the typhus group, and a transitional group, with additional basal lineages unclassified (1). Rickettsia rickettsii is a member of the SFG rickettsiae and the causative agent of Rocky Mountain spotted fever (RMSF), the most severe of the SFG rickettsioses. Even today, RMSF is a potentially life-threatening disease with mortality rates reaching 20% if not treated appropriately (2). Strains of R. rickettsii differ markedly, however, in the severity of human disease that they cause as well as in their virulence in animal model systems (3, 4). Since its earliest recognition, it was observed that cases in the Bitterroot Valley of western Montana were much more severe than in surrounding areas, with case fatality rates upward of 80% before the advent of antibiotics (5). This contrasts with fatality rates near 5% in nearby Idaho. Recent genomic comparisons of the highly virulent R. rickettsii Sheila Smith strain with the avirulent Iowa strain (6) and strains of intermediate virulence (4) identified differences in several genes, which might represent potential virulence determinants (4, 6).
One such factor is an ankyrin repeat-containing protein (ARP), the Rickettsia ankyrin repeat protein 2 (RARP-2) (7) (A1G_05165 in Sheila Smith strain, RrIowa_1113 in Iowa strain). While RARP-2 homologs are present in all R. rickettsii strains, both the avirulent Iowa strain and the moderately virulent R strain share a 588-bp deletion in this gene, leading to a lesser number of ankyrin repeats in the mature proteins (4, 6).
The ankyrin domain is the most common protein-protein interaction motif in nature (8), and evidence points to ankyrin repeat-containing proteins (ARPs) as key virulence factors of intracellular bacteria (9). Due to high flexibility and sequence degeneracy, ankyrin domains allow for interaction with a diversity of cellular targets (10). Each ARP characterized from the intracellular pathogens Anaplasma phagocytophilum, Ehrlichia chaffeensis, Legionella pneumophila, and Orientia tsutsugamushi performs a unique task, such as directly influencing gene transcription, hijacking vesicular trafficking, interrupting signaling, or disrupting organelles (11–17). Despite the occurrence of ARPs in all rickettsial genomes, their roles during rickettsial pathogenesis remain unstudied.
Given the capacity of ARPs to act as virulence factors, it has been proposed that differences in RARP-2 may contribute to the increased virulence of the Sheila Smith strain (4, 6). Here, we demonstrate that RARP-2 is a type IV secreted effector, but only the Sheila Smith RARP-2 homolog (SS-RARP-2) colocalizes with endoplasmic reticulum (ER) membranes during infection. Expression of the virulent Sheila Smith RARP-2 homolog (SS-RARP-2) in the avirulent Iowa strain restores the lytic plaque phenotype characteristic of virulent R. rickettsii strains but does not change virulence in a guinea pig model.
RESULTS
Conservation of RARP-2 in the genus Rickettsia.
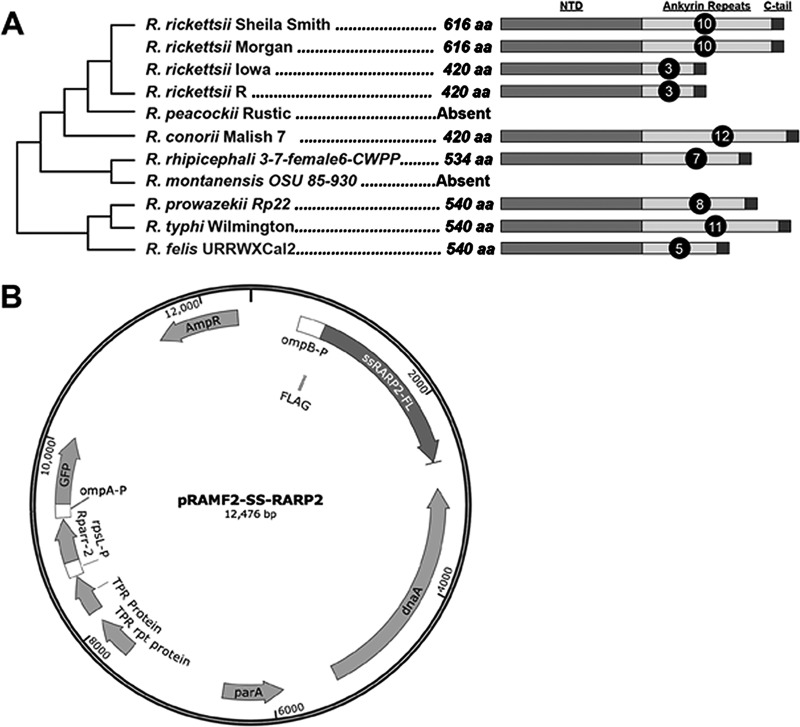
A genomic comparison of strains of R. rickettsii that differ in virulence identified a limited number of genes that uniquely diverged in the avirulent Iowa strain (4). One of these genes, RrIowa_1113, is annotated as RARP-2 in Rickettsia typhi (7). Architecturally, RARP-2 proteins contain a highly conserved N-terminal domain of unknown function, a variable ankyrin domain comprised of 3 to 12 ankyrin repeat units, and a distinct C-terminal tail (Fig. 1A; see also Fig. S1 in the supplemental material). In addition to a single nucleotide polymorphism (SNP) that distinguishes R. rickettsii Iowa RARP-2 from the other R. rickettsii strains, Iowa also has a deletion of 588 bp relative to the highly virulent Sheila Smith strain that deletes 7 of the 10 ankyrin repeat units. This deletion is also present in the moderately virulent R. rickettsii R strain (Fig. 1A), another western Montana isolate that is closely related to Sheila Smith but of reduced virulence (4).
FIG 1 .
RARP-2 organization and expression vector description. (A) Conservation of RARP-2 in R. rickettsii and selected species of the spotted fever and typhus groups of rickettsiae. Note that the number of ankyrin repeats differs in strains of R. rickettsii. See Fig. S1 for additional species and information. NTD, N-terminal domain; aa, amino acids. (B) pRAMF2 plasmid engineered for expression in rickettsiae. The ompB promoter expresses N-terminal FLAG-tagged proteins of interest; the ompA promoter expresses GFP to identify transformants.
RARP-2 homologs are conserved in pathogenic rickettsiae and harbor variable ankyrin repeats. In silico analysis of RARP-2 homologs across 49 Rickettsia genomes. Proteins were retrieved from NCBI using R. typhi strain Wilmington RARP-2 (locus tag RT0600) as the query in a blastp search against the NR (All GenBank+RefSeq nucleotides + EMBL + DDBJ + PDB) database, coupled with a search against the Conserved Domains Database (A. Marchler-Bauer et al., Nucleic Acids Res 39:D225–D229, 2011, https://doi.org/10.1093/nar/gkq1189). Searches were performed with composition-based statistics across Rickettsia. No filter was used. Default matrix parameters (BLOSUM62) and gap costs (existence, 11; extension, 1) were implemented, with an inclusion threshold of 0.005. Subjects were aligned using MUSCLE with default parameters (R. C. Edgar, Nucleic Acids Res 32:1792–1797, 2004, https://doi.org/10.1093/nar/gkh340). The alignment was manually adjusted to delineate predicted ankyrin repeats using a consensus ankyrin repeat model (L. K. Mosavi, T. J. Cammett, D. C. Desrosiers, and Z.-Y. Peng, Protein Sci 13:1435–1448, 2004, https://doi.org/10.1110/ps.03554604). These analyses indicated that RARP-2 homologs consist of a highly conserved N-terminal domain (NTD) of unknown function, an ankyrin domain composed of 3 to 12 repeats of 28 residues, and a C-terminal tail containing a predicted T4SS signal. Phylogeny at left was estimated as previously described (T. Driscoll, J. J. Gillespie, E. K. Nordberg, A. F. Azad, and B. W. Sobral, Genome Biol Evol 5:621–645, 2013, https://doi.org/10.1093/gbe/evt036), with additional genomes annotated using RAST (R. K. Aziz et al., BMC Genomics 9:75, 2008, https://doi.org/10.1186/1471-2164-9-75). The Rickettsia classification scheme follows previous studies: red, ancestral lineages; blue, transitional group; aquamarine, typhus group; and brown, spotted fever group (J. J. Gillespie et al., PLoS One 2:e266, 2007, https://doi.org/10.1371/journal.pone.0000266). Rickettsia helvetica is unclassified according to recent recommendations (T. Driscoll, J. J. Gillespie, E. K. Nordberg, A. F. Azad, and B. W. Sobral, Genome Biol Evol 5:621–645, 2013, https://doi.org/10.1093/gbe/evt036; J. J. Gillespie et al., PLoS One 3:e2018, 2008, https://doi.org/10.1371/journal.pone.0002018). Note that divergent RARP-2 homologs (found only in R. bellii and R. felis) share less than ~37% identity with RARP-2 proteins in the N-terminal domain, with very different ankyrin repeat sequences that do not match RARP-2 ankyrin repeats in reciprocal blastp searches. Download FIG S1, TIF file, 23.5 MB (24.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
pRAMF2 rickettsial expression vector
To evaluate whether SS-RARP-2 contributes to the increased virulence in strain Sheila Smith, we expressed SS-RARP-2 in the Iowa strain and evaluated changes in phenotype. RARP-2 homologs were cloned into a rickettsial expression plasmid, pRAMF2 (Fig. 1B), a modified version of the pRAM18dRGA plasmid (18). This plasmid was modified to allow for recombinant expression of N- and C-terminal FLAG-tagged proteins in rickettsiae using the strong ompB promoter. The ompB promoter was chosen to drive the expression cassette due to high levels of expression in the transformed bacteria (data not shown). The promoter rpsL is used to drive rifampin resistance, and the promoter ompA was used to drive green fluorescent protein (GFP) in order to identify positive transformants. The rpsL-rparr2-ompA-gfp cassette is located on a section of the plasmid backbone separate from the multiple cloning site (MCS) and associated FLAG tag to prevent readthrough.
Expression of SS-RARP-2 in an avirulent strain restores a lytic plaque phenotype.
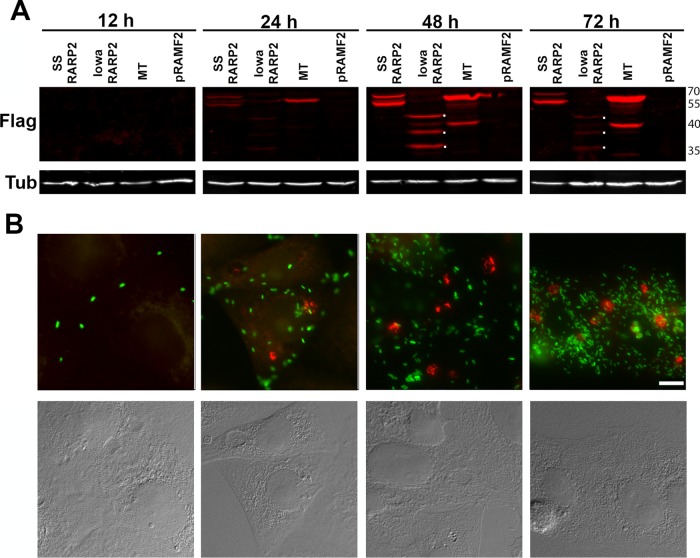
Strains of R. rickettsii differing in virulence have been shown to form distinct plaque phenotypes (6, 19). The avirulent R. rickettsii strain Iowa demonstrates characteristic nonlytic, opaque plaques, compared to the lytic, clear plaque phenotype of virulent Sheila Smith. Both SS-RARP-2 and Iowa-RARP-2 were expressed from pRAMF2 as N-terminal FLAG-tagged fusion proteins in R. rickettsii Iowa. Expression of SS-RARP-2 in Iowa restored the lytic plaque phenotype characteristic of the virulent strain (Fig. 2A). Expression of Iowa-RARP-2 in R. rickettsii Iowa did not change the plaque phenotype, indicating that the lytic phenotype was not due simply to overexpression of the SS-RARP-2 homolog.
FIG 2 .
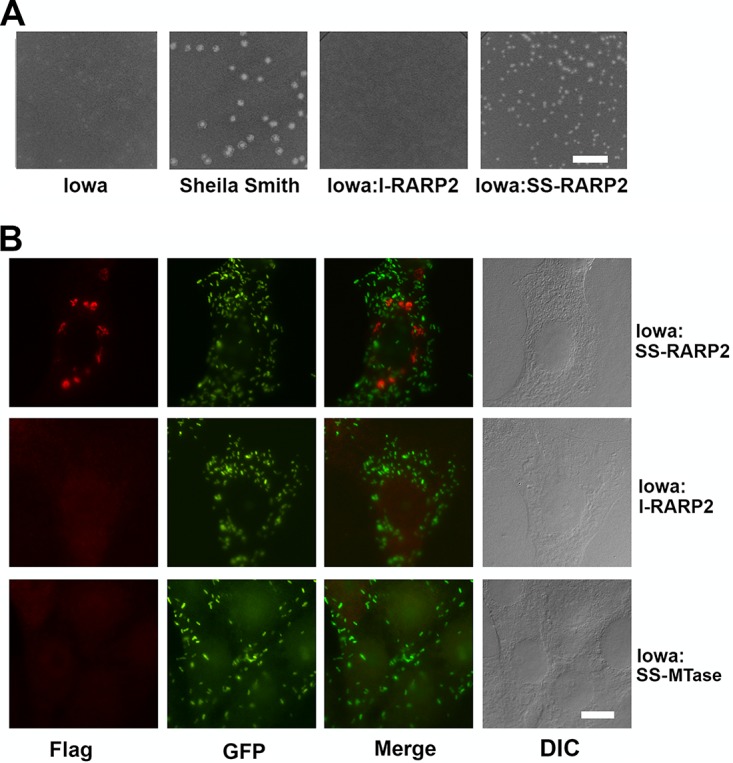
Expression of RARP-2 homologs in the R. rickettsii Iowa strain. (A) Plaque morphologies of R. rickettsii Iowa, R. rickettsii Sheila Smith, and R. rickettsii Iowa expressing Iowa RARP-2 (Iowa:I-RARP-2) or Sheila Smith RARP-2 (Iowa:SS-RARP-2). Expression of the SS-RARP-2 homolog in the Iowa strain restores a lytic plaque phenotype. Bar, 10 mm. (B). Extrarickettsial structures are formed following expression of SS-RARP-2 from R. rickettsii Iowa but not expression of Iowa-RARP-2 or MTase. FLAG-tagged RARP-2 was detected using an anti-FLAG antibody (red). Rickettsiae are expressing GFP (green). Corresponding Nomarski differential interference contrast (DIC) images are provided. Bar, 10 µm.
Vesicular structures are formed by SS-RARP-2 but not Iowa-RARP-2.
The localization of the RARP-2 homologs expressed in R. rickettsii Iowa during infection of eukaryotic cells was examined (Fig. 2B). The SS-RARP-2 construct displayed pleomorphic structures in the cytosol of infected cells and was not observed in association with the rickettsiae. Expression of SS-RARP-2 from the virulent R strain yielded similar vesicular structures (Fig. S2). Iowa-RARP-2 did not exhibit such structures, nor did a Sheila Smith control fusion protein, A1G_06690, annotated as a methyltransferase (MTase) predicted to be cytosolically localized. We therefore examined expression of these proteins during infection over a 72-h interval postinfection (Fig. 3). Expression of all constructs was detectable by immunoblotting at 24 h postinfection (hpi). The cytosolic structures formed by SS-RARP-2 were detected by 24 hpi and at each time point thereafter. Both SS-RARP-2 and the MTase showed some evidence of processing. However, the Iowa-RARP-2 exhibited a more extensive and marked proteolytic breakdown with maximal expression observed at 48 hpi. At no time did Iowa RARP-2 or MTase display extrarickettsial cytosolic structures (not shown). To confirm the expression and internal localization of the MTase in rickettsiae, a lysozyme treatment step was included in the immunofluorescent staining to confirm expression and localization within rickettsiae (Fig. S3).
FIG 3 .
Expression of RARP-2 during infection. (A) FLAG-tagged SS-RARP-2, Iowa-RARP-2, MTase, and pRAMF2 empty vector control were overexpressed from R. rickettsii Iowa in Vero76 cells (MOI of 1) and sampled at 12, 24, 48, and 72 hpi for immunoblotting with an anti-FLAG antibody. Dots to the right of the bands indicate the major breakdown products of Iowa-RARP-2 at 48 and 72 hpi. Reduced signal at 72 h is presumably due to further proteolysis of the products. Tubulin was used as a loading control. (B) Cultures infected with SS-RARP-2-FLAG were fixed at times corresponding to panel A and stained with anti-FLAG antibodies (red) for observation by immunofluorescence assay. GFP-expressing rickettsiae are green. Bar, 10 µm. Corresponding Nomarski differential interference contrast images are provided.
Expression of SS-RARP-2 from the virulent R strain yields similar vesicular structures as those expressed from the avirulent Iowa strain. R::SS-RARP-2-infected cells at 48 hpi were fixed and stained with an anti-FLAG antibody (red) and merged with images showing GFP-rickettsia (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Download FIG S2, TIF file, 1.8 MB (1.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
FLAG-MTase is expressed and is internally localized in rickettsiae. MTase was expressed from pRAM2 as an N-terminal FLAG fusion in R. rickettsii Iowa in Vero cells. Fixed cells were incubated with 5 mg/ml lysozyme (Sigma-Aldrich) in 10 mM Tris-HCl (pH 7.5) for 1 h at 37°C (+lysozyme) or not (-lysozyme) and stained for immunofluorescence as described in Materials and Methods. Bar, 10 µm. Download FIG S3, TIF file, 7.6 MB (7.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
A rabbit polyclonal antipeptide antibody was prepared to examine native RARP-2 and tested for reactivity against RARP-2 in Sheila Smith- and Iowa-infected cells. The antibody recognized overexpressed recombinant RARP-2 but did not detect specific antigen from parental R. rickettsii (Fig. S4A), suggesting that RARP-2 may be of low abundance. RARP-2 from Sheila Smith and Iowa appeared to be transcribed equivalently and at approximately the same level as dnaK (Fig. S4B).
An anti-RARP-2 antiserum fails to detect native RARP-2 from wild-type R. rickettsii Sheila Smith and Iowa. (A) A rabbit polyclonal antipeptide antibody recognized overexpressed recombinant RARP-2 but did not detect specific antigen from parental rickettsiae, suggesting that RARP-2 may be of low abundance. Anti-FLAG staining of the recombinant proteins is shown in green. Anti-RARP-2 is shown in red. A panel showing the merged images is also provided. Dots to the right of the bands indicated RARP-2 fragments recognized by the antiserum. (B) Reverse transcriptase quantitative PCR (RT-qPCR) showing equivalent transcription of RARP-2 from Sheila Smith and Iowa. Three Vero cell culture flasks per strain were infected with rickettsiae and harvested at 48 hpi. Medium was removed, and cells were lysed in 6 ml Trizol. Two hundred microliters 1-bromo-3-chloropropane/ml Trizol was added, and samples were centrifuged at 16,000 × g for 15 min. RNA was extracted from the aqueous phase using the New England BioLabs (NEB) Monarch total RNA miniprep kit. After extraction, an additional DNA removal step was performed using the Turbo DNA-free kit (Thermo Fisher). RNA quality was checked on 1% Tris-borate-EDTA (TBE) gels. Primers were designed using the IDT PrimerQuest tool (RARP2_F, CTGATGAAGGTACAACTCCTGTATTA; RARP2_R, CGGCTCCTGAATGACAAGAA; DnaK_F, CCAAGAGGTTTGCCACAAATAG; DnaK_R, GCTCTTTACCGCTTGCTTTATC). Gene fragments of DnaK and RARP-2 were cloned into TopoTA, and plasmids were used to establish standard curves and calculate qPCR efficiencies and copy numbers. RT-qPCR was performed using 1 ng of purified RNA with the Luna universal one-step RT-qPCR kit (NEB) on a Roche Light Cycler 480 II. Efficiencies were 2.00 for RARP-2 and 1.96 for DnaK. No-RT controls were included for all samples and did not show any amplification. All reactions were performed in triplicate on biological triplicate samples. Download FIG S4, TIF file, 13.7 MB (14MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
To better resolve the organization of these structures, immunoelectron microscopy using horseradish peroxidase-conjugated secondary antibodies with diaminobenzidine detection was performed on R. rickettsii Iowa expressing each of these constructs. At an ultrastructural level, SS-RARP-2 was observed in association with membranous material. Consistent with the results by immunofluorescence, the structures were also found to be pleomorphic but with multilamellar structures frequently observed (Fig. 4).
FIG 4 .
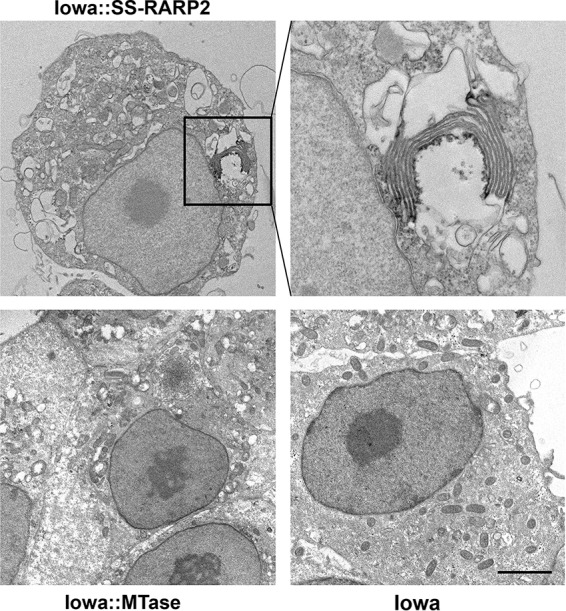
Immunoelectron microscopy of SS-RARP-2-induced formation of multilamellar structures in the host cytosol. FLAG-tagged SS-RARP-2, MTase, and pRAMF2 empty vector control were expressed from R. rickettsii Iowa in Vero76 cells (MOI of 1; 48 hpi). Cultures were fixed and stained with anti-FLAG antibody with an anti-mouse-horseradish peroxidase-conjugated secondary antibody. Specimens were developed with the Pierce diaminobenzidine (DAB) metal-enhanced substrate kit prior to embedding and sectioning for transmission electron microscopy. Bar, 5 µm.
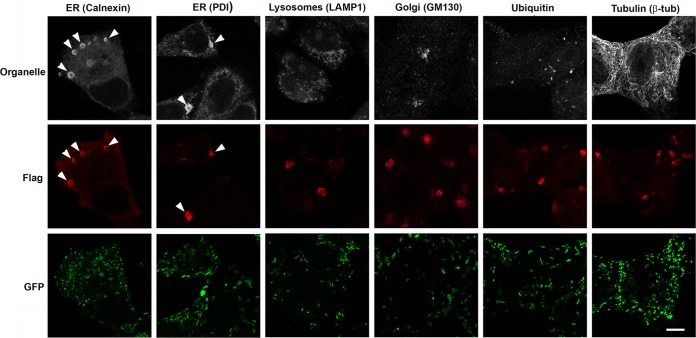
RARP-2 structures colocalize with ER markers.
The source of the membranes associated with SS-RARP-2 is unknown. Therefore, we screened for colocalization of these structures with markers for various cellular organelles by immunofluorescence (Fig. 5). SS-RARP-2 was observed in association with the ER markers calnexin and protein disulfide isomerase (PDI). No colocalization was observed with markers for the Golgi apparatus (GM130), lysosomes (Lamp1), ubiquitinated proteins (ubiquitin), or microtubules (β-tubulin). Association with autophagosomes was assessed by examination for colocalization of the SS-RARP-2 structures with expressed GFP-LC3, and no colocalization was observed (Fig. S5). These structures were associated with either calnexin or PDI 97.3% of the time. These data suggest that SS-RARP-2 structures are derived from ER membrane.
FIG 5 .
RARP-2 structures colocalize with ER markers. R. rickettsii SS-RARP-2 was expressed from R. rickettsii Iowa in Vero76 cells (MOI of 1). Cultures were stained with anti-FLAG antibody (red) and observed by immunofluorescence assay after 48 hpi. Pleomorphic structures were observed in the cytosol of infected cells. Cultures were counterstained for various organelles: ER, lysosomes, Golgi apparatus, ubiquitin, and β-tubulin (white). RARP-2 structures colocalize only with the ER markers calnexin and PDI. Rickettsiae expressing GFP are green. Arrowheads identify multiple RARP-2 vesicular structures colocalizing with ER markers. Bar, 10 µm.
RARP-2 does not associate with autophagosomes. Potential association with autophagosomes was assessed by examination of the SS-RARP-2 structures with coexpressed GFP-LC3. GFP was used as a negative control. No association with LC3 was observed. Bar, 10 µm. Download FIG S5, TIF file, 3.9 MB (4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RARP-2 is secreted from R. rickettsii during infection.
The structures formed by expressed SS-RARP-2 are clearly not associated with the intracellular rickettsiae. RARP-2 has previously been predicted to be a type IV secretion system (T4SS) effector (7). To confirm secretion from rickettsiae and exposure to the cytosol, we transformed R. rickettsii Iowa with reporter plasmids encoding N-terminally glycogen synthase kinase (GSK)-tagged SS-RARP-2, Iowa RARP-2, or GFP control proteins. GSK-tagged fusion proteins are phosphorylated only upon exposure to host cytoplasmic Ser/Thr kinases and are commonly used to evaluate secretion of various bacterial effectors into the host cytoplasm (20–22). In a time course experiment, SS-GSK-RARP-2 was expressed and phosphorylated by 24 hpi, whereas GSK-GFP was expressed but not phosphorylated (Fig. 6A).
FIG 6 .
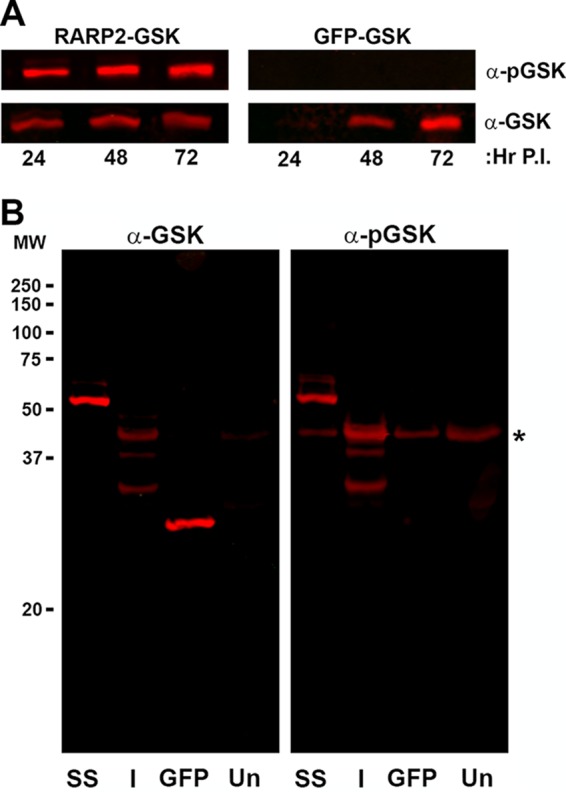
SS-RARP-2 and Iowa-RARP-2 homologs are secreted when expressed from R. rickettsii during host cell infection. (A) GSK-tagged SS-RARP-2 and GSK-tagged GFP were expressed from R. rickettsii Iowa, and samples were collected at 24, 48, and 72 hpi. Specimens were immunoblotted for detection with an anti-GSK epitope tag antibody and a phospho-GSK-specific antibody to detect tagged protein exposure to host cytosolic kinases. GFP-GSK was used as a nonsecreted control protein and does not show reactivity with the anti-phospho-GSK probe. (B) GSK-tagged SS-RARP-2, GSK-tagged Iowa-RARP-2, and GSK-tagged GFP were expressed from R. rickettsii Iowa. Whole-cell lysates were prepared from infected cultures and uninfected cultures (Un) at 48 hpi and probed for phosphorylation of the GSK epitope as described above. Despite proteolysis of GSK-Iowa-RARP-2, multiple fragments were phosphorylated, indicating secretion and exposure to the cytosol. Host cellular phospho-GSK is indicated by an asterisk.
Examination of SS-RARP-2 and Iowa-RARP-2 at 48 hpi indicated that both are translocated and exposed to the cytosol (Fig. 6B). As was observed with the FLAG-tagged Iowa-RARP-2, Iowa-GSK-RARP-2 showed evidence of proteolytic degradation.
Ectopic expression.
To evaluate RARP-2 effects on host cells in the absence of additional rickettsial proteins, we ectopically expressed full-length GFP-SS-RARP-2 in Vero cells (Fig. 7) and visualized the transfected cells by microscopy. The SS-RARP-2 construct formed large pleomorphic structures morphologically identical to those formed after secretion from rickettsiae. Neither Iowa-RARP-2 nor the MTase formed similar structures even when expressed within the cytosol of eukaryotic cells. Collectively, the results suggest that Iowa-RARP-2 is secreted from rickettsiae but, even when present in the cytosol, does not form membranous structures as does RARP-2 from the virulent Sheila Smith strain.
FIG 7 .
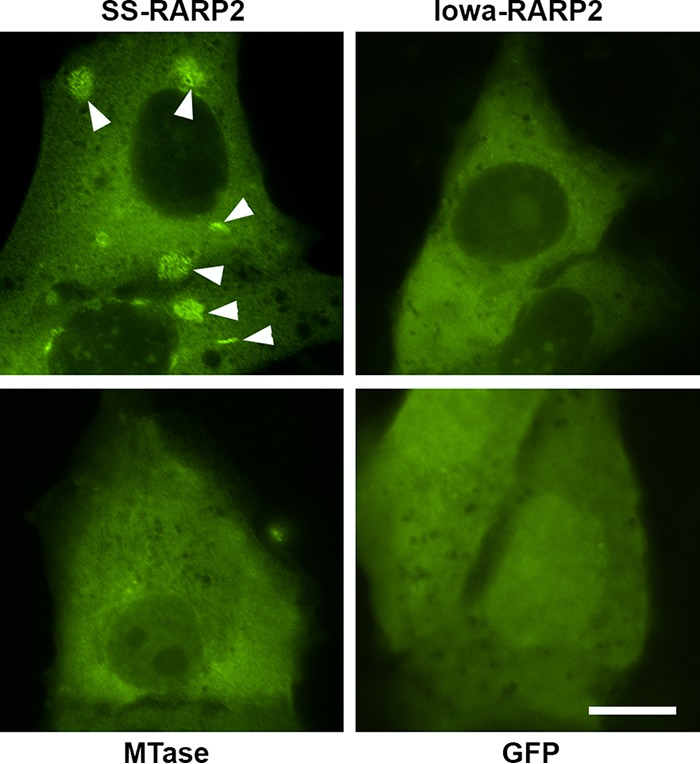
Ectopic expression of RARP-2 in Vero cells. Pleomorphic vesicular structures are formed by R. rickettsii Sheila Smith RARP-2 during ectopic expression (arrowheads). GFP-tagged RARP-2 homologs or an empty GFP expression vector was transfected into Vero cells, fixed after 18 h of expression, and visualized by confocal microscopy. Shown are R. rickettsii Sheila Smith RARP-2 (SS-RARP2), R. rickettsii Iowa RARP-2 (Iowa-RARP2), R. rickettsii Sheila Smith methyltransferase A1G_06690 (MTase), and pEGFP-C1 empty vector control (GFP). Bar, 10 µm.
RARP-2 is a type IV secreted effector.
The T4SS coupling protein VirD4 regulates effector entry into the secretion channel (23). The Rickettsia VirD4 homolog (RvhD4) has been shown to directly interact with the T4SS effector RalF (24) as well as those of bacteria closely related to Rickettsia, Ats1 of Anaplasma phagocytophilum and ECH0825 of Ehrlichia chaffeensis (25, 26).
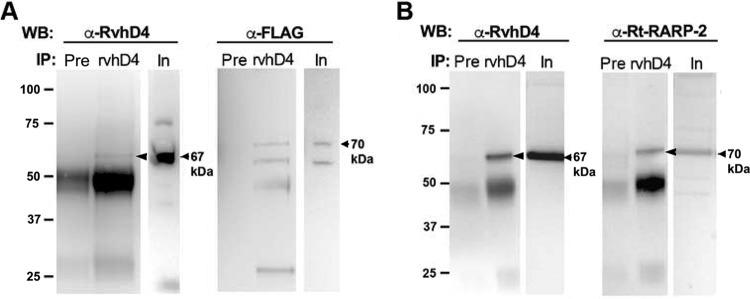
To evaluate the potential role of RARP-2 as a T4SS effector, we tested for an interaction of R. rickettsii and R. typhi RARP-2 with RvhD4 by coimmunoprecipitation. FLAG-tagged SS-RARP-2 and R. typhi RARP-2 were coimmunoprecipitated using anti-R. typhi RvhD4 antibody or preimmune serum as a negative control. Following immunoprecipitation by anti-RvhD4 antibody, RARP-2 was detected by immunoblotting using anti-R. typhi RARP-2 or anti-FLAG antibody (Fig. 8A and B). RARP-2-FLAG was not precipitated by the preimmune serum. These results indicate an interaction of RARP-2 with the T4SS effector coupling protein RvhD4, implying that RARP-2 is a T4SS effector.
FIG 8 .
Type IV secretion of RARP-2. (A) FLAG-tagged SS-RARP-2 expressed in R. rickettsii Iowa was coimmunoprecipitated using anti-RvhD4 antibody or preimmune serum (Pre). Input (In) and immunoprecipitated proteins (IP) were detected by Western blotting (WB) using anti-RvhD4 antibody (left) or anti-FLAG antibody (right). The expected size for SS-RvhD4 is 67 kDa (arrow), and the expected size for SS-RARP-2 is 70 kDa (arrow). (B) Endogenous R. typhi RARP-2 (Rt-RARP-2) homolog was coimmunoprecipitated using anti-RvhD4 antibody or preimmune serum (Pre). Input (In) and precipitated proteins (IP) were detected by Western blotting (WB) using anti-RvhD4 antibody or anti-Rt-RARP-2 antibody. The expected size for Rt-RvhD4 is 67 kDa (arrow), and the expected size for Rt-RARP-2 is 70 kDa (arrow). Bands near 50 kDa represent immunoglobulin heavy chain.
The association of R. typhi RARP-2 with RvhD4 was corroborated by a bacterial two-hybrid assay (Fig. S6). Collectively, these data indicate that RARP-2 interacts with the T4SS effector coupling protein RvhD4 and that RARP-2 is a T4SS effector.
Association of R. typhi RARP-2 with RvhD4 was corroborated by a bacterial two-hybrid assay. (A) The bacterial two-hybrid assay was performed by transforming codon-optimized R. typhi RvhD4 bait and R. typhi RARP-2 prey vectors into Escherichia coli BacterioMatch II reporter electrocompetent cells. Growth on dual selective medium indicates that the RARP-2 C-terminal tail interacts with RvhD4 (FL), and this interaction is abolished when 37 residues of the Rt-RARP-2 C-terminal tail are deleted (ΔCT). (B) Quantification of panel A. Percent growth was calculated from cotransformed bacterial CFUs on dual selective screening medium relative to CFU obtained on nonselective medium. Error bars represent mean ± standard deviation (SD) from three independent experiments (Student’s two-sided t test). Download FIG S6, TIF file, 1.7 MB (1.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Functional domain analysis of RARP-2.
ARP effector domain architectures frequently coexist with various functional modules. RARP-2 might act similarly to other known microbial ARPs that use the ankyrin repeats for substrate binding, while the N terminus possesses the functional domain that acts on the associated host protein (10). While the C-terminal region of RARP-2 has been predicted to contain ankyrin repeats, the highly conserved N-terminal region has no predicted functional domains. However, structural modeling of the N-terminal region using Phyre2 (27) revealed similarities to clan CD cysteine proteases (Fig. S7) (27, 28). We therefore generated FLAG-tagged constructs with the predicted catalytic cysteine mutated to an alanine (C109A). Constructs with the C-terminal domain deleted from C109A were created (C109A-ΔCT). In addition, wild-type RARP-2 (with the cysteine intact) but with the ankyrin repeat domain deleted (ΔAnk) was also expressed.
RARP-2 N-terminal domain harbors a putative clan CD cysteine protease active site. Using Phyre2 (L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass, and M. J. Sternberg, Nat Protoc 10:845–858, 2015, https://doi.org/10.1038/nprot.2015.053), we modeled several RARP-2 homologs to clan CD cysteine endopeptidases (data not shown). Accordingly, we aligned several RARP-2 homologs with proteases from selected clan CD families: legumain (C13), caspase 1 (C14), clostripain (C11), and gingipain R (C25). The sequence segments containing the experimentally identified or predicted catalytic His and Cys residues (pink and yellow) in each of the four families and RARP-2 homologs were subsequently adjusted based on a previous study [J. M. Chen, N. D. Rawlings, R. A. Stevens, and A. J. Barrett, FEBS Lett 441:361–365, 1998, https://doi.org/10.1016/S0014-5793(98)01574-9]. Upstream of the catalytic residues, there is a block of four predominantly hydrophobic residues (green). Other residues that tend to be conserved between the families are printed in white on black. Blue numbers indicate amino acid coordinates flanking the active site. Accession numbers from UniProt are as follows: Q99538, human legumain; P49046, jack bean (Canavalia ensiformis) asparaginyl endopeptidase; P42665, Schistosoma japonicum hemoglobinase; P49048, Caenorhabditis elegans hypothetical protein T05E11.6; Q92643, human GPI8 protein; P42574, human caspase 3; P55210, human caspase 7; P55212, human caspase 6; O01382, Drosophila melanogaster caspase; P29466, human caspase 1; P42573, Caenorhabditis elegans CED3 protein; P09870, Clostridium histolyticum α-clostripain; Q8A866, Bacteroides thetaiotaomicron clostripain-related protein; Q9WYY6, Thermotoga maritima clostripain-related protein; Q51816, Porphyromonas gingivalis gingipain R; B0VHP1, Cloacimonas acidaminovorans putative gingipain R; F2I9M7, Fluviicola taffensis gingipain R; A0A075MRD2, endosymbiont of Acanthamoeba sp. strain UWC8 uncharacterized protein; Q1RID2, Rickettsia bellii putative ankyrin repeat protein RBE_0801; Q4UKP4, Rickettsia felis uncharacterized protein; Q1RI97, Rickettsia bellii uncharacterized protein; Q68WC7, Rickettsia typhi uncharacterized protein; A0A0H3AY85, Rickettsia rickettsii strain Sheila Smith uncharacterized protein; B0BUJ3, Rickettsia rickettsii Iowa uncharacterized protein. Download FIG S7, TIF file, 1 MB (1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
These constructs were transformed into R. rickettsii Iowa, and transformants were analyzed for changes in SS-RARP-2 localization and differences in plaque phenotypes (Fig. 9). No transformants were recovered for SS-RARP-2 lacking the C terminus (SS-RARP-2-ΔCT) in three attempts. The SS-RARP-2-C109A construct was secreted and formed vesicular structures, although these tended to be smaller and showed a more perinuclear localization. Association with ER markers was retained (Fig. S8). However, the SS-RARP-2-ΔCT-C109A mutant did not form structures and produced opaque plaques. T4SS secretion signals are typically localized to the C terminus of effector proteins (29–32); thus, this may signify a failure of secretion. Interestingly, the plaque phenotype was also opaque for SS-RARP-2-C109A, suggesting that even though this construct is secreted and associates with membrane, the cysteine is essential for the lytic phenotype of SS-RARP-2. Both the putative catalytically active cysteine residue and appropriate localization conferred by the ankyrin repeats therefore seem necessary for the change in plaque phenotype.
FIG 9 .
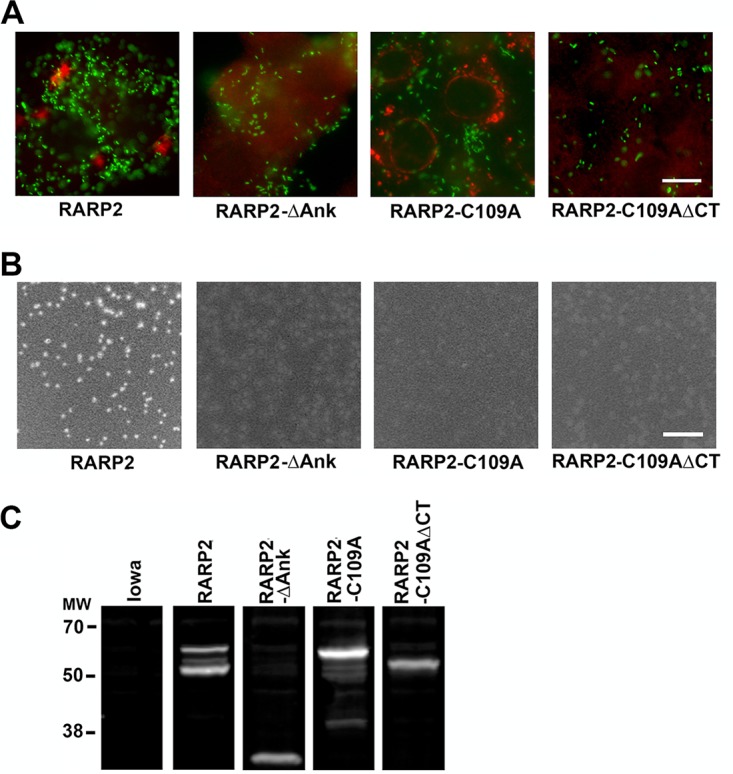
The C-terminal tail and ankyrin repeats are required for the formation of RARP-2 vesicular structures. (A) Deletion constructs were overexpressed from R. rickettsii Iowa infecting Vero76 cells (MOI of 1). Constructs examined were SS-RARP-2 as an N-terminal FLAG fusion (RARP-2); RARP-2 with the ankyrin repeat domain deleted (RARP-2-ΔAnk); RARP-2 with the predicted catalytic cysteine mutated to an alanine (RARP-2-C109A); and RARP-2-N with the C-terminal domain deleted from C109A (RARP-2-C109A-ΔCT). Cultures were stained with anti-FLAG antibody (red) and observed by immunofluorescence assay after 48 hpi. GFP-expressing rickettsiae are green. Bar, 10 μm. (B) Plaque morphologies of R rickettsii Iowa expressing each of the constructs above. Bar, 10 mm. (C) Immunoblot probed with anti-FLAG antibody demonstrating expression of FLAG-tagged recombinant proteins above and the absence of signal from parental R rickettsii Iowa. All panels are from a single gel and immunoblot taken at the same exposure. Lanes were separated for presentation.
The SS-RARP-2-C109A construct is secreted and forms perinuclear vesicular structures but retains association with ER markers. Iowa::SS-RARP-2-C109A-infected cells at 48 hpi were fixed and stained with an anti-FLAG antibody (red) and anticalnexin or PDI (white). GFP-expressing rickettsiae are green. Images were merged and include DAPI staining (blue). Bar, 10 µm. Download FIG S8, TIF file, 1.9 MB (2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
In transformants lacking the ankyrin repeat domain (ΔAnk), no cytosolic structures were observed and the plaque phenotype remained opaque. Because the C-terminal tail of the ΔAnk construct is intact, we would predict that it is secreted but may not function normally. This conclusion is reinforced by the observation that Iowa-RARP-2, which contains only three ankyrin repeats compared with 10 in SS-RARP-2, is secreted but does not form cytosolic vesicular structures. Collectively, these domain analysis data indicate that the C-terminal tail and ankyrin repeat domains are necessary for vesicular structure formation and that the C-terminal tail is required for secretion.
Expression of SS-RARP-2 homolog in avirulent Iowa strain does not restore virulence in guinea pig model.
The guinea pig model of infection was used to determine the effect of RARP-2 complementation on R. rickettsii Iowa virulence. Guinea pigs were inoculated with 100 PFU of R. rickettsii Sheila Smith, R. rickettsii Iowa, R. rickettsii Iowa expressing SS-RARP-2, or R. rickettsii Iowa expressing MTase. Temperatures were monitored for 16 days. Only those animals infected with R. rickettsii Sheila Smith developed fevers; thus, complementation of R. rickettsii Iowa with Sheila Smith RARP-2 did not restore virulence to the avirulent Iowa strain (Fig. 10). All animals showed seroconversion, indicating that they were infected. This level of inoculum does not provide sufficient antigenic mass to cause seroconversion without replication (6).
FIG 10 .
Complementation of RARP-2 in R. rickettsii Iowa does not restore virulence. Guinea pigs were infected with approximately 100 PFU of R. rickettsii Sheila Smith (SS), Iowa, Iowa:SS-RARP-2, or Iowa:SS-MTase, and temperatures were monitored for 16 days.
DISCUSSION
Strains of R. rickettsii differ dramatically in the severity of disease in humans as well as in animal model systems despite being genetically very similar (4, 33). R. rickettsii has a small genome of approx 1.3 Mbp that is predicted to encode approximately 1,350 proteins (6). Comparative genomics has identified relatively few differentially encoded proteins as putative virulence factors (4, 6), but thus far, no molecular basis for the strain-level differences in virulence has been discerned. One of these putative virulence factors is the ankyrin repeat-containing protein RARP-2, which in the avirulent Iowa strain contains a large internal deletion relative to the virulent Sheila Smith strain and possesses one nonsynonymous SNP not observed in other R. rickettsii strains (4). Here, we have complemented the avirulent Iowa strain with RARP-2 from virulent Sheila Smith. Using epitope-tagged versions of RARP-2 expressed from rickettsiae or ectopically in eukaryotic cells, we show that RARP-2 is secreted into the cytosol of the host and that RARP-2 from the virulent strain associates with host ER to form membranous structures. RARP-2 from the avirulent Iowa strain was also secreted into the cytosol but did not exhibit demonstrable association with membrane. Interestingly, a critical cysteine residue from a predicted protease catalytic active site on RARP-2 was required for the lytic plaque phenotype but not for association with host membrane. Interaction with a rickettsial VirD4 homolog indicates that secretion is via a T4SS. Although expression of Sheila Smith RARP-2 in R. rickettsii Iowa altered the phenotype, it was unable to restore virulence in a guinea pig model of infection.
Rickettsia genomes encode variable numbers of predicted ankyrin repeat-containing proteins. Genomes showing high incidences of horizontal gene transfer, such as Rickettsia felis, Rickettsia bellii, and Rickettsia massiliae, are most abundant in ARPs (34). The most conserved ARPs are RARP-1 and RARP-2 (7). The ubiquitous RARP-1 was recently characterized as a Sec-TolC secreted effector (35), while RARP-2 is a predicted rickettsial effector conserved in most, but not all, Rickettsia species (7). RARP-2, when present, consists of an N-terminal domain of unknown function, an ankyrin domain composed of 3 to 12 repeats, and a distinct C-terminal tail. The reduced number of ankyrin repeats in RARP-2 of the attenuated R. rickettsii strains is suggestive of possible reductive evolution, although the impact on virulence is unclear. Alignment of RARP-2 from the R. rickettsii virulent strain Sheila Smith and the avirulent strain Iowa shows the deletion of internal ankyrin repeats from Iowa but conservation of the first two and last repeats, as well as the C-terminal tail. Interestingly, this conservation of the first two and last repeats occurs across all rickettsial RARP-2 homologs. It is possible that the ankyrin repeats play a role in targeting of RARP-2 in the host cell. Neither the Iowa-RARP-2, containing only three ankyrin repeats, nor the SS-RARP-2-ΔAnk construct formed vesicular structures. It is possible that with a diminished or deleted ankyrin repeat domain, the protein is aberrantly targeted. A greater number of ankyrin repeats may increase the avidity of RARP-2 for its target.
Structural modeling of the N-terminal domain suggests that RARP-2 may possess a cysteine protease active site similar to other clan CD cysteine proteases such as eukaryotic legumains and caspases and bacterial gingipains and clostripains, which are secreted effectors from highly pathogenic bacteria (28, 36, 37). This may be related to the loss of the lytic plaque phenotype after mutation of the potential catalytic cysteine residue in SS-RARP-2. Interestingly, no SS-RARP-2-ΔCT transformants could be recovered; yet after mutating the putative protease catalytic cysteine 109 to an alanine, ΔCT transformants were readily obtained and appeared to not be secreted compared to SS-RARP-2-C109A. If the N-terminal domain indeed harbors a protease activity, this could contribute to toxicity when overexpressed without a secretion signal. It is unknown whether endogenous autocatalytic protease activity occurs in an incorrectly targeted Iowa RARP-2 and contributes to the proteolytic breakdown observed.
RARP-2 homologs from members of both the spotted fever group and the typhus group directly interact with the T4SS coupling protein RvhD4. This suggests that, despite having some phylogenetic group-level variation in their C-terminal-tail secretion signals, RARP-2 homologs are T4SS effectors. All sequenced Rickettsia genomes encode a T4SS (23), and several putative rickettsial T4SS effectors have been predicted (7). RARP-2 appears to be a cytosolically localized T4SS effector. Clearly, RARP-2 from the virulent Sheila Smith strain targets different sites within the host cell once secreted, likely due to the presence of additional ankyrin repeat units.
Previous studies indicate that RARP-2 gene expression increases during rickettsial transition to a mammalian host environment. Transcription of R. rickettsii R strain RARP-2 showed a ≥3-fold increase during Vero cell infection compared to infection of an Ixodes scapularis tick embryo-derived cell line, ISE6 (38). Similarly, Galletti et. al. showed a 2-fold increase in R. rickettsii Taiaçu RARP-2 expression in ticks with both simultaneous blood-feeding and temperature upshift compared to unfed, room-temperature ticks (39). Whether RARP-2 is necessary or functional during rickettsial growth in arthropods remains to be determined.
The lytic plaque phenotype has classically been correlated with virulent strains of R. rickettsii, while opaque plaques were associated with avirulent strains incapable of lysing the host cell (40–43). The restoration of the lytic phenotype in Iowa expressing SS-RARP-2 suggested potentially higher virulence than the avirulent parental strain. Surprisingly, there was no change in the ability to cause fever in the guinea pig model of infection. The Iowa strain has several genomic differences from the virulent strains (4). It is likely that factors other than RARP-2 prevent complementation of virulence in the guinea pig model. We had previously observed that reconstitution of another potential rickettsial virulence factor, RelA/SpoT, in the Iowa and R strains also influenced lytic versus opaque plaque phenotypes but did not alter virulence in the guinea pig model (44). Plaque phenotype alone may therefore not be a reliable marker for virulence in R. rickettsii.
The availability of multiple complete genome sequences along with recent advances in rickettsial genetics (45) now offers the opportunity to more fully characterize the molecular basis for rickettsial pathogenesis. Although genomic comparisons provide some guidance as to certain genes that may be involved in pathogenesis, bioinformatics alone will not suffice for associating genotype with phenotype. For example, some rickettsial proteins, such as the autotransporter Sca2, are present and functional in many avirulent SFG rickettsiae and yet their disruption in R. rickettsii decreases virulence (46). A comprehensive inventory of rickettsial virulence determinants will therefore necessitate a number of complementary approaches.
MATERIALS AND METHODS
Bacterial strains, growth, and purification.
R. rickettsii strains Sheila Smith (47) and Iowa (6, 48) were grown and propagated at 34°C in Vero cells in M199 medium plus 2% fetal bovine serum and purified by Renografin density gradient centrifugation (49) with modifications (50). Infected cells were lysed by Dounce homogenization and partially purified by differential centrifugation followed by centrifugation through a 30% Renografin pad. The rickettsiae were washed twice in 250 mM sucrose for use in plasmid transformations or infections. Numbers of viable rickettsiae were determined by plaque assay on Vero cell monolayers as previously described (46, 51). Procedures for R. typhi growth and purification are found in Text S1 in the supplemental material.
Supplemental materials and methods. Download TEXT S1, DOCX file, 0.02 MB (19.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Transformation of rickettsiae.
Purified R. rickettsii was transformed with plasmids as previously described (46, 52) Clonal transformants were obtained by 4 repetitions of picking individual plaques, expanding the plaques in Vero cell monolayers with M199 and 200 ng/ml rifampin for PCR verification, and then recloning as previously described (46).
Plasmid construction.
The shuttle vector pRAMF2 was modified from the original rickettsial shuttle vector pRAM18dRGA vector developed by Burkhardt et al. (18). Specifically, pRAM18dRGA was digested with KpnI to remove the multiple cloning site (MCS). A synthetic MCS was ordered from Integrated DNA Technologies (IDT; Coralville, IA) which contained restriction enzyme sites (PspOMI and RsrII) to insert a promoter of interest followed by an ATG start codon and in-frame FLAG tag sequence. Immediately following the FLAG sequence are restriction sites (BsiWI/BssHII) to allow in-frame insertion of a gene of interest for recombinant expression followed by an in-frame stop codon. The synthetic MCS was designed as two separate ~60-bp single-stranded cDNA fragments which, when annealed, contained overhangs that simulated KpnI digestion. The MCS was then cloned into the KpnI-digested pRAM18dRGA. The pRAMF2-SS-RARP-2-ΔAnk construct was created by dual digestion of the pRAMF2 vector as described above but with InFusion insertion of two fragments: (i) residues 1 to 279 and (ii) residues 560 to 616. Site-directed mutagenesis (QuikChange Lightning site-directed mutagenesis kit; Agilent) was used to create the C109A mutants. pAHG was constructed by replacing the N-terminal FLAG tag of pRAMF2 with GSK (MSGRPRTTSFAES).
Enhanced GFP (EGFP) fusions to the N terminus of RARP-2 were constructed using pEGFP-C1 (Clontech, Mountain View, CA). Genes were amplified from R. rickettsii strains as indicated using forward primers (IDT, Coralville, IA) that incorporated an XhoI or BamHI site and reverse primers (IDT) that incorporated an EcoRI site.
The plasmid expressing GFP-LC3 (53) was a kind gift of T. Yoshimori.
Ectopic expression.
Plasmids were used to transfect Vero cells on glass coverslips in 24-well plates using Lipofectamine reagents (Invitrogen) according to the manufacturer’s instructions.
Antibodies.
Monoclonal antibodies (MAbs) 13-2 and 13-3 have been previously described (54). Anti-FLAG was from Sigma. Monoclonal antibody to Lamp1 was obtained from the Developmental Studies Hybridoma Bank. Anticalnexin and anti-GM130 were from Abcam. Antimonoubiquitin and antipolyubiquitin were from Enzo Life Sciences, and anti-β-tubulin was from BD Pharmingen. Secondary antibodies [anti-mouse Alexa Fluor 488, anti-rabbit Alexa Fluor 594, or peroxidase-conjugated F(ab′)2 donkey anti-mouse IgG] were from Jackson ImmunoResearch.
Immunofluorescence.
Vero cells were infected with rickettsia at a multiplicity of infection (MOI) of 5 overnight at 37°C in M199 medium. Monolayers were fixed in 3.7% paraformaldehyde and permeabilized with phosphate-buffered saline (PBS) with 0.01% Triton X-100 and 0.05% sodium dodecyl sulfate (SDS). Fixed coverslips were stained with primary antibodies as described above, washed, and detected with secondary anti-mouse Alexa Fluor 594 or anti-rabbit Alexa Fluor 594 antibodies. Rickettsiae were detected based upon GFP expression. Images were acquired on a Nikon Eclipse 80i microscope with a 60× 1.4-numerical-aperture oil immersion objective and a Nikon DS-Qi1Mc camera. Confocal microscopy was performed on a Zeiss LSM-880 microscope.
SDS-PAGE and immunoblotting.
R. rickettsii lysates in Laemmli buffer were separated by electrophoresis on a 10% sodium dodecyl sulfate-polyacrylamide gel for 1 h at 150 V. Protein was transferred at 100 V for 1 h to a polyvinylidene difluoride (PVDF) membrane for immunoblotting with primary antibody for 1 h in Odyssey blocking buffer (Li-Cor). Blots were washed with Tris-buffered saline–0.1% Tween 20 (TBST) and incubated with appropriate secondary antibody. Blots were processed and imaged using the Li-Cor Odyssey CLx infrared imaging system according to the manufacturer’s instructions.
Procedures for the coimmunoprecipitations with anti-RvhD4 and immunoblotting with anti-Rt-RARP-2 or anti-FLAG are found in Text S1 in the supplemental material.
GSK secretion assay.
The glycogen synthase kinase (GSK) tag was fused to RARP-2 as described above. Secreted GSK-tagged protein was detected by immunoblotting with a phosphospecific GSK-3β antibody (Cell Signaling Technology). A GSK-3β tag antibody (Cell Signaling Technology) was used to detect total (nonphosphorylated and phosphorylated) GSK.
Transmission electron microscopy.
Vero cells were grown on Thermanox coverslips (Nunc) and infected at an MOI of approximately 1 with R. rickettsii Iowa expressing SS-RARP-2-FLAG, SS-MTase-FLAG, or parental Iowa for 48 h. Cells were rinsed twice with Hanks balanced salt solution (HBSS) and fixed with periodate-lysine-paraformaldehyde (PLP fixative; 75 mM lysine, 37 mM sodium phosphate, 10 mM sodium periodate, 2% paraformaldehyde) plus 0.25% glutaraldehyde for 2 h at room temperature. Specimens were processed for transmission electron microscopy as previously described (55). Micrographs were acquired using a Hitachi 7500 transmission electron microscope (TEM) (Hitachi High Technologies America, Inc.) at 80 kV and recorded on a bottom-mount AMT camera system (Advanced Microscopy Techniques Corp.).
Guinea pig inoculations.
Female Hartley strain guinea pigs (350 g) were purchased from Charles River Laboratories (MA) and housed in an animal biosafety level 3 laboratory under a protocol approved by the Rocky Mountain Laboratories Animal Care and Use Committee. Guinea pigs were implanted with transponders (Bio Medic Data Systems, Inc., Seaford, DE) to monitor temperatures. R. rickettsii strains Sheila Smith, Iowa, Iowa::pRAMF2-SS-RARP-2, and Iowa::pRAMF2-MTase were inoculated intradermally with 100 PFU. Temperatures were monitored for 16 days after infection. Animals were euthanized on day 28, and sera were collected for antibody titration.
ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the NIAID/NIH (1ZAAI000977-12 to T.H.) and grants (R01AI017828 and R01AI126853 to A.F.A. and R21AI26108 to J.J.G. and M.S.R.). S.S.L. is supported in part by NIH/NIAID T32AI007540.
We thank Uli Munderloh for her generous sharing of plasmid pRAM18dRGA. We also thank Magda Beier-Sexton for administrative and technical support to the UMB authors.
Footnotes
Citation Lehman SS, Noriea NF, Aistleitner K, Clark TR, Dooley CA, Nair V, Kaur SJ, Rahman MS, Gillespie JJ, Azad AF, Hackstadt T. 2018. The rickettsial ankyrin repeat protein 2 is a type IV secreted effector that associates with the endoplasmic reticulum. mBio 9:e00975-18. https://doi.org/10.1128/mBio.00975-18.
REFERENCES
- 1.Gillespie JJ, Williams K, Shukla M, Snyder EE, Nordberg EK, Ceraul SM, Dharmanolla C, Rainey D, Soneja J, Shallom JM, Vishnubhat ND, Wattam R, Purkayastha A, Czar M, Crasta O, Setubal JC, Azad AF, Sobral BS. 2008. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One 3:e2018. doi: 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiedeman C, McQuiston J, Ngo TH, Lancaster MJ, McElroy K, Carpenter LR, Mosites E, Dunn JR. 2009. Knowledge, attitudes, and practices regarding Rocky Mountain spotted fever among health care providers, Tennessee. Am J Trop Med Hyg 88:162–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anacker RL, List RH, Mann RE, Wiedbrauk DL. 1986. Antigenic heterogeneity in high- and low-virulence strains of Rickettsia rickettsii revealed by monoclonal antibodies. Infect Immun 51:653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark TR, Noriea NF, Bublitz DC, Ellison DW, Martens C, Lutter EI, Hackstadt T. 2015. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun 83:1568–1576. doi: 10.1128/IAI.03140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ricketts HT. 1991. Some aspects of Rocky Mountain spotted fever as shown by recent investigations [1909]. Rev Infect Dis 13:1227–1240. doi: 10.1093/clinids/13.6.1227. [DOI] [PubMed] [Google Scholar]
- 6.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Porcella SF, Hackstadt T. 2008. Genomic comparison of virulent Rickettsia rickettsii Sheila Smith and avirulent Rickettsia rickettsii Iowa. Infect Immun 76:542–550. doi: 10.1128/IAI.00952-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillespie JJ, Kaur SJ, Rahman MS, Rennoll-Bankert K, Sears KT, Beier-Sexton M, Azad AF. 2015. Secretome of obligate intracellular Rickettsia. FEMS Microbiol Rev 39:47–80. doi: 10.1111/1574-6976.12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosavi LK, Minor DL Jr, Peng ZY. 2002. Consensus-derived structural determinants of the ankyrin repeat motif. Proc Natl Acad Sci U S A 99:16029–16034. doi: 10.1073/pnas.252537899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Al-Khodor S, Price CT, Kalia A, Abu Kwaik Y. 2010. Functional diversity of ankyrin repeats in microbial proteins. Trends Microbiol 18:132–139. doi: 10.1016/j.tim.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jernigan KK, Bordenstein SR. 2014. Ankyrin domains across the Tree of Life. PeerJ 2:e264. doi: 10.7717/peerj.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol 9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- 13.Price CT, Jones SC, Amundson KE, Kwaik YA. 2010. Host-mediated post-translational prenylation of novel Dot/Icm-translocated effectors of Legionella pneumophila. Front Microbiol 1:131. doi: 10.3389/fmicb.2010.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rikihisa Y, Lin M. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis type IV secretion and Ank proteins. Curr Opin Microbiol 13:59–66. doi: 10.1016/j.mib.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VieBrock L, Evans SM, Beyer AR, Larson CL, Beare PA, Ge H, Singh S, Rodino KG, Heinzen RA, Richards AL, Carlyon JA. 2014. Orientia tsutsugamushi ankyrin repeat-containing protein family members are type 1 secretion system substrates that traffic to the host cell endoplasmic reticulum. Front Cell Infect Microbiol 4:186. doi: 10.3389/fcimb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Stevenson HL, Scott MJ, Ismail N. 2015. Type I interferon contributes to noncanonical inflammasome activation, mediates immunopathology, and impairs protective immunity during fatal infection with lipopolysaccharide-negative ehrlichiae. Am J Pathol 185:446–461. doi: 10.1016/j.ajpath.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW. 2009. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect Immun 77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burkhardt NY, Baldridge GD, Williamson PC, Billingsley PM, Heu CC, Felsheim RF, Kurtti TJ, Munderloh UG. 2011. Development of shuttle vectors for transformation of diverse Rickettsia species. PLoS One 6:e29511. doi: 10.1371/journal.pone.0029511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackstadt T, Messer R, Cieplak W, Peacock MG. 1992. Evidence for the proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect Immun 60:159–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauler LD, Hackstadt T. 2014. Expression and targeting of secreted proteins from Chlamydia trachomatis. J Bacteriol 196:1325–1334. doi: 10.1128/JB.01290-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brodsky IE, Medzhitov R. 2008. Reduced secretion of YopJ by Yersinia limits in vivo cell death but enhances bacterial virulence. PLoS Pathog 4:e1000067. doi: 10.1371/journal.ppat.1000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia JT, Ferracci F, Jackson MW, Joseph SS, Pattis I, Plano LR, Fischer W, Plano GV. 2006. Measurement of effector protein injection by type III and type IV secretion systems by using a 13-residue phosphorylatable glycogen synthase kinase tag. Infect Immun 74:5645–5657. doi: 10.1128/IAI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillespie JJ, Ammerman NC, Dreher-Lesnick SM, Rahman MS, Worley MJ, Setubal JC, Sobral BS, Azad AF. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. doi: 10.1371/journal.pone.0004833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rennoll-Bankert KE, Rahman MS, Gillespie JJ, Guillotte ML, Kaur SJ, Lehman SS, Beier-Sexton M, Azad AF. 2015. Which way in? The RalF Arf-GEF orchestrates rickettsia host cell invasion. PLoS Pathog 11:e1005115. doi: 10.1371/journal.ppat.1005115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu H, Bao W, Lin M, Niu H, Rikihisa Y. 2012. Ehrlichia type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol 14:1037–1050. doi: 10.1111/j.1462-5822.2012.01775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu H, Kozjak-Pavlovic V, Rudel T, Rikihisa Y. 2010. Anaplasma phagocytophilum Ats-1 is imported into host cell mitochondria and interferes with apoptosis induction. PLoS Pathog 6:e1000774. doi: 10.1371/journal.ppat.1000774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JM, Rawlings ND, Stevens RA, Barrett AJ. 1998. Identification of the active site of legumain links it to caspases, clostripain and gingipains in a new clan of cysteine endopeptidases. FEBS Lett 441:361–365. doi: 10.1016/S0014-5793(98)01574-9. [DOI] [PubMed] [Google Scholar]
- 29.Hohlfeld S, Pattis I, Püls J, Plano GV, Haas R, Fischer W. 2006. A C-terminal translocation signal is necessary, but not sufficient for type IV secretion of the Helicobacter pylori CagA protein. Mol Microbiol 59:1624–1637. doi: 10.1111/j.1365-2958.2006.05050.x. [DOI] [PubMed] [Google Scholar]
- 30.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schröder G, Vergunst AC, Carena I, Dehio C. 2005. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci U S A 102:856–861. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simone M, McCullen CA, Stahl LE, Binns AN. 2001. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol Microbiol 41:1283–1293. doi: 10.1046/j.1365-2958.2001.02582.x. [DOI] [PubMed] [Google Scholar]
- 32.Vergunst AC, van Lier MC, den Dulk-Ras A, Stüve TA, Ouwehand A, Hooykaas PJ. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci U S A 102:832–837. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anacker RL, Philip RN, Williams JC, List RH, Mann RE. 1984. Biochemical and immunochemical analysis of Rickettsia rickettsii strains of various degrees of virulence. Infect Immun 44:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Merhej V, Raoult D. 2011. Rickettsial evolution in the light of comparative genomics. Biol Rev Camb Philos Soc 86:379–405. doi: 10.1111/j.1469-185X.2010.00151.x. [DOI] [PubMed] [Google Scholar]
- 35.Kaur SJ, Rahman MS, Ammerman NC, Beier-Sexton M, Ceraul SM, Gillespie JJ, Azad AF. 2012. TolC-dependent secretion of an ankyrin repeat-containing protein of Rickettsia typhi. J Bacteriol 194:4920–4932. doi: 10.1128/JB.00793-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pantaléon V, Soavelomandroso AP, Bouttier S, Briandet R, Roxas B, Chu M, Collignon A, Janoir C, Vedantam G, Candela T. 2015. The Clostridium difficile protease Cwp84 modulates both biofilm formation and cell-surface properties. PLoS One 10:e0124971. doi: 10.1371/journal.pone.0124971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamatake K, Maeda M, Kadowaki T, Takii R, Tsukuba T, Ueno T, Kominami E, Yokota S, Yamamoto K. 2007. Role for gingipains in Porphyromonas gingivalis traffic to phagolysosomes and survival in human aortic endothelial cells. Infect Immun 75:2090–2100. doi: 10.1128/IAI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellison DW, Clark TR, Sturdevant DE, Virtaneva K, Hackstadt T. 2009. Limited transcriptional responses of Rickettsia rickettsii exposed to environmental stimuli. PLoS One 4:e5612. doi: 10.1371/journal.pone.0005612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galletti MF, Fujita A, Rosa RD, Martins LA, Soares HS, Labruna MB, Daffre S, Fogaça AC. 2016. Virulence genes of Rickettsia rickettsii are differentially modulated by either temperature upshift or blood-feeding in tick midgut and salivary glands. Parasit Vectors 9:331. doi: 10.1186/s13071-016-1581-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anacker RL, McCaul TF, Burgdorfer W, Gerloff RK. 1980. Properties of selected rickettsiae of the spotted fever group. Infect Immun 27:468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eremeeva ME, Dasch GA, Silverman DJ. 2001. Quantitative analyses of variations in the injury of endothelial cells elicited by 11 isolates of Rickettsia rickettsii. Clin Diagn Lab Immunol 8:788–796. doi: 10.1128/CDLI.8.4.788-796.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hackstadt T. 1996. The biology of rickettsiae. Infect Agents Dis 5:127–143. [PubMed] [Google Scholar]
- 43.Johnson JW, Pedersen CE Jr.. 1978. Plaque formation by strains of spotted fever rickettsiae in monolayer cultures of various cell types. J Clin Microbiol 7:389–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clark TR, Ellison DW, Kleba B, Hackstadt T. 2011. Complementation of Rickettsia rickettsii RelA/SpoT restores a nonlytic plaque phenotype. Infect Immun 79:1631–1637. doi: 10.1128/IAI.00048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood DO, Wood RR, Tucker AM. 2014. Genetic systems for studying obligate intracellular pathogens: an update. Curr Opin Microbiol 17:11–16. doi: 10.1016/j.mib.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kleba B, Clark TR, Lutter EI, Ellison DW, Hackstadt T. 2010. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun 78:2240–2247. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bell EJ, Pickens EG. 1953. A toxic substance associated with the rickettsias of the spotted fever group. J Immunol 70:461–472. [PubMed] [Google Scholar]
- 48.Cox HR. 1941. Cultivation of Rickettsiae of the Rocky Mountain spotted fever, typhus and Q fever groups in the embryonic tissues of developing chicks. Science 94:399–403. doi: 10.1126/science.94.2444.399. [DOI] [PubMed] [Google Scholar]
- 49.Weiss E, Coolbaugh JC, Williams JC. 1975. Separation of viable Rickettsia typhi from yolk sac and L cell host components by Renografin density gradient centrifugation. Appl Microbiol 30:456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark TR, Lackey AM, Kleba B, Driskell LO, Lutter EI, Martens C, Wood DO, Hackstadt T. 2011. Transformation frequency of a mariner-based transposon in Rickettsia rickettsii. J Bacteriol 193:4993–4995. doi: 10.1128/JB.05279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cory J, Yunker CE, Ormsbee RA, Peacock M, Meibos H, Tallent G. 1974. Plaque assay of rickettsiae in a mammalian cell line. Appl Microbiol 27:1157–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu ZM, Tucker AM, Driskell LO, Wood DO. 2007. Mariner-based transposon mutagenesis of Rickettsia prowazekii. Appl Environ Microbiol 73:6644–6649. doi: 10.1128/AEM.01727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anacker RL, Mann RE, Gonzales C. 1987. Reactivity of monoclonal antibodies to Rickettsia rickettsii with spotted fever and typhus group rickettsiae. J Clin Microbiol 25:167–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mital J, Miller NJ, Dorward DW, Dooley CA, Hackstadt T. 2013. Role for chlamydial inclusion membrane proteins in inclusion membrane structure and biogenesis. PLoS One 8:e63426. doi: 10.1371/journal.pone.0063426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RARP-2 homologs are conserved in pathogenic rickettsiae and harbor variable ankyrin repeats. In silico analysis of RARP-2 homologs across 49 Rickettsia genomes. Proteins were retrieved from NCBI using R. typhi strain Wilmington RARP-2 (locus tag RT0600) as the query in a blastp search against the NR (All GenBank+RefSeq nucleotides + EMBL + DDBJ + PDB) database, coupled with a search against the Conserved Domains Database (A. Marchler-Bauer et al., Nucleic Acids Res 39:D225–D229, 2011, https://doi.org/10.1093/nar/gkq1189). Searches were performed with composition-based statistics across Rickettsia. No filter was used. Default matrix parameters (BLOSUM62) and gap costs (existence, 11; extension, 1) were implemented, with an inclusion threshold of 0.005. Subjects were aligned using MUSCLE with default parameters (R. C. Edgar, Nucleic Acids Res 32:1792–1797, 2004, https://doi.org/10.1093/nar/gkh340). The alignment was manually adjusted to delineate predicted ankyrin repeats using a consensus ankyrin repeat model (L. K. Mosavi, T. J. Cammett, D. C. Desrosiers, and Z.-Y. Peng, Protein Sci 13:1435–1448, 2004, https://doi.org/10.1110/ps.03554604). These analyses indicated that RARP-2 homologs consist of a highly conserved N-terminal domain (NTD) of unknown function, an ankyrin domain composed of 3 to 12 repeats of 28 residues, and a C-terminal tail containing a predicted T4SS signal. Phylogeny at left was estimated as previously described (T. Driscoll, J. J. Gillespie, E. K. Nordberg, A. F. Azad, and B. W. Sobral, Genome Biol Evol 5:621–645, 2013, https://doi.org/10.1093/gbe/evt036), with additional genomes annotated using RAST (R. K. Aziz et al., BMC Genomics 9:75, 2008, https://doi.org/10.1186/1471-2164-9-75). The Rickettsia classification scheme follows previous studies: red, ancestral lineages; blue, transitional group; aquamarine, typhus group; and brown, spotted fever group (J. J. Gillespie et al., PLoS One 2:e266, 2007, https://doi.org/10.1371/journal.pone.0000266). Rickettsia helvetica is unclassified according to recent recommendations (T. Driscoll, J. J. Gillespie, E. K. Nordberg, A. F. Azad, and B. W. Sobral, Genome Biol Evol 5:621–645, 2013, https://doi.org/10.1093/gbe/evt036; J. J. Gillespie et al., PLoS One 3:e2018, 2008, https://doi.org/10.1371/journal.pone.0002018). Note that divergent RARP-2 homologs (found only in R. bellii and R. felis) share less than ~37% identity with RARP-2 proteins in the N-terminal domain, with very different ankyrin repeat sequences that do not match RARP-2 ankyrin repeats in reciprocal blastp searches. Download FIG S1, TIF file, 23.5 MB (24.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of SS-RARP-2 from the virulent R strain yields similar vesicular structures as those expressed from the avirulent Iowa strain. R::SS-RARP-2-infected cells at 48 hpi were fixed and stained with an anti-FLAG antibody (red) and merged with images showing GFP-rickettsia (green) and 4′,6-diamidino-2-phenylindole (DAPI) (blue). Download FIG S2, TIF file, 1.8 MB (1.9MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
FLAG-MTase is expressed and is internally localized in rickettsiae. MTase was expressed from pRAM2 as an N-terminal FLAG fusion in R. rickettsii Iowa in Vero cells. Fixed cells were incubated with 5 mg/ml lysozyme (Sigma-Aldrich) in 10 mM Tris-HCl (pH 7.5) for 1 h at 37°C (+lysozyme) or not (-lysozyme) and stained for immunofluorescence as described in Materials and Methods. Bar, 10 µm. Download FIG S3, TIF file, 7.6 MB (7.8MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
An anti-RARP-2 antiserum fails to detect native RARP-2 from wild-type R. rickettsii Sheila Smith and Iowa. (A) A rabbit polyclonal antipeptide antibody recognized overexpressed recombinant RARP-2 but did not detect specific antigen from parental rickettsiae, suggesting that RARP-2 may be of low abundance. Anti-FLAG staining of the recombinant proteins is shown in green. Anti-RARP-2 is shown in red. A panel showing the merged images is also provided. Dots to the right of the bands indicated RARP-2 fragments recognized by the antiserum. (B) Reverse transcriptase quantitative PCR (RT-qPCR) showing equivalent transcription of RARP-2 from Sheila Smith and Iowa. Three Vero cell culture flasks per strain were infected with rickettsiae and harvested at 48 hpi. Medium was removed, and cells were lysed in 6 ml Trizol. Two hundred microliters 1-bromo-3-chloropropane/ml Trizol was added, and samples were centrifuged at 16,000 × g for 15 min. RNA was extracted from the aqueous phase using the New England BioLabs (NEB) Monarch total RNA miniprep kit. After extraction, an additional DNA removal step was performed using the Turbo DNA-free kit (Thermo Fisher). RNA quality was checked on 1% Tris-borate-EDTA (TBE) gels. Primers were designed using the IDT PrimerQuest tool (RARP2_F, CTGATGAAGGTACAACTCCTGTATTA; RARP2_R, CGGCTCCTGAATGACAAGAA; DnaK_F, CCAAGAGGTTTGCCACAAATAG; DnaK_R, GCTCTTTACCGCTTGCTTTATC). Gene fragments of DnaK and RARP-2 were cloned into TopoTA, and plasmids were used to establish standard curves and calculate qPCR efficiencies and copy numbers. RT-qPCR was performed using 1 ng of purified RNA with the Luna universal one-step RT-qPCR kit (NEB) on a Roche Light Cycler 480 II. Efficiencies were 2.00 for RARP-2 and 1.96 for DnaK. No-RT controls were included for all samples and did not show any amplification. All reactions were performed in triplicate on biological triplicate samples. Download FIG S4, TIF file, 13.7 MB (14MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RARP-2 does not associate with autophagosomes. Potential association with autophagosomes was assessed by examination of the SS-RARP-2 structures with coexpressed GFP-LC3. GFP was used as a negative control. No association with LC3 was observed. Bar, 10 µm. Download FIG S5, TIF file, 3.9 MB (4MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Association of R. typhi RARP-2 with RvhD4 was corroborated by a bacterial two-hybrid assay. (A) The bacterial two-hybrid assay was performed by transforming codon-optimized R. typhi RvhD4 bait and R. typhi RARP-2 prey vectors into Escherichia coli BacterioMatch II reporter electrocompetent cells. Growth on dual selective medium indicates that the RARP-2 C-terminal tail interacts with RvhD4 (FL), and this interaction is abolished when 37 residues of the Rt-RARP-2 C-terminal tail are deleted (ΔCT). (B) Quantification of panel A. Percent growth was calculated from cotransformed bacterial CFUs on dual selective screening medium relative to CFU obtained on nonselective medium. Error bars represent mean ± standard deviation (SD) from three independent experiments (Student’s two-sided t test). Download FIG S6, TIF file, 1.7 MB (1.7MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
RARP-2 N-terminal domain harbors a putative clan CD cysteine protease active site. Using Phyre2 (L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass, and M. J. Sternberg, Nat Protoc 10:845–858, 2015, https://doi.org/10.1038/nprot.2015.053), we modeled several RARP-2 homologs to clan CD cysteine endopeptidases (data not shown). Accordingly, we aligned several RARP-2 homologs with proteases from selected clan CD families: legumain (C13), caspase 1 (C14), clostripain (C11), and gingipain R (C25). The sequence segments containing the experimentally identified or predicted catalytic His and Cys residues (pink and yellow) in each of the four families and RARP-2 homologs were subsequently adjusted based on a previous study [J. M. Chen, N. D. Rawlings, R. A. Stevens, and A. J. Barrett, FEBS Lett 441:361–365, 1998, https://doi.org/10.1016/S0014-5793(98)01574-9]. Upstream of the catalytic residues, there is a block of four predominantly hydrophobic residues (green). Other residues that tend to be conserved between the families are printed in white on black. Blue numbers indicate amino acid coordinates flanking the active site. Accession numbers from UniProt are as follows: Q99538, human legumain; P49046, jack bean (Canavalia ensiformis) asparaginyl endopeptidase; P42665, Schistosoma japonicum hemoglobinase; P49048, Caenorhabditis elegans hypothetical protein T05E11.6; Q92643, human GPI8 protein; P42574, human caspase 3; P55210, human caspase 7; P55212, human caspase 6; O01382, Drosophila melanogaster caspase; P29466, human caspase 1; P42573, Caenorhabditis elegans CED3 protein; P09870, Clostridium histolyticum α-clostripain; Q8A866, Bacteroides thetaiotaomicron clostripain-related protein; Q9WYY6, Thermotoga maritima clostripain-related protein; Q51816, Porphyromonas gingivalis gingipain R; B0VHP1, Cloacimonas acidaminovorans putative gingipain R; F2I9M7, Fluviicola taffensis gingipain R; A0A075MRD2, endosymbiont of Acanthamoeba sp. strain UWC8 uncharacterized protein; Q1RID2, Rickettsia bellii putative ankyrin repeat protein RBE_0801; Q4UKP4, Rickettsia felis uncharacterized protein; Q1RI97, Rickettsia bellii uncharacterized protein; Q68WC7, Rickettsia typhi uncharacterized protein; A0A0H3AY85, Rickettsia rickettsii strain Sheila Smith uncharacterized protein; B0BUJ3, Rickettsia rickettsii Iowa uncharacterized protein. Download FIG S7, TIF file, 1 MB (1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The SS-RARP-2-C109A construct is secreted and forms perinuclear vesicular structures but retains association with ER markers. Iowa::SS-RARP-2-C109A-infected cells at 48 hpi were fixed and stained with an anti-FLAG antibody (red) and anticalnexin or PDI (white). GFP-expressing rickettsiae are green. Images were merged and include DAPI staining (blue). Bar, 10 µm. Download FIG S8, TIF file, 1.9 MB (2MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Supplemental materials and methods. Download TEXT S1, DOCX file, 0.02 MB (19.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.