Abstract
Background
The green peach aphid (GPA), Myzus persicae, is economically one of the most threatening pests in pepper cultivation, which not only causes direct damage but also transmits many viruses. Breeding aphid resistant pepper varieties is a promising and environmentally friendly method to control aphid populations in the field and in the greenhouse. Until now, no strong sources of resistance against the GPA have been identified. Therefore the main aims of this study were to identify pepper materials with a good level of resistance to GPA and to elucidate possible resistance mechanisms.
Results
We screened 74 pepper accessions from different geographical areas for resistance to M. persicae. After four rounds of evaluation we identified one Capsicum baccatum accession (PB2013071) as highly resistant to M. persicae, while the accessions PB2013062 and PB2012022 showed intermediate resistance. The resistance of PB2013071 resulted in a severely reduced uptake of phloem compared to the susceptible accession, as determined by Electrical Penetration Graph (EPG) studies. Feeding of M. persicae induced the expression of callose synthase genes and resulted in callose deposition in the sieve elements in resistant, but not in susceptible plants.
Conclusions
Three aphid resistant pepper accessions were identified, which will be important for breeding aphid resistant pepper varieties in the future. The most resistant accession PB2013071 showed phloem-based resistance against aphid infestation.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1340-3) contains supplementary material, which is available to authorized users.
Keywords: Aphid resistance screening, Capsicum baccatum, EPG, Callose, Real-time PCR
Background
Pepper (Capsicum spp.) belongs to the Solanaceae family and is one of the economically most important and widely cultivated vegetable crops. The annual global production area and yield of pepper are 3.7 million hectares and 37 million tons, respectively (FAOSTAT, 2015). The genus Capsicum originates from Central and South America and 25 distinct species have been reported [1], among which five are domesticated: C. annuum, C. chinense, C. frutescens, C. baccatum, and C. pubescens [2].
Aphids (Aphididae) are the most wide-spread pest insects. More than 100 aphid species are reported as economically important pests and most crops suffer from one or more species [3]. The green peach aphid (GPA), Myzus persicae, is one of the most threatening pests in pepper and many other crops. It is a generalist that causes many types of damages in pepper, including chlorosis, necrosis, wilting, defoliation and flower and fruit abortion. It produces honeydew when feeding on plants, which may affect fruit quality and reduce photosynthetic capacity by stimulating mold development. However the most serious damage is done indirectly by the viruses that GPA may vector, including Potato virus Y, Pepper mottle virus, Pepper severe mosaic virus, Pepper yellow mosaic virus, and Peru tomato mosaic virus [4].
As phloem-feeding insects, aphids use their specialized mouthparts, the stylets, to penetrate plant tissue and to take up nutrients without inflicting serious damage [5, 6]. To study aphid probing and feeding behaviour, the electrical penetration graph (EPG) technique can be used [7]. In the EPG technique an aphid and a plant are wired into an electrical circuit, and aphid activity on the plant is recorded as waveforms that are specific for different probing and feeding activities [8, 9]. The EPG technique can be applied to explore the nature of the differences in aphid behaviour on resistant and susceptible plants, for instance to determine where in the leaf an aphid encounters a specific plant resistance factor [5, 10–12].
In several cases it has been observed that aphids show a significantly shorter period of phloem feeding on resistant than on susceptible plants [11, 12]. One possible explanation is occlusion of the phloem vessels in response to feeding [13, 14], which may be caused by callose deposition [13, 15]. Callose, a ß-1,3-glucan, is an important component in the defense response to mechanical wounding, pathogen infection and insect infestation [16–18]. In Arabidopsis thaliana callose deposition was induced and the expression of related synthase genes was enhanced in response to whitefly infestation [19]. In rice, callose deposition was suggested as an important resistance factor against the brown plant hopper [15].
Callose is produced by callose synthases (CalS), which are encoded by a family of callose synthase genes. Twelve, ten, six, nine and eight synthase genes were identified and characterized in A. thaliana [20, 21], rice [22], barley [23], wheat [24] and grapevine [25], respectively. These genes were studied in detail in A. thaliana. The CalS7 gene was reported to be expressed specifically in the phloem vessels and was responsible for callose deposition induced by mechanical wounding [26]. The CalS12 was mainly shown to be required for wound and papillary callose formation in response to pathogen attack [27, 28] and to aphid feeding [29]. The expression of CalS1 was found to be up-regulated after infestation with aphids and whiteflies [19, 30]. Besides the role of callose formation and deposition in plant resistance, the breakdown of callose might be another factor. Callose degradation, which is governed by some ß-1,3-glucanases, was shown to cause susceptibility in the interaction between the brown plant hopper and rice [15] as well as in the interaction between bird cherry-oat aphid and barley [31].
Due to the severe negative effects of aphids on crop yield and quality, chemical pesticides have been widely used to control aphids. However, with more and more reports on aphids developing resistance to pesticides [32, 33] and growing concern about the environmental impact of insecticides, breeding aphid resistant pepper varieties is a desirable alternative which will become an indispensable part of integrated pest management. Plant resistance mechanisms against insects, including aphids, are classified as antixenosis, antibiosis and tolerance [34–37]. Antixenosis, or non-preference, affects insect settling or feeding through repellence or deterrence [38]. Antibiosis-based resistance impairs insect survival, growth, development and fecundity, caused by chemical or morphological adaptations of the plant [36, 39, 40]. Tolerance reduces damage to the plant after insect feeding, in spite of the presence of insect population densities similar to those on susceptible plants [34, 40]. A number of genes conferring resistance to aphids have been identified in crops, including among others in wheat [41], soybean [42], lettuce [43] and cowpea [44]. However, only two genes have been cloned, the tomato Mi-1.2 gene which confers resistance to the potato aphid Macrosiphum euphorbiae, to the whitefly Bemisia tabaci and to three species of root-knot nematodes [45–47], and the melon Vat gene, which confers resistance to the cotton aphid Aphis gossypii, as well as to non-persistent viruses when vectored by A. gossypii [48]. Both genes are of the NBS-LRR type [45, 48] and work according to the gene-for-gene principle which means that the R gene in the plant recognizes an effector secreted by the aphid, and activates an aphid-specific defense response [35]. Until now only a few studies to identify donors of resistance genes that may be used in pepper breeding have been published [49, 50]. One C. pubescens plant showed antixenosis rather than antibiosis resistance to the GPA [49], but detailed information on this accession was not provided, and no hybridization between C. pubescens and C. annuum has been reported yet. Franz et al. detected significant differences among 50 pepper accessions in choice tests with GPA, however no strong resistance was found [50]. De Costa et al. identified a pepper cultivar which was resistant against the A. gossypii, but it is unknown if it is also resistant to GPA [51]. Therefore, there is still an urgent need for pepper accessions resistant to GPA.
This research was carried out to identify accessions with a good level of resistance to GPA and to shed light on the possible resistance mechanism. We evaluated a collection of C. annuum, C. chinense, C. frutescens and C. baccatum accessions for GPA resistance and identified resistant accessions in C. baccatum. The resistance, mainly affecting aphid reproduction, is most likely phloem based and accompanied by callose deposition.
Results
Selection of pepper accessions resistant to GPA
Evaluation of 50 accessions, representing 4 Capsicum species, for GPA resistance showed large and highly significant differences (Additional file 1: Table S1) for the two resistance parameters used: survival of the original nymphs and the number of next generation nymphs produced. Survival rate ranged from 6 to 97%, while the average number of new nymphs produced by each living adult during infestation varied from 0 to 0.8.
After transferring the GPA rearing from Chinese cabbage to C. annuum accession CGN19226, ten selected accessions (Additional file 1: Table S1 and Fig. 1) were re-tested with the GPA colony that had been adapted to pepper. These included seven accessions showing a low aphid survival and also a low production of second generation nymphs in the first experiment. The accessions C. annuum CGN19226 and C. frutescens PB2012045 were chosen as susceptible standards as they are from different species and origins. Accession C. annuum CGN19194 was selected as no second generation nymphs were produced on it, while the number of surviving adults was high, suggesting that this accession may possess a resistance mechanism affecting reproduction only. In this second experiment the two susceptible standards were again completely susceptible. Accession CGN19194 was also highly susceptible; the reduced reproduction observed in the first test was not confirmed in the second one using the aphids adapted to pepper. Among the seven accessions selected as resistant in the first experiment, the five C. chinense accessions respectively showed varying levels of resistance based on the two resistance parameters between the two experiments (T-test, P < 0.01). However, the two C. baccatum accessions (PB2012022 and PB2012024) continued to show an impaired reproduction in the second experiment, which was the same as that in the first experiment.
Fig. 1.
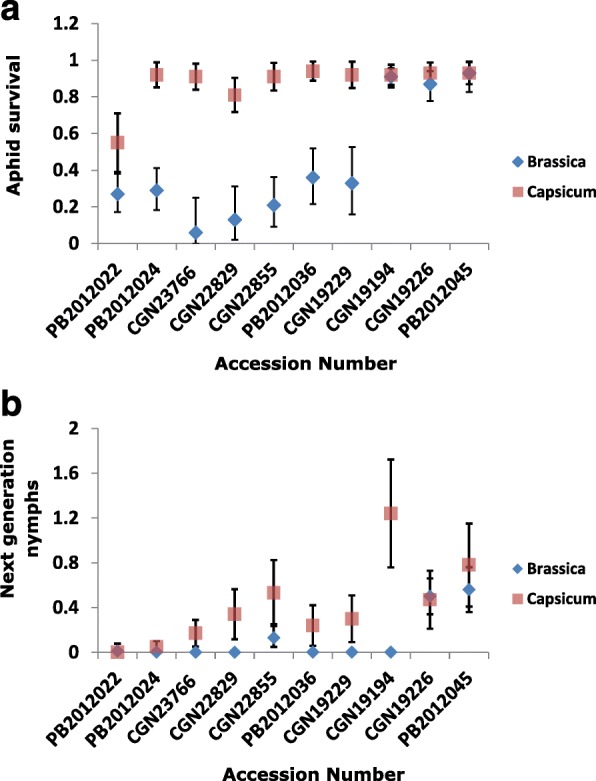
Performance of M. persicae after adaptation to different host plants. Ten selected accessions were infested with aphids reared on Chinese cabbage (Brassica rapa cv. Granaat; blue icon) or pepper (C. annuum CGN19226; red icon). Performance parameters used: survival of the original nymphs (a) and the number of next generation nymphs produced (b). Survival was determined by dividing the number of living aphids by the total number of aphids (dead and alive) in the clip cage. The number of next generation nymphs was divided by the average number of living aphids present, calculated as (2*living aphids + dead aphids)/2. Each bar represents the mean values ± SD. More details on the statistics can be found in Additional file 1: Table S1
Based on the results of the initial screening, we decided to focus further efforts on the screening of C. baccatum accessions (Additional file 2: Table S2). In the third experiment accession C. annuum CGN19226 was used as susceptible standard. Evaluation of 38 accessions showed significant variation for aphid survival and aphid fecundity: survival of original nymphs varied from 0.49 to 0.98 and the number of new nymphs produced per aphid ranged from 0 to 0.89. The accessions PB2013071, PB2013062 and CGN23260 were among the most resistant although they were not significantly different from a number of others, based on aphid survival and next generation nymphs produced. Accession PB2012022 showed a slightly higher nymph survival, but no next generation nymphs, confirming previous results. The accession C. baccatum PB2013046 was as susceptible as the susceptible standard C. annuum accession CGN19226. For this reason we transferred the GPA rearing to PB2013046 and re-tested eight accessions for resistance using GPA reared on this susceptible C. baccatum accession (Additional file 2: Table S2 and Fig. 2). In this fourth experiment, we classified PB2013071, PB2013062, CGN23260 (no reproduction, relatively low survival: < 0.7) together with PB2012022 and CGN22834 (also no reproduction, somewhat higher survival: > 0.7) as resistant, CGN22858 (some reproduction, low survival) as an intermediate resistant, and PB2013046 together with CGN19226 (high reproduction, high survival) as susceptible accessions. In this experiment, the accession C. baccatum PB2013071 was again the most resistant, as it continued showing the lowest survival and no reproduction. The accession C. baccatum PB2013046 was again as susceptible as C. annuum accession CGN19226. The correlation coefficient between the number of new nymphs produced by C. annuum and C. baccatum adapted aphids (third and fourth experiment) was 0.83, which was calculated on the basis of the eight accessions tested with both populations.
Fig. 2.
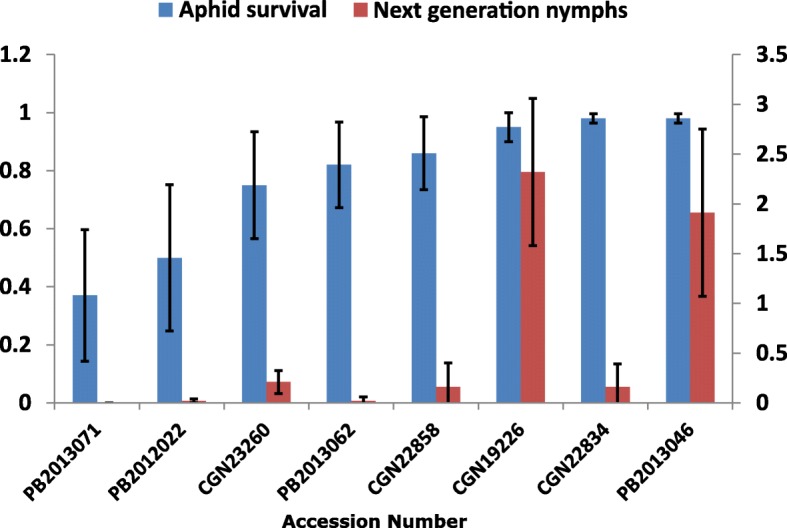
Performance of M. persicae on eight accessions after adaptation on C. baccatum. Aphids were reared on accession PB2013046. Performance parameters used: survival of the original nymphs (blue column) and the number of next generation nymphs produced (red column). Survival was determined by dividing the number of living aphids by the total number of aphids (dead and alive) in the clip cage. The number of next generation nymphs was divided by the average number of living aphids present, calculated as (2* living aphids + dead aphids)/2. Each bar represents the mean values ± SD. More details on the statistics can be found in Additional file 2: Table S2
GPA population development on selected accessions
The three selected resistant C. baccatum accessions (PB2013071, PB2013062 and PB2012022), the susceptible C. baccatum accession (PB2013046) and the susceptible C. annuum accession (CGN19226) were used for further confirmation of resistance and susceptibility using a population development experiment. Results are shown in Table 1. PB2013046 is confirmed as a susceptible accession on which aphids show a high survival rate and strong fecundity, which was even higher than the C. annuum susceptible standard (CGN19226). Accession PB2013071 showed the highest level of resistance, while the accessions PB2013062 and PB2012022 were intermediate.
Table 1.
Population development of the aphid M. persicae on five Capsicum accessions
| Accession number | Adults | Nymphsa | |
|---|---|---|---|
| PB2013071 | 33 | a | 0.7 |
| PB2012022 | 158 | b | 1.2 |
| PB2013062 | 337 | c | 1.6 |
| PB2013046 | 2655 | d | 2.0 |
| CGN19226 | 1633 | d | 2.0 |
aAverage number of nymphs according to visual scale: 0 = none, 1 = few (< 50), 2 = many (> 50)
Mean values of adult count followed by the same letter are not significantly different (LSD- test on log-transformed scale at P < 0.05)
EPG analysis on accessions PB2013071 and PB2013046
Results for the parameters extracted from the EPG recordings are presented in Table 2. No significant difference was found between the resistant accession PB2013071 and the susceptible accession PB2013046 for parameters related with non-probing, pathway phase, derailed stylet mechanics and xylem phase. However, significant differences were seen during the phloem phase E1 (salivation into the phloem) and E2 (phloem sap ingestion) (T-test, P < 0.05). The total duration of E1 on PB2013071 was more than two times as long as on PB2013046, while the total duration of E2 on PB2013071 was only about one-eighth of that on PB2013046. However, there was no significant difference in the number of aphids that successfully reached phloem ingestion E2: 75% on PB2013046 and 47% on PB2013071 (Fisher exact test, P = 0.101). The total number of individual cell punctures (potential drops) and average number of potential drops per minute of pathway phase were both more on PB2013071 than PB2013046 (T-test, P < 0.01).
Table 2.
M. persicae EPG parameters measured on a susceptible (PB2013046) and a resistant (PB2013071) C. baccatum accession
| Class | Trait Definitiona | 2,013,046 | 2,013,071 | P-value |
|---|---|---|---|---|
| Non-probing (NP) | Number of NP | 15.7 | 17.3 | 0.5645 |
| Total duration of NP (min) | 21.3 | 21.3 | 0.9879 | |
| Probes | Number of Probes | 14.8 | 16.3 | 0.5870 |
| Total duration of Probes (min) | 338.6 | 338.6 | 0.9874 | |
| Pathway phase (C) | Number of C (pathway periods) | 24.4 | 27.8 | 0.3528 |
| Total duration of pathway period (min) | 125.0 | 155.1 | 0.1206 | |
| Derailed stylet (F) | Number of periods with F form | 3.3 | 1.5 | 0.0650 |
| Total duration of F period (min) | 72.9 | 59.8 | 0.6230 | |
| Xylem phase (G) | Number of periods with G form | 2.1 | 1.7 | 0.5651 |
| Total duration of G period (min) | 32.9 | 33.4 | 0.9686 | |
| Time to first G phase (min) | 162.8 | 153.2 | 0.8326 | |
| Phloem phase (E) | Number of salivation periods (E1) | 6.1 | 9.4 | 0.0265 |
| Time to first E1 (min) | 105.1 | 82.2 | 0.4575 | |
| Total duration of E1 (min) | 28.6 | 80.0 | 0.0001 | |
| Total duration of phloem uptake (E2, min) | 78.9 | 10.5 | 0.0032 | |
| Time to first E2 (min) | 258.5 | 324.0 | 0.0414 | |
| Number of E1 followed by E2 | 1.2 | 0.3 | 0.0049 | |
| Total duration of E1E2 (min) | 86.6 | 5.5 | 0.0008 | |
| Time to first E1E2 (min) | 225.4 | 324.4 | 0.0037 | |
| Potential drops (Pd) | Number of potential drops | 83.7 | 156.4 | 0.0000 |
| Number of Pd per min of Pathway C | 0.7 | 1.0 | 0.0025 | |
| Aphids reaching E2 | Percentage of aphids reaching E2 | 75% | 47% | 0.1010 |
Data are based on 20 and 17 aphids tested on PB2013046 and PB2013071, respectively. Mean values are shown
Callose deposition
Callose deposition is considered important for plant resistance against pathogens and insects [15, 52]. We studied the accumulation of callose in resistant and susceptible plants after GPA feeding. Detached leaves were infested with GPA for 24 h, after which three or four leaf disks were prepared for the callose deposition study. Representative results are shown in Fig. 3 and additional images can be found in Additional file 3: Figure S1. Callose signals were detected in the vascular tissue of all sampled leaf disks from accession PB2013071, but not in accession PB2013046 treated by GPA or in leaf disks of both accessions without aphids infestation.
Fig. 3.
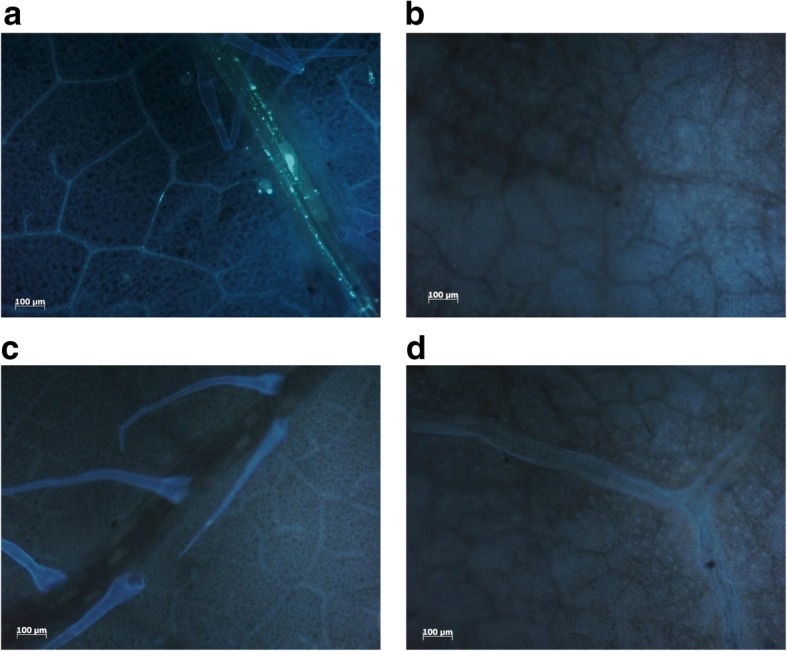
Histochemical staining of callose in the GPA-infested leaves (a, b) and GPA-free leaves (c, d). Resistant accession PB2013071 (a, c); susceptible accession PB2013046 (b, d). Staining was carried out 24 h after the start of the infestation
Identification and expression of callose related genes
Nine putative Callose Synthase (CalS) genes were identified in the C. annuum sequences and named with reference to the most homologous gene in Arabidopsis, CaCalS1, CaCalS3, CaCalS5, CaCalS7, CaCalS8, CaCalS9, CaCalS10, CaCalS11 and CaCalS12. The length of open reading frames (ORFs) and gene IDs in both pepper genome sequences are listed in Additional file 4: Table S3. A neighbour-joining tree of CalS proteins among pepper, Arabidopsis and grapevine is shown in Additional file 5: Figure S2.
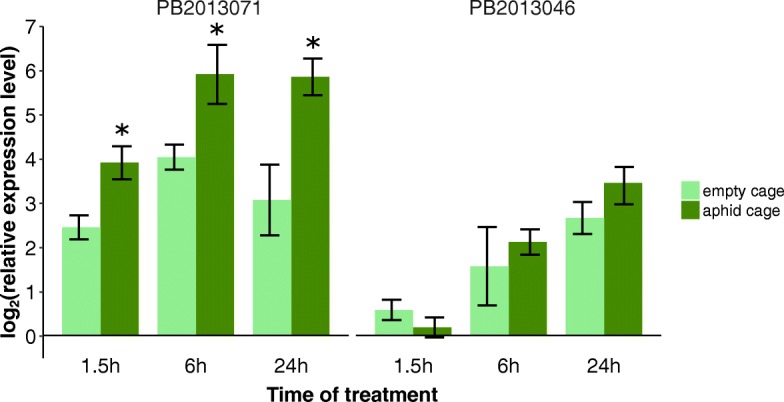
To shed light on the regulation of the callose deposition we compared the expression of callose synthase genes (CalS family genes) and the basic ß-1,3-glucanase gene (BGLU) in GPA-infested leaves with those of non-infested leaves. Nine putative CalS family genes were analyzed by real-time PCR. Among these nine genes, only CalS1 (Fig. 4a) and CalS7 (Fig. 4b) showed a clear change in transcript accumulation upon aphid infestation. In the leaves of PB2013071 infested with GPA, no difference in expression was detected for both genes after 1.5 h, but expression was significantly up-regulated at 6 h and 24 h after the start of the infestation compared to empty cages (T-test, P < 0.05). The expression level of CalS1 increased 5.6-fold (T-test, P = 0.0004) and that of CalS7 increased 3.9-fold (T-test, P = 0.0088) 24 h post-infestation compared to empty cages. In the leaves of PB2013046 infested with GPA, expression of the CalS1 and CalS7 genes remained stable during 24 h, except that CalS7 after 1.5 h showed significantly lower expression level in GPA infested leaves compared to GPA free leaves (T-test, P = 0.0017). The expression of the CalS1 and CalS7 gene in leaves of both accessions after 1.5 h, 6 h, 24 h with empty clip cages remained constant (ANOVA, P > 0.05).
Fig. 4.
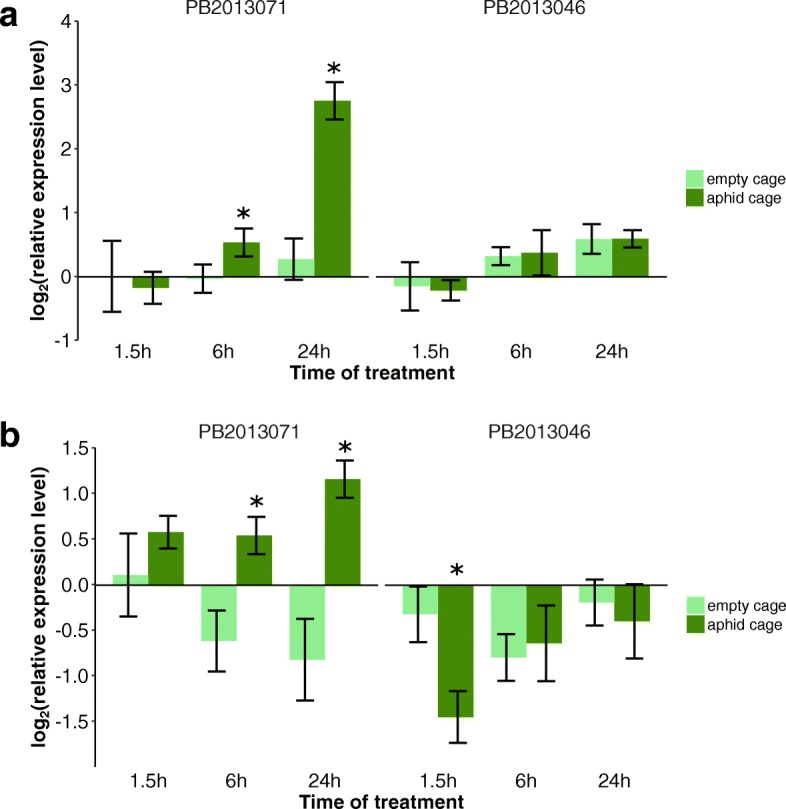
Expression analysis of Callose synthase genes CalS1 (a) and CalS7 (b) after aphid infestation. Gene expression was quantified relative to the value obtained from leaf samples without clip cage or aphid infestation (time point 0 h). Data was log2-transformed. Each bar represents the mean values of three or four biological replicates, each with two technical replicates. The actin gene was used as the reference gene. * indicates a significant difference in level of gene expression between the GPA treated sample and the GPA-free (empty clip cage) sample at that time points (T-test, P < 0.05). Each bar represents the mean values ± SD
The BGLU gene was up-regulated in PB2013071 at all three time-points during the 24 h of aphid infestation compared to empty cages (Fig. 5) (T-test, P < 0.05). The ratio of transcripts with and without aphid infestation increased to 2.3 at 1.5 h (T-test, P = 0.0070), to 3.9 at 6 h (T-test, P = 0.0062) and to 6.4 at 24 h (T-test, P = 0.0141) after the start of the infestation. In contrast, there was no significant difference in expression of the BGLU gene in PB2013046 between plants with GPA treatment for 1.5 h, 6 h and 24 h and plants with empty cages at the same time points. In leaves that received empty clip cages, the expression of the BGLU gene increased after 1.5 h, 6 h, 24 h, in both accessions (ANOVA, P < 0.05).
Fig. 5.
Expression analysis of the BGLU gene after aphid infestation. Gene expression was quantified relative to the value obtained from leaf samples without clip cage or aphid infestation (time point 0 h). Data was log2-transformed. Each bar represents the mean values of three or four biological replicates, each with two technical replicates. The actin gene was used as the reference gene. * indicates a significant difference in level of gene expression between the GPA treated sample and the GPA-free (empty clip cage) sample at that time points (T-test, P < 0.05). Each bar represents the mean values ± SD
Discussion
Importance of rearing history during evaluation of GPA performance
When the initial evaluations were performed using GPA reared on cabbage or pepper, large differences were seen in aphid survival: GPA survival was relatively low when cabbage reared GPA were used and high when pepper reared GPA were used. The effect of GPA rearing history varied among Capsicum accessions. There was hardly any effect on C. annuum accessions, whereas on C. chinense accessions the effect of the rearing was pronounced. It has been reported before that the host plant on which an aphid colony is reared can affect the performance of aphids. For example, the grain aphid Sitobion avenae reared on wheat performed less well on the cocksfoot than on wheat [53], and A. gossypii that adapted to cotton or cucumber could not survive and reproduce after reciprocal host transfer [54].
About the background of host adaptation in our test system we can only speculate. (1) As there are differences in metabolite content between cabbage and pepper, aphids may have to develop/adjust their detoxification system to adapt to the host plant, which may take several generations. For instance, the enzymatic detoxification system, a family of glutathione S-transferases, was reported to be involved in adaptation of GPA to different species containing different glucosinolates [55]. (2) Another hypothesis involves a change in endosymbiont composition after transferring from one plant species to the other. Mutualistic symbionts play an instrumental role in plant-insect interactions [56]. Host plant specialization of pea aphid Acyrthosiphon pisum was reported to be influenced by the facultative pea aphid U-type symbiont (PAUS) [57]. Also, the abundance of Buchnera aphidicola, the primary endosymbiotic bacterium of GPA, was found to affect GPA host acceptance and stylet penetration on host plants [58]. In our case, the rearing on C. annuum may have changed the aphid metabolism, introduced a new endosymbiont species or increased the abundance of an already present symbiont species, improving their performance on C. chinense. Based on the observations made, it is highly recommended that evaluations of germplasm are carried out using insect populations that are adapted to the species, or that re-testing is conducted with adapted aphids to confirm results of resistance screenings especially when aphids are reared on evolutionary distant plant materials, as on Chinese cabbage in our case.
A wide diversity in GPA resistance among Capsicum accessions
The high multiplication rate of aphids makes them a pest in many crops [59]. Even in the presence of natural enemies (predators and parasitoids) it is often difficult to control the growth of aphid populations. Varieties that are highly or even partly resistant to aphids can make a big difference by reducing the multiplication rate of the aphids and thus give natural enemies more chance to control them [60]. To develop such varieties, resistance sources need to be identified in crossable species and in this paper we describe the identification of such sources. Accessions from four inter-crossable Capsicum species were evaluated for resistance against the GPA and considerable variation was observed. After four rounds of evaluation, we identified a number of C. baccatum accessions with a relatively high and stable level of aphid resistance. A GPA population development experiment among five selected accessions confirmed their resistance. Resistance primarily seems to affect the production of next generation nymphs and to a lesser extent the survival of the aphid itself. Accession C. baccatum PB2013046 showed susceptibility with the highest GPA survival rate and fecundity while C. baccatum PB2013071 showed the strongest resistance, with a significantly lower GPA survival than on the susceptible accession and a severely impaired population development. Accessions C. baccatum PB2013062 and C. baccatum PB2012022 showed intermediate levels of resistance. These three accessions are the first C. baccatum accessions in which resistance to GPA is demonstrated and may be used for breeding resistant varieties in the other Capsicum species as well. The species C. baccatum has been used for pepper breeding as donor of anthracnose [61, 62] and powdery mildew resistance [63]. With respect to insect resistance, two C. baccatum accessions were reported as a good source for thrips (Thrips parvispinus and Frankliniella occidentalis) resistance [64] and three C. baccatum accessions were identified as tolerant but not resistant to cotton aphid (A. gossypii) [50]. To our knowledge, this is the first report of a strong antibiosis type of resistance to GPA in Capsicum.
Impaired phloem uptake on a resistant accession
The Electrical Penetration Graph (EPG) technique allows an in-depth study of the feeding behaviour of piercing-sucking insects [7] and is able to reveal possible constraint encountered by such insects when trying to feed on plants [5, 9]. The EPG analysis revealed significant differences in parameters related with the phloem phase of GPA feeding on the resistant versus susceptible pepper accession. In comparison to the susceptible accession PB2013046, on the resistant PB2013071 the phloem salivation periods were longer and more frequent, and the phloem uptake periods were much shorter, suggesting that the resistance is most likely located in the phloem. In other words, aphids feeding on resistant accession PB2013071 have difficulties to initiate and sustain phloem sap ingestion. Aphids feeding on accessions containing a phloem based resistance are likely to grow more slowly, have lower fecundity and are more likely to die early due to the problems they experience with taking up sufficient nutrition. This is in line with our observations. Besides the possibility to control aphid population, phloem based resistance may reduce the transmission of persistent viruses because generally aphids cannot acquire persistent viruses during short-time feeding [65]. It is likely that the percentage of plants infested with persistent viruses will also decrease when the number of aphids carrying virus is low [66].
No significant differences were observed in the pre-phloem phase, with the exception of the number of potential drops. Potential drops indicate that the aphid’s stylets puncture cells along the pathway to the phloem [67]. The number of potential drops was much higher on the resistant accession PB2013071 than on the susceptible accession PB2013046. One biotype of soybean aphid (Glycine max) was also shown to have a higher number of potential drops when feeding on resistant genotypes than on susceptible genotypes [11]. It has been reported that potential drops are related with aphid transmission of non-persistently transmitted viruses [68, 69]. However, it is unknown if they are indicative for a specific plant resistance component. In spite of the difference in number of potential drops, the total duration of the pathway phase was not different between the two accessions. We examined the number of cell layers between the epidermis and the phloem in the two accessions, which might have a relation with the number of cells punctured while passing to the phloem; however we did not observe a difference between the two accessions in this respect (results not shown). Therefore, it remains unclear if the higher number of potential drops on the resistant plant is important for resistance.
Induced callose deposition in the resistant accession
One possible mechanism of phloem-based resistance might be occlusion of the phloem vessels in response to aphid feeding, which may result from callose deposition. Callose induction and formation is a defense response to phloem-sucking pests that plugs the sieve element to obstruct feeding [15, 30, 70–72]. Our data clearly show callose deposition 24 h after the start of the aphid infestation on detached leaves from the resistant accession PB2013071, but not on the susceptible accession PB2013046 and also not on non-infested leaves of either accession. This suggests that callose deposition may be one of the mechanisms behind the phloem-based resistance. The fact that callose deposition was studied on detached leaves and not on intact plants may have resulted in a weaker callose response. We did not assess the resistance on detached leaves, but studies on lettuce with the aphid Nasonovia ribisnigri [73] suggest that the expression of resistance may be partially reduced in detached leaves compared to intact plants. It is also reported that callose deposition is observed in epidermal and mesophyll cell walls in the interaction of A. gossypii with melon plants carrying resistance gene Vat [74].
As a strong callose signal was found in leaf veins of resistant pepper plants after GPA feeding and not in susceptible plants, it was hypothesized that one or several CalS family genes or ß-1,3-glucanase gene(s) might be involved in this difference between resistant and susceptible plants after GPA infestation. We carried out quantitative real-time PCR to examine whether callose deposition could be due to increased CalS gene expression upon aphid attack. Among the nine putative CalS family genes, the CalS1 gene was found to be significantly up-regulated at 6 h and 24 h post-infestation of GPA feeding in the leaves of PB2013071, while the level of gene transcripts remained constant in the leaves of PB2013046 during the initial 24 h of aphid infestation. The CalS1 gene has been reported in Arabidopsis to accumulate after whitefly and aphid infestation [19, 30]. Besides the CalS1 gene, we detected that transcripts of CalS7 in the infested leaves of resistant accession PB2013071 also significantly increased after 6 h and 24 h compared to non-infested leaves, but less than the transcripts of CalS1. The CalS7 gene is the only phloem-specific callose synthase gene and it is responsible for callose biosynthesis in developing sieve elements as well as for callose deposition after mechanical wounding in mature phloem [26]. Here we report for the first time an induction of CalS7 transcription upon infestation with a phloem-feeding insect. The expression of the two CalS genes increased after aphid attack in leaves of the resistant accession but not in leaves of the susceptible accession. We speculate that the CalS1 and/or CalS7 genes are responsible for callose deposition in leaves of the resistant accession PB2013071 after GPA feeding. As in A. thaliana CalS1 also can be induced by other phloem-feeding insects like the whitefly B. tabaci [19] and cabbage aphid Brevicoryne brassicae [30], the CalS1 gene might have a common role in callose deposition induced by phloem-feeders. As the CalS7 gene is expressed specifically in phloem vessels [75], the sampling of entire leaf disks rather than just leaf veins for real-time PCR may lead to an underestimation of the level of induction in phloem punctured by the insect. The role of these two CalS genes in callose deposition needs to be further studied. As transformation of pepper is difficult [76], it may not be so easy to do this by silencing the two CalS genes. It may be more effective to carry out a genetic (fine) mapping study to identify genes involved in the resistance.
The BGLU protein, also known as pathogenesis-related (PR) protein 2, is responsible for hydrolyzing callose (ß-1,3-glucan) in order to destabilize the cell wall of pathogens as well as to activate some immunity elicitors which can stimulate defense responses against pathogen attack [77]. In pepper plants BGLU has been reported to play an important role during defense against pathogens [78–80]. The BGLU protein or BGLU gene transcript has also been found to accumulate in leaves of wheat [81] and Arabidopsis [82] after aphid infestation. The BGLU gene is considered as a marker of the salicylic acid (SA)-dependent defense response in plants [83, 84]. Also, some ß-1,3-glucanases of the same family as BGLU were proposed as susceptibility factors in the interaction between brown plant hopper and rice [15] as well as between bird cherry-oat aphid and barley [31]. It is thought that the feeding barrier for insects caused by callose deposition can be weakened in susceptible plants due to accumulation of ß-1,3-glucanase, while callose deposition can be maintained in resistant plants when the expression of ß-1,3-glucanase gene is low. However, in contrast to this hypothesis we found that expression of the BGLU gene increased during the 24 h of GPA feeding in the leaves of resistant accession PB2013071, but not in leaves of susceptible accession PB2013046. There may be a delicate balance between the expression level of the callose synthesis and callose degrading genes. The BGLU accumulation might be caused by the plant’s need to degrade callose in the phloem, as callose deposition may affect the transport of assimilates. The fact that also under the empty clip cages the expression of the BGLU gene increased may be related to the involvement of the BGLU gene in the general defense response [85–87]. Putting a clip cage on a leaf may inflict such a response.
The accumulation of the CalS1 and CalS7 gene transcripts seems not to coincide with impaired phloem uptake as recorded by EPG. The gene expression increased after 6 h infestation whereas aphids already show difficulty in phloem feeding before that time. One possible explanation is that callose deposition is regulated at the protein level in the early stage of the defense response. In bean, callose can be induced within 5–10 min after injury through the activation of proteases [28]. We found that aphids tried to start phloem probing after about 1.5 h on resistant as well as susceptible plants (EPG parameter: time to first E1). However, no callose deposition was detected 1.5 h after the start of the aphid infestation (results not shown), which suggests that callose deposition is not involved in the early response of PB2013071 to aphid feeding. Another possible mechanism of phloem vessel occlusion is plugging by phloem proteins (P-proteins), which can block sieve tubes of higher-level plants rapidly [88–92]. P-proteins based occlusion is thought to be a faster and earlier response than callose deposition [90]. It may be speculated that specific P-proteins are involved in the early response to aphids on the resistant accessions, while callose deposition is induced later to prevent aphid feeding in a more stable and long-lasting way.
Conclusion
In conclusion, we identified three C. baccatum accessions that are resistant to the green peach aphid and one C. baccatum accession that is susceptible. Accession PB2013071 shows the highest aphid resistance, which seems to be phloem based according to the EPG recordings. The resistance is accompanied by callose deposition in the sieve elements, which may be at least partially causal. The up-regulated expression of the CalS1 and CalS7 genes in the resistant accession is in line with this observation.
Methods
Plant materials and growing condition
The plant materials used consisted of accessions of C. annuum, C. chinense, C. frutescens and C. baccatum that were obtained from the Centre for Genetic Resources, the Netherlands (CGN) and from the collection of Wageningen University & Research. Based on the results of an initial evaluation of about 50 accessions, additional material from C. baccatum were screened. The accession codes, names, and species of all materials used can be found in the Additional file 1: Tables S1 and Additional file 2: Table S2.
Two weeks after sowing, plants were transplanted into 14 cm pots with potting compost and grown in a standard greenhouse at 19–21 °C, 60–70% relative humidity and a 16–8 h light–dark photoperiod at Wageningen University & Research, Wageningen, NL. Plants were watered every other day and no aphid control was applied during growth and testing.
Aphid population
The GPA (M. persicae) population used originated from the population used by [93]. Initially it was reared on Chinese cabbage (B. rapa) cv. Granaat; later the rearing was transferred to C. annuum accession CGN19226 and subsequently to C. baccatum accession PB2013046. The aphid rearing was maintained in a standard greenhouse under the same conditions as the pepper plants.
Evaluation of Capsicum accessions for GPA resistance in a clip cage test
All evaluations were carried out in the greenhouses of Wageningen University & Research, Wageningen, NL and were performed in four experiments during summer and autumn. The first experiment, including 50 accessions (Additional file 1: Table S1), was done when plants were eight weeks old. Plants were tested in a complete block design with four blocks in the same glasshouse compartment, with one plant of each accession per block and two clip cages containing per cage 10 1-day-old GPA nymphs that were obtained from a rearing on Chinese cabbage. The clip cages were placed on the abaxial side of the top two fully expanded leaves of the plants. After seven days the numbers of surviving and dead aphids as well as new nymphs produced in each clip cage were counted. The second experiment was conducted similarly to the first with the following changes. Ten accessions were selected from the 50 tested in the first experiment (Additional file 1: Table S1). They were re-tested in a complete block design with 10 blocks when they were seven weeks old, one plant per accession in each block, again per plant with two clip-on cages with 10 1-day-old nymphs, originating from a rearing on C. annuum accession CGN19226.
In the third experiment only C. baccatum accessions were evaluated, together with C. annuum CGN19226 as susceptible control (Additional file 2: Table S2) in a complete block design with four blocks under conditions similar to the first two experiments. They were evaluated with two clip cages per plant, containing 5 1-day-old GPA nymphs per cage obtained from a rearing on CGN19226, when plants were seven weeks old. During the fourth experiment, eight selected accessions from the third experiment (including the susceptible C. annuum CGN19226) were re-tested in a complete block design with five blocks (Additional file 2: Table S2). Similar to the third experiment plants were evaluated with two clip cages containing 5 1-day-old GPA nymphs originated from a rearing on the susceptible C. baccatum accession PB2013046, when the plants were seven weeks old. After eight days all clip cages were observed.
For statistical analysis, the observations from two clip cages per plant were combined. Survival was determined by dividing the number of living aphids by the total number of aphids (dead and alive) in the clip cage. The number of new nymphs was divided by the average number of living aphids present, calculated as (2*living aphids + dead aphids)/2. Additionally, data used for ANOVA analysis were transformed to obtain a more or less constant residual variance: survival as arcsin(sqrt(x)) and nymphs as sqrt(x). Significance of differences in the means was evaluated using the LSD test (P < 0.05) on the transformed data.
Population development
A population development experiment was used to further confirm resistance/susceptibility of the accessions. Ten plants of each selected accession were randomized in one greenhouse compartment. Approx. 40 days after sowing each plant was infested with 5 wingless GPA adults and 10 nymphs and enclosed in an aphid-proof sleeve. After 19 days, the number of adult aphids was counted and the number of nymphs was estimated according to a visual scale (0 = none, 1 = few (< 50), 2 = many nymphs (> 50)). For ANOVA analysis, the number of adults per plant was transformed to log(x). Significance of differences of means was tested by LSD test (P < 0.05).
Electrical Penetration Graph
The Electrical Penetration Graph (EPG) technique was used to monitor GPA probing and feeding behaviour on the most resistant (PB2013071) and a susceptible (PB2013046) C. baccatum accession. For each accession, 10 seven-week-old plants were each probed with two adult aphids placed on the abaxial side of the top two fully expanded leaves. Experimental setup was as described by [94]. Recording lasted for six hours at 20 ± 2 °C under constant light. The EPG patterns were transformed into waveforms using the Stylet+a software version 1.20 (http://www.epgsystems.eu/). Extraction of resistance parameters from the waveforms was carried out using EPG-Calc 6.1.3 [95]. T-tests were used to determine the significance of the differences between the accessions for various EPG parameters. The Fisher exact test was used to determine the significance of the difference in percentage of aphids that reached E2 during six hours’ recording.
Callose deposition
Histological analysis of in situ callose deposition was performed essentially as described by [96] on the resistant (PB2013071) and susceptible (PB2013046) C. baccatum accession. The second fully expanded leaf with petiole was cut with scissors from each plant and immediately put into a 6 cm-diameter petri dish with 1.5% water-agar medium. Twenty randomly selected wingless aphids were put gently into the petri dish, which was sealed by Parafilm M (Bemis NA, USA). Four plants/replicates were used for each treatment or control. After 24 h, three to four leaf disks (1.3 cm in diameter) containing highest number of aphids were sampled from the detached leaf and directly placed in 96% ethanol with their abaxial side up to remove chlorophyll. After washing in 0.07 M K2HPO4 (pH = 9), leaf disks were stained for 2 h in 0.1% (w/v) aniline blue in 0.07 M K2HPO4 (pH = 9) at room temperature. Samples were subsequently mounted on glass slides with 70% glycerol. Callose fluorescence was observed qualitatively under UV light, and photos were taken using the Zeiss Axiophoto digital imaging microscope (Carl Zeiss AG, Germany). Control leaf samples without aphids were treated in the same way; leaf disks were taken from areas comparable to the areas taken from the infested leaves. In total 12 leaf disks were observed for accession PB2013071 and 14 for accession PB2013046 after 24 h GPA treatment; and 12 leaf disks were observed for both accessions as control.
Gene expression analysis
The expression level of callose related genes was analyzed by quantitative real-time PCR. Seven-week-old plants received three clip cages containing 15 randomly selected wingless aphids per cage. Leaf disks were collected from the clip cage areas 1.5, 6 and 24 h after the start of aphid infestation. After gently brushing aphids away, disks were flash-frozen in liquid nitrogen and stored at − 80 °C until use. Leaf disks under an empty clip cage were also collected after 1.5, 6 and 24 h and used as reference. Additionally, leaf disks without clip cage and aphid infestation were collected just before the infestation stated (time point 0 h). Four biological replicates were used per treatment with aphid infestation and three per treatment with empty clip cages. For the reference without clip cages (time point 0 h) also three biological replicates were used. In all cases, two plants were pooled together as one biological replicate.
The sequences of CalS family genes were obtained from the Pepper Genome Platform (http://peppergenome.snu.ac.kr/) [97] and the Pepper Genome Database (http://peppersequence.genomics.cn/page/species/index.jsp) [98] through BlastP queries [99] referring to the sequences from Arabidopsis (https://www.arabidopsis.org/index.jsp). Genes were identified and named according to phylogenetic tree of CalS family genes among Arabidopsis, grapevine and pepper which was constructed by MEGA5 [100]. Besides the CalS family genes, the basic ß-1,3-glucanase gene (CA03g30020, BGLU) was obtained from the Pepper Genome Platform (http://peppergenome.snu.ac.kr/). The pepper actin gene (CA12g08730) was used as an internal reference for normalization of gene expression [101]. Gene specific primers were designed using Primer3Plus (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi) and are listed in Additional file 6: Table S4.
Total RNA was isolated with the RNeasy plant mini kit (Qiagen, USA) according to the suppliers’ recommendations. After treatment with DNase I (Invitrogen, USA), 1 μg RNA template was reversely transcribed into cDNA using the iScript™ cDNA Synthesis Kit (Bio-Rad, USA). Quantitative real-time PCR was conducted using the iQ™ SYBR Green Supermix (Bio-Rad, USA) and the CFX96 Touch™ Real-Time system (Bio-Rad, USA).
The PCR mix contained 5 μl 2× iQ™ SYBR GREEN Supermix, 0.3 μl forward primer (10 μM), 0.3 μl reverse primer (10 μM) and 2 μl cDNA template with 10-time dilution, into a final volume of 10 μl. Quantitative RT-PCR was performed in duplicate using the following program: 95 °C for 3 min followed by 40 cycles of 95 °C for 15 s, and 60 °C for 1 min. As the primers were designed on the gene sequences from C. annuum, the QPCR products were sequenced to validate the region of amplification in C. baccatum. Relative expression was calculated with the 2-ΔΔCt method [102]. Independent-samples t-tests on log2-transformed data were used to determine the significance of the differences between certain time points after GPA infestation and no GPA infestation (P < 0.05).
Additional files
Table S1. Evaluation of Capsicum accessions for resistance against the aphid M. persicae. (DOCX 21 kb)
Table S2. Evaluation of C. baccatum accessions for resistance against the aphid M. persicae. (DOCX 19 kb)
Figure S1. Histochemical staining of callose in 24 h GPA-infested leaves. Resistant accession PB2013071 (A, C, E, G); susceptible accession PB2013046 (B, D, F, H). Bars = 100 μm. (PDF 275 kb)
Table S3. Callose synthase (CalS) genes in C. annuum. (DOCX 15 kb)
Figure S2. Phylogenetic analysis of pepper (Ca), Arabidopsis (At) and grapevine (Vv) CalS proteins, using the MEGA [100] neighbour-joining algorithm. (PDF 10 kb)
Table S4. Primer sequences used in real-time PCR. (DOCX 14 kb)
Acknowledgements
We would like to thank Dr. Freddy Tjallingii for valuable discussions on the EPG data, Sean Geurts and Maarten Peters for taking care of the plants.
Funding
Funding for this project was provided by a grant (PPS 1409–029) from the Ministry of Economic Affairs of the Netherlands as well as the breeding companies Syngenta B.V., the Netherlands and Bayer Crop Science B.V., the Netherlands. The resistance evaluations were also co-funded by Enza Zaden Research & Development B.V., the Netherlands, and Rijk Zwaan Zaadteelt en Zaadhandel B.V., the Netherlands. MS is supported by a grant from the China Scholarship Council.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article (and its Additional files). The accessions used are available from the Centre for Genetic resources the Netherlands or from Wageningen University and Research under an MTA for non-commercial applications.
Authors’ contributions
Resistance evaluations were carried out by GS and WvW. The EPG experiment was carried out by WvW. The callose deposition and real-time PCR experiments were carried out by MS. MS, RV and BV carried out the data analysis and drafted the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1340-3) contains supplementary material, which is available to authorized users.
Contributor Information
Mengjing Sun, Email: mengjing.sun@wur.nl.
Roeland E. Voorrips, Email: roeland.voorrips@wur.nl
Greet Steenhuis-Broers, Email: greet.steenhuis@upcmail.nl.
Wendy van’t Westende, Email: wendy.vantwestende@wur.nl.
Ben Vosman, Phone: +31 317 480838, Email: ben.vosman@wur.nl.
References
- 1.Baral J, Bosland P. An update synthesis of the Capsicum genus. Capsicum Eggplant Newsletter. 2002:11–21.
- 2.Pickersgill B. Cytogenetics and evolution of Capsicum L. Chromosome engineering in plants: genetics, breeding, evolution, part B Elsevier, Amsterdam. 1991:139–60.
- 3.Blackman RL, Eastop VF. 2 Volume Set: John Wiley & Sons. 2008. Aphids on the World's Herbaceous Plants and Shrubs. [Google Scholar]
- 4.Kenyon L, Kumar S, Tsai WS, Hughes J. Virus diseases of peppers (Capsicum spp.) and their control. Adv Virus Res. 2014;90:297–354. doi: 10.1016/B978-0-12-801246-8.00006-8. [DOI] [PubMed] [Google Scholar]
- 5.Tjallingii WF. Salivary secretions by aphids interacting with proteins of phloem wound responses. J Exp Bot. 2006;57(4):739–745. doi: 10.1093/jxb/erj088. [DOI] [PubMed] [Google Scholar]
- 6.Dedryver C-A, Le Ralec A, Fabre F. The conflicting relationships between aphids and men: a review of aphid damage and control strategies. C R Biol. 2010;333(6):539–553. doi: 10.1016/j.crvi.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Tjallingii W. Electrical recording of stylet penetration activities. In: Aphids, their biology, natural enemies and control. Elsevier Science Publishers; 1988. p. 95–108.
- 8.Tjallingii W. Electrical nature of recorded signals during stylet penetration by aphids. Entomologia Experimentalis et Applicata. 1985;38(2):177–186. doi: 10.1111/j.1570-7458.1985.tb03516.x. [DOI] [Google Scholar]
- 9.Tjallingii WF, Garzo E, Fereres A. New structure in cell puncture activities by aphid stylets: a dual-mode EPG study. Entomologia experimentalis et applicata. 2010;135(2):193–207. doi: 10.1111/j.1570-7458.2010.00983.x. [DOI] [Google Scholar]
- 10.Alvarez A, Tjallingii W, Garzo E, Vleeshouwers V, Dicke M, Vosman B. Location of resistance factors in the leaves of potato and wild tuber-bearing Solanum species to the aphid Myzus persicae. Entomologia Experimentalis et Applicata. 2006;121(2):145–157. doi: 10.1111/j.1570-8703.2006.00464.x. [DOI] [Google Scholar]
- 11.Chandran P, Reese JC, Khan SA, Wang D, Schapaugh W, Campbell LR. Feeding behavior comparison of soybean aphid (Hemiptera: Aphididae) biotypes on different soybean genotypes. J Econ Entomol. 2013;106(5):2234–2240. doi: 10.1603/EC13126. [DOI] [PubMed] [Google Scholar]
- 12.Khan SA, Marimuthu M, Predeesh C, Aguirre-Rojas LM, Reese JC, Smith CM. Electrical Penetration Graph Recording of Russian Wheat Aphid (Hemiptera: Aphididae) Feeding on Aphid-Resistant Wheat and Barley. J Econ Entomol. 2015;108(5):2465–2470. doi: 10.1093/jee/tov183. [DOI] [PubMed] [Google Scholar]
- 13.Will T, Tjallingii WF, Thönnessen A, van Bel AJ. Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci. 2007;104(25):10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaupels F, Knauer T, van Bel AJ. A combinatory approach for analysis of protein sets in barley sieve-tube samples using EDTA-facilitated exudation and aphid stylectomy. J Plant Physiol. 2008;165(1):95–103. doi: 10.1016/j.jplph.2007.07.023. [DOI] [PubMed] [Google Scholar]
- 15.Hao P, Liu C, Wang Y, Chen R, Tang M, Du B, Zhu L, He G. Herbivore-induced callose deposition on the sieve plates of rice: an important mechanism for host resistance. Plant Physiol. 2008;146(4):1810–1820. doi: 10.1104/pp.107.111484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donofrio NM, Delaney TP. Abnormal callose response phenotype and hypersusceptibility to Peronospora parasitica in defense-compromised Arabidopsis nim1-1 and salicylate hydroxylase-expressing plants. Mol Plant-Microbe Interact. 2001;14(4):439–450. doi: 10.1094/MPMI.2001.14.4.439. [DOI] [PubMed] [Google Scholar]
- 17.Saheed SA, Cierlik I, Larsson KA, Delp G, Bradley G, Jonsson L, Botha CE. Stronger induction of callose deposition in barley by Russian wheat aphid than bird cherry-oat aphid is not associated with differences in callose synthase or β-1, 3-glucanase transcript abundance. Physiol Plant. 2009;135(2):150–161. doi: 10.1111/j.1399-3054.2008.01180.x. [DOI] [PubMed] [Google Scholar]
- 18.Luna E, Pastor V, Robert J, Flors V, Mauch-Mani B, Ton J. Callose deposition: a multifaceted plant defense response. Mol Plant-Microbe Interact. 2011;24(2):183–193. doi: 10.1094/MPMI-07-10-0149. [DOI] [PubMed] [Google Scholar]
- 19.Kempema LA, Cui X, Holzer FM, Walling LL. Arabidopsis transcriptome changes in response to phloem-feeding silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 2007;143(2):849–865. doi: 10.1104/pp.106.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richmond TA, Somerville CR. The cellulose synthase superfamily. Plant Physiol. 2000;124(2):495–498. doi: 10.1104/pp.124.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verma DPS, Hong Z. Plant callose synthase complexes. Plant Mol Biol. 2001;47(6):693–701. doi: 10.1023/A:1013679111111. [DOI] [PubMed] [Google Scholar]
- 22.Hazen SP, Scott-Craig JS, Walton JD. Cellulose synthase-like genes of rice. Plant Physiol. 2002;128(2):336–340. doi: 10.1104/pp.010875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schober MS, Burton RA, Shirley NJ, Jacobs AK, Fincher GB. Analysis of the (1, 3)-β-D-glucan synthase gene family of barley. Phytochemistry. 2009;70(6):713–720. doi: 10.1016/j.phytochem.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Fu M, Xu M, Zhou T, Wang D, Tian S, Han L, Dong H, Zhang C. Transgenic expression of a functional fragment of harpin protein Hpa1 in wheat induces the phloem-based defence against English grain aphid. J Exp Bot. 2014;65(6):1439–1453. doi: 10.1093/jxb/ert488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu Y, Jiao L, Fu S, Yin L, Zhang Y, Lu J. Callose Synthase Family Genes Involved in the Grapevine Defense Response to Downy Mildew Disease. Phytopathology. 2016;106(1):56–64. doi: 10.1094/PHYTO-07-15-0166-R. [DOI] [PubMed] [Google Scholar]
- 26.Xie B, Wang X, Zhu M, Zhang Z, Hong Z. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011;65(1):1–14. [DOI] [PubMed]
- 27.Jacobs AK, Lipka V, Burton RA, Panstruga R, Strizhov N, Schulze-Lefert P, Fincher GB. An Arabidopsis callose synthase, GSL5, is required for wound and papillary callose formation. Plant Cell. 2003;15(11):2503–2513. doi: 10.1105/tpc.016097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakashima J, Laosinchai W, Cui X, Brown RM. New insight into the mechanism of cellulose and callose biosynthesis: proteases may regulate callose biosynthesis upon wounding. Cellulose. 2003;10(4):369–389. doi: 10.1023/A:1027336605479. [DOI] [Google Scholar]
- 29.Lü B, Sun W, Zhang S, Zhang C, Qian J, Wang X, Gao R, Dong H. HrpNEa-induced deterrent effect on phloem feeding of the green peach aphid Myzus persicae requires AtGSL5 and AtMYB44 genes in Arabidopsisthaliana. J Biosci (Bangalore) 2011;36(1):123–137. doi: 10.1007/s12038-011-9016-2. [DOI] [PubMed] [Google Scholar]
- 30.Kuśnierczyk A, Winge P, Jørstad TS, Troczyńska J, Rossiter JT, Bones AM. Towards global understanding of plant defence against aphids–timing and dynamics of early Arabidopsis defence responses to cabbage aphid (Brevicoryne brassicae) attack. Plant Cell Environ. 2008;31(8):1097–1115. doi: 10.1111/j.1365-3040.2008.01823.x. [DOI] [PubMed] [Google Scholar]
- 31.Mehrabi S, Åhman I, Jonsson LM. The constitutive expression and induction of three β-1, 3-glucanases by bird cherry-oat aphid in relation to aphid resistance in 15 barley breeding lines. Arthropod Plant Interact. 2016;10(2):101–111. doi: 10.1007/s11829-016-9415-2. [DOI] [Google Scholar]
- 32.Silver AR, Van Emden HF, Battersby M. A biochemical mechanism of resistance to pirimicarb in two glasshouse clones of Aphis gossypii. Pestic Sci. 1995;43(1):21–29. doi: 10.1002/ps.2780430104. [DOI] [Google Scholar]
- 33.Wang K-Y, Liu T-X, Yu C-H, Jiang X-Y, Yi M-Q. Resistance of Aphis gossypii (Homoptera: Aphididae) to fenvalerate and imidacloprid and activities of detoxification enzymes on cotton and cucumber. J Econ Entomol. 2002;95(2):407–413. doi: 10.1603/0022-0493-95.2.407. [DOI] [PubMed] [Google Scholar]
- 34.Painter RH. Insect resistance in crop plants. Soil Sci. 1951;72(6):481. doi: 10.1097/00010694-195112000-00015. [DOI] [Google Scholar]
- 35.Smith CM, Boyko EV. The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomologia Experimentalis et Applicata. 2007;122(1):1–16. doi: 10.1111/j.1570-7458.2006.00503.x. [DOI] [Google Scholar]
- 36.Van Emden HF, Harrington R: Aphids as crop pests: Cabi; 2007.
- 37.Züst T, Agrawal AA. Mechanisms and evolution of plant resistance to aphids. Nature plants. 2016;2:15206. doi: 10.1038/nplants.2015.206. [DOI] [PubMed] [Google Scholar]
- 38.Goggin FL. Plant–aphid interactions: molecular and ecological perspectives. Curr Opin Plant Biol. 2007;10(4):399–408. doi: 10.1016/j.pbi.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Williams W, Kennedy GG, Yamamoto RT, Thacker J, Bordner J. 2-Tridecanone: a naturally occurring insecticide from the wild tomato Lycopersicon hirsutum f. glabratum. Science. 1980;207(4433):888–889. doi: 10.1126/science.207.4433.888. [DOI] [PubMed] [Google Scholar]
- 40.Smith CM. Plant resistance to arthropods: molecular and conventional approaches: Springer Science & Business Media; 2005.
- 41.Boyko E, Starkey S, Smith M. Molecular genetic mapping of Gby, a new greenbug resistance gene in bread wheat. Theor Appl Genet. 2004;109(6):1230–1236. doi: 10.1007/s00122-004-1729-2. [DOI] [PubMed] [Google Scholar]
- 42.Kim K-S, Bellendir S, Hudson KA, Hill CB, Hartman GL, Hyten DL, Hudson ME, Diers BW. Fine mapping the soybean aphid resistance gene Rag1 in soybean. Theor Appl Genet. 2010;120(5):1063–1071. doi: 10.1007/s00122-009-1234-8. [DOI] [PubMed] [Google Scholar]
- 43.Eenink A, Dieleman F, Groenwold R. Resistance of lettuce (Lactuca) to the leaf aphid Nasonovia ribis nigri 2. Inheritance of the resistance. Euphytica. 1982;31(2):301–304. doi: 10.1007/BF00021644. [DOI] [Google Scholar]
- 44.Githiri S, Ampong-Nyarko K, Osir E, Kimani P. Genetics of resistance to Aphis craccivora in cowpea. Euphytica. 1996;89(3):371–376. [Google Scholar]
- 45.Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M. The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat Biotechnol. 1998;16(13):1365–1369. doi: 10.1038/4350. [DOI] [PubMed] [Google Scholar]
- 46.Rossi M, Goggin FL, Milligan SB, Kaloshian I, Ullman DE, Williamson VM. The nematode resistance gene Mi of tomato confers resistance against the potato aphid. Proc Natl Acad Sci. 1998;95(17):9750–9754. doi: 10.1073/pnas.95.17.9750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nombela G, Williamson VM, Muñiz M. The root-knot nematode resistance gene Mi-1.2 of tomato is responsible for resistance against the whitefly Bemisia tabaci. Mol Plant-Microbe Interact. 2003;16(7):645–649. doi: 10.1094/MPMI.2003.16.7.645. [DOI] [PubMed] [Google Scholar]
- 48.Pauquet J, Burget E, Hagen L, Chovelon V, Menn Al, Valot N, Desloire S, Caboche M, Rousselle P, Pitrat M et al.: Map-based cloning of the Vat gene from melon conferring resistance to both aphid colonization and aphid transmission of several viruses. In Olomouc: Palacký University in Olomouc; 2004: 325–329.
- 49.Bosland PW, Ellington JJ. Comparison of Capsicum annuum and C. pubescens for antixenosis as a means of aphid resistance. HortScience. 1996;31(6):1017–1018. [Google Scholar]
- 50.Frantz JD, Gardner J, Hoffmann MP, Jahn MM. Greenhouse screening of Capsicum accessions for resistance to green peach aphid (Myzus persicae) HortScience. 2004;39(6):1332–1335. [Google Scholar]
- 51.Da Costa JG, Pires EV, Riffel A, Birkett MA, Bleicher E, Sant’Ana AEG. Differential preference of Capsicum spp. cultivars by Aphis gossypii is conferred by variation in volatile semiochemistry. Euphytica. 2011;177(3):299–307. doi: 10.1007/s10681-010-0250-8. [DOI] [Google Scholar]
- 52.Ton J, Mauch-Mani B. β-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. 2004;38(1):119–130. doi: 10.1111/j.1365-313X.2004.02028.x. [DOI] [PubMed] [Google Scholar]
- 53.De Barro P, Sherratt T, David O, Maclean N. An investigation of the differential performance of clones of the aphid Sitobion avenae on two host species. Oecologia. 1995;104(3):379–385. doi: 10.1007/BF00328374. [DOI] [PubMed] [Google Scholar]
- 54.Liu XD, Zhai BP, Zhang XX. Specialized host-plant performance of the cotton aphid is altered by experience. Ecol Res. 2008;23(5):919–925. doi: 10.1007/s11284-007-0458-9. [DOI] [Google Scholar]
- 55.Francis F, Vanhaelen N, Haubruge E. Glutathione S-transferases in the adaptation to plant secondary metabolites in the Myzus persicae aphid. Arch Insect Biochem Physiol. 2005;58(3):166–174. doi: 10.1002/arch.20049. [DOI] [PubMed] [Google Scholar]
- 56.Sugio A, Dubreuil G, Giron D, Simon J-C. Plant–insect interactions under bacterial influence: ecological implications and underlying mechanisms. J Exp Bot. 2014;66(2):467–478. doi: 10.1093/jxb/eru435. [DOI] [PubMed] [Google Scholar]
- 57.Tsuchida T, Koga R, Fukatsu T: Host plant specialization governed by facultative symbiont. Science 2004, 303(5666):1989–1989. [DOI] [PubMed]
- 58.Machado-Assefh CR, Lopez-Isasmendi G, Tjallingii WF, Jander G, Alvarez AE. Disrupting Buchnera aphidicola, the endosymbiotic bacteria of Myzus persicae, delays host plant acceptance. Arthropod Plant Interact. 2015;9(5):529–541. doi: 10.1007/s11829-015-9394-8. [DOI] [Google Scholar]
- 59.Leather S, Dixon A. Aphid growth and reproductive rates. Entomologia experimentalis et applicata. 1984;35(2):137–140. doi: 10.1111/j.1570-7458.1984.tb03373.x. [DOI] [Google Scholar]
- 60.Thomas M, Waage J. Integration of biological control and host plant resistance breeding: a scientific and literature review: CTA. 1996. [Google Scholar]
- 61.Yoon JB, Yang DC, Lee WP, Ahn SY, Park HG. Genetic resources resistant to anthracnose in the genus Capsicum. Horticulture Environment and Biotechnology. 2004;45(6):318–323. [Google Scholar]
- 62.Park S-K, Kim SH, Park HG, Yoon JB. Capsicum germplasm resistant to pepper anthracnose differentially interact with Colletotrichum isolates. Horticulture Environment and Biotechnology. 2009;50(1):17–23. [Google Scholar]
- 63.De Souza V, Café-Filho A. Resistance to Leveillula taurica in the genus Capsicum. Plant Pathol. 2003;52(5):613–619. doi: 10.1046/j.1365-3059.2003.00920.x. [DOI] [Google Scholar]
- 64.Maharijaya A, Vosman B, Steenhuis-Broers G, Harpenas A, Purwito A, Visser RG, Voorrips RE. Screening of pepper accessions for resistance against two thrips species (Frankliniella occidentalis and Thrips parvispinus) Euphytica. 2011;177(3):401–410. doi: 10.1007/s10681-010-0277-x. [DOI] [Google Scholar]
- 65.Gray SM, Banerjee N. Mechanisms of arthropod transmission of plant and animal viruses. Microbiol Mol Biol Rev. 1999;63(1):128–148. doi: 10.1128/mmbr.63.1.128-148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Radcliffe EB, Ragsdale DW. Aphid-transmitted potato viruses: the importance of understanding vector biology. American J Potato Res. 2002;79(5):353–386. doi: 10.1007/BF02870173. [DOI] [Google Scholar]
- 67.Tjallingii W, Esch TH. Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiol Entomol. 1993;18(3):317–328. doi: 10.1111/j.1365-3032.1993.tb00604.x. [DOI] [Google Scholar]
- 68.Mart B, Collar J, Tjallingii W, Fereres A. Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non-persistently transmitted plant viruses. J Gen Virol. 1997;78(10):2701–2705. doi: 10.1099/0022-1317-78-10-2701. [DOI] [PubMed] [Google Scholar]
- 69.Symmes E, Walker G, Perring T. Stylet penetration behavior of Myzus persicae related to transmission of Zucchini yellow mosaic virus. Entomologia experimentalis et applicata. 2008;129(3):258–267. doi: 10.1111/j.1570-7458.2008.00784.x. [DOI] [Google Scholar]
- 70.Botha C, Matsiliza B, Bornman C. Reduction in transport in wheat (Triticum aestivum) is caused by sustained phloem feeding by the Russian wheat aphid (Diuraphis noxia) S Afr J Bot. 2004;70(2):249–254. doi: 10.1016/S0254-6299(15)30242-8. [DOI] [Google Scholar]
- 71.Saheed S, Larsson K, Delp G, Botha C, Jonsson L, Bradley G. Wound callose synthesis in response to Russian wheat aphid and bird cherry-oat aphid feeding on barley cv Clipper. S Afr J Bot. 2007;73(2):310. doi: 10.1016/j.sajb.2007.02.110. [DOI] [Google Scholar]
- 72.Mondal HA, Louis J, Archer L, Patel M, Nalam VJ, Sarowar S, Sivapalan V, Root DD, Shah J. Arabidopsis ACTIN-DEPOLYMERIZING FACTOR3 is required for controlling aphid feeding from the phloem. Plant Physiol. 2017:01438.02017. [DOI] [PMC free article] [PubMed]
- 73.Broeke CJ, Dicke M, Loon JJ. Feeding behavior and performance of Nasonovia ribisnigri on grafts, detached leaves, and leaf disks of resistant and susceptible lettuce. Entomologia Experimentalis et Applicata. 2016;159(1):102–111. doi: 10.1111/eea.12430. [DOI] [Google Scholar]
- 74.Villada ES, González EG, López-Sesé AI, Castiel AF, Gómez-Guillamón ML. Hypersensitive response to Aphis gossypii Glover in melon genotypes carrying the Vat gene. J Exp Bot. 2009;60(11):3269–3277. doi: 10.1093/jxb/erp163. [DOI] [PubMed] [Google Scholar]
- 75.Xie B, Wang X, Hong Z. Precocious pollen germination in Arabidopsis plants with altered callose deposition during microsporogenesis. Planta. 2010;231(4):809–823. doi: 10.1007/s00425-009-1091-3. [DOI] [PubMed] [Google Scholar]
- 76.Kothari S, Joshi A, Kachhwaha S, Ochoa-Alejo N. Chilli peppers—a review on tissue culture and transgenesis. Biotechnol Adv. 2010;28(1):35–48. doi: 10.1016/j.biotechadv.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 77.Linthorst HJ, Van Loon L. Pathogenesis‐related proteins of plants. Crit Rev Plant Sci. 1991;10(2):123–50.
- 78.Jung HW, Hwang BK. Pepper gene encoding a basic β-1, 3-glucanase is differentially expressed in pepper tissues upon pathogen infection and ethephon or methyl jasmonate treatment. Plant Sci. 2000;159(1):97–106. doi: 10.1016/S0168-9452(00)00334-4. [DOI] [PubMed] [Google Scholar]
- 79.Zheng H-Z, Kim Y-W, Lee H-J, Park R-D, Jung W-J, Kim Y-C, Lee S-H, Kim T-H, Kim K-Y. Quantitative changes of PR proteins and antioxidative enzymes in response to Glomus intraradices and Phytophthora capsici in pepper (Capsicum annuum L.) plants. J Microbiol Biotechnol. 2004;14(3):553–562. [Google Scholar]
- 80.Wang J-E, Li D-W, Zhang Y-L, Zhao Q, He Y-M, Gong Z-H. Defence responses of pepper (Capsicum annuum L.) infected with incompatible and compatible strains of Phytophthora capsici. Eur J Plant Pathol. 2013;136(3):625–638. doi: 10.1007/s10658-013-0193-8. [DOI] [Google Scholar]
- 81.Van der Westhuizen A, Qian XM, Botha AM. β-1, 3-Glucanases in wheat and resistance to the Russian wheat aphid. Physiol Plant. 1998;103(1):125–131. doi: 10.1034/j.1399-3054.1998.1030115.x. [DOI] [Google Scholar]
- 82.De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux J-P, Van Loon L, Dicke M. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant-Microbe Interact. 2005;18(9):923–937. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 83.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. Acquired resistance in Arabidopsis. Plant Cell. 1992;4(6):645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moran PJ, Thompson GA. Molecular responses to aphid feeding in Arabidopsis in relation to plant defense pathways. Plant Physiol. 2001;125(2):1074–1085. doi: 10.1104/pp.125.2.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sardesai* N, Subramanyam* S, Nemacheck J, Williams CE. Modulation of defense-response gene expression in wheat during Hessian fly larval feeding. J Plant Interact. 2005;1(1):39–50. doi: 10.1080/17429140500309498. [DOI] [Google Scholar]
- 86.Xue M, Wang C-X, Bi M-J, Li Q-L, Liu T-X. Induced defense by Bemisia tabaci biotype B (Hemiptera: Aleyrodidae) in tobacco against Myzus persicae (Hemiptera: Aphididae) Environ Entomol. 2010;39(3):883–891. doi: 10.1603/EN09307. [DOI] [PubMed] [Google Scholar]
- 87.De Zutter N, Audenaert K, Ameye M, De Boevre M, De Saeger S, Haesaert G, Smagghe G. The plant response induced in wheat ears by a combined attack of Sitobion avenae aphids and Fusarium graminearum boosts fungal infection and deoxynivalenol production. Mol Plant Pathol. 2017;18(1):98–109. doi: 10.1111/mpp.12386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cronshaw J, Sabnis DD. Phloem proteins. In: Sieve elements: Springer; 1990. p. 257–83.
- 89.Kehr J. Phloem sap proteins: their identities and potential roles in the interaction between plants and phloem-feeding insects. J Exp Bot. 2006;57(4):767–774. doi: 10.1093/jxb/erj087. [DOI] [PubMed] [Google Scholar]
- 90.Furch AC, Hafke JB, Schulz A, van Bel AJ. Ca2+−mediated remote control of reversible sieve tube occlusion in Vicia faba. J Exp Bot. 2007;58(11):2827–2838. doi: 10.1093/jxb/erm143. [DOI] [PubMed] [Google Scholar]
- 91.Zhang C, Shi H, Chen L, Wang X, Lü B, Zhang S, Liang Y, Liu R, Qian J, Sun W. Harpin-induced expression and transgenic overexpression of the phloem protein gene AtPP2-A1 in Arabidopsis repress phloem feeding of the green peach aphid Myzus persicae. BMC Plant Biol. 2011;11(1):11. doi: 10.1186/1471-2229-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ernst AM, Jekat SB, Zielonka S, Müller B, Neumann U, Rüping B, Twyman RM, Krzyzanek V, Prüfer D, Noll GA. Sieve element occlusion (SEO) genes encode structural phloem proteins involved in wound sealing of the phloem. Proc Natl Acad Sci. 2012;109(28):1980–9. [DOI] [PMC free article] [PubMed]
- 93.Chen X, Vosman B, Visser RG, van der Vlugt RA, Broekgaarden C. High throughput phenotyping for aphid resistance in large plant collections. Plant Methods. 2012;8(1):1. doi: 10.1186/1746-4811-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alvarez AE, Broglia VG, D’Amato AM A, Wouters D, van der Vossen E, Garzo E, Tjallingii WF, Dicke M, Vosman B. Comparative analysis of Solanum stoloniferum responses to probing by the green peach aphid Myzus persicae and the potato aphid Macrosiphum euphorbiae. Insect Sci. 2013;20(2):207–227. doi: 10.1111/j.1744-7917.2012.01505.x. [DOI] [PubMed] [Google Scholar]
- 95.Giordanengo P. EPG-Calc: a PHP-based script to calculate electrical penetration graph (EPG) parameters. Arthropod Plant Interact. 2014;8(2):163–169. doi: 10.1007/s11829-014-9298-z. [DOI] [Google Scholar]
- 96.Kissoudis C, Sunarti S, van de Wiel C, Visser RG, van der Linden CG, Bai Y. Responses to combined abiotic and biotic stress in tomato are governed by stress intensity and resistance mechanism. J Exp Bot. 2016;67(17):5119–5132. doi: 10.1093/jxb/erw285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim S, Park M, Yeom S-I, Kim Y-M, Lee JM, Lee H-A, Seo E, Choi J, Cheong K, Kim K-T. Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nat Genet. 2014;46(3):270–278. doi: 10.1038/ng.2877. [DOI] [PubMed] [Google Scholar]
- 98.Qin C, Yu C, Shen Y, Fang X, Chen L, Min J, Cheng J, Zhao S, Xu M, Luo Y. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc Natl Acad Sci. 2014;111(14):5135–5140. doi: 10.1073/pnas.1400975111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McGinnis S, Madden TL. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(suppl 2):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bin WS, Wei LK, Ping DW, Li Z, Wei G, Bing LJ, Gui PB, Jian WH, Feng CJ. Evaluation of appropriate reference genes for gene expression studies in pepper by quantitative real-time PCR. Mol Breed. 2012;30(3):1393–1400. doi: 10.1007/s11032-012-9726-7. [DOI] [Google Scholar]
- 102.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Evaluation of Capsicum accessions for resistance against the aphid M. persicae. (DOCX 21 kb)
Table S2. Evaluation of C. baccatum accessions for resistance against the aphid M. persicae. (DOCX 19 kb)
Figure S1. Histochemical staining of callose in 24 h GPA-infested leaves. Resistant accession PB2013071 (A, C, E, G); susceptible accession PB2013046 (B, D, F, H). Bars = 100 μm. (PDF 275 kb)
Table S3. Callose synthase (CalS) genes in C. annuum. (DOCX 15 kb)
Figure S2. Phylogenetic analysis of pepper (Ca), Arabidopsis (At) and grapevine (Vv) CalS proteins, using the MEGA [100] neighbour-joining algorithm. (PDF 10 kb)
Table S4. Primer sequences used in real-time PCR. (DOCX 14 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article (and its Additional files). The accessions used are available from the Centre for Genetic resources the Netherlands or from Wageningen University and Research under an MTA for non-commercial applications.


