Abstract
A number of reagents have been used to define the sequence-specific protein–DNA contacts by footprinting analysis. We report a new in vivo technique using the complex of 1,10-phenanthroline and copper [(OP2)Cu] as a probe to study various intracellular DNA–protein interactions in whole cells. The versatility of the protocol is demonstrated by applying the technique to address various processes. The protocol is applied to (i) detect structural alterations in DNA as a result of single base substitution, (ii) footprint site-specific DNA-binding proteins, (iii) analyze promoter occupancy by RNA polymerase and (iv) analyze molecular interactions during transcription initiation. The results demonstrate that in vivo (OP)2Cu probing is a useful tool in studying important cellular processes involving DNA–protein interactions and has potential applications in post-genomic research.
INTRODUCTION
Understanding protein–DNA interactions is necessary to get an insight into various cellular processes such as transcription, replication, DNA repair and recombination. Among the various techniques developed to study molecular interactions, footprinting is most commonly used as it provides valuable information on how a protein recognizes DNA. A number of enzymatic and chemical probes are used that cleave or modify DNA differentially as a result of protein binding to its site. Enzymatic probes include DNase I (1,2), exonuclease III (3,4), P1 nuclease (5) and micrococcal nuclease (6). Dimethylsulfate (DMS) (7,8), ethyl nitrosourea (9), potassium permanganate (10), bis(1,10-phenanthroline)–copper (I) (11), methidiumpropyl-EDTA.iron (II) [MPE.Fe(II)] (12), diethyl pyrocarbonate (13,14), peroxonitrous acid (15) and osmium tetroxide (16) are examples of chemical probes used. While the list of reagents that are used for in vitro footprinting reactions is extensive, the agents that are applied to in vivo probing reactions are limited. DMS (7,8) and potassium permanganate (10) have proved useful in studying DNA–protein interactions in vivo. In addition to these two chemicals, DNase I has been used in ethanol permeabilized Escherichia coli cells in situ (17). In the post-genomic scenario of functional genomics, it has become important to develop new tools for in vivo probing of molecular interactions.
The chemical nuclease activity of 1,10-phenanthroline–copper [(OP)2Cu] was demonstrated in 1978 while studying the mechanism of inhibition of E.coli DNA polymerase by 1,10-phenanthroline (OP). The nucleolytic activity of (OP)2Cu has many advantages as a footprinting reagent when compared to other commonly used reagents: (i) the (OP)2Cu chelate is a small molecule that gives a high resolution footprint; (ii) (OP)2Cu cuts at almost every sequence position, though the intensity of cutting may depend on local primary sequence (18,19); (iii) since (OP)2Cu binds to the minor groove of DNA, it reveals minor groove interactions. Any alterations in the major groove width due to protein binding and distortion will also be reflected as a change in the reactivity pattern of (OP)2Cu in the minor groove. (iv) (OP)2Cu has the ability to detect protein induced changes in DNA (20,21). The chemistry of the DNA cleavage reaction of (OP)2Cu has not previously been developed as an in vivo footprinting technique to study DNA–protein interactions. In this study, the potential of (OP)2Cu as a tool for in vivo footprinting assays is considered. The procedure we have developed using the reagent is applied successfully to probe DNA–protein interactions in vivo.
MATERIALS AND METHODS
Strains, plasmids, enzymes and chemicals
Escherichia coli DH10B was used for propagating the various plasmids. Plasmids pLW4, pVN184 and ptin7 (22), pBM2 (B.D.Paul and V.Nagaraja, unpublished), pACMK (23) and pVR7 (24) were used for different footprinting reactions. Klenow fragment of E.coli DNA polymerase and T4 polynucleotide kinase were from New England Biolabs. [γ-32P]ATP was purchased from NEN. The chemicals used for the footprinting reactions, OP, neocuproine, 3-mercaptopropionic acid (3-MPA) and copper sulfate, were purchased from Sigma. DNA manipulation techniques (plasmid isolation, end-labeling of primers with [γ-32P]ATP etc.) were carried out as described (25).
In vivo 1,10-phenanthroline–copper [(OP)2Cu] footprinting reaction
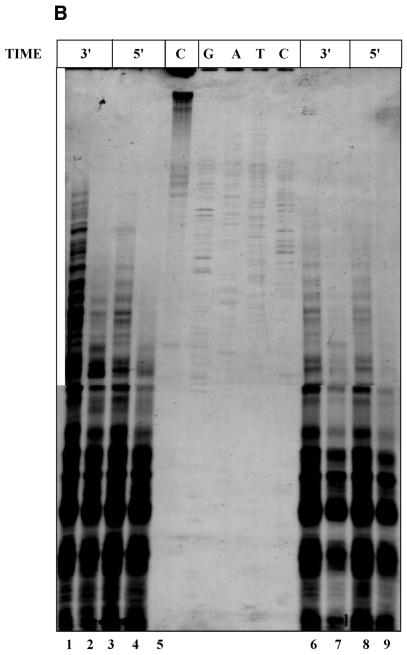
Overnight-grown E.coli cells harboring various plasmid constructs (described in Results and figure legends) were used as pre-inoculum (1%) and the cultures were grown in 4.0 ml of Luria–Bertani medium to an OD600 of 0.6. Various concentrations of OP and CuSO4 were used in order to standardize the conditions of single hit kinetics, and representative data are shown in Figure 1A. A representative autoradiogram for the pattern of cleavage with respect to (OP)2Cu concentration and time is depicted in Figure 1B. Copper sulfate, OP or 3-MPA alone did not cleave the plasmid DNA. Single hit pattern would depend on the size of the plasmid, copy number and also number of different plasmids present in the cell. Accordingly, different concentrations of the reagents were used for different plasmids after appropriate standardization. For plasmids pLW4, ptin7 and T2G, 200 µl of a mixture of 5 mM OP and 0.375 mM CuSO4 and 200 µl of 580 mM 3-MPA were added to 4.0 ml of culture and shaken vigorously at 37°C for 1 min. The reaction was quenched by adding 200 µl of 0.1 M neocuproine (2,9-dimethyl-1,10-phenanthroline) to the culture and mixing thoroughly for another 10 s. The culture tubes were chilled in ice and processed immediately for plasmid isolation by the boiling method (25). For the pBM2 and pVR7 as well as pUC18 and pACMK plasmid systems, 100 µl of 5 mM OP and 0.375 mM CuSO4 and 100 µl of 580 mM 3-MPA were used and the reaction was carried out for 30 s to get single hit conditions. Primer extension reactions were carried out using end-labeled primers (5′-GGAATCCGCCTTAAATAACA-3′ and 5′-TGACCGGCAGCAAAATG-3′) as described earlier (7). Samples were electrophoresed in a 6% denaturing PAGE and autoradiographed. Using the same end-labeled primers Sanger’s dideoxy sequencing was carried out.
Figure 1.
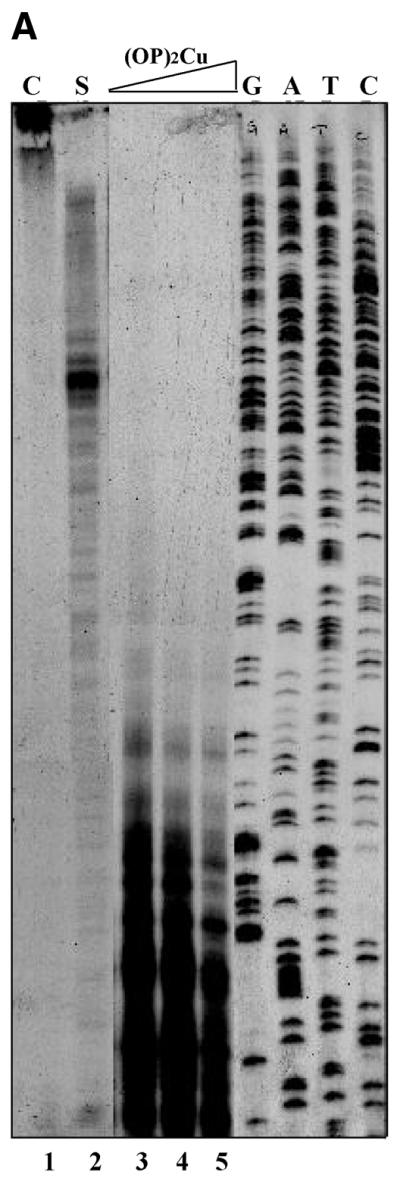
Standardization of in vivo (OP)2Cu probing. (A) Single hit conditions. Aliquots of 4.0 ml of culture of E.coli DH10B cells harboring plasmid pLW4 were subjected to treatment with 200 µl of increasing concentrations of mixture of OP and copper sulfate (lane 2, 5 mM OP/0.375 mM CuSO4; lane 3, 80 mM OP/6 mM CuSO4; lane 4, 160 mM OP/12 mM CuSO4; lane 5, 400 mM OP/30 mM CuSO4) and 200 µl of 580 mM 3-MPA. The reaction was terminated after 1 min by the addition of 200 µl of 0.1 M neocuproine to the reaction mix as described in Materials and Methods. Primer extension products of the plasmid from untreated culture (C) and near single hit conditions (S) are shown in lanes 1 and 2, respectively. G, A, T and C refer to sequencing lanes with the same primer. (B) Time and concentration dependence. In vivo (OP)2Cu reaction was carried out with cells containing plasmid pLW4 alone (lanes 1, 3, 6 and 8) or in combination with pVN184 (lanes 2, 4, 7 and 9) for different time points as indicated in the figure. Lanes 1–4 and 6–9 were treated with 200 µl of a mixture of 20 mM OP/1.5 mM CuSO4 and 40 mM OP/3 mM CuSO4, respectively, and 200 µl of 580 mM 3-MPA as described in Materials and Methods. Primer extension product of plasmid DNA isolated from cells treated with CuSO4 alone is shown in lane 5 (C).
RESULTS
Detection of differences in the DNA conformation
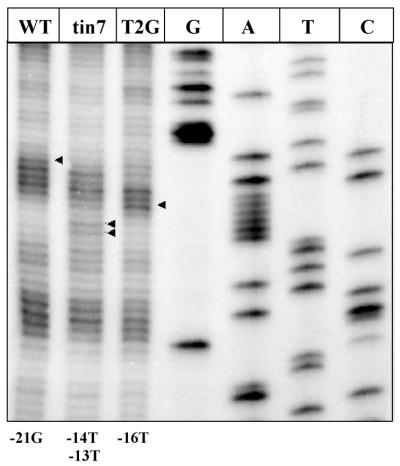
(OP)2Cu cleaves the phosphodiester backbone of nucleic acids at physiological pH and temperatures. Though the reagent cuts at every base, the intensity of the cleavage is dependent on the structure of DNA due to a particular base sequence (18,19). We have probed the variation in DNA structure by assessing the sensitivity pattern to single hit cleavage by (OP)2Cu for three different DNA sequences that differ from each other by a single base in the promoter spacer region (Fig. 2). In the top strand, –21G was hypersensitive in the wild-type (WT) promoter spacer region whereas in T4G (tin7), –14T and –13T were cleaved more often. T2G mutant showed a hypersensitive band at position –16T, while –19T and –20A were less susceptible to cleavage with (OP)2Cu as compared to WT and T4G. We have observed this difference in the cleavage pattern in DNA sequences in in vitro (OP)2Cu analysis and correlated to promoter strength of WT and mutant spacer sequences (26). The results presented in Figure 2 show the utility of the technique for in vivo probing to assess structural changes in DNA arising as a result of base substitutions.
Figure 2.
In vivo (OP)2Cu cleavage to detect changes in DNA structure. Escherichia coli cells harboring plasmid pLW4 having wild-type sequence in the spacer region of the promoter (WT) or two other substitution mutations (tin7 and T2G, respectively) in the spacer region of the promoter were used for (OP)2Cu cleavage reaction as described in Materials and Methods. G, A, T and C refer to sequencing lanes using Sanger’s dideoxy method. Arrowheads indicate the hypersensitive residues upon (OP)2Cu cleavage.
Probing DNA–protein interactions involved in different cellular events
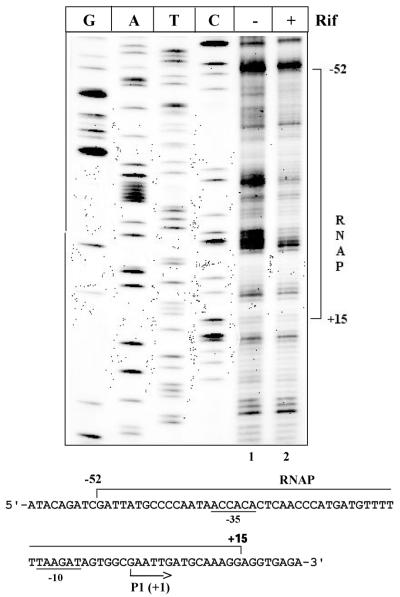
Binding of RNA polymerase (RNAP) to promoter sequences. Binding of RNAP at a promoter is the first step in the transcription initiation process. Expression of a large number of genes is regulated at this step. Consequently, RNAP contacts at different promoters have been studied extensively in vitro to delineate the features of promoter–polymerase interactions. Other than KMnO4 probing, which assesses open complex formation at the promoter, there are no other convenient tools to evaluate in vivo promoter occupancy by RNAP. Use of (OP)2Cu as an in vivo probe for RNAP binding to a promoter is demonstrated in Figure 3. Escherichia coli DH10B cells harboring the ptin7 plasmid were treated with rifampicin (200 µg/ml) to arrest RNAP at the promoter, followed by treatment with (OP)2Cu as described in Materials and Methods. A region of 67 bp was protected upon rifampicin treatment of the cells from –52 to +15 with respect to the +1 transcription start site of the promoter (Fig. 3). DNase I footprinting with the promoter of the tin7 mutant revealed a protected region from –56 to +21 (22). The larger footprint obtained by DNase I could be because of steric hindrance between RNAP and DNase I. (OP)2Cu being a small molecule, a better resolved RNAP footprint on the same promoter was obtained.
Figure 3.
RNAP binding to the promoter. Absence (–) or presence (+) of rifampicin is indicated. G, A, T and C refers to sequencing lanes. The sequence below shows the Mu mom regulatory region. The protected region is indicated with a bracket. The –10 and –35 elements are underlined.
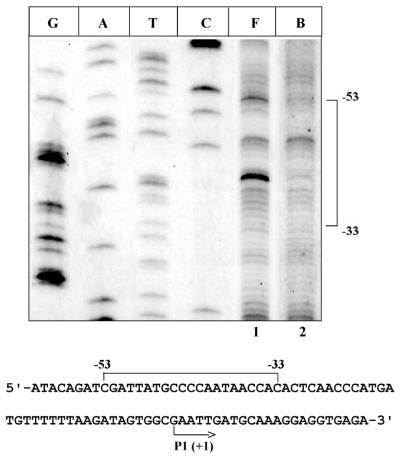
Footprinting of cis elements with trans factors. Site-specific interactions of a large number of DNA-binding proteins at their respective cis site serve as determinants of diverse molecular events. A large number of regulatory proteins influence gene expression by binding to their recognition sequence. A wealth of information is available on in vitro footprinting with various trans factors at their binding sites near or distant from the promoter region. The utility of (OP)2Cu as an in vivo tool to probe the specific binding is addressed by using regulatory protein C from bacteriophage Mu. The C binding site has been identified by in vitro analysis using a variety of footprinting reagents (27). Plasmids ptin7 (harboring the C binding sequence) and pVN184 (C protein producing plasmid) were used to map the C protein binding site. A footprint from –53 to –33 (with respect to the transcription start site of the mom gene) was obtained in the presence of C protein (Fig. 4, lanes 1 and 2). The location of the binding site lies within the region of in vitro DNase I footprint (–57 to –28) with purified C protein (28).
Figure 4.
Footprinting of site-specific DNA-binding protein. ‘F’ indicates free DNA where cells containing ptin7 plasmid have been treated with (OP)2Cu as described in Materials and Methods. ‘B’ indicates C protein bound to its site in the regulatory region of the mom operon (as shown by a bracket in the sequence below). The transcription start site is indicated with an arrow. Sequencing lanes are marked as G, A, T and C.
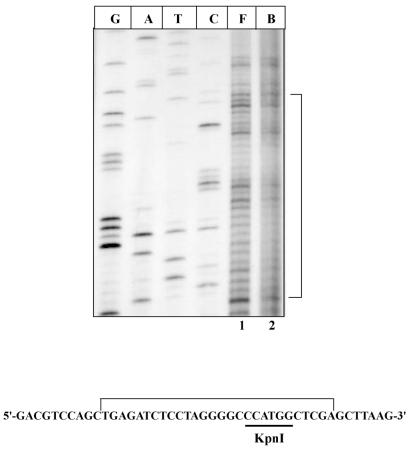
In order to assess the versatility of the technique, we extended the footprinting analysis to map the DNA binding by other types of DNA recognition proteins. DNA methyltransferases and their corresponding restriction enzymes recognize specific DNA sequences in a site-specific fashion. Application of the in vivo (OP)2Cu footprinting technique to assess the interaction of KpnI methyltransferase with its recognition site present in pUC18 plasmid is shown in Figure 5. The plasmid pACMK (23) was used for the expression of KpnI methyltransferase. An overnight-grown (16 h) culture (4.0 ml) of both pUC18 and pACMK were subjected to (OP)2Cu probing as described in Materials and Methods. A region of 28 bp was protected encompassing the KpnI recognition sequence (Fig. 5, lane 2). These results demonstrate the usefulness of the technique to probe diverse site-specific interactions.
Figure 5.
Footprinting of KpnI methyltransferase. Free (F) and bound (B) lanes contain pUC18 and pACMK plasmids, respectively, after treatment with (OP)2Cu and extension with DNA polymerase I (Klenow) as described in Materials and Methods. The bracket indicates the protected region both in the figure and in the sequence of the multiple cloning site of pUC18 plasmid. The recognition sequence for KpnI methyltransferase is underlined. G, A, T and C are the sequencing lanes.
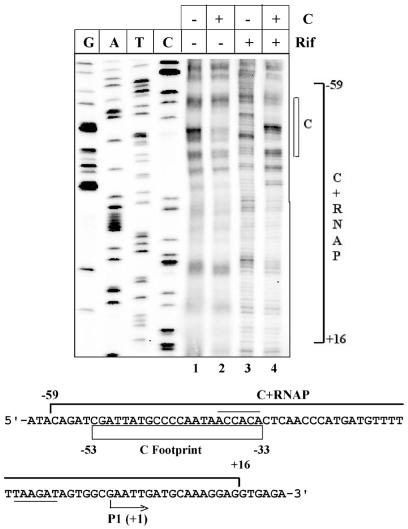
Multi-protein complex formation: activation of transcription initiation. The majority of molecular events are triggered as a result of the interaction of a number of proteins with DNA. In most cases, complexes of several proteins establish a molecular communication network. The potential of in vivo footprinting with (OP)2Cu to study the processes involving interplay of factors is assessed in Figure 6 by analyzing the transcription activation process. Binding of the transactivator protein C is a prerequisite step for the recruitment of RNAP to the momP1 promoter of bacteriophage Mu. When E.coli cells harboring the pBM2 plasmid (containing the momP1 promoter) are arrested with rifampicin, RNAP fails to show any detectable footprint (Fig. 6, lane 3). However, in the presence of C protein, there is an extended region of protection from –59 to +16 (top strand) indicative of binding of RNAP to the promoter. These results essentially reproduce the earlier in vitro footprinting experiments with purified C protein and RNAP (28).
Figure 6.
Activator-mediated RNAP binding to weak promoter. Absence (–) or presence (+) of C protein (C) or rifampicin (Rif) are indicated. C footprint is shown as an open rectangle in the sequence of the mom regulatory region below the figure. The –10 and –35 hexamers are underlined and overlined, respectively. The transcription start site is indicated with an arrow. The bracket indicates the extended footprint obtained due to binding of both C protein and RNAP. G, A, T and C are sequencing lanes.
DISCUSSION
Here, we demonstrate the use of (OP)2Cu as a versatile probe to assess the in vivo interaction of protein(s) with DNA. The method is successfully applied to study (i) structural differences in DNA, (ii) interaction of RNAP with promoter, (iii) interaction of specific DNA-binding proteins with their cognate sites and (iv) recruitment of transcription machinery.
Amongst the wide range of probes available to define DNA–protein interactions in vitro, very few reagents have been used to analyze in vivo interactions. The small size of the molecule, sequence-independent recognition, backbone cleavage chemistry and the ability to bind to minor groove, are the features of (OP)2Cu that makes it an attractive in vivo probe. In contrast, both DMS and KMnO4 are specific base modifiers revealing A, G or T contacts (29,30). Thus, (OP)2Cu footprinting provides complementary information to the data obtained with DMS and KMnO4. Though DNase I has been used for in vivo footprinting (17), the cells require pre-treatment with ethanol for the entry of DNase I into the cells. The (OP)2Cu footprinting technique, on the other hand, does not require permeabilization and gives better resolution of interaction of the protein with its specific site as compared to the DNase I footprinting method. In our protocol of in vivo footprinting with (OP)2Cu there is no need for any pre-treatment of cells. Also, no post-treatment of the DNA is required as opposed to the piperidine cleavage reaction required for DMS and KMnO4 footprinting reactions. Sequence-dependent structural variability can also be detected using (OP)2Cu. Although the scission reaction of (OP)2Cu is not specific for any particular base, the rate of cutting the DNA backbone does rely on the nucleotide sequence (18,19). We have utilized this property of (OP)2Cu for comparing in vivo scission pattern to reveal differences in DNA conformation of mutant promoters. Another important feature is that arduous protein purification steps are eliminated. Thus, the technique is readily applied to study the interactions of single protein or multi-protein–DNA complexes as there is no need to purify and reconstitute all the protein components to study their effect.
The present protocol could be applied to study the in vivo DNA–protein interactions in diverse biological systems. In organisms where permeability to the reagents could pose a problem, pre-treatment of cells with chemicals such as toluene or alcohol would aid the entry of (OP)2Cu complex. Reagents such as EDTA and SDS could be used to alter the permeability of the cells. We have shown previously that low concentrations of SDS (0.01%), which do not affect growth of cells, also lead to permeabilization of cells (31). (OP)2Cu could also be used in vivo to study DNA–protein interactions at the chromosomal level when combined appropriately with a PCR-based amplification method of primer extension to increase its sensitivity further. It should, however, be mentioned that for each plasmid or chromosomal system (depending on the size of the plasmid/chromosome) the concentration of (OP)2Cu, 3-MPA and neocuproine to be used has to be standardized accordingly to get a single hit pattern.
The in vivo footprinting technique presented here appears to be a powerful probe to study the dynamics of various cellular molecular processes involving DNA–protein interactions. For example, interplay of regulatory factors influencing gene regulation at the transcription level, replication initiation, repair pathways, recombination reactions or functional interaction in nucleo–protein complexes could be analyzed. Another obvious application is in in vivo promoter mapping to identify novel promoters. As the present protocol does not require purified species-specific components, it has significant application potential for post-genomic research for functional probing of a large number of proteins whose DNA-binding property is unknown. Thus, the method developed in this study using (OP)2Cu has many potential applications in analysing DNA–protein interactions occurring in intracellular processes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Bindu Diana Paul for providing the plasmid pBM2 and S. Chandrashekaran for the plasmid pACMK. S.B. is supported by Jawaharlal Nehru Center for Advanced Scientific Research, Bangalore. The work was supported by a grant (to V.N.) from the Department of Science and Technology, Government of India.
References
- 1.Schmitz A. and Galas,D.J. (1979) The interaction of RNA polymerase and lac repressor with the lac control region. Nucleic Acids Res., 6, 111–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buchanan R.L. and Gralla,J.D. (1987) Factor interactions at simian virus 40 GC-box promoter elements in intact nuclei. Mol. Cell Biol., 7, 1554–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger W., Schickor,P. and Heumann,H. (1989) A cinematographic view of Escherichia coli RNA polymerase translocation. EMBO J., 8, 2745–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pavco P.A. and Steege,D.A. (1990) Elongation by Escherichia coli RNA Polymerase is blocked in vitro by a site specific DNA binding protein. J. Biol. Chem., 265, 9960–9969. [PubMed] [Google Scholar]
- 5.Bhaduri T., Basak,S., Sikder,D. and Nagaraja,V. (2000) Inhibition of Mycobacterium smegmatis topoisomerase I by specific oligonucleotides. FEBS Lett., 486, 126–130. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L. and Gralla,J.D. (1989) Micrococcal nuclease as a probe for bound and distorted DNA in lac transcription and repression complexes. Nucleic Acids Res., 17, 5017–5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gralla J.D. (1985) Rapid ‘footprinting’ on supercoiled DNA. Proc. Natl Acad. Sci. USA, 82, 3078–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borowiec J.A. and Gralla,J.D. (1986) High-resolution analysis of lac transcription complexes inside cells. Biochemistry, 25, 5051–5057. [DOI] [PubMed] [Google Scholar]
- 9.Siebenlist U. and Gilbert,W. (1980) Contacts between Escherichia coli RNA polymerase and an early promoter of phage T7. Proc. Natl Acad. Sci. USA, 77, 122–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sasse-Dwight S. and Gralla,J.D. (1989) KMnO4 as a probe for lac promoter DNA melting and mechanism in vivo. J. Biol. Chem., 264, 8074–8081. [PubMed] [Google Scholar]
- 11.Sigman D.S., Graham,D.R., D’Aurora,V. and Stern,A.M. (1979) Oxygen-dependent cleavage of DNA by the 1,10-phenanthroline. cuprous complex. Inhibition of Escherichia coli DNA polymerase I. J. Biol. Chem., 254, 12269–12272. [PubMed] [Google Scholar]
- 12.van Dyke M.W. and Dervan,P.B. (1983) Chromomycin, mithramycin, and olivomycin binding sites on heterogeneous deoxyribonucleic acid. Footprinting with (methidiumpropyl-EDTA) iron (II). Biochemistry, 22, 2373–2377. [DOI] [PubMed] [Google Scholar]
- 13.Herr, W. (1985) Diethyl pyrocarbonate: a chemical probe for secondary structure in negatively supercoiled DNA. Proc. Natl Acad. Sci. USA, 82, 8009–8013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston B.H. and Rich,A. (1985) Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell, 42, 713–724. [DOI] [PubMed] [Google Scholar]
- 15.King P.A., Jamison,E., Strahs,D., Anderson,V.E. and Brenowitz,M. (1993) ‘Footprinting’ proteins on DNA with peroxonitrous acid. Nucleic Acids Res., 21, 2473–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galazka G., Palecek,E., Wells,R.D. and Klysik,J. (1986) Site-specific OsO4 modification of the B-Z junctions formed at the (dA-dC)32 region in supercoiled DNA. J. Biol. Chem., 261, 7093–7098. [PubMed] [Google Scholar]
- 17.Cassler M.R., Grimwade,J.E., McGarry,K.C., Mott,R.T. and Leonard,A.C. (1999) Drunken-cell footprints: nuclease treatment of ethanol-permeabilized bacteria reveals an initiation-like nucleoprotein complex in stationary phase replication origins. Nucleic Acids Res., 27, 4570–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Veal J.M. and Rill,R.L. (1989) Sequence specificity of DNA cleavage by bis(1,10-phenanthroline)copper(I): effects of single base pair transitions on the cleavage of preferred pyrimidine-purine-pyrimidine triplets. Biochemistry, 28, 3243–3250. [DOI] [PubMed] [Google Scholar]
- 19.Yoon C., Kuwabara,M.D., Spassky,A. and Sigman,D.S. (1990) Sequence specificity of the deoxyribonuclease activity of 1,10-phenanthroline-copper ion. Biochemistry, 29, 2116–2121. [DOI] [PubMed] [Google Scholar]
- 20.Spassky A. and Sigman,D.S. (1985) Nuclease activity of 1,10-phenanthroline-copper ion. Conformational analysis and footprinting of the lac operon. Biochemistry, 24, 8050–8056. [DOI] [PubMed] [Google Scholar]
- 21.Spassky A. (1986) Visualization of the movement of the Escherichia coli RNA polymerase along the lac UV5 promoter during the initiation of the transcription. J. Mol. Biol., 188, 99–103. [DOI] [PubMed] [Google Scholar]
- 22.Balke V., Nagaraja,V., Gindlesperger,T. and Hattman,S. (1992) Functionally distinct RNA polymerase binding sites in the phage Mu mom promoter region. Nucleic Acids Res., 20, 2777–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandrashekaran S., Babu,P. and Nagaraja,V. (1999) Characterization of DNA binding activities of over-expressed KpnI restriction endonuclease and modification methylase. J. Biosci., 24, 269–277. [Google Scholar]
- 24.Ramesh V., De,A. and Nagaraja,V. (1994) Engineering hyperexpression of bacteriophage Mu C protein by removal of secondary structure at the translational initiation region. Protein Eng., 7, 1053–1057. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 26.Basak S., Lars,O., Hattman,S. and Nagaraja,V. (2001) Intrinsic DNA distortion of the bacteriophage Mu momP1 promoter is a negative regulator of its transcription. A novel mode of toxic gene expression. J. Biol. Chem., 276, 19836–19844. [DOI] [PubMed] [Google Scholar]
- 27.Ramesh V. and Nagaraja,V. (1996) Sequence-specific DNA binding of the phage Mu C protein: footprinting analysis reveals altered DNA conformation upon protein binding. J. Mol. Biol., 260, 22–33. [DOI] [PubMed] [Google Scholar]
- 28.Gindlesperger T.L. and Hattman,S. (1994) In vitro transcriptional activation of the phage Mu mom promoter by C protein. J. Bacteriol., 176, 2885–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxam A.M. and Gilbert,W. (1980) Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol., 65, 499–560. [DOI] [PubMed] [Google Scholar]
- 30.Sasse-Dwight S. and Gralla,J.D. (1991) Footprinting protein-DNA complexes in vivo. Methods Enzymol., 208, 146–168. [DOI] [PubMed] [Google Scholar]
- 31.Nagaraja V. (1981) Interaction of Mycobacteriophage I3 with its host Mycobacterium smegmatis. Ph.D. Thesis, Microbiology and Cell Biology Laboratory, Indian Institute of Science, Bangalore, India.