Abstract
We report a top-down proteomic analysis of the membrane-bound peripheral light-harvesting complex LH2 isolated from the purple photosynthetic bacterium Rhodobacter (Rb.) sphaeroides. The LH2 complex is coded for by the puc operon. The Rb. sphaeroides genome contains two puc operons, designated puc1BAC and puc2BA. Although previous work has shown consistently that the LH2 β polypeptide coded by the puc2B gene was assembled into LH2 complexes, there are contradictory reports whether the Puc2A polypeptides are incorporated into LH2 complexes. Furthermore, post-translational modifications (PTM) of this protein offer the prospect that it could coordinate bacteriochlorophyll a (Bchl a) by a modified N-terminal residue. Here we describe the LH2-complex components on the basis of electron-capture dissociation (ECD) fragmentation to confirm the identity and sequence of the protein subunits. We found that both gene products of the β polypeptides are expressed and assembled in the mature LH2 complex, but only the Puc1A-encoded polypeptide α is observed here. The methionine of the Puc2B-encoded polypeptide is missing, and a carboxyl group is attached to the threonine at the N terminus. Surprisingly, one amino acid encoded as an isoleucine in both the puc2B gene and the mRNA is found as valine in the mature LH2 complex, suggesting an unexpected and unusual post-translational modification or a specific tRNA recoding of this one amino acid.
TOC image
1. Introduction
The capture and utilization of solar energy is one of the most fundamental processes on Earth. Anoxygenic photosynthesis can occur in the absence of air without producing oxygen.1, 2 The photosynthetic complexes of purple phototrophic bacteria have a rather simple modular construction system that often utilizes two basic types of light-harvesting complexes, called light-harvesting complex 1 (LH1) and light-harvesting complex 2 (LH2). These function to absorb light energy and to transfer that energy rapidly and efficiently to the photochemical reaction centers where it is trapped by photochemistry. LH2 is composed of heterodimeric units, consisting of α, β apoprotein pairs that serve as a scaffold to bind Bchl a and a carotenoid (Car) for optimal energy transfer. Those heterodimers aggregate to produce circular ring structures containing eight or nine heterodimeric units. The LH1 complex from Rb. sphaeroides contains an LH1 dimer with 28 heterodimer unit.3 LH2 complexes are adjacent to the LH1-reaction center (RC) core complex, and together, the two complexes effectively capture the light energy that sustains growth of the organism. The ratio of LH2 complexes to RC is variable and depends on growth conditions.4, 5
X-ray crystallography revealed structural data to atomic resolution of two types of LH2 complexes from Rhodopseudomonas (Rps.) acidophila6 and from Rhodospirillum (Rsp.) molischianum7. The structure at 100 K of LH2 from Rps. acidophila was refined to 2.0 Å resolution by Papiz et al.6 The crystal structure of LH2 from Rs. molischianum was determined by molecular replacement at 2.4 Å resolution by using x-ray diffraction.7 In both structures, the modular α, β-heterodimers form a circular ring structure. LH2 from Rs. molischianum forms octamers instead of the nonamers observed in the Rps. acidophila structure. Both types of LH2 complexes contain relatively isolated Bchl a molecules parallel to the plane of the membrane that absorb light at 800 nm (B800) and closely coupled Bchl a dimers that absorb at 850 nm (B850). One of the major differences of the two crystal structures lies in the nature of B800 orientation: aspartate is the Mg ligand in Rs. molischianum as opposed to carboxyl-methionine in Rps. acidophila.
Rhodobacter (Rb.) sphaeroides, a member of the α-3 subclass of proteobacteria, is an exemplary model organism for the creation and study of novel protein expression systems as its genome is sequenced, genetic systems are available, and its metabolism is well characterized.7 Like many purple phototrophic bacteria, the photosynthetic apparatus of Rb. sphaeroides is composed of three multimeric transmembrane protein complexes: the LH2 light-harvesting complex, the LH1-reaction-center complex (RC-LH1), and the cytochrome (cyt) bc1 complex.9 The 3D structure of a dimeric RC-LH1-PufX complex was determined to 8 Å by x-ray crystallography, and a model was built.3 Although there is no atomic resolution structure of LH2 from Rb. sphaeroides available to date, a projection map of this LH2 clearly shows the nonameric organization of the ring.10 This is also observed for LH2 from Rps. acidophila, but different from the octamer-ring structure of LH2 from Rs. molischianum.
Theiler et al.11 sequenced the apoproteins of LH2 from Rb. sphaeroides in 1984 and found a degree of heterogeneity at the N-terminus of β subunit, with some chains starting with threonine and others having an additional methionine residue at the N-terminal position. DNA sequence of the photosynthesis region of Rhodobacter sphaeroides 2.4.1 is described by Choudhary et al.12 and the amino acid sequence predicted by the genome is consistent with the previous protein sequencing result. A few years later, a new operon (designated the puc2BA operon), displaying a high degree of similarity to the original pucBA genes of Rb. sphaeroides, was identified and studied genetically and biochemically by Zeng et al in 2003.13 (See Fig.S1 in supplemental information) Employing genetic and biochemical approaches, they obtained evidence that the Puc2B-encoded polypeptide is able to enter into LH2 complex formation, but neither the full-length Puc2A-encoded polypeptide nor its N-terminal 48-amino-acid derivative is able to enter into LH2 complex formation. In contrast, Wang et al.14 isolated LH2 from mutated strains and found Puc2B and both the N-terminal version and the intact version of the Puc2A-encoded polypeptides. They suggested that the transcription of puc2BA and the assembly of the LH2 complex is independent of the expression of puc1BA, and is only dependent upon the expression of pucC. According to the result from SDS-PAGE, either the first 54 amino-acid residues of the N-terminus or the one containing a 251 residue C-terminal extension of Puc2A encoded polypeptide can be assembled into the LH2 complex. It is possible that the manipulations of genome affect the assembly of LH2 complex, leading to the contradictory results. Later, Woronowicz et al.15 described a proteomic analysis of the expression levels of the various Puc1BA and Puc2BA operon-encoded polypeptides in the LH2 complexes assembled in Rb. sphaeroides. Surprisingly, the Puc2A polypeptide containing a 251 residue C-terminal extension is of major abundance. It was also reported that genomes of Rps. acidophila and Rhodopseudomonas (R.) palustris contain additional, highly homologous copies of the puc operon encoding the α, β polypeptides of the LH2 complexes.16, 17 All five copies of the puc operon in R. palustris were expressed and regulated by incident light intensity, whereas only two copies of the puc operon products were detected in LH2 complexes from Rps. acidophila.
We applied mass spectrometry (MS) to this problem because MS is now playing a role in intrinsic-membrane protein (IMPs) analyses, and high-throughput proteomics technology can accelerate the understanding of membrane protein structure/function relationships. Precise characterization of whole intrinsic membrane protein (IMPs), however, remains a challenge despite their essential roles in cell biology. The hydrophobicity of IMPs makes them difficult to be analyzed by traditional bottom-up mass spectrometry owing to its bias toward soluble, hydrophilic peptides that are easily recovered during sample processing and chromatography, and that ionize and dissociate well during mass spectrometry.18 For example, many membrane proteins are insoluble under the conditions for enzyme digestion, and subsequent steps in analysis could further lead to precipitation.
In this paper, we report a top-down MS study of the intact LH2 from wide type Rb. sphaeroides to identify and sequence this peripheral antenna system. IMP solubility also challenges the “top down” approach in which intact proteins are introduced directly to the mass spectrometer. In early work from Whitelegge et al.19, they studied the seven-transmembrane helix protein bacteriorhodopsin and the Dl and D2 reaction-center subunits from spinach thylakoids and demonstrated the potential of top-down analysis of IMPs. Later, they described using top-down high-resolution Fourier transform mass spectrometry with collision-induced dissociation (CID) to study post-translationally modified integral membrane proteins with polyhelix bundles and transmembrane porin motifs.20 Whereas CID fragmentation occurs by increasing the internal energy of peptide/protein ions and causing protons to move, electron-capture dissociation (ECD) generally breaks bonds near the location of a protonated site that can attract the electrons.21, 22 The top-down analysis on the c-subunit of ATP synthase (AtpH) shows that thermal activation concomitant with electron delivery increased coverage in the transmembrane domain compared to CID fragmentation.23
Materials and methods
2.1 LH2 preparation
Rb. sphaeroides wild-type strain ATCC 2.4.1 was grown photosynthetically at RT in 1 L bottles. The membrane-enriched pellet obtained from ultracentrifugation of the sonicated cells was re-suspended in 20 mM Tris (pH = 8.0) to a final concentration of OD (850) = 50, solubilized by the addition of lauryldimethylamine N-oxide (LDAO) to a concentration of 1% (w/v), and allowed to incubate for 30 min at room temperature. Solubilization was stopped by dilution of the mixture with 20 mM Tris (pH = 8.0) to a final LDAO concentration of 0.1%. This mixture was ultracentrifuged once again at 200,000×g for 1 h to remove insoluble debris. The supernatant was collected and loaded onto an anion-exchange column (QSHP resin, GE Healthcare, Uppsala, Sweden) that had been equilibrated with 20 mM tris-HCl, 0.1% (w/v) LDAO (pH = 8.0). After washing extensively, LH2 was then eluted with a linear gradient from 100 mM to 500 mM NaCl. Fractions with the highest A850nm:A280nm ratios (greater than 3.0) were pooled, and the accumulated sample applied to a HiLoad™ Superdex™ 200 prep grade column (GE Healthcare). The further purified LH2 was precipitated with acetone and then solubilized with 20% formic acid before being infused into the MS spectrometer.
2.2 Top-down LC-MS analysis of LH2
Resins (PLRP/S, 5 μm, 1000 Å) were packed into 100 μm IntegraFrit capillary (Waters Inc., Milford, MA). A NanoAcuity UPLC (Waters Inc., Milford, MA) was used to separate protein subunits. The gradient was delivered by a NanoAcuity UPLC (0–5 min, 15% solvent B; 5–35 min, 15–90% solvent B. Solvent A: water, 0.1% formic acid; Solvent B: acetonitrile, 0.1% formic acid) at a flow rate 1 μL/min. Two mass spectrometers, a hybrid ion-mobility quadrupole ToF (Synapt G2, Waters Inc., Milford, MA) and a 12 T FTICR mass spectrometer (Solarix, BrukerDaltonics, Bremen, Germany) were operated under normal ESI conditions (capillary voltage 1-2 kV, source temperature ~ 100 °C). The typical ECD pulse length was 0.4 s, ECD bias 0.4 V, and ECD lens 10 V. The ECD hollow cathode heater current was 1.6 A. MS parameters were slightly modified for each individual sample to obtain an optimized signal. For introduction to give ECD fragmentation, an Advion Triversa Nanomate sample robot infused the sample into the 12 T FTICR. Precursor ions were each isolated over a 10 m/z window. Data were processed by using Bruker Daltonics BioTools and Protein Prospector (from the University of California-San Francisco MS Facility web site). Manual data interpretations combined with software tools were adapted to achieve improved sequence coverage. The mass tolerance for fragment ions assignment was 0.02 Da.
2.3 puc2B gene identification
Rb. sphaeroides genomic DNA was extracted by Qiagen® DNeasy Plant Mini Kit. puc2B genes were PCR-amplified by left primer GCTCCGAGCCCTGATAGTAG and right primer AAGCTGAGCAGAGGGGTCTT. The purified PCR product was cloned and sequenced.
2.4 puc2B mRNA identification
Rb. sphaeroides genomic RNA was extracted by TRIzol® Reagent (Life Technologies, Grand Island, NY). Briefly, the cells were broken by ultrasonification in TRIzol® Reagent. After phase separation by chloroform and precipitation by isopropyl alcohol, RNA precipitates were washed by 70% ethanol. TURBO DNA-free™ Kit (Life Technologies, Grand Island, NY) was used to eliminate any remaining DNA contamination. The first strand cDNA was synthesized by RevertAid First Strand cDNA Synthesis Kit (Life Technologies, Grand Island, NY). After synthesis of the first-strand, primer-1 and primer-2 were used to PCR-amplify the puc2B sequence. The purified PCR product was cloned and sequenced.
2.5 Homology modelling
Homology models of the two subunits were generated by using the Phyre224 online modeling suite. The two subunits were combined, and energy minimization was performed by Phenix.25 The top model was aligned to the crystal structure of LH2 from Rps. acidophila5 by Pymol (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC.).
2. Results and discussion
3.1 Composition of mature LH2 complexes
To study the composition of mature LH2 complexes, we purified the whole complex from a photosynthetically cultured wild type strain ATCC 2.4.1 of Rb. sphaeroides. There are three major protein components as seen in the chromatogram of the denatured LH2 protein; these components were later identified as the Puc1B-, Puc2B-, and Puc1A-encoded polypeptides by top-down MS (Fig. 1). The Puc2A α polypeptide was not detected. The β subunits were eluted earlier than the α subunit as the latter is more hydrophobic. Because there is 94% sequence identity of the two β subunits, the elution times of the two β copies are nearly identical. This result is in accord with those of Zeng et al.13, who also found that only the Puc1A-encoded α subunit exists in the mature LH2 complex. Wang et al.14 isolated LH2 from mutated strains and found Puc2B- and both the N-terminal version and the intact versions of the Puc2A-encoded polypeptides. Later, Woronowicz et al.15 found that the Puc2A-encoded polypeptide containing a 251 residue C-terminal extension is a highly abundant protein of the LH2 complex. The large protein fragment they detected does not contain any apparent membrane-spanning regions. They suggested that this peptide is not part of the functional complex and instead arises from in vivo enzymatic cleavage, representing an adventitious co-eluent of CNE and readily detected because mass spectrometers have a bias toward detecting soluble peptides. The existence of certain peptides from Puc2A-encoded polypeptide indicates, however, it may have some assembly role in the complex. The differences between these results are presently not fully understood. It is possible that the growth conditions and the light intensity can affect the expression and assembly of those polypeptides. Nevertheless, the Puc2A-encoded polypeptide is not a major component according to our results. The experimental MW of each subunit were deconvoluted by Bruker Compass DataAnalysis software and shown in Table 1.
Figure 1.

Liquid chromatogram of denatured LH2
Table 1.
Molecular weight of each subunit from LH2
| Subunit | Theoretical MW(mono) | Experimental MW(mono) |
|---|---|---|
| Puc1A polypeptide | 5595.0692 Da | 5595.0512 Da |
| Puc1B polypeptide | 5456.8854 Da | 5456.8706 Da |
| Puc2B polypeptide | 5355.8191 Da | 5355.8082 Da |
3.2 Sequence and post translational modifications
Natural photosynthetic organisms have developed a large variety of light-harvesting strategies that allow them to live nearly everywhere where sunlight can penetrate.26 Most of the antenna systems are pigment-containing, integral membrane proteins. Detailed sequence information of some of those proteins is still not fully known except when there is a high-resolution crystal structure available. Although the MS-based proteomics characterization of the Rb. sphaeroides intra-cytoplasmic membrane assembly was reported by several groups,15, 27, 28 the entire sequence was not identified because 100% coverage was not achieved, which is usually the case for membrane proteins. Those missing regions may play an important role of the function of antenna systems.
We used top-down mass spectrometry to determine the sequence and post-translational modifcations (PTM) information of the LH2 complex. Although not every bond of the polypeptides fragmented when we submitted the protein to ECD on a 12 T FTICR mass spectrometer, we found ~ 70% coverage of the sequence (the mass spectra are in Fig. S4 in supplementary information). Furthermore, many complementary ions shown in the spectrum and their accurate mass measurements (within a few ppm) provided by the instrument provide high confidence for the results. Unlike CID fragmentation, which occurs by increasing the internal energy of peptide/protein ions until peptide-bond cleavage occurs, electron-based fragmentation (ECD) breaks different bonds near the sites of positive charge where the electron capture occurs, preserving PTM information. The ECD-based top-down sequencing identifies not only the sequence information of the protein but also the location of the PTMs.
The PTM information of LH2 complex has been of interest for some time. Papiz et al.5 reported the high-resolution crystal structure of Rps. acidophila, and they found that a carboxyl-modified Met1 of the α subunit is ligated to Mg2+ of B800. Our top-down MS investigation of the structure of LH2 and the possible coordination of B800 BChl-a shows that the experimental molecular weight of the α subunit is consistent with that predicted from the gene sequence (3 ppm accuracy), clearly indicating there is no carboxylation modification on Met in Rb. sphaeroides LH2. We observed some oxidation of methionine, and this was probably introduced during the sample handling. MS/MS with ECD fragmentation provided further evidence that the predicted sequence is correct (Fig. 3).
Figure 3.
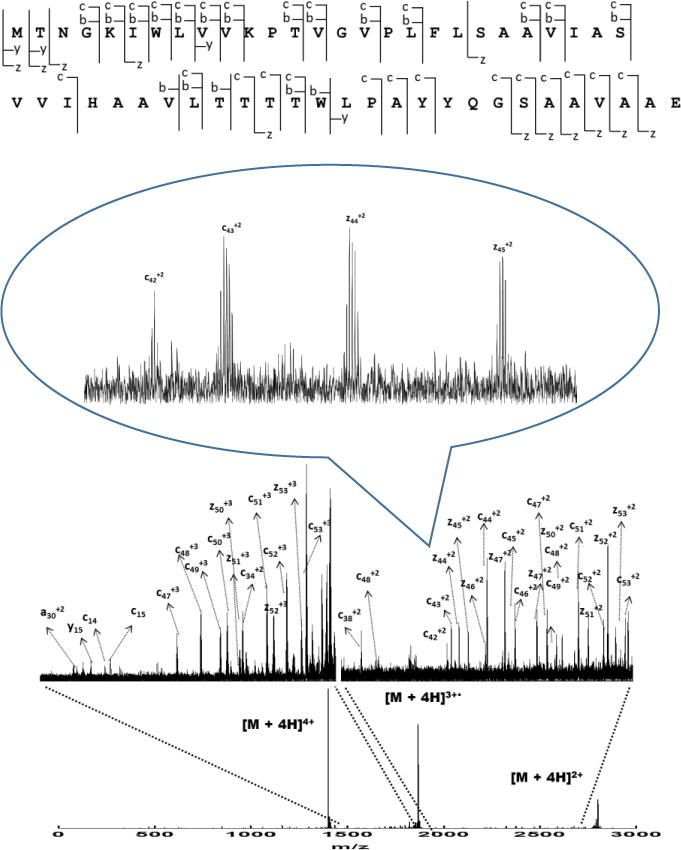
Sequence coverage and ECD product-ion spectrum of Puc1A-encoded polypeptides
The central Mg2+ ion chelation in the core of the Bchl a macrocycle helps preserve the planar conformation of the pigment molecule.29 In principle, oxygen or nitrogen atoms on amino acid side chains (e.g., aspartate, glutamate, asparagine, glutamine, serine, threonine, histidine) or even water can interact with this central Mg atom of BChl-a. In the LH2 complex of Rps. acidophila, the ligation of B800 BChl a is accomplished in part by a carboxyl modification on methionine whereas in Rs. molischianum, the corresponding ligand is aspartate (Asp-6). For Rb. sphaeroides, there is no modification of the α-Met, indicating that the N-terminal Met is not the ligand. To obtain a better understanding of the structure, 52 residues (96% of sequence) were modelled with 100% confidence by the single highest scoring template (Fig. 3A),24 suggesting that the N terminal region of α subunit of Rb. sphaeroides is quite similar to that of Rps. acidophila (Fig. 3C). The reason they have similar structures but different coordination schemes is not clear. To identify similar regions that may be a consequence of structural relationships, we aligned the sequence of Rs. molischianum and Rps. acidophila from RCSB protein data bank to the sequence of Rb. sphaeroides we identified. From the sequence alignment result from ClustalW2, the amino acid in Rb. sphaeroides is Asp-6 whereas in Rs. molischianum, it is asparagine; the former is likely the Mg2+ ligand in the LH2 complex (Fig. 4A).
Figure 4.

Sequence alignment of LH2 subunits. (a) α subunits from different LH2 complexes: (1), Rps. acidophila (2), Rb. sphaeroides (3), Rs. molischianum (b) Puc1B- and Puc2B-encoded polypeptides from Rb. sphaeroides.
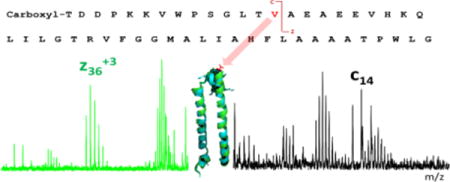
Similarly, the molecular weight obtained from the mass spectrum is consistent with the theoretical MW predicted from the amino-acid sequence of the Puc1B-encoded peptide without any post-translational modifications (3 ppm accuracy) (Fig. 5). The MW observed for the Puc2B-encoded peptide, however, is 101.0589 Da less than predicted. The mass difference doesn’t match a simple modification with or without removal of the N terminal methionine. To address this discrepancy, we analyzed the fragmentation patterns and found that the methionine on the N terminus is removed and a carboxyl group is attached to threonine. In addition, the fourteenth amino acid counting from the N terminus, is a valine instead of the isoleucine that is coded in the gene sequence (Fig. 6). The fragment ions (C13+, C14+, Z363+) displayed on the spectrum confirm this assignment.
Figure 5.

Sequence coverage and ECD product-ion spectrum of Puc1B -encoded polypeptides.
Figure 6.

Sequence coverage and ECD product-ion spectrum of Puc2B-encoded polypeptides.
We were surprised to find carboxylation on the threonine residue at the N terminus. The high mass fragments (Fig. 7) clearly show that losses of OH, COOH, COONH, and then threonine from N terminus, consistent with this modification. The simulated isotopic pattern of fragments on the N terminus also corresponds well with the experimental patterns (See Fig. S2 in supplemental information). To verify that the dissociation spectrum is interpreted correctly, we undertook bottom-up sequencing. The N terminal peptide has a COOH modification on threonine according to CID (See Fig.S3 in supplemental information).
Figure 7.
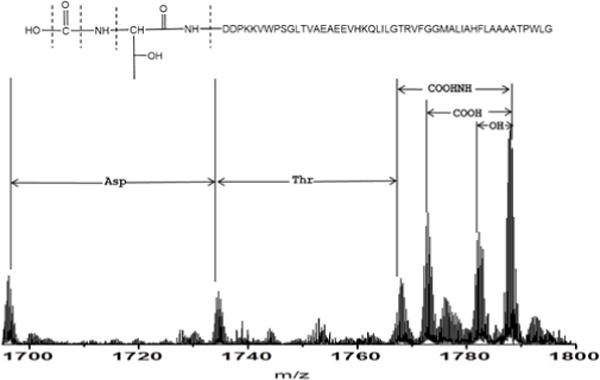
High mass fragments in the ECD product-ion spectrum of Puc2B-encoded polypeptides.
Proteins can carry several PTMs, and some proteins may display large numbers of different modifications.30 New modifications31 and unexpectedly extensive PTMs32 can occur, and they are poorly accounted for in existing databases. There are several reports about PTMs of light-harvesting proteins. The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry revealed 40 gene products with variable post-translational modifications.33 Michael et al.34 found acetylation and phosphorylation on spinach light-harvesting chlorophyll protein II aside from the removal of methionine at the N terminus. A proteomics study of the green alga Chlamydomonas reinhardtii light-harvesting proteins shows the presence of differentially N-terminally processed forms of Lhcbm3 and phosphorylation of a threonine residue at the N terminus.35 For Rps. acidophila, the ligation of B800 BChl a is accomplished in part by a carboxyl-methionine on the N terminus.5
Carboxylation generally happens on glutamate residues, which is required for function of factors II, VII, IX, and X, protein C, protein S, and some bone proteins.36, 37 Although our study identified the carboxylation of a threonine residue at the N terminus of Puc2B, the functional role of this PTM is not clear. The assignments of the N terminal fragments further confirm the sequence and PTM information of this polypeptide (Fig. 7). Zeng et al.13 found that the ratio of B800 to B850 of the LH2-2 complex in mutant ∆PUC2BA (pUC2ASPhoA) is greater than that of the LH2-1 complex in mutant ∆PUC2BA (0.75 and 0.67, respectively). This result suggests that the Bchl a moieties have a slightly altered binding environment in the Puc2B-encoded complex compared to that of the Puc1B-encoded peptide. For the β subunit, 38 residues (75% of sequence) were modelled with 99.9% confidence by the single highest scoring template.21 The first twelve amino acids are not covered in the homology modeling because there is high sequence discrepancy of the N terminal region of β subunit (Fig. 3A). It is possible, however, that the carboxyl group serves to coordinate the Mg of B800 in the LH2 complex, increasing the ratio of B800 to B850.
3.3 Substitution of Valine for Isoleucine
Initially, we could not match the experimental molecular weight of Puc2B to the molecular weight predicted by the genome, even after removal of methionine and attachment of a carboxyl group on the N terminal threonine. As discussed above, an analysis of the C and Z ions that are produced upon ECD shows that the 14th amino acid from N terminal is valine, not the isoleucine predicted from the gene sequence. The experimental MW matches the theoretical value within 2 ppm, confirming this assignment. Interestingly, we observed two polypeptides, mostly as the “valine version”, but there is also a small amount of the “isoleucine version” in a roughly 10:1 ratio (Fig. 8). The sequence chemically determined by Theiler et al.11 matches the predicted sequence of the Puc1B protein. It is likely that their samples also contained the Puc2B protein but did not obtain sequence from it due to the blocked N-terminus.
Figure 8.
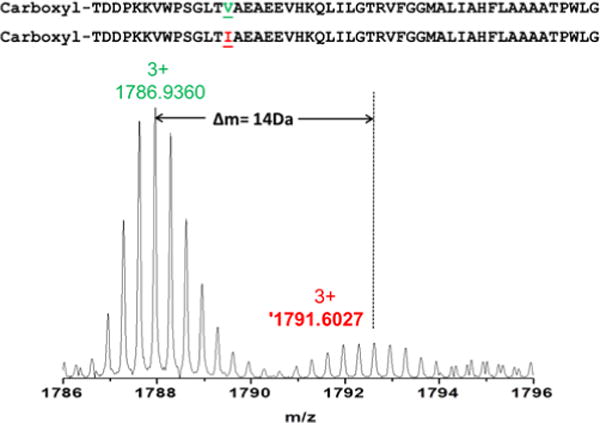
Precursor ions of puc2B-encoded polypeptides.
Mass spectrum of parent ions of Puc2B-encoded polypeptides.
The question now arises as to the origin of this isoleucine to valine conversion. The spinach chloroplast genome reports a codon for Ser at position 2 whereas Phe was detected at position 2 of PetL of chloroplast-encoded subunits.38 The authors believe there might be either a DNA sequencing error or a RNA editing event. To eliminate the possibility of a mutation happening during culturing of cells over many generations, we sequenced the puc2B gene, and we found it to be the same as in the NCBI database (i.e., the 14th codon is ATC, which codes for isoleucine). Another possibility to explain the substitution is RNA editing in which the codon is changed after transcription but before translation. To study the Puc2B-encoded peptide at the transcriptional level, we sequenced the mRNA that is encoded for this region. The result shows that there is no variation at the mRNA level, and the mRNA also indicates isoleucine (See Fig.S5 in supplemental information). One possibility is a mis-sense error, which results in the substitution of one amino acid for another probably by mischarging the isoleucine tRNA with valine. However, all other isoleucines in the protein, all of which are coded for by the ATC isoleucine codon, appear to be correctly inserted, so the only way that this could be the case is for a context-specific change to be made in just this one place. Another possibility is that the amino acid residue is not valine but its isomer, norvaline, which is sometimes abundant.39, 40 It is also likely that it is a PTM process. The existence of two forms of the polypeptides also suggests that the demethylation is not 100% complete. Thus, we suggest the isoleucine-to-valine conversion is either a posttranslational modification in which a methyl group is cleaved from isoleucine or a norvaline substituion.
Although there are very few protein demethylation cases reported, one well-known example is histone demethylation mediated by the nuclear amine oxidase homolog LSD1.41 To the best of our knowledge, the post-translational demethylation process of isoleucine to valine has not been reported previously. Norvaline is usually found in place of leucine instead of isoleucine.42 According to the homology modelling structure, the valine is located in the loop region of the β subunit. (Fig. 3D) The biological significance of this conversion of the Puc2B-encoded polypeptide is not clear at this stage, but given the unprecedented nature of this substitution, more study is needed. After this conversion, three amino acids on the N terminus are different and the rest of the amino acids are the same for the two copies of the β polypeptides. (Fig. 4B)
3. Conclusions
The composition and PTMs of LH2 of purple phototrophic bacteria likely play important roles in absorbing light energy effectively and in allowing the organism to adapt to a changing environment (i.e., light intensity). In this study, a top-down proteomic analysis of the membrane-bound peripheral light harvesting complex LH2 isolated from WT Rb. sphaeroides confirms the identity and sequence of these protein subunits. We showed that polypeptide encoded by puc1A is the sole source of the α subunit in the LH2 complex. Consistent with previous reports, this work also shows that both LH2 β polypeptides coded by the puc1B and puc2B gene are assembled into LH2 complexes. Overall, the complex has a similar structure to those in other purple bacteria (Fig. 2B). Unusual PTMs occur for this protein. For example, a carboxyl group is attached to the N-terminal Thr along with removal of the Met on the Puc2B-encoded polypeptide. The carboxyl group, instead of the carboxyl-methionine in Rps. acidophila, likely coordinates Bchl a in Rb. sphaeroides. The unexpected substitution of valine to isoleucine in Puc2B-encoded polypeptide is likely to be a PTM or a norvaline substitution. The biological significance of this conversion is currently not clear and will be the subject of future studies.
Figure 2.
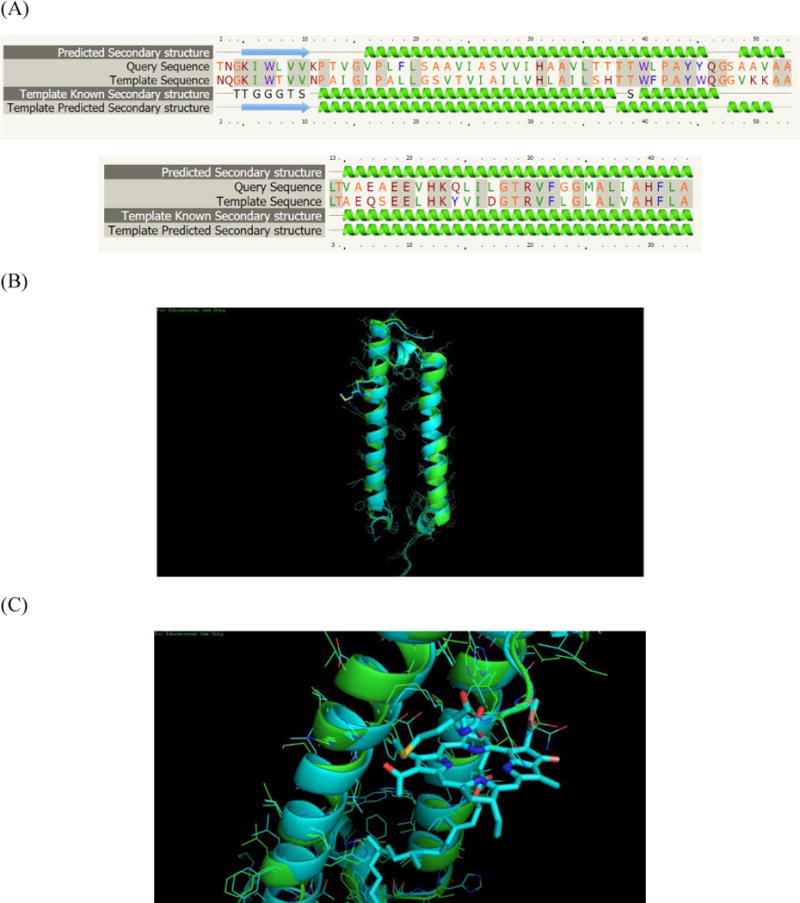
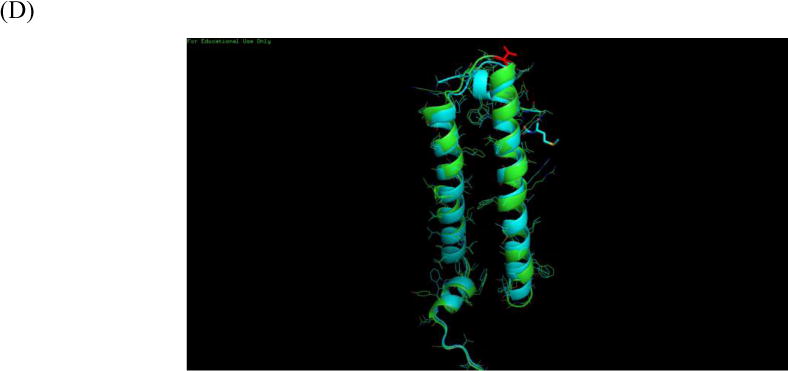
Homology model of LH2 from Rb. sphaeroides. (A) Sequence alignment to the template (B) LH2 from Rb. sphaeroides is shown in green, LH2 from Rps. acidophila is shown in cyan (C) N-terminus structure of two LH2. B800 is coordinated by carboxyl-Met from Rps. acidophila. (D) Valine from β subunit of Rb. sphaeroides is shown in red
Supplementary Material
Acknowledgments
We want to thank Dr. Philip C. Andrews for discussion about the nonverline substitution.
Experiments confirm the peptide carrying carboxyl modification by bottom-up approach; sequence alignment of Puc1A- and Puc2A-encoded polypeptides; comparison of simulated and experimental isotopic pattern of parents and fragment ions on N-teriminus of Puc2B-encoded polypeptide; isolation spectrums of Puc1A, Puc1B and Puc2B encoded polypeptides; DNA and RNA Puc2B operon sequencing result and trace files around 14th valine region.
Funding
This research is funded by the Photosynthetic Antenna Research Center (PARC), an Energy Frontier Research Center funded by the DOE, Office of Science, Office of Basic Energy Sciences under Award Number DE-SC 0001035 and National Institute of General Medical Science under Grant Number: 8 P41 GM103422. All sample preparation and molecular biology work was supported by the DOE grant, while top-down mass spectrometry was supported by the NIH grant. YL, HZ, RS, and HL were supported by the DOE grant.
Footnotes
Supporting Information is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pullerits T, Sundstrom V. Photosynthetic light-harvesting pigment-protein complexes: Toward understanding how and why. Acc Chem Res. 1996;8:381–389. [Google Scholar]
- 2.Blankenship RE. Molecular Mechanisms of Photosynthesis. Wiley-Blackwell 2013 May; [Google Scholar]
- 3.Qian P, Rapiz MZ, Jackson PJ, Brindley AA, Ng IW, Olsen JD, Dickman MJ, Bullough PA, Hunter N. Three-Dimensional Structure of the Rhodobacter sphaeroides RC-LH1-PufX Complex: Dimerization and Quinone Channels Promoted by PufX. Biochemistry. 2013;52:7575–7585. doi: 10.1021/bi4011946. [DOI] [PubMed] [Google Scholar]
- 4.Codgell RJ, Southall J, Gardiner AT, Law CJ, Gall A, Roszak AW, Isaacs NW. How purple photosynthetic bacteria harvest solar energy. J Bacteriol. 2006;9:201–206. [Google Scholar]
- 5.Jaw CJ, Roszak AW, Southall J, Gardiner AT, Isaacs NW, Codgell RJ. The structure and function of bacterial light-harvesting complexes. Mol Membr Biol. 2004;21:183–191. doi: 10.1080/09687680410001697224. [DOI] [PubMed] [Google Scholar]
- 6.Rapiz MZ, Prince SM, Howard T, Cogdell RJ, Isaacs NW. The Structure and Thermal Motion of the B800–850 LH2 Complex from Rps. acidophila at 2.0 A° Resolution and 100 K: New Structural Features and Functionally Relevant Motions. J Mol Biol. 2003;326:1523–1538. doi: 10.1016/s0022-2836(03)00024-x. [DOI] [PubMed] [Google Scholar]
- 7.Koepke J, Hu X, Muenke C, Schulten K, Michel H. The crystal structure of the light-harvesting complex II (B800–850) from Rhodospirillum molischianum. Structure. 1996;4(5):581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 8.Jaschke PR, Saer RG, Noll S, Beatty JT. Modification of the genome of Rhodobacter sphaeroides and construction of synthetic operons. Method enzymol. 2009;497:519–538. doi: 10.1016/B978-0-12-385075-1.00023-8. [DOI] [PubMed] [Google Scholar]
- 9.Scheuring S, Nevo R, Liu L, Mangenot S, Charuvi D, Boudier T, Prima V, Hubert P, Sturgis JN, Reich Z. The architecture of Rhodobacter sphaeroides chromatophores. Biochim Biophys Acta. 2014;1837(8):1263–1270. doi: 10.1016/j.bbabio.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 10.Walz T, Jamieson SJ, Bowers CM, Bullough PA, Hunter CN. Projection Structures of Three Photosynthetic Complexes from Rhodobacter sphaeroides: LH2 at 6 Å, LH1 and RC-LH1 at 25 Å. J Mol Biol. 1998;282:833–845. doi: 10.1006/jmbi.1998.2050. [DOI] [PubMed] [Google Scholar]
- 11.Theiler R, Suter F, Zuber H, Cogdell RJ. A comparison of the primary structures of the two B800-850-apoproteins from wild-type Rhodopseudomonas sphaeroides strain 2.4.1 and a carotenoidless mutant strain R26.1. FEBS. 1984;175(2):231–237. [Google Scholar]
- 12.Choudhary M, Kaplan S. DNA sequence analysis of the photosynthesis region of Rhodobacter sphaeroides 2.4.1. Nucleic Acids Res. 2000;28:862–867. doi: 10.1093/nar/28.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng X, Choudhary M, Kaplan S. A Second and Unusual pucBA Operon of Rhodobacter sphaeroides 2.4.1: Genetics and Function of the Encoded Polypeptides. J Bacteriol. 2003;185:6171–6184. doi: 10.1128/JB.185.20.6171-6184.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang W, Hu Z, Li J, Chen G. Expression, characterization and actual function of the second pucBA in Rhodobacter sphaeroides. Biosci Rep. 2009;29:165–172. doi: 10.1042/BSR20080061. [DOI] [PubMed] [Google Scholar]
- 15.Woronowicz K, Olubanjo OB, Sung HC, Lamptey JL, Niederman RA. Differential assembly of polypeptides of the light-harvesting 2 complex encoded by distinct operons during acclimation of Rhodobacter sphaeroides to low light intensity. Photosynth Res. 2012;111:125–138. doi: 10.1007/s11120-011-9707-4. [DOI] [PubMed] [Google Scholar]
- 16.Tadrosl MH, Waterkampl K. Multiple copies of the coding regions for the light-harvesting B800-850 alpha- and beta-polypeptides are present in the Rhodopseudomonas palustris genome. The EMBO Journal. 1989;8:1303–1308. doi: 10.1002/j.1460-2075.1989.tb03509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardiner AT, MacKenzie RC, Barrett SJ, Kaiser K, Cogdell RJ. The purple photosynthetic bacterium Rhodopseudomonas acidophila contains multiple puc peripheral antenna complex (LH2) genes: Cloning and initial characterisation of four β/α pairs. Photosynth Res. 1996;49:223–235. doi: 10.1007/BF00034783. [DOI] [PubMed] [Google Scholar]
- 18.Whitelegge JP. Integral Membrane Proteins and Bilayer Proteomics. Anal Chem. 2013;85:2558–2568. doi: 10.1021/ac303064a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitelegge JP, Gundersen CB, Faully KE. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998;7:1423–1430. doi: 10.1002/pro.5560070619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan CM, Souda P, Bassilian S, Ujwal R, Zhang J, Abramson J, Ping P, Durazo A, Bowie JU, Hasan SS, Baniulis D, Cramer WA, Faull KF, Whitelegge JP. Post-translational Modifications of Integral Membrane Proteins Resolved by Top-down Fourier Transform Mass Spectrometry with Collisionally Activated Dissociation. Mol Cell Proteomics. 2010;9:791–803. doi: 10.1074/mcp.M900516-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zubarev RA, Kelleher NL, Mclafferty FW. Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. J Am Chem Soc. 1998;120:3265–3266. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 22.Cui W, Rohrs HW, Gross ML. Top-down mass spectrometry: Recent developments, applications and perspectives. Analyst. 2011;136:3854–3864. doi: 10.1039/c1an15286f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zabrouskov V, Whitelegge JP. Increased Coverage in the Transmembrane Domain with Activated-Ion Electron Capture Dissociation for Top-Down Fourier- Transform Mass Spectrometry of Integral Membrane Proteins. J Proteome Res. 2007;6(6):2205–2210. doi: 10.1021/pr0607031. [DOI] [PubMed] [Google Scholar]
- 24.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 25.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Huang LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croce R, Amerongen HV. Natural strategies for photosynthetic light harvesting. Nat Chem Biol. 2014;10:492–501. doi: 10.1038/nchembio.1555. [DOI] [PubMed] [Google Scholar]
- 27.Jackson RJ, Lewis HJ, Tucker JD, Hunter CN, Dickman MJ. Quantitative proteomic analysis of intracytoplasmic membrane development in Rhodobacter sphaeroides. Mol Microbiol. 2012;84:1062–1078. doi: 10.1111/j.1365-2958.2012.08074.x. [DOI] [PubMed] [Google Scholar]
- 28.Woronowicz K, Harrold JW, Kay JM, Niederman RA. Structural and Functional Proteomics of Intracytoplasmic Membrane Assembly in Rhodobacter sphaeroides. J Mol Microbiol Biotechnol. 2013;23:48–62. doi: 10.1159/000346520. [DOI] [PubMed] [Google Scholar]
- 29.Fiedora L, Kaniab A, Mysliwa-Kurdziela B, Orzelb L, Stochelb G. Understanding chlorophylls: Central magnesium ion and phytyl as structural determinants. Biochim Biophys Acta. 2008;1777:1491–1500. doi: 10.1016/j.bbabio.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 31.Medzihradszky KF, Darula Z, Perlson E, Fainzilber M, Chalkley RJ, Ball H, Greenbaum D, Bogyo M, Tyson DR, Bradshaw A, Burlingame AL. O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes. Mol Cell Proteomics. 2004;3:429–440. doi: 10.1074/mcp.M300140-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Liao G, Xie L, Li X, Cheng Z, Xie J. Unexpected extensive lysine acetylation in the trump-card antibiotic producer Streptomyces roseosporus revealed by proteome-wide profiling. J Proteomics. 2014;106:260–269. doi: 10.1016/j.jprot.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 33.Gómez SM, Nishio JN, Faull KF, Whitelegge JP. The Chloroplast Grana Proteome Defined by Intact Mass Measurements from Liquid Chromatography Mass Spectrometry. Mol Cell Proteomics. 2002;1:46–59. doi: 10.1074/mcp.m100007-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Michel H, Griffin PR, Shabanowitz J, Hunt DF, Bennett J. Tandem mass spectrometry identifies sites of three post-translational modifications of spinach light-harvesting chlorophyll protein II. Proteolytic cleavage, acetylation, and phosphorylation. J Biol Chem. 1991;266:17584–17591. [PubMed] [Google Scholar]
- 35.Stauber EJ, Fink A, Markert C, Kruse O, Johanningmeier U, Hippler MC. Proteomics of Chlamydomonas reinhardtii Light-Harvesting Proteins. Eukaryot Cell. 2003;2:978–994. doi: 10.1128/EC.2.5.978-994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkner KL, Runge KW. The physiology of vitamin K nutriture and vitamin K-dependent protein function in atherosclerosis. J Thromb Haemost. 2009;4:363–371. doi: 10.1111/j.1538-7836.2004.00968.x. [DOI] [PubMed] [Google Scholar]
- 37.Lee T, Lu C, Chen S, Bretana NA, Cheng T, Su M, Huang K. Investigation and identification of protein γ-glutamyl carboxylation sites. BMC Bioinformatics. 2011;12(Suppl 13):S10. doi: 10.1186/1471-2105-12-S13-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitelegge JP, Zhang H, Aguilera R, Taylor RM, Cramer WA. Full subunit coverage liquid chromatography electrospray ionization mass spectrometry (LCMS+) of an oligomeric membrane protein: cytochrome b6f complex from spinach and the cyanobacterium Mastigocladus laminosus. Mol Cell Proteomics. 2002;1:816–27. doi: 10.1074/mcp.m200045-mcp200. [DOI] [PubMed] [Google Scholar]
- 39.Kisumi M, Sugiura M, Chibata I. Biosynthesis of norvaline, norleucine, and homoisoleucine in Serratia marcescens. J biochem. 1976;80(2):333–9. doi: 10.1093/oxfordjournals.jbchem.a131281. [DOI] [PubMed] [Google Scholar]
- 40.Alvarez-Carreno C, Becerra A, Lazcano A. Norvaline and Norleucine May Have Been More Abundant Protein Components during Early Stages of Cell Evolution. Orig Life Evol Biosph. 2013;43:365–375. doi: 10.1007/s11084-013-9344-3. [DOI] [PubMed] [Google Scholar]
- 41.Shi Y, Matson C, Muligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Apotol I, Levine J, Lippicott J, Leach J, Hess E, Glascock CB, Weickert MJ, Blackmore R. Incorporation of Norvaline at Leucine Positions in Recombinant Human Hemoglobin Expressed in Escherichia coli. J Biol Chem. 1997;272:28980–28988. doi: 10.1074/jbc.272.46.28980. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


