Abstract
Natural and human-related landscape features influence the ecology and water quality of lakes. Summarizing these features in a hydrologically meaningful way is critical to understanding and managing lake ecosystems. Such summaries are often done by delineating watershed boundaries of individual lakes. However, many technical challenges are associated with delineating hundreds or thousands of lake watersheds at broad spatial extents. These challenges can limit the application of analyses and models to new, unsampled locations. We present the Lake-Catchment (LakeCat) Dataset (https://www.epa.gov/national-aquatic-resource-surveys/lakecat) of watershed features for 378,088 lakes within the conterminous USA. We describe the methods we used to: 1) delineate lake catchments, 2) hydrologically connect nested lake catchments, and 3) generate several hundred watershed-level metrics that summarize both natural (e.g., soils, geology, climate, and land cover) and anthropogenic (e.g., urbanization, agriculture, and mines) features. We illustrate how this data set can be used with a random forest model to predict the probability of lake eutrophication by combining LakeCat with data from US Environmental Protection Agency’s National Lakes Assessment (NLA). This model correctly predicted the trophic state of 72% of NLA lakes, and we applied the model to predict the probability of eutrophication at 297,071 unsampled lakes across the conterminous USA. The large suite of LakeCat metrics could be used to improve analyses of lakes at broad spatial extents, improve the applicability of analyses to unsampled lakes, and ultimately improve the management of these important ecosystems.
Keywords: lakes, watersheds, National Hydrography Dataset (NHD), StreamCat Dataset, National Lakes Assessment (NLA), landscape descriptors, chlorophyll a, eutrophication, macrosystems ecology
Environmental and ecological conditions within lakes are the result of complex processes that interact at several spatial and temporal scales (Soranno et al. 2014). Understanding these processes, their interactions, and the scales across which they interact, a focus of macrosystems ecology (Fei et al. 2016), can improve the management and conservation of these important ecosystems (Heffernan et al. 2014). A critical component of characterizing lakes within a macrosystems framework is characterizing lake watersheds. The ecology and water quality of lakes can be understood and effectively managed only in the context of their surrounding landscapes (Likens 1984, Foster and Dearing 1987, Soranno et al. 2017). For example, Read et al. (2015) used a macrosystems approach to explore the relative roles of local and regional drivers on lake water quality across the conterminous US (CONUS) and found that basin-scale (henceforth, watershed) metrics of land use (e.g., % of the lake watershed used for agriculture) can account for a large percentage of the variance (45–62%) of lake water-quality metrics (i.e., turbidity, total P, total N, dissolved organic C, and conductivity). Likewise, harmful effects of human-related alterations to lake watersheds on water quality can be mitigated by the presence of natural land covers, such as wetlands or forests (Fergus et al. 2011, Read et al. 2015). However, producing watershed metrics of landscape features at large spatial scales (e.g., regional to continental) can prove challenging. We describe a data set called the Lake-Catchment Dataset (LakeCat) that was developed at the US Environmental Protection Agency’s (USEPA) Western Ecology Division. LakeCat contains data that characterize a large suite of landscape features for 378,088 lakes and their associated watersheds across the CONUS.
In recent years, several large, often publicly available data sets have been developed that characterize the biota or water quality of aquatic ecosystems, such as lakes, at various spatial scales (e.g., USEPA 2009, Moe et al. 2013, Soranno et al. 2017). Use of these data sets has the potential to improve our understanding and management of these ecosystems (Hering et al. 2013, Soranno and Schimel 2014, Soranno et al. 2017), but researchers are faced with the challenge of developing indicators of landscape features and processes that influence lakes and that can explain the variation in physical and ecological features among these systems (e.g., Hollister and Milstead 2010, Hollister et al. 2011, Soranno et al. 2017). Doing so requires specialized geospatial techniques that account for the directional flow of water over land surfaces in a geographic information system (GIS). Such data often are acquired via delineation of individual lake-watershed boundaries to summarize landscape features in a hydrologically meaningful way (e.g., Hollister and Milstead 2010, Soranno et al. 2017). Furthermore, application of analytical results to new locations requires the same process of watershed delineation and summarization. However, doing so can be difficult if hundreds or thousands of lakes are to be analyzed or if lake watersheds are nested within each other.
We developed LakeCat to provide geospatial data for researchers and managers to conduct regional-to-national analyses of lakes. In this way, LakeCat complements the USEPA’s Stream–Catchment (StreamCat) Dataset (Hill et al. 2016). StreamCat contains watershed-level summaries of >500 geospatial metrics for ~2.65 million stream segments within the CONUS (https://www.epa.gov/national-aquatic-resource-surveys/streamcat) and has been used successfully to predict the biological condition at >1 million stream segments across the CONUS (Fox et al. 2017, Hill et al. 2017).
The objectives of our paper are to: 1) describe the development and main features of the LakeCat Dataset, and 2) demonstrate its utility by modeling and predicting national-scale eutrophication. We start by defining the geospatial framework and the terminology used in LakeCat. We then describe the algorithms we used to generate the LakeCat geospatial framework, including lake catchment delineation, generation of FROM–TO relationships among nested catchments (i.e., network topology), and linking information from multiple, nested catchments to produce full, hydrologically complete summaries of landscape features for each lake. This description includes the quality-assurance procedures we used to ensure the integrity of LakeCat data. We then provide a summary of the main landscape metrics that are available from LakeCat. Last, we illustrate how LakeCat can be applied to address ecological and management questions. Specifically, we linked LakeCat data with field samples of chlorophyll a (Chl a) concentration from the USEPA’s 2012 National Lakes Assessment (NLA) (USEPA 2017) to model and predict the probability of eutrophication at 297,071 lakes across the CONUS. In addition to the documentation provided here, all code and processing steps are available in a public GitHub repository (https://github.com/USEPA/LakeCat).
LAKECAT DEVELOPMENT
Geospatial framework and terminology
The National Hydrography Dataset (NHD) is a geospatial data set that is available at several scales and depicts waterbodies across the USA (USGS 2001). For LakeCat, we used the medium-resolution NHD (1:100,000 scale) that is available for download as part of the National Hydrography Dataset Plus version 2.1 (NHDPlusV2), a value-added version of the 1:100,000-scale NHD (McKay et al. 2012). We used the NHDPlusV2 because LakeCat was developed as a complementary data set to StreamCat, which was also built on the NHDPlusV2 framework (Hill et al. 2016). In addition, the NHDPlusV2 contains hydrologic rasters that depict the flow of water across landscape that we used to develop the LakeCat framework (McKay et al. 2012).
The NHDPlusV2 contains 448,512 waterbody polygons that include estuaries, ice masses, playas, and others within the CONUS. Of these polygons, 379,097 are classified as lakes, ponds, or reservoirs, and these polygons form the basis of LakeCat (minimum size = 0.1 ha). Through our process, we were able to calculate and provide metrics for 378,088 lakes (Fig. 1). About ⅓ of these lakes (125,993) intersect with line features that represent the national stream network of the medium-resolution NHD (on-network lakes; e.g., Fig. 2). The remaining 252,095 lakes are geographically isolated from NHD stream lines (off-network lakes; Fig. 3). Off-network lakes required special processing to produce lake catchments and associated metrics (see Off-network lakes below). We used a Python script (NHDtblMerge function in LakeCat_functions.py; available at https://github.com/USEPA/LakeCat) to distinguish between on- and off-network lakes.
Fig. 1.
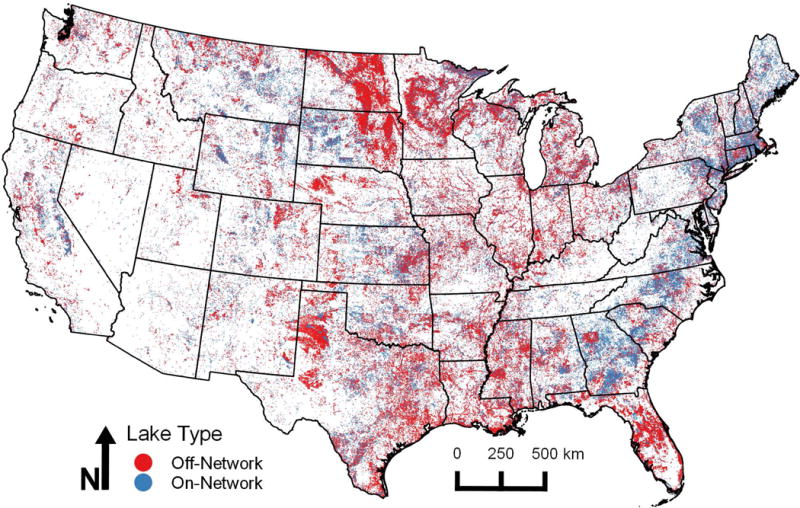
Map of 379,095 lakes within the National Hydrography Dataset version 2 (NHDPlusV2). Blue lakes intersect with lines of the NHDPlusV2 stream network and are designated as ‘on-network’ lakes (n = 125,993). Red lakes (n = 252,095) are geographically isolated from stream lines and are designated as ‘off-network’ lakes within LakeCat.
Fig. 2.
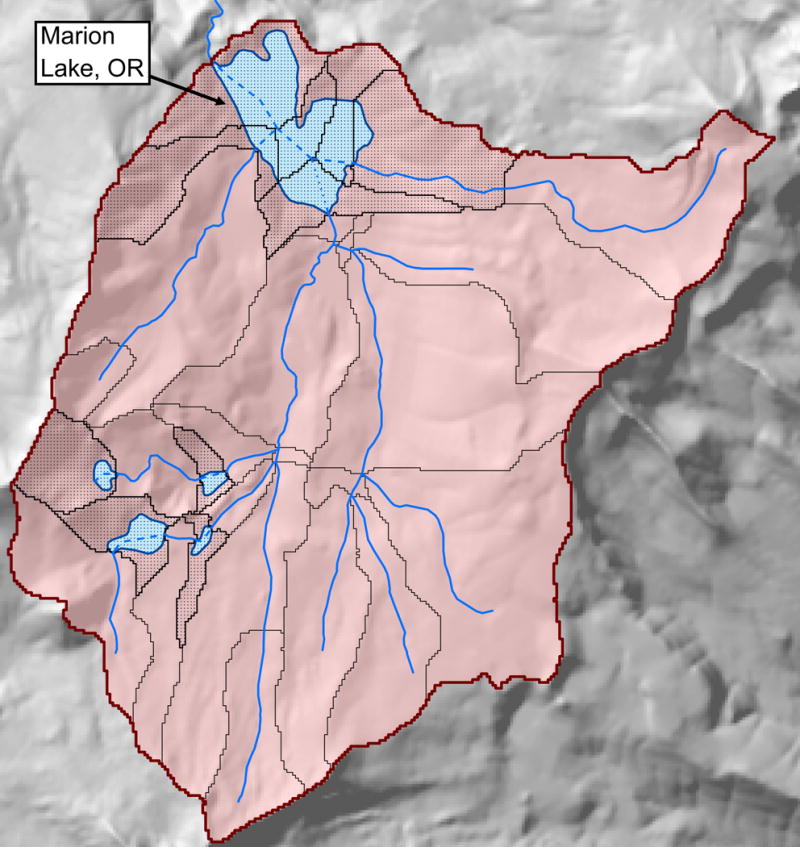
A detailed map of a set of on-network lakes in the state of Oregon. Lake polygons are light blue. National Hydrography Dataset (NHD) Plus version 2 catchments are outlined in black. Catchments that intersect with lake waterbodies have black stippling. The watershed of Lake Marion is composed of several, hydrologically linked catchments (pink area). Solid blue lines are NHD rivers and streams, and dashed blue lines are artificial paths used by the NHD to provide connectivity of rivers and streams across lakes. OR = Oregon.
Fig. 3.
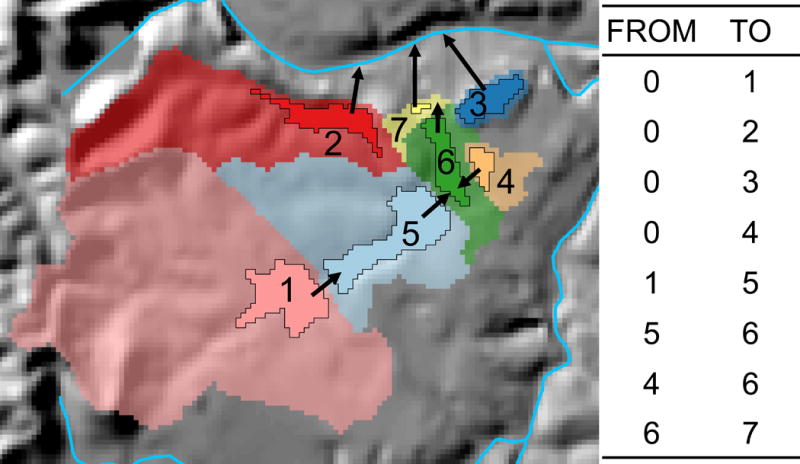
A series of nested and nonnested off-network lakes. Lakes are opaque with black borders and their local catchments are of matching, partially transparent colors. Arrows indicate the direction of hydrologic connection from each lake and its associated catchment. Numbers indicate unique lake identifiers (IDs) and the table shows the FROM-TO connection of each lake ID within the network of lakes. In this example, lakes 1–4 are headwater lakes, i.e., they have no upslope lakes. Lake 5 is downslope of lake 1, lake 6 is downslope of lakes 4 and 5, and lake 7 is downslope of lake 6. Thus, lake 7 is directly and indirectly connected to lakes 1, 4, 5, and 6 and the aggregation of these catchments is its watershed.
On-network lakes
Each on-network lake intersects with ≥1 NHDPlusV2 stream lines (blue lines in Fig. 2). With stream lines, the NHDPlusV2 also distributes a GIS polygon layer that represents the area of the landscape that drains directly to each stream segment, excluding upstream contributions. These smaller units are called catchments in the NHDPlusV2 and StreamCat (black polygons in Fig. 2). The NHDPlusV2 catchments were used to produce statistical summaries of geospatial layers (e.g., land cover). These catchment summaries were hydrologically aggregated to produce metrics for the full upstream contributing areas of each stream segment (Hill et al. 2016). For example, the red area in Fig. 2 is the aggregation of all upstream catchments of Marion Lake, Oregon. The NHDPlusV2 and StreamCat use the term watershed to refer to a set of hydrologically aggregated catchments, and we used this term in LakeCat for consistency. On-network lakes already were included as part of the StreamCat Dataset, and we drew from StreamCat to populate catchment and watershed data for on-network lakes in LakeCat. On-network lakes can have several catchments that feed directly into a lake, such as Marion Lake in Fig. 2. In LakeCat, metrics for these multipart lake catchments are reported as a single, aggregated value. For example, all catchments that overlap with Marion Lake in Fig. 2 are aggregated and reported as the local catchment for this lake (i.e., black stippled area).
Off-network lakes
To maintain consistency with the terminology used in the NHDPlusV2 and StreamCat, we also called areas that were hydrologically connected to off-network lakes catchments (i.e., colored areas surrounding lakes 1–7 in Fig. 3). Likewise, we used the term watershed to denote a set of nested catchments that represent the full contributing area to a downslope lake. In Fig. 3, catchments 1, 4, 5, and 6 contribute flow directly and indirectly to lake-catchment 7, and the integration of these catchments represent its full watershed. For off-network lakes, we delineated the local contributing areas for each, excluding nested contributions from upslope lakes (Fig. 3). We hydrologically linked these catchments to produce watershed metrics for each off-network lake (see Linking off-network lake catchments below).
Generating off-network lake catchments
To delineate catchments of off-network lakes, we first converted lake polygons to GIS rasters, where the values of lake pixels were their numerical identifiers (unique identifiers [IDs] called COMIDs) in the NHDPlusV2 (numbered polygons in Fig. 3). We overlaid these lake rasters onto rasters that depict the direction of flow across land surfaces. These flow direction rasters are distributed as part of the NHDPlusV2 (McKay et al. 2012). We then used the lake rasters as pour points in the ArcGIS Watershed tool (ESRI 2016)) to delineate lake-specific catchments, where the values of pixels within the delineated catchments are unique IDs that can be associated back to the lake on which it was based. For lakes with no additional upslope lakes (i.e., a headwater lake), a catchment and watershed are the same geographic unit (e.g., lakes 1–4 in Fig. 3). For a series of hydrologically connected lakes, this delineation process results in a series of nested catchments, where the border of the downslope lake’s catchment ends at the initiation of the next upslope lake’s catchment, usually at the edge of the upslope lake itself (e.g., lakes 1, 4–7 in Fig. 3). Each local catchment represents the contribution of flow from the surrounding landscape, excluding the areas of the upslope, hydrologically connected catchments (see makeBasins function in LakeCat_functions.py https://github.com/USEPA/LakeCat).
The catchments produced with our approach include the areas under each lake as part of their contributing area. In contrast, Soranno et al. (2017) removed lake areas from catchment areas to produce LAGOS-NE, a data set of ~50,000 lakes in the midwestern and northeastern USA. Our approach produced catchments similar to those used by the USEPA to calculate geospatial metrics for the 2007 and 2012 lake assessments (https://www.epa.gov/national-aquatic-resource-surveys) in that they also included lake areas. Inclusion of lake areas in the denominator of watershed calculations can reduce the proportion of certain land uses within watersheds (e.g., urbanization) when compared to the approach of Soranno et al. (2017). On the other hand, exclusion of lake areas from watersheds is complicated by the fact that lake areas can vary from year to year (e.g., drought vs nondrought years). Furthermore, the exclusion of lake areas also may exclude important features that underlie lakes, such as geology. We chose to include lake areas in this version of LakeCat to be consistent with the NLA.
Linking off-network lake catchments
We created a topological table that depicts the hydrologic relationships of off-network lakes. This table contains ‘FROM’ column and ‘TO’ columns. IDs of catchments that connect hydrologically to a downslope catchment are in the FROM column. Likewise, the downslope catchment’s ID is in the TO column. For example, catchments 1 to 4 in Fig. 3 are headwater catchments and are listed in rows of the TO column and have FROM-column values of 0, i.e., 0 in the FROM column denotes headwater catchments. Catchments 2 and 3 in Fig. 3 do not connect hydrologically to any downslope lakes and eventually flow to the nearby stream segment (arrows indicate hydrologic connections in Fig. 3), Thus, catchments 2 and 3 are listed in the TO column but are absent from the FROM column. In contrast, catchments 1 and 4 connect to downslope catchments 5 and 6, respectively. Thus, their values also appear in the FROM column, whereas 5 and 6 appear in the TO column. In Fig. 3, we depict lakes and catchments as sharing unique IDs. In practice, the COMIDs of lakes can be very long numerical values and we found that GIS processes were far more efficient if the unique IDs of catchments were represented with shorter values. Therefore, we gave catchments shorter IDs and linked processed values back to lake COMIDs through a relational table.
To create the topology table, we used the NHDPlusV2 flow-direction raster and the raster of catchments that was produced with the ArcGIS Watershed tool (see Generating off-network lake catchments). This process required that we identify flow connections between adjacent catchments. First, we compared each pixel in the catchment raster to each of its 8 cardinal and ordinal neighbors. If a neighboring pixel had a different catchment ID than the focal pixel, we flagged it as a potential flow connection between the focal pixel and the adjacent catchment. Next, we determined whether the direction of flow of the focal pixel was toward the neighboring catchment. If both criteria were met, the focal pixel represented a flow connection to a downslope catchment and was recorded in the topological flow table in the FROM column. Likewise, the downslope catchment ID was recorded in the TO column of the flow table. In practice, we shifted the catchment raster with open source Python tools and conducted a series of mathematical operations on the shifted and original catchment rasters to implement this logic and conduct pixel-by-pixel comparisons. To illustrate our approach, we provide the steps used to determine that catchments 6 and 7 in Fig. 3 have a south-to-north flow connection in Fig. 4. In this figure, we used a 3 × 3 window that is fixed in space to demonstrate the process of connecting the center pixel (catchment 6) to its downslope neighbor in catchment 7, i.e., the 3 × 3 window is for illustrative purposes only. In practice, the shifting and subsequent mathematical operations were done on the entire raster at once, which made the operation efficient and simple to implement with existing GIS tools. The process to connect these catchments was as follows:
Shift the catchment raster (Cat) southward by 1 pixel (shifted raster = Catʹ in Fig. 4). Note that the entire raster Cat is shifted southward to produce Catʹ, but only the area under the fixed 3 × 3 window is depicted in Fig. 4.
Apply a logical query to identify where Cat ≠ Catʹ and set the output raster (Query1) equal to 1 where true (Fig. 4). Pixels with value = 1 represent south-to-north borders between adjacent catchments in Cat, with these pixels falling on the south side of this border.
Apply a 2nd logical query to identify values in the flow direction raster that indicate northward flow (ESRI flow-direction rasters use value = 64 to represent northward flow), and set output raster (Query2) equal to 1 where true (Fig. 4).
Multiply rasters from steps 1 to 3: Query1 × Query2 × Catʹ.
Fig. 4.
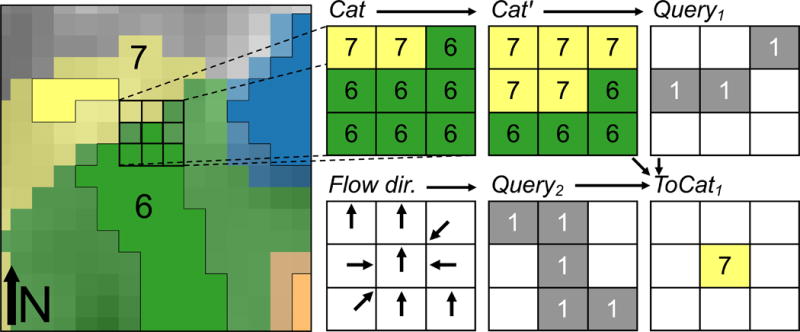
Diagram of the connection between lakes 6 and 7 from Fig. 3. The connection between lakes 6 and 7, and their associated catchments, is identified through a series of spatial shifts of the underlying rasters and a set of queries (see text). The 3 × 3 window that provides detail in this figure is fixed in space and the catchment raster, Cat, is shifted underneath this fixed position to produce Catʹ. However, this 3 × 3 window does not exist when the process is implemented. Instead, shifting and subsequent queries and mathematical operations are conducted on the entire raster data set at once to improve efficiency and to make implementation easier with existing geographic information system tools.
The result of step 4 is a raster (ToCat1) (Fig. 4) with pixels at the northern edge of the south-to-north border that meet the criteria of steps 2 and 3, but now have values of Catʹ rather than Cat. That is, the pixel values in this raster are the TO COMIDs of downslope, south-to-north connected catchments but the locations of these pixels are now shifted upslope to the FROM-pixel locations. Steps 1 to 4 were repeated in each of the remaining 7 cardinal and ordinal directions to identify catchment connections in each direction. The results were then joined to produce a single raster (ToCat1–8) of all flow connections. We then combined the ToCat1–8 and Cat rasters to identify all unique pairwise combinations of pixel values and produced a table of these combinations. The column in this table that lists Cat pixels is the FROM column and the column for ToCat1–8 is the TO column (see rollArray and makeFlows functions in LakeCat_functions.py at https://github.com/USEPA/LakeCat for Python code to connect lake catchments).
Landscape layers
LakeCat was designed to parallel StreamCat in the numbers and types of landscape metrics calculated for each catchment and watershed. The original set of StreamCat metrics included summaries of both natural and anthropogenic landscape layers (Hill et al. 2016). These metrics were selected based on 3 papers that identified these layers as being of ecological relevance to streams (i.e., Carlisle et al. 2009, Falcone et al. 2010, Wang et al. 2011). However, others have shown that several of these layers have similar relevance to lake conditions (e.g., Fergus et al. 2011, Read et al. 2015, Hollister et al. 2016). Variables originally included in StreamCat were derived from STATSGO soils (USDA 2006), lithology (Cress et al. 2010), runoff (McCabe and Wolock 2011), 2006 land cover and imperviousness of human-made surfaces (Homer et al. 2007), mines (USGS 2003), and roads (USCB 2014) among others (see Hill et al. 2016). Since publication, several hundred more metrics have been added to the original set in StreamCat. These new metrics include 2011 land cover and imperviousness of artificial surfaces (Homer et al. 2007), forest loss by year (2001–2013) (Hansen et al. 2013), and the geochemical and geophysical characteristics of underlying lithology (Olson and Hawkins 2014) (see the StreamCat website for a list of updates since publication; https://www.epa.gov/national-aquatic-resource-surveys/streamcat). We summarized all of these layers for LakeCat, and like StreamCat, more metrics may be added as geospatial data become available. However, We excluded some summaries if no analog existed for lakes. For example, StreamCat includes summaries of some landscape layers within a 100-m riparian buffer of streams and rivers. We did not include summaries of landscapes within a buffer of lakes but future versions of LakeCat might include such metrics.
Catchment and watershed summaries
To generate watershed metrics for each lake, we first overlaid catchments onto a suite of natural and anthropogenic landscape layers (see Landscape layers) and calculated statistical summaries of features within the boundaries (ArcGIS Zonal Statistics tool). For continuous-variable data sets, such as climate rasters, we calculated the sum of pixel values and the count of pixels within each catchment. For categorical data sets, such as land-cover classes, we calculated the sum of each pixel type and the sum of all pixels within each catchment. We then accumulated these local catchment summaries to produce watershed-level metrics. To accumulate the summaries of nested catchments, we developed a Python routine that created a list of all upslope catchments for each lake catchment (also see Hill et al. 2016). In the example of Fig. 3, the accumulation list for lake 7 includes lakes 1, 4, 5, and 6 (Table 1). Likewise, the accumulation list for lake 6 includes only lakes 1, 4, and 5 (Fig. 3). We then associated the catchment summaries with the catchment IDs in these lists and calculated full watershed averages for continuous variables with:
| (Eq. 1) |
in which Lw is the mean of the continuous landscape layer for watershed w, xc is the sum of values for all pixels in catchment c with data, and nc is the number of pixels with data in catchment c, where catchment c is a member of the list of catchments that make up watershed w (Hill et al. 2016). Equation 1 is applied to pixels with data, i.e., missing data are ignored in calculations. Some catchments may cross international boundaries, and pixels within the catchment that are outside of the CONUS are assigned a missing data value. We used similar calculations were used to calculate watershed percentages of categorical rasters (e.g., land cover type) as:
| (Eq. 2) |
in which Pw,i represents the percent of watershed w composed of class i, xc,i is the count of pixels of class i in catchment c, and nc is the number of all pixels in catchment c, with catchment c being a member of a list of catchments within watershed w (Hill et al. 2016). As with StreamCat, watershed metrics for point data were calculated as densities within the watershed, such as the density of dams. For these metrics, we used:
| (Eq. 3) |
where Vw is the density of point features within watershed w, nc is the count of points within catchment c, and πc is the proportion of area (Ac) of catchment c within the CONUS. Equation 3 also can be used to calculate the density of line features (e.g., roads) within watersheds by summing the length of features within each catchment (i.e., nc becomes the total length of line features in catchment c) (Hill et al. (2016). Other characteristics of point or line features also can be summarized for watersheds by replacing nc in Eq. 3 with layer attributes:
| (Eq. 4) |
where xc represents attribute of interest (e.g., dam volume) for each point or line feature in catchment c. We used πc to account for the fact that most of the point and line features used in LakeCat stop at international borders. Not accounting for the proportion of the catchment and watershed within the CONUS would down-weight these densities and averages by areas for which data are unavailable (Hill et al. 2016). In LakeCat, we also documented the percentage of each catchment and watershed area that overlapped with data. That is, we accounted for areas of missing values arising from international borders and other reasons. We recommend that users of LakeCat examine these percentages to determine the appropriateness of data completeness for analyses. The Python scripts LakeCat.py, LakeCat_functions.py, and MakeFinalTables_LakeCat.py at https://github.com/USEPA/LakeCat contain code to produce catchment and watershed summaries.
Table 1.
Accumulation lists for lake catchments in Fig. 3. Catchments 1–4 have no upslope, contributing catchments. Catchment 5 receives flow from catchment 1 (brackets ‘[]’ indicate a list of upslope catchments). Catchment 6 receives flow from catchments 4 and 5 directly, and from catchments 1–5. The list of contributing catchments is linked to statistical summaries (e.g., sum and count) of landscape layers for these catchments and used to calculate full watershed metrics for LakeCat.
| Focal catchment COMID | List of contributing upstream catchments | |||
|---|---|---|---|---|
| 1 | ||||
| 2 | ||||
| 3 | ||||
| 4 | ||||
| 5 | [1] | |||
| 6 | [5 | 4 | 1] | |
| 7 | [6 | 5 | 4 | 1] |
Data validation
We conducted extensive quality assurance at each step to ensure the integrity of LakeCat data. Many of these procedures were originally conducted during the development of the StreamCat Dataset, such as checking and documenting the geographic projection, extent, and data values of landscape layers that were used to produce watershed metrics (Hill et al. 2016). We visually examined several hundred catchment delineations and calculated several metrics by hand to ensure that our algorithms were accurately producing watershed-level accumulations, i.e., those produced by Eqs 1–3. In addition, we compared the areas of off-network lake catchments to the areas of the NHDPlusV2 catchments they fell within. Off-network lakes, in general, had small watershed areas and should not be larger than the NHDPlusV2 catchments that contained them (see makeBasins function in LakeCat_functions.py at https://github.com/USEPA/LakeCat). We found 326 cases where lake catchments were larger than the NHDPlusV2 catchments within which they fell. Upon examination, we determined that these off-network lakes did indeed cross NHDPlusV2 catchment boundaries. In all cases, the lake catchment appeared to be hydrologically correct despite the lake crossing NHDPlusV2 catchment boundaries, and we retained these records in LakeCat. The identities of these lakes were stored in a table that is available for download from the LakeCat website (https://www.epa.gov/national-aquatic-resource-surveys/lakecat). We were unable to generate LakeCat metrics for 1009 of the 379,097 NHD lakes. Of these 1009 lakes, 939 were outside the boundary of the NHDPlusV2 geospatial layers we used as the basis for LakeCat; 44 had boundaries that extended beyond their NHDPlusV2 processing regions, which resulted in inaccurate catchment delineations; 8 were duplicated in the NHDPlusV2 (i.e., same lake area and unique ID), 9 had polygons that overlapped with other lakes (i.e., similar or the same lake area with differing unique IDs), and 9 were very small and generated errors when we attempted to delineate catchment boundaries with the ArcGIS Watershed tool. See documentation available at the LakeCat website (https://www.epa.gov/national-aquatic-resource-surveys/lakecat) for additional details on the quality assurance procedures used to identify and remove these lakes.
We conducted an independent evaluation by comparing LakeCat metrics with 1156 watershed summaries of land cover that were produced by the USEPA as part of the 2007 NLA (USEPA 2010). The NLA watershed summaries were produced with an independent method and an earlier version of the NHDPlus (version 1), but both data sets used the same land-cover layer, i.e., the 2006 National Land Cover Dataset (NLCD) (Homer et al. 2007). In general, we found a high degree of correspondence between the 2 sets of summaries. For example, a regression of the % of the watershed composed of cropland from each data set had an r2 = 0.91 and a slope = 0.96 (Fig. 5). We examined several cases where substantial disagreement occurred between the 2 sets of summaries. For many cases of large differences, the NLA either combined 2 adjacent lakes or split a single lake to create a custom lake boundary for watershed delineation if doing so better represented a lake’s state at the time of sampling (e.g., during wet or dry years, respectively). However, the vast majority of disagreements were caused by differences between versions 1 and 2 of the NHDPlus, and their associated hydrologic flow rasters. NHDPlusV2 is a substantial improvement in the representation of hydrologic features and their connections over version 1 (McKay et al. 2012). Which of 2 watershed delineations is more valid is not always possible to determine. However, in all of the cases we examined, we determined that LakeCat produced delineations that were as defensible as the 2007 NLA and, in most cases, were an improvement.
Fig. 5.
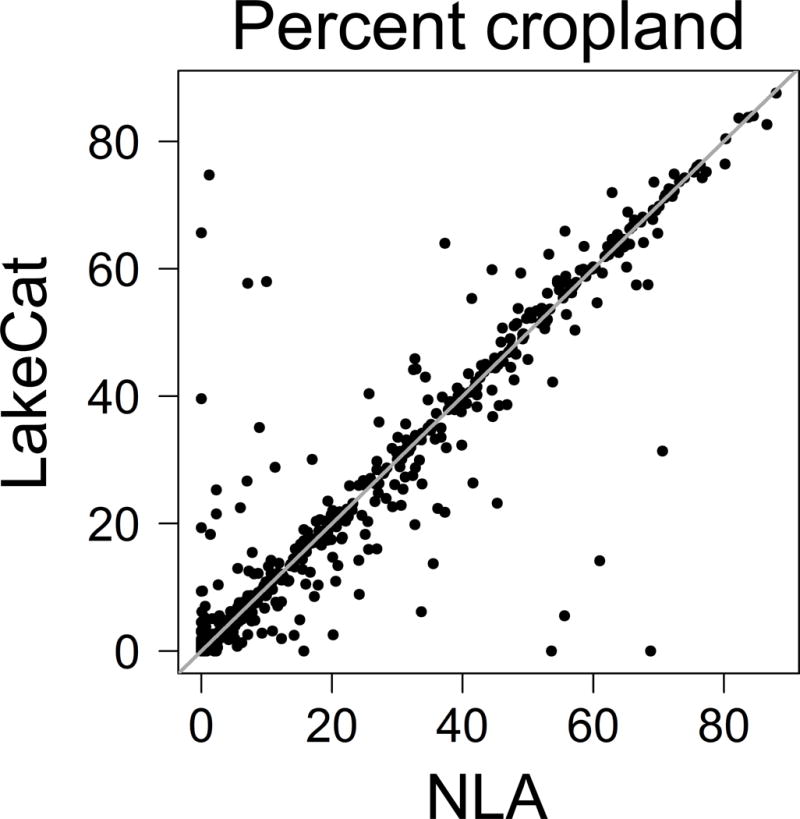
Plot of LakeCat vs 2007 National Lake Assessment (NLA) estimates of the % lake watersheds consisting of cropland (2006 National Land Cover Dataset).
ACCESSING LAKECAT DATA
LakeCat data are available for download through the USEPA’s National Aquatic Resource Survey website (https://www.epa.gov/national-aquatic-resource-surveys/lakecat) as comma-delimited text files. At the time of publication, LakeCat contained 135 metrics at each of the catchment and watershed scales, i.e., 270 total metrics when including both scales (see Appendix S1 for a list of LakeCat variables at the time of publication). Each table is based on the source data used to produce them, such as the 2006 NLCD (Homer et al. 2007) or STATSGO soils data (USDA 2006). LakeCat has far fewer records than StreamCat, which enabled production of a single CONUS-wide table for each metric set. The first 6 columns in each table contain ancillary information regarding each lake: the unique ID of the lake (i.e., the COMID that can be linked to the NHDPlusV2), the areas of the lake catchment and full contributing watershed, and the % of each lake catchment or watershed that overlapped with the landscape layer used to produce the metric (e.g., lake watersheds that cross into Canada and overlap with areas of missing data will have % completeness values <100%), and a column (inStreamCat) that indicates whether the lake was on the NHDPlusV2 stream network (inStreamCat = 1) or was geographically isolated from this network (inStreamCat = 0). The remaining columns contain the set of local catchment and watershed summaries for the landscape layer used to produce the table. Local-catchment and watershed metrics have ‘Cat’ and ‘Ws’ appended to the end of their names, respectively. The format of these data should facilitate the use of LakeCat data with analytical and geospatial software, such as R (see Appendix S2 for example R code linking sample coordinates to the NHDPlusV2).
In addition to the data set, the geospatial framework we developed and used to make LakeCat also is available for download (https://www.epa.gov/national-aquatic-resource-surveys/lakecat). This framework includes lake catchments as rasters and ESRI shapefiles (ESRI 2016), and FROM-TO topology tables that connect nested off-network lakes. It also includes a set of Python vectors (i.e., NumPy arrays; van der Walt et al. 2011) that were used to map catchment values to upslope COMIDs for each lake in the NHDPlusV2 and provide full watershed accumulations. All code used to develop LakeCat is available on GitHub (https://github.com/USEPA/LakeCat). However, this code was developed specifically for our computer systems, and no effort has been made to make the LakeCat algorithms and code more generically accessible to users. This code is available in the spirit of transparency. We do not have the resources to support those attempting to run or modify LakeCat code or algorithms.
MODELING ILLUSTRATION
Model development
A major benefit of extensive, nationally consistent data sets, such as LakeCat, is the ability to use the data to model and predict conditions at locations that have not yet been sampled (Hill et al. 2017). Doing so could provide states, tribes, and other management entities with an important tool to identify lakes that may be suffering from impaired conditions and to focus assessment and monitoring resources. To illustrate the utility of LakeCat for such applications, we used LakeCat data to model and predict the probability of lake eutrophication across the CONUS. First, we tied the geographic coordinates of lakes from the USEPA’s 2012 NLA to LakeCat data (see Appendix S2 for R code). Next, we used random forests (Breiman 2001) to model eutrophic and noneutrophic sampling sites as a binary response variable (Fig. 6A) with the randomForest package in R (version 3.4.0) (Liaw and Wiener 2002, R Development Core Team 2017). Hollister et al. (2016) developed a similar model to predict the trophic state of lakes with data from the 2007 NLA and a combination of GIS and in-lake biophysical measurements collected at the time of sampling. However, the use of field-collected biophysical measurements to model lake eutrophication would limit our ability to predict to new, unsampled lakes within LakeCat. Therefore, we used only geospatial predictors from LakeCat to develop our model. The 2012 NLA sampled Chl a at lakes across the CONUS and used a numerical criterion of >7 μg/L to designate lakes as being in a eutrophic state (USEPA 2017).
Fig. 6.
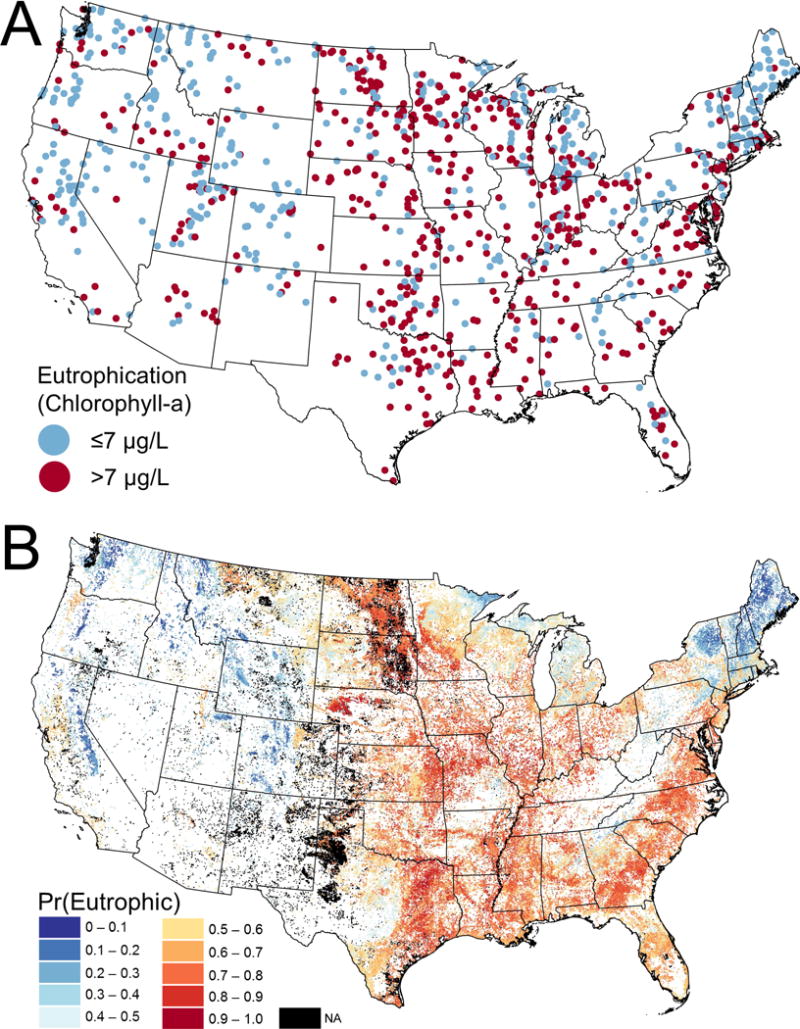
A.—A binary classification of lake samples into eutrophic and noneutrophic conditions based on chlorophyll a concentrations. The USEPA used a criterion of >7 μg/L to designate lakes as eutrophic (USEPA 2017). B.—Predicted probabilities of lake eutrophication [Pr(Eutrophication)] based on a random forest model that used LakeCat metrics as predictor variables. The map represents each lake as a single point. Black points are lakes that were excluded from the National Lakes Assessment sampling frame (see text).
Land use (e.g., agriculture) can influence the trophic state of lakes (Smith et al. 1999, Taranu and Gregory-Eaves 2008, Hollister et al. 2016), and we expected that the LakeCat estimates of these land uses would be important in predicting eutrophication. To model eutrophication as a binary response, we followed the approach of Fox et al. (2017) and Hill et al. (2017). We used all LakeCat variables (Appendix S1) as possible predictors in the model with no selection. We balanced the binary responses in each tree within the random forest to reduce biases that can occur with unbalanced response data (Hill et al. 2017). We also generated several new variables for modeling by combining a subset of LakeCat variables. For example, we summed the watershed percentages of the two 2011 NLCD agricultural types to produce a composite metric of all agriculture. We also produced composite metrics of NLCD 2011 urbanization and natural land covers.
We report the model performance as % sites correctly classified (PCC) as being eutrophic or noneutrophic and the area under the receiver operating curve (AUC). AUC compares the true positive and false positive rates of classification. An AUC = 0.5 implies that a model can predict no better than random, and an AUC = 1 indicates a perfect classifier (Hosmer and Lemeshow 2004). These performance metrics were based on random forest’s out-of-bag data. Random forest withholds ~⅓ of the observations at the time each tree is constructed. Performance metrics based on these data are considered a reliable estimate of true model performance (Cutler et al. 2007). Random forest also can rank predictor variables in terms of their contribution to model performance and plot the partial dependencies of the response variable to a variable while accounting for the effect of all other variables (Cutler et al. 2007).
We predicted and mapped the probability of eutrophication for all NHD lakes that met the 2012 NLA sampling criteria framework (n = 297,071). These lakes were not sampled but rather were part of the set of lakes that the NLA selected from for sampling. This sampling frame is designed to allow EPA scientists to make inferences regarding the condition of lakes regionally and nationally and excludes lakes with surface areas <1 ha and lakes that are designated within the NHD as intermittent (Peck et al. 2013). To facilitate mapping of the 297,071 lakes that were candidates for NLA sampling, we mapped the predicted probabilities at the centroid of each lake and excluded lake polygons from the map.
Model performance and map
The random forest model correctly predicted eutrophication at 73% of sites and had an AUC = 0.80 based on the out-of-bag evaluations. In addition, the PCC was balanced between true eutrophic and true noneutrophic sites (PCC = 72 and 74%, respectively), indicating the model was an unbiased predictor of eutrophication. The map of predicted probabilities of eutrophication [Pr(Eutrophication)] showed higher probabilities throughout the midwestern and southeastern states, excluding portions of the southern Appalachian Mountains (Fig. 6B). Lower Pr(Eutrophication) values occurred in most of the western states, the northern portions of the Great Lakes states, and some northeastern states. However, in western states, slightly higher Pr(Eutrophication) was predicted in valleys with large amounts of agriculture, such as Central Valley, California; Willamette Valley, Oregon; and Cache and Salt Lake Valleys, Utah.
Metrics of agricultural activity constituted 7 of the top 10 variables when ranked by their contribution to model accuracy (Fig. 7), and larger values of each agricultural metric were positively related to higher Pr(Eutrophication), as we had hypothesized. These metrics included % watershed composed of agriculture in 2011 (PctAg2011Ws), 1997 pesticide usage (Pestic97Ws and Pestic97Cat, i.e., pesticides in the watershed and catchment, respectively), watershed fertilizer usage (FertWs), erodibility of soils on agricultural land in the watershed (AgKffactWs), and watershed biological N2 fixation of cultivated crops (CBNFWs). Pesticides are not known to cause eutrophication directly, but this metric may be especially good for representing the intensity of agricultural activity within lake catchments and watersheds. In general, the watershed version of each metric (i.e., Ws variables in Fig. 7) ranked higher than its local catchment counterpart (Cat in Fig. 7), indicating the importance of understanding the aggregated watershed context in water-quality management. Three predictors (BFIWs, BFICat, and PctConif2011Ws) had negative relationships with Pr(Eutrophication). The BFI metrics are based on the USGS base-flow index raster (Wolock 2003) and were originally developed to indicate the % stream flow consisting of ground water relative to event flow. Lakes with BFIWs and BFICat values >50 were less likely to be eutrophic, all else being equal. The USGS BFI is negatively related to lake nutrients, especially total P (Cheruvelil et al. 2013), and may offer an explanation for the importance of this variable in our model of eutrophication. Likewise, lake watersheds with >10% conifer forest in 2011 were also less likely to be eutrophic; a pattern that is consistent with the negative association found by Read et al. (2015) between watershed forest cover and lake nutrients.
Fig. 7.
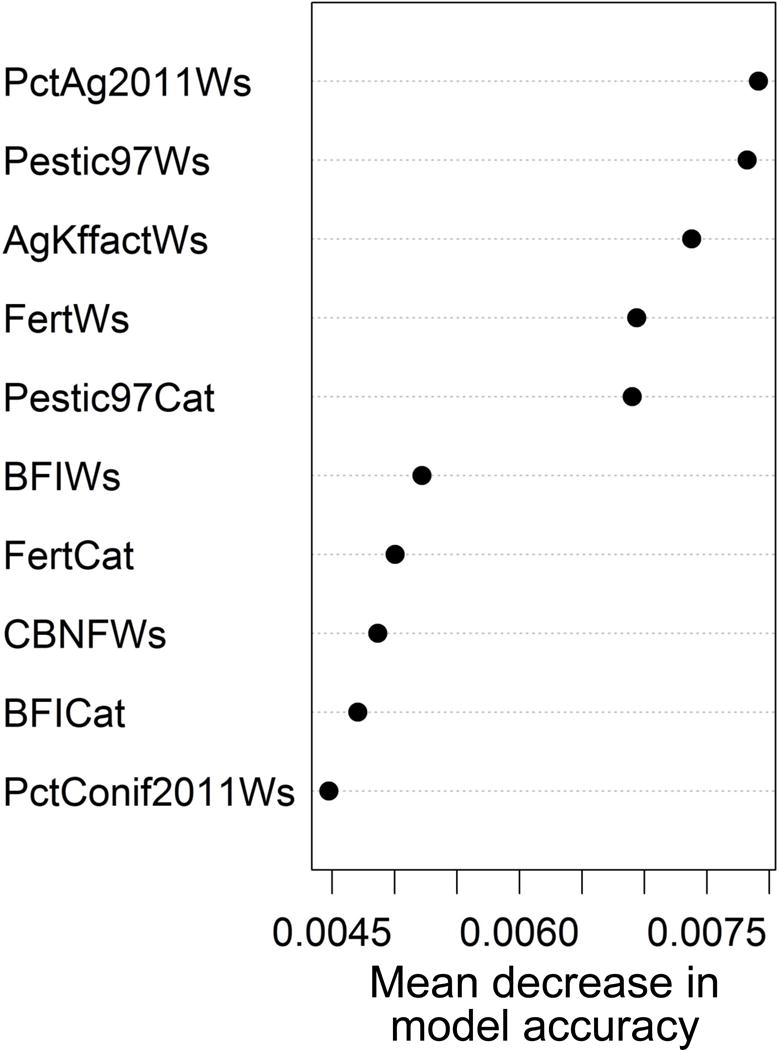
Top 10 variables ranked by their importance to the model (mean decrease in model accuracy). PctAg2011Ws = % of the watershed consisting of agriculture (cropland and hay classes of 2011 National Land Cover Dataset); Pestic97Cat and Pestic97Ws = mean 1997 pesticide use (kg/km2) within catchment and watershed, respectively; FertCat and FertWs = mean rate of synthetic N fertilizer application to agricultural land (kg N ha–1 y–1) within catchment and watershed, respectively; AgKffactWs = mean erodibility factor (unitless) of soils on agricultural land within the watershed; BFICat and BFIWs = mean of US Geological Survey base-flow raster within catchment and watershed, respectively; PctConif2011Ws = % of the watershed composed of coniferous forest (2011 NLCD); CBNFWs = mean rate of biological N2 fixation from the cultivation of crops (kg N ha–1 y–1) within the watershed.
DISCUSSION
LakeCat provides a nationally consistent data set of natural and anthropogenic geospatial metrics for lakes across the CONUS. To our knowledge, LakeCat is the first data set of such an extensive suite of catchment and watershed metrics for this number of lakes. To date, linking lake samples to watershed-based geospatial information has been a time-consuming task that required specialized expertise, a process that LakeCat will shorten substantially. As with StreamCat, additional metrics may be added to LakeCat as they become available. These metrics may not be limited to catchment summaries of landscape layers. For example, the position of a lake within a network of lakes, the connectivity among lakes, and their connectivity to downslope waters can play important roles in determining the physical, chemical, and ecological characteristics of these systems (Martin and Soranno 2006, Sadro et al. 2012, Guimarães et al. 2014). The methods and tools we developed for LakeCat are flexible and could be adapted to characterize the connectivity among freshwater ecosystems (e.g., Fergus et al. 2017).
As illustrated by our model of eutrophication, LakeCat can easily provide covariates to model the physical and ecological phenomena of lakes at broad spatial extents. Furthermore, the application of this model to all LakeCat sites in the NLA sampling frame produced a spatially explicit prediction of the probability of eutrophication for 297,071 lakes across the CONUS. To our knowledge, no such map has been produced previously. This task was accomplished with ~60 lines of code in R (Appendix S3), demonstrating the way in which LakeCat can facilitate analysis and mapping of physical and ecological phenomena of lake ecosystems. These types of models and maps could provide managers with an important tool for prioritizing limited financial and personnel resources for monitoring and assessment of lakes. However, the utility of LakeCat is not limited to development of such models. Our illustration was intended mainly to highlight an analysis that would not have been possible before LakeCat. For example, simpler queries of landuse metrics within LakeCat can identify lakes with minimal human-related alterations to their watershed and help prioritize these lakes for conservation or for potential use as regional reference lakes (Stoddard et al. 2006, Soranno et al. 2011). Likewise, a query of lakes with large amounts of human-related alteration could identify systems that are at particular risk of water quality and biological impairment.
The objective of developing LakeCat was to provide researchers and managers with high-quality geospatial data for a large number of lakes and their associated catchments and watersheds for analyses at various spatial extents (e.g., regional to national). Our hope is to facilitate the studies and analyses that are needed to manage these important ecosystems effectively. In addition, LakeCat fits well within the emerging framework of macrosystems ecology (Heffernan et al. 2014) and the use of large databsets to inform freshwater research (Soranno and Schimel 2014, Isaak et al. 2017). We think that LakeCat will be an important contribution to these efforts.
Supplementary Material
Acknowledgments
We thank Pat Soranno and Nicole Smith of the Michigan State University Continental Limnology group, and C. Emi Fergus and James Markwiese of the USEPA Western Ecology Division, and 2 anonymous referees for comments that greatly improved the article. The information in this document has been funded entirely by the US Environmental Protection Agency, in part by an appointment to the Internship/Research Participation Program at the Office of Research and Development, USEPA, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the US Department of Energy and EPA. The views expressed in this journal article are those of the authors and do not necessarily reflect the views or policies of the USEPA. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Author contributions: RAH contributed to the project design, algorithm development, and wrote most of the manuscript. MHW contributed to the projected design and algorithm development. RMD contributed to algorithm development and quality-assurance procedures. SGL and ARO contributed to the project design. All authors edited and contributed to the manuscript.
LITERATURE CITED
- Breiman L. Random forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- Carlisle D, Falcone J, Meador M. Predicting the biological condition of streams: use of geospatial indicators of natural and anthropogenic characteristics of watersheds. Environmental Monitoring and Assessment. 2009;151:143–160. doi: 10.1007/s10661-008-0256-z. [DOI] [PubMed] [Google Scholar]
- Cheruvelil KS, Soranno PA, Webster KE, Bremigan MT. Multi-scaled drivers of ecosystem state: quantifying the importance of the regional spatial scale. Ecological Applications. 2013;23:1603–1618. doi: 10.1890/12-1872.1. [DOI] [PubMed] [Google Scholar]
- Cress J, Soller D, Sayre R, Comer P, Harumi W. Scientific Investigations Map 3126. US Geological Survey; Denver, Colorado: 2010. Terrestrial ecosystems—surficial lithology of the conterminous United States. [Google Scholar]
- Cutler DR, Edwards TC, Beard KH, Cutler A, Hess KT, Gibson J, Lawler JJ. Random forests for classification in ecology. Ecology. 2007;88:2783–2792. doi: 10.1890/07-0539.1. [DOI] [PubMed] [Google Scholar]
- Falcone JA, Carlisle DM, Weber LC. Quantifying human disturbance in watersheds: variable selection and performance of a GIS-based disturbance index for predicting the biological condition of perennial streams. Ecological Indicators. 2010;10:264–273. [Google Scholar]
- Fei S, Guo Q, Potter K. Macrosystems ecology: novel methods and new understanding of multi-scale patterns and processes. Landscape Ecology. 2016;31:1–6. [Google Scholar]
- Fergus CE, Lapierre JF, Oliver SK, Skaff NK, Cheruvelil KS, Webster K, Scott C, Soranno P. The freshwater landscape: lake, wetland, and stream abundance and connectivity at macroscales. Ecosphere. 2017;8:e01911–n/a. [Google Scholar]
- Fergus CE, Soranno PA, Cheruvelil KS, Bremigan MT. Multiscale landscape and wetland drivers of lake total phosphorus and water color. Limnology and Oceanography. 2011;56:2127–2146. [Google Scholar]
- Foster IDL, Dearing JA. Lake-catchments and environmental chemistry: a comparative study of contemporary and historical catchment processes in midland England. GeoJournal. 1987;14:285–297. [Google Scholar]
- Fox EW, Hill RA, Leibowitz SG, Olsen AR, Thornbrugh DJ, Weber MH. Assessing the accuracy and stability of variable selection methods for random forest modeling in ecology. Environmental Monitoring and Assessment. 2017;189:316. doi: 10.1007/s10661-017-6025-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães TdFR, Hartz SM, Becker FG. Lake connectivity and fish species richness in southern Brazilian coastal lakes. Hydrobiologia. 2014;740:207–217. [Google Scholar]
- Hansen MC, Potapov PV, Moore R, Hancher M, Turubanova SA, Tyukavina A, Thau D, Stehman SV, Goetz SJ, Loveland TR, Kommareddy A, Egorov A, Chini L, Justice CO, Townshend JRG. High-resolution global maps of 21st-century forest cover change. Science. 2013;342:850–853. doi: 10.1126/science.1244693. [DOI] [PubMed] [Google Scholar]
- Heffernan JB, Soranno PA, Angilletta MJ, Buckley LB, Gruner DS, Keitt TH. Macrosystems ecology: understanding ecological patterns and processes at continental scales. Frontiers in Ecology and the Environment. 2014;12:5–14. [Google Scholar]
- Hering D, Borja A, Carvalho L, Feld CK. Assessment and recovery of European water bodies: key messages from the WISER project. Hydrobiologia. 2013;704:1–9. [Google Scholar]
- Hill RA, Fox EW, Leibowitz SG, Olsen AR, Thornbrugh DJ, Weber MH. Predictive mapping of the biotic condition of conterminous-USA rivers and streams. Ecological Applications. 2017;27:18. doi: 10.1002/eap.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RA, Weber MH, Leibowitz SG, Olsen AR, Thornbrugh DJ. The Stream-Catchment (StreamCat) Dataset: a database of watershed metrics for the conterminous United States. Journal of the American Water Resources Association. 2016;52:120–128. [Google Scholar]
- Hollister J, Milstead WB. Using GIS to estimate lake volume from limited data. Lake and Reservoir Management. 2010;26:194–199. [Google Scholar]
- Hollister JW, Milstead WB, Kreakie BJ. Modeling lake trophic state: a random forest approach. Ecosphere. 2016;7:e01321–n/a. [Google Scholar]
- Hollister JW, Milstead WB, Urrutia MA. Predicting maximum lake depth from surrounding topography. PLoS ONE. 2011;6:e25764. doi: 10.1371/journal.pone.0025764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer C, Dewitz J, Fry J, Coan M, Hossain N, Larson C, Herold N, McKerrow A, VanDriel JN, Wickham J. Completion of the 2001 National Land Cover Database for the conterminous United States. Photogrammetric Engineering and Remote Sensing. 2007;73:337–341. [Google Scholar]
- Hosmer DWJ, Lemeshow S. Applied logistic regression. 2nd. John Wiley and Sons; New York: 2004. [Google Scholar]
- Isaak DJ, Wenger SJ, Young MK. Big biology meets microclimatology: defining thermal niches of ectotherms at landscape scales for conservation planning. Ecological Applications. 2017;27:977–990. doi: 10.1002/eap.1501. [DOI] [PubMed] [Google Scholar]
- Liaw A, Wiener M. Classification and regression by randomForest. R News. 2002;2:18–22. [Google Scholar]
- Likens GE. Beyond the shoreline: a watershed-ecosystem approach. Verhandlungen der Internationalen Vereinigung für theoretische und angewandte Limnologie. 1984;22:22. [Google Scholar]
- Martin SL, Soranno PA. Lake landscape position: relationships to hydrologic connectivity and landscape features. Limnology and Oceanography. 2006;51:801–814. [Google Scholar]
- McCabe GJ, Wolock DM. Independent effects of temperature and precipitation on modeled runoff in the conterminous United States. Water Resources Research. 2011;47:W11522. [Google Scholar]
- McKay L, Bondelid T, Dewald T, Johnston J, Moore R, Reah A. NHDPlus Version 2: user guide. US Geological Survey; Reston, Virginia: 2012. (Available from: http://www.horizon-systems.com/NHDPlus/NHDPlusV2_home.php) [Google Scholar]
- Moe SJ, Schmidt-Kloiber A, Dudley BJ, Hering D. The WISER way of organising ecological data from European rivers, lakes, transitional and coastal waters. Hydrobiologia. 2013;704:11–28. [Google Scholar]
- Olson JR, Hawkins CP. Geochemical Characteristics of the Conterminous United States: US Geological Survey (USGS) data release. US Geological Survey; Reston, Virginia: 2014. (Available from: http://dx.doi.org/10.5066/F7X0653P) [Google Scholar]
- Peck DV, Olsen AR, Weber MH, Paulsen SG, Peterson C, Holdsworth SM. Survey design and extent estimates for the National Lakes Assessment. Freshwater Science. 2013;32:1231–1245. [Google Scholar]
- Read EK, V, Patil P, Oliver SK, Hetherington AL, Brentrup JA, Zwart JA, Winters KM, Corman JR, Nodine ER, Woolway RI, Dugan HA, Jaimes A, Santoso AB, Hong GS, Winslow LA, Hanson PC, Weathers KC. The importance of lake-specific characteristics for water quality across the continental United States. Ecological Applications. 2015;25:943–955. doi: 10.1890/14-0935.1. [DOI] [PubMed] [Google Scholar]
- Sadro S, Nelson CE, Melack JM. The influence of landscape position and catchment characteristics on aquatic biogeochemistry in high-elevation lake-chains. Ecosystems. 2012;15:363–386. [Google Scholar]
- Smith VH, Tilman GD, Nekola JC. Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environmental Pollution. 1999;100:179–196. doi: 10.1016/s0269-7491(99)00091-3. [DOI] [PubMed] [Google Scholar]
- Soranno PA, Bacon LC, Beauchene M, Bednar KE, Bissell EG, Boudreau CK, Boyer MG, Bremigan MT, Carpenter SR, Carr JW, Cheruvelil KS, Christel ST, Claucherty M, Collins SM, Conroy JD, Downing JA, Dukett J, Fergus CE, Filstrup CT, Funk C, Gonzalez MJ, Green LT, Gries C, Halfman JD, Hamilton SK, Hanson PC, Henry EN, Herron EM, Hockings C, Jackson JR, Jacobson-Hedin K, Janus LL, Jones WW, Jones JR, Keson CM, King KBS, Kishbaugh SA, Lapierre JF, Lathrop B, Latimore JA, Lee Y, Lottig NR, Lynch JA, Matthews LJ, McDowell WH, Moore KEB, Neff BP, Nelson SJ, Oliver SK, Pace ML, Pierson DC, Poisson AC, Pollard AI, Post DM, Reyes PO, Rosenberry DO, Roy KM, Rudstam LG, Sarnelle O, Schuldt NJ, Scott CE, Skaff NK, Smith NJ, Spinelli NR, Stachelek JJ, Stanley EH, Stoddard JL, Stopyak SB, Stow CA, Tallant JM, Tan PN, Thorpe AP, Vanni MJ, Wagner T, Watkins G, Weathers KC, Webster KE, White JD, Wilmes MK, Yuan S. LAGOS-NE: a multi-scaled geospatial and temporal database of lake ecological context and water quality for thousands of US lakes. GigaScience. 2017;6:1–22. doi: 10.1093/gigascience/gix101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranno PA, Cheruvelil KS, Bissell EG, Bremigan MT, Downing JA, Fergus CE. Cross-scale interactions: quantifying multi-scaled cause-effect relationships in macrosystems. Frontiers in Ecology and the Environment. 2014;12:65–73. [Google Scholar]
- Soranno PA, Schimel DS. Macrosystems ecology: big data, big ecology. Frontiers in Ecology and the Environment. 2014;12:3. [Google Scholar]
- Soranno PA, Wagner T, Martin SL, McLean C, Novitski LN, Provence CD, Rober AR. Quantifying regional reference conditions for freshwater ecosystem management: a comparison of approaches and future research needs. Lake and Reservoir Management. 2011;27:138–148. [Google Scholar]
- Stoddard JL, Larsen DP, Hawkins CP, Johnson RK, Norris RH. Setting expectations for the ecological condition of streams: the concept of reference condition. Ecological Applications. 2006;16:1267–1276. doi: 10.1890/1051-0761(2006)016[1267:seftec]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Taranu ZE, Gregory-Eaves I. Quantifying relationships among phosphorus, agriculture, and lake depth at an inter-regional scale. Ecosystems. 2008;11:715–725. [Google Scholar]
- USCB (US Census Bureau) TIGER/Line Shapefiles (machine-readable data files) US Census Bureau; Washington, DC: 2014. (Available from: http://www.census.gov/geo/maps-data/data/tiger.html) [Google Scholar]
- USDA (US Department of Agriculture) US General Soil Map (STATSGO) National Geospatial Center of Excellence, Natural Resource Conservation Service, US Department of Agriculture; Washington, DC: 2006. (Available from: http://www.ncgc.nrcs.usda.gov/products/datasets/statsgo/) [Google Scholar]
- USEPA (US Environmental Protection Agency) National Lakes Assessment 2007:a collaborative survey. Office of Water and Office of Research and Development, US Environmental Protection Agency; Washington, DC: 2009. (EPA/841/R-09/001). [Google Scholar]
- USEPA (US Environmental Protection Agency) National Lakes Assessment 2007 Technical Appendix. Office of Water and Office of Research and Development, US Environmental Protection Agency; Washington, DC: 2010. (EPA 841-R-09-001a). [Google Scholar]
- USEPA (US Environmental Protection Agency) National Lakes Assessment 2012:Technical Report. Office of Water and Office of Research and Development, US Environmental Protection Agency; Washington, DC: 2017. (EPA841-R-16-114). [Google Scholar]
- USGS (US Geological Survey) National Hydrography Dataset (NHD) US Geological Survey; Reston, Virginia: 2001. (Available from: https://nhd.usgs.gov/) [Google Scholar]
- USGS (US Geological Survey) Active mines and mineral processing plants. US Geological Survey; Reston, Virginia: 2003. (Available from: http://tin.er.usgs.gov/metadata/mineplant.faq.html) [Google Scholar]
- van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: a structure for efficient numerical computation. Computing in Science and Engineering. 2011;13:22–30. [Google Scholar]
- Wang L, Infante D, Esselman P, Cooper A, Wu D, Taylor W, Beard D, Whelan G, Ostroff A. A hierarchical spatial framework and database for the National River Fish Habitat Condition Assessment. Fisheries. 2011;36:436–449. [Google Scholar]
- Wolock DM. Open-File Report 03–263, digital data set. US Geological Survey; Reston, Virginia: 2003. Base-flow index grid for the conterminous United States. (Available from: http://water.usgs.gov/lookup/getspatial?bfi48grd) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


